Abstract
The F1c10 subcomplex of the yeast F1F0-ATP synthase includes the membrane rotor part c10-ring linked to a catalytic head, (αβ)3, by a central stalk, γδϵ. The Saccharomyces cerevisiae yF1c10·ADP subcomplex was crystallized in the presence of Mg·ADP, dicyclohexylcarbodiimide (DCCD), and azide. The structure was solved by molecular replacement using a high resolution model of the yeast F1 and a bacterial c-ring model with 10 copies of the c-subunit. The structure refined to 3.43-Å resolution displays new features compared with the original yF1c10 and with the yF1 inhibited by adenylyl imidodiphosphate (AMP-PNP) (yF1(I–III)). An ADP molecule was bound in both βDP and βTP catalytic sites. The αDP-βDP pair is slightly open and resembles the novel conformation identified in yF1, whereas the αTP-βTP pair is very closed and resembles more a DP pair. yF1c10·ADP provides a model of a new Mg·ADP-inhibited state of the yeast F1. As for the original yF1 and yF1c10 structures, the foot of the central stalk is rotated by ∼40 ° with respect to bovine structures. The assembly of the F1 central stalk with the F0 c-ring rotor is mainly provided by electrostatic interactions. On the rotor ring, the essential cGlu59 carboxylate group is surrounded by hydrophobic residues and is not involved in hydrogen bonding.
Keywords: ATP Synthase, Bioenergetics, Crystal Structure, Membrane Proteins, Mitochondria
Introduction
The F1F0-ATP synthase is an essential membrane rotary motor that uses transmembrane electrochemical ion gradients to synthesize ATP. To date, only cryo-electron microscopy has provided a complete view of the yeast ATP synthase (1). The structure of the Saccharomyces cerevisiae F1c10-ATP synthase subcomplex provided the first model at 3.9-Å resolution of the molecular assembly between the membrane rotor ring and the central stalk of a F1F0-ATP synthase (2). Both adenylyl imidodiphosphate (AMP-PNP)3 and ADP were added to the crystallization medium, a structure that will be referred to as yF1c10. It was solved by molecular replacement using the Cα coordinates of the crystal structures of the AMP-PNP-inhibited bovine F1-ATPase containing (αβ)3γ-subunits, which is considered as the bovine reference structure bF1·AMP-PNP (3), and of the Escherichia coli ϵ-subunit, the homologous of the mitochondrial δ-subunit (4), as models. A decameric ring of c-subunits (also named subunit 9 in yeast) was afterward found in the residual electron densities and the NMR solution structure of the E. coli c-monomer (5), which shares only 18% of identity with the yeast subunit, was used to build the c10-ring in the maps. The smallest subunit (ϵ-subunit) was indistinguishable in electron densities but was present as shown by a SDS-PAGE analysis of the crystal (2). Unfortunately and unexpectedly, the peripheral stalk of the enzyme dissociated during the crystallization process. Owing to the lack of crystals that diffracted at high enough resolution and lack of crystal structures of yeast subunits or subcomplexes, refinement of the yF1c10 was impossible a few years ago. Although the model deposited in the Protein Data Bank is a mix of x-ray and NMR unrefined Cα atom coordinates of bovine and E. coli homologous proteins, the structure was fitted successfully in the envelope of the electron microscopy map of F1F0-ATP synthase to obtain the envelope of the peripheral stalk (1, 6).
The first F1-ATPase x-ray structure (3) supports Boyer's binding change mechanism for catalysis (7). It was proposed that (i) the empty and open catalytic site βE was the open site with low affinity for nucleotides, (ii) the βTP site filled by ATP (or AMP-PNP) was the loose conformation, and (iii) the βDP site filled by ADP was the tight conformation where synthesis occurs (8). The opening of catalytic sites can be estimated by means of the buried area at the α-β interface. The loose and tight conformations were further identified in the structures of the ground state (9) and of an intermediate state analog (10), but they remained debatable because the βTP site has also been proposed as the high affinity catalytic site (11).
The inconsistent presence of ADP in the βDP site of the bovine reference structure (3) has been explained by the Mg·ADP-inhibited state of the enzyme. Dicyclohexylcarbodiimide (DCCD) inhibits both the H+ transport and the ATPase activity (12). It has been used to block the enzyme for crystallization (13). It reacts preferentially with a single c-subunit of the F0 rather than a single β-subunit of the F1. During the past decade, the detailed structures of the bovine F1-sector inhibited by Mg·ADP (bF1·ADP, Protein Data Bank (PDB) code 1w0k) (10) or covalently inhibited on βDPGlu199 by DCCD (bF1·DCCD) (13) and of the yeast enzyme in the presence of AMP-PNP and ADP at 2.8-Å resolution (yF1) (14) have been refined. There are three independent copies of the enzyme in the yF1 asymmetric unit, yF1(I) and yF1(III) were similar, and yF1(II) was different. In the nucleotide-free yeast F1 structure (yF1apo(I–III)), the DP and TP pairs adopt “closed” conformations despite the absence of bound nucleotides (15).
Concerning the membrane rotors, the structure solved at 2.4-Å resolution of the Na+-F-ATPase c-ring from Ilyobacter tartaricus contains 11 monomers (16). The K-subunit (NtpK) of the Na+-V-ATPase of Enterococcus hirae with four transmembrane helices forms decameric rings (17). In the E. hirae DCCD-inhibited K10-ring structure (PDB code 2DB4), dicyclohexyl-N-acylurea (DCU) was found to be linked to Glu139 equivalent to the yeast cGlu59, with DCU groups protruding outside the ring.
Here, we report the x-ray structure of the F1c10 subcomplex from S. cerevisiae inhibited by Mg·ADP and DCCD (yF1c10·ADP) refined to a resolution of 3.43 Å. However, DCU modifications on both sectors could not be ascertained on the electron density maps. The βDP and βTP catalytic sites adopt closed conformations and are both occupied by ADP. This structure represents the first view of the Mg·ADP-inhibited state of the yeast enzyme. The structure of the F1-sector was compared with the structures of the bovine enzyme and with all independent copies found in the yeast F1 with bound AMP-PNP and without bound nucleotides. Compared with the bovine enzyme, the yeast central stalk is twisted. In the F0 rotor ring, the essential cGlu59 carboxylate group is only surrounded by apolar residues. Its closest hydrogen bond acceptor, the cLeu57 carbonyl oxygen of the adjacent c-subunit, is too far away to make a direct hydrogen bond. The proton binding has specific features compared with the bacterial Na+ transporting (16) or the cyanobacterial (19) and chloroplastic (20) H+-transporting F-type ATP synthase rotor structures.
EXPERIMENTAL PROCEDURES
yF1F0 Enzyme Purification and yF1c10 Crystallization
All purification steps were performed as described previously (21) except that the nickel-nitrilotriacetic acid elution fraction was concentrated and subjected to a purification step on a Superdex 200 gel filtration column (HR 10/30). The gel filtration buffer contained 0.64 mm n-dodecyl β-d-maltoside, 100 mm NaCl, 2 mm MgCl2, 25 mm trehalose, 0.5 mm EDTA, 3 mm sodium azide, 20 mm Tris-HCl, pH 8.0. Fractions corresponding to the major peak were analyzed by SDS-PAGE electrophoresis. These fractions contained all the known subunits of the monomeric F1F0 (subunits α, β, γ, 4, 6, OSCP, d, δ, h, f, ϵ, i, 8, and 9) (supplemental Fig. S1). Assembly states were asserted by nondenaturing Blue native PAGE. The purified F1F0-ATP synthase is active, migrated as a fully assembled monomeric form (21). Neither ADP nor ATP was added during the purification. The protein solution was concentrated with a Centricon concentrator unit (YM-100, Millipore). The final enzyme concentration (9 mg/ml) was assessed by the Lowry method. The enzyme was incubated for 1 h at 20 °C with 660 μm ADP and then inhibited for 1 h with 100 μm DCCD in the presence of 2.5 mm DTT and 0.5 mm PMSF.
Crystallizations were carried out at 20 °C, by the sitting drop vapor diffusion method, after mixing equal volumes of protein and reservoir solutions (100 mm NaCl, 12% PEG MME 5000, 3 mm sodium azide, 100 mm Hepes, pH 7.5). Crystals grew in a few days as flattened rods of typically 150 to 400 μm in length and 20 μm in thickness. Crystals were cryo-protected with mother liquor containing 15% (v/v) glycerol and flash-frozen in liquid nitrogen.
Structure Determination
X-ray diffraction data were collected on beamline ID29 (European Synchrotron Radiation Facility). Many crystals mounted from the same drop diffracted at an ∼4-Å resolution, with the best one diffracting at 3.2-Å resolution. Severe anisotropic diffraction and radiation damage limited the quality of the data sets. The best data set (77% complete at 3.43 Å) was merged with one of lower resolution at 3.67 Å. All data were processed with IMOSFLM (22) and the CCP4 suite (23). The cell parameters of yF1c10·ADP are close to those of the original yF1c10 structure (2). The current structure was solved by molecular replacement with the PHASER program (24). The three independent conformations yF1(I–III) in which nucleotides were removed (PDB code 2HLD) were used successively as search model (14). The solutions obtained with yF1(I) and yF1(III) were better (Z-score, 50; Rwork, 0.44) than with yF1(II) (Z-score, 30; Rwork, 0.47). Because the central stalk of yF1(I) was more complete than that of yF1(III), the yF1(I) solution was retained. The asymmetric unit contains one molecule resulting in a solvent content of 67% (Vm = 3.7 Å3/Da). Rigid body refinement of individual subunits resulted in Rwork and Rfree equal to 0.38 using 3.7 Å data. In the F1-sector, the electron density maps are mainly continuous along the backbones and are good enough to rule out the possibility of statistical disorder.
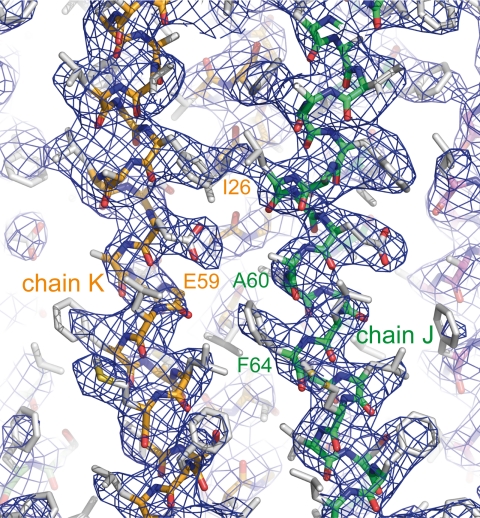
A c-monomer template was built using the coordinates of the I. tartaricus rotor c11-ring (PDB code 1YCE) (16) by removing six and seven residues at the N and C termini, respectively. To ascertain the number of c-subunits in the crystal, ring models containing from 8 to 12 copies (N) of the c-subunit were constructed as described below. The main axis of the I. tartaricus ring was oriented along the Z-axis and then rotated to place the center of the template on the X-axis. Rings were finally generated by successive (N−1) rotations of (2π/N) degrees of the template about the Z-axis. Optimization of the ring radii was based on an analysis of intersubunit contacts. With the fixed current F1 model as input, molecular replacement searches were run using each ring model with PHASER (24). The best solution was clearly obtained for the decameric ring, i.e. a Z-score of 24, instead of a Z-score <15 with the other ring models. Rigid body and overall temperature factor refinement of individual subunits resulted in Rwork and Rfree equal to 0.33. In the c-ring, the side chains were added to the model with the help of the I. tartaricus model (32% of identity, 59% of similarity with S. cerevisiae). The backbone electron density was better defined in a third and in the lower half of the ring where the sequence could be assigned with certainty. When no side chain information was present in the template or no density visible in the map, the most favored rotamer with acceptable van der Waals exclusion test was added to the model. Strong noncrystallographic symmetry restraints were applied separately on helices H1 and H2 but not on the loop. The chains of the c-ring were labeled from J to S in the direction of the hydrolysis (25), starting with the chain having the lowest overall temperature factor. The assignment was checked on 2Fo − Fc unaveraged and Non-crystallographic Symmetry-averaged (calculated using the DM program) electron density maps on chains with the lowest temperature factors (Fig. 1) using COOT (26).
FIGURE 1.
View around the proton binding site. The 10-fold Non-crystallographic Symmetry-averaged 2Fo − Fc electron density map at 3.43-Å resolution with the refined coordinates superimposed is shown. The contour level is 1.5σ.
Recently, modifications of standard refinement protocols were developed to refine low resolution structures for which a structural knowledge of components is available at higher resolution (27). F1- and c10-chains were divided into several groups according to the structural domains: the N-terminal β-barrel domain (α26–95 or β6–82), the central nucleotide binding domain (α96–381 or β83–357) and the C-terminal helical domain (α382–509 or β358–475) of each catalytic head subunit, the coiled-coil helical domain (γ1–59, γ201–278) and the globular domain (γ71–200) of the γ-subunit, the β-sandwich domain (δ9–90) and the two C-terminal helices (δ92–97, δ99–138) of the δ-subunit and finally the outer (c1–41) and inner (c42–76) helices of each c-subunit. Rigid body, individual coordinates and atomic displacement parameter refinements were further performed with PHENIX (28). Extensive torsion angle simulated annealing refinement with harmonic restraints applied on atomic coordinates was carried out using the maximum likelihood amplitude target in CNS (version 1.2) (29). The final model refined to 3.43 Å resolution had Rwork and Rfree of 0.312 and 0.319, respectively. Data and refinement statistics are supplied in Table 1. The refined structure was subjected to validation using PROCHECK (30). The secondary structures were assigned using STRIDE (31). The r.m.s.d. between equivalent Cα atoms and the calculation of rotation angles were obtained with LSQKAB from the CCP4 package (23). Protein structures were illustrated by using the PyMOL program (32). The electrostatic potentials were calculated with APBS (33). supplemental Fig. S2 was drawn with ESPRIPT (34). To facilitate comparison with the bovine sequence, the bovine numbering is indicated in parentheses.
TABLE 1.
Data processing and refinement statistics
| Data set |
|||
|---|---|---|---|
| Crystal 1 | Crystal 2 | Merged data | |
| Data collection | |||
| Space group | P21 | P21 | P21 |
| a, b, c (Å) | 134.7, 174.0, 137.2 | 136.1, 173.7, 136.3 | 135.0, 173.9, 137.0 |
| β | 92.6° | 92.9° | 92.7° |
| Resolution range (Å) | 40–3.43 | 40–3.67 | 40–3.43 |
| Highest resolution bin | 3.62–3.43 | 3.87–3.67 | 3.62–3.43 |
| No. of unique reflectionsa | 64,957 (8463) | 26,999 (4220) | 74,433 (7927) |
| Multiplicitya | 2.6 (2.4) | 1.5 (1.4) | 2.8 (2.4) |
| Completenessa (%) | 77.2 (69.0) | 39.9 (42.2) | 88.1 (64.4) |
| Rsym, Rmergea,b | 0.19 (0.72) | 0.15 (0.73) | 0.19 (0.72) |
| 〈I/σ(I)〉a,b | 4.2 (1.3) | 3.0 (1.3) | 4.3 (1.3) |
| B-factor Wilson (Å2) | 69 | 83 | 73 |
| Rwork, Rfreec | 0.312, 0.319 | ||
| Refinement statistics | |||
| No. of protein atoms | 30,671 | ||
| No. of ligand atoms | 152 | ||
| Protein B-factors (Å2) | 106 | ||
| r.m.s.d. bond lengths (Å) | 0.005 | ||
| r.m.s.d. bond angles | 0.89° | ||
| Ramachandran plot (%)d | 91.6, 7.7, 0.7 | ||
a Statistics for the highest resolution bin are shown in parentheses.
b Rsym and Rmerge were calculated by ΣhΣj|Ih,j − 〈Ih〉|/ΣhΣjIh,j, where h is the index for unique reflections, and j is the index for symmetry redundant reflections. Rsym and Rmerge are the reliability factors for single crystal (1 and 2) and merged data sets, respectively. Ih is the mean weighted intensity after rejection of outliers.
c Rwork and Rfree were calculated by Σ‖Fobserved| − k|Fcalculated‖/Σ|Fobserved|. Rfree was calculated using 5% random data (3767 reflections) omitted from refinement.
d Percentage of residues in most favored regions, allowed regions and disfavored regions, respectively.
Intermolecular Lattice Contacts
The buried surface areas (Å2) of interfaces in yF1c10·ADP, yF1, and bF1-ATPase were calculated with the PDBePISA server.
RESULTS
Overall Description of the Structure
The structure was solved by molecular replacement using the coordinates of the yeast yF1 (14) and of the I. tartaricus rotor ring (16). These high resolution structures provided accurate models to phase and then refine the yeast F1c10 subcomplex structure. According to the functional α- and β-subunits reference scheme, as in Ref. 3, chains A, B, and C were named αE-, αTP-, αDP-subunits, and chains D, E, and F were named βDP-, βE-, and βTP-subunits, respectively (supplemental Fig. S2). The final model contains all subunits of both the F1-sector (αE26–510, αTP25–509, αDP26–510, βE6–478, βTP7–478, βDP6–475, and γ1–59 and γ71–278, δ9–138, and ϵ1–59) and the F0 membrane rotor composed of a ring of 10 c-subunits (c1–76) (Fig. 2). No densities that could be attributed to any peripheral stalk subunits were visible. The overall structure of the F1-sector of yF1c10·ADP is more similar to the three yeast yF1(I–III) structures (root mean square deviation (r.m.s.d.) < 1.7 Å) than to those of bovine bF1 structures (r.m.s.d. > 4.5 Å) (Table 2). yF1(I), the most complete model, and yF1(III) were found to be more similar (r.m.s.d. < 1.47 Å). Thus, yF1(I) will be considered as the yeast reference structure.
FIGURE 2.
Ribbon diagram of the overall structure of the yeast F1c10·ADP complex. The α-, β-, γ-, δ- and ϵ-subunits are shown in red, blue, green, yellow, and cyan, respectively, whereas the c-ring is multicolored. The nucleotides and magnesium ions are drawn in a space-filling model. The symmetry axes of the F1-stator and F0-rotor are drawn in black.
TABLE 2.
r.m.s.d. values (Å) in Cα positions between α- and β-subunits, E, TP and DP pairs, and γ-, δ- and ϵ-subunits of yF1c10·ADP with yF1(I) (PDB code 2HLD, chains A–I), yF1(II) (PDB code 2HLD, chains J–R) (13), and bF1 (bF1·DCCD, PDB code 1E79) (12)
| yF1(I) | yF1(II) | bF1 | |
|---|---|---|---|
| αE/αE | 0.42 | 1.21 | |
| αDP/αDP | 0.50 | 1.12 | |
| αTP/αTP | 1.18 | 1.88 | |
| αTP/αDP | 1.29 | 1.27 | |
| γδϵ/γδϵ | 1.47 | 3.05 | |
| δ/δ | 1.05 | 2.16 | |
| βE/βE | 0.50 | 0.87 | |
| βDP/βDP | 0.54 | 0.87 | |
| βTP/βTP | 0.53 | 0.89 | |
| βTP/βDP | 0.71 | 0.73 | |
| γ/γ | 0.91 | 2.60 | |
| ϵ/ϵ | 0.70 | 2.64 | |
| E/E | 0.54 | 0.89 | 1.17 |
| DP/DP | 0.67 | 1.45 | 1.76 |
| TP/TP | 1.64 | 1.43 | 2.46 |
| TP/DP | 1.90 | 1.02 | 1.14 |
| F1/F1 | 1.47 | 1.68 | 4.53 |
| (αβ)3 | 1.22 | 1.43 | 2.13 |
Structure of the F0 Rotor Ring
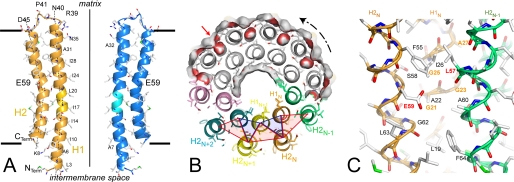
The c-subunit folds as a helical hairpin with the N-terminal helix c2–38 (helix H1) connected by a polar loop c39–45 containing an R(N/Q)P motif to the C-terminal helix c46–74 (helix H2) (Fig. 3A). A decameric c-ring displays an hourglass shape with a central pore. The ring is 60 Å in height and has a diameter of 57 Å at the top, 51 Å at the bottom, and 43 Å in the middle. The above-mentioned distances are between local helical axes on the cGly21-Cα atoms plane. As shown in Fig. 3B, the neighboring helices H1 are tightly packed with 7.5 Å between their local axes. One outer helix H2N interacts with two inner helices, H1N at 7.4 Å and H1N+1 at 9.0 Å to form a tightly packed three-helix bundle (open angle ∼50 °) turned inwards. Furthermore, one inner helix H1N interacts with two outer helices, H2N and H2N−1, to form a more open three-helix bundle (open angle ∼80 °) turned outwards that harbors the proton binding site. The neighboring helices H2 are further away at 11.5 Å (Fig. 3B). In helix H1, a 14 ° kink in the cLeu20 region follows a two-turn long 310 helix (c16–20) (Fig. 3A). A conserved GxGxGx(G/A) motif in the middle of the helix H1 (c21–27) (supplemental Fig. S3) participates in this close packing (50). In helix H2, a more pronounced 29 ° kink is located below cGlu59 in the region opposite the glycine-rich motif (Fig. 3A). The central constriction is caused by the bend of both inner and outer helices. The pore has a diameter of 14–16 Å in the middle and 23 Å at the bottom and is lined by Ile10, Ile14, Ile17, Ile24, Ile28, and Leu20 residues (Fig. 3A). There are some narrow clefts between the outer helices, and one of these clefts (Fig. 3B) is lined by Leu19, Gly21, Ala22, Gly23, and Ile26 from H1N, Glu59, and Gly62 from H2N and Ser58, Ala60, Thr61, Phe64, from H2N−1. There is no intrinsic channel lined by polar groups in the upper and lower parts of the c10-ring. The cGlu59 side chains with the lowest temperature factors (J–M) display well defined electron densities (Fig. 1). Nearby, small residual densities are not sufficiently high to model likely DCU modifications. One of oxygen atoms of the cGlu59 side chain is accessible from the outer surface, whereas the second atom is turned toward the center of the outer three-helix bundle and is not hydrogen bonded (Fig. 3, B and C). Large rotations of the cGlu59 side chains are sterically hindered by cIle26, cPhe55, cAla56, and cLeu63 of the same c-subunit.
FIGURE 3.
A, the c-subunit adopts a helical hairpin structure. Local helical axes of the H1 (c5–19, c23–37) and H2 (c47–59, c60–74) helices and the ring axis are drawn as dashed lines and full lines, respectively. Short horizontal lines indicate the position of assumed limits of the inner mitochondrial membrane. Two diametrically opposed c-subunits are shown. The two-turn long 310 helix is marked in light colors. Hydrophobic residues lining the pore and significant residues are labeled. B, section of the c10-ring viewed from F1 at the level of proton binding sites. The molecular surface of a half-ring is colored in gray and red for apolar and acidic residues, respectively. One cleft located slightly below the proton binding site is indicated by the red arrow. The inner and outer three-helix bundles are outlined by blue and red triangles, respectively. The rotation direction of the ATP synthesis is indicated. C, the essential cGlu59 carboxylate group is surrounded by hydrophobic residues and is not involved in hydrogen bonding. The conserved residues of the GxGxGx(G/A) motif are labeled in orange.
In yF1c10·ADP, TP Pair Resembles the DP Pair of Yeast and Bovine F1
Despite the limited data resolution, residual electron densities appeared clearly in five nucleotide binding sites. These densities were sufficiently defined to be able to distinguish between di- and triphosphate nucleotides (supplemental Fig. S4). Mg·ATP molecules were modeled in the three noncatalytic sites, whereas Mg·ADP molecules were modeled in both the βDP and βTP catalytic sites. The refined temperature factors of nucleotides were compatible with fully occupied binding sites. No densities were observed in the βE catalytic site. The mean overall temperature factor of the αE-, αDP-, βDP-, and βTP-subunits (95 Å2) is lower than that of the αTP- and βE-subunits (125 Å2).
When α- and β-subunits were individually compared with those of the yF1(I) model (14) (Table 2), there were no significant differences in any subunits (r.m.s.d. values 0.42–0.54 Å), with the exception of the αTP-subunit (r.m.s.d. 1.18 Å). When the αTP-subunit was split into subdomains I (αTP25–147 and αTP162–351) and II (αTP148–161 and αTP352–509), the deviations were decreased to 0.58 Å and 0.71 Å, respectively (supplemental Fig. S5). The change can be described as a rigid body rotation of 7.7 ° of subdomain II. When E, DP, and TP pairs are compared with yF1(I) pairs (Table 2), no significant differences are observed for the E and DP pairs (r.m.s.d. values 0.54 and 0.67 Å, respectively), whereas the TP pair appears quite different (r.m.s.d. 1.64 Å). The E pair is largely open (buried area of 1820 Å2, supplemental Table S1) and similar to the yF1(I) E pair (r.m.s.d. 0.54 Å), but slightly less open than the E pair in the bF1·DCCD structure (1780 Å2, r.m.s.d. 1.17 Å). The DP interface is moderately open (2180 Å2) and is similar to the novel conformation of the DP pair identified in yF1(I) (2010 Å2, r.m.s.d. 0.67 Å) (Fig. 4A and see Fig. 5, A–C), whereas the DP pairs of yF1(II) (2740 Å2, r.m.s.d. 1.45 Å) and of bF1·DCCD (2940 Å2, r.m.s.d. 1.76 Å) are 6.3 ° more closed.
FIGURE 4.
The (α/β)DP pair of yF1c10·ADP resembles the (α/β)DP pair of yF1(I), whereas the (α/β)TP of yF1c10·ADP resembles the (α/β)DP of yF1(II). Superimposition of the DP pair of yF1c10·ADP with the DP pair of yF1(I) (A) and the TP pair of yF1c10·ADP with the DP pair of yF1(II) (B). The α- and β-subunits are superimposed. The αTP- and βTP-subunits of yF1c10·ADP are shown in red and blue, respectively. The yF1(I) and yF1(II) (13) subunits are shown in green. The αArg375 side chains and nucleotides are drawn as ball and sticks.
FIGURE 5.
Four serial cross-sections perpendicular to the pseudo 3-fold axes (▴) are viewed from the top of F1. Superimposition was performed using α-subunits. The (αβ)3 catalytic heads of yF1c10·ADP (multicolored) are shown with bF1·DCCD (blue) (or with yF1(I) on supplemental Fig. S6). The nucleotides of yF1c10·ADP are drawn as a ball-and-stick diagram, and Mg2+ is drawn as a sphere. The central stalks are shown using a schematic except for the coiled-coil of γ-subunits (γ1–50 and γ205–276), which are drawn as linked Cα. Blue, bF1·DCCD (12); green, yF1(I) (13); and red, yF1c10·ADP. The yF1c10·ADP ring (multicolored surface) and the rotor axis (yellow decagon) are drawn. A, the C-terminal helices γ256–278 are represented as sticks in the proline-rich collar (αPro290, αPro291, αGly292, and βPro276). The conserved γGly273(268) is near the αDP- and βDP-subunits. B, at the end of the coiled-coil γ1–17 and γ230–259, the Tyr255(250) backbone is more twisted in yeast. C, near the hinge residues, γ18–25 and γ235–242, the yeast and bovine N-terminal helices clearly deviate. The arrow marks the deviation of ∼8 Å of the tip αTP407–411 toward the βTP-subunit. D, the foot of the yeast yF1c10·ADPcentral stalk is rotated 43 ° relative to the bF1·DCCD stalk in the ATP direction of the hydrolysis.
The TP interface is highly buried (2960 Å2) similar to the TP interface in yF1apo(III) (2840 Å2, r.m.s.d. 0.90 Å) (15). Interestingly, the TP pair resembles the DP pairs of yF1(II) (2740 Å2, r.m.s.d. 1.02 Å) (Fig. 4B) and bF1·DCCD (2940 Å2, r.m.s.d. 1.14 Å). It is definitely more closed than the TP interface of yF1(I) (2280 Å2, r.m.s.d. 1.64 Å) (Fig. 5C). This closure can be described as an overall 5.7° rotation of the αTP- toward the βTP-subunit and results in the ∼8 Å displacement of the tip (αTP407–411) of the helix-turn-helix structure of the C-terminal domain toward the adjacent βTP-subunit, tangentially to the C-terminal helix of the γ-subunit (Fig. 5C and supplemental Figs. S5 and S6C). As a probable consequence, the tip is visible in the 2Fobs − Fcalc electron density maps contoured at 1.5σ, whereas it is absent in the Fobs − Fcalc electron density maps of yF1 contoured at 2.0σ. In the crystal, neighboring molecules are arranged to form infinite columns along the [101̄] direction of the unit cell by a coaxial association with the collar protruding from c10-ring that plugs the cavity lined by the six N-terminal β-barrels of (αβ)3. In the orthogonal plane, neighboring columns are in the opposite direction and display a V-shaped arrangement (supplemental Fig. S2). They interact through the central nucleotide-binding domains. Four connecting loops (α103–108, α120–126, α224–229, and α193–195) of αE- and αDP-subunits interact tightly, whereas the outer surfaces of βDP- and βTP-subunits face each other more loosely. The outer surfaces of αTP- and βE-subunits are widely solvent-accessible. The TP closure cannot be attributed to direct lattice contacts of the α-subunit.
Faint residual densities were observed in the hydrophobic binding pocket around the βDP- and not βTP-Glu200 side chain without structural changes in the neighborhood when the βDP-subunit was compared with yF1(I) and βTP-subunit to yF1apo(III). Attempts to model DCU modification were unsuccessful. Indeed, no structural changes were observed previously between bF1·DCCD (13) and bF1·ADP (11) structures. Finally, the yF1c10·ADP structure represents an Mg·ADP-inhibited state of the yeast F1 enzyme, which is relatively different from those observed in bovine structures.
Central Stalk
A whole γδϵ central stalk was observed in yeast F1 structures (2, 14, 15) and in bF1·DCCD (13). The γ-, δ- and ϵ-subunits of yF1c10·ADP are individually similar to those of yF1(I) (r.m.s.d. < 1.05 Å, Table 2). Overall, the γδϵ central stalks of the yeast F1 structures (yF1c10·ADP, yF1(I), yF1(II), yF1c10) are also similar (r.m.s.d. < 1.7 Å), whereas they differ significantly from the bovine one (r.m.s.d. = 3.05 Å, Table 2). The overall temperature factors of γ-, δ- and ϵ-subunits are 120 Å2, 147 Å2, and 149 Å2, respectively. In F1 complexes, the orientation of the γ-subunit relative to the (αβ)3 component determines the catalytic state of the enzyme and, according to the binding change mechanism, a 120 ° rotation of the γ-subunit during the ATP hydrolysis cycle results in E → TP, TP → DP, and DP → E interconversions. The three α-subunits were used to superimpose the central pseudo 3-fold axes. The upper part (γ256–278) of the C-terminal helix of the γ-subunit are well superimposed (Fig. 5A). This helical segment is constrained to lie inside the narrowest part of the α3β3 inner channel lined by the proline-rich collar (αPro290(288), αPro291(289), αGly292 and βPro276(276)) (3). A conserved γGly273(268) allows this helix to nestle against the αDP- and βDP-subunit interface. Below, the N- and C-terminal helices interact to form a coiled coil (Fig. 5B). From a tight turn of the yeast backbone at residue γTyr255(250), the stalk deviates increasingly as it becomes closer to the c-ring rotor, with ∼18-Å shifts relative to bF1·DCCD at the periphery of the foot (Fig. 5D). Accordingly, the long C-terminal helix is more curved in yeast (75 Å of curvature radius) than in bovine structures (100 Å of curvature radius). The γ-subunit twist is definitely an intrinsic property of the yeast enzyme. In yF1c10·ADP, the region γ235–242 of the C-terminal helix interacts with the helical domains of αTP- and βTP-subunits (Fig. 5C), and the TP closure results in a significant rotation/translation of this helix. In yeast, the region γ18–25 of the N-terminal helix interacts with the C-terminal helical domains of αE- and βDP-subunits, and the partial opening of the DP interface in the yeast structures could result from these interactions. The αDP-subunit interacts with the globular domain of the γ-subunit, whereas the helical domain of βE-subunit does not interact directly with the γ-subunit (Fig. 5C). The closure of the TP pair and the subtle differences in the relative orientation and position of the γ-subunit result from the Mg·ADP inhibition. At the F1-F0 rotor interface, the foot of the yeast central stalk is rotated by ∼40 ° (36 ° for yF1(I), 41 ° for yF1c10 or 43 ° for yF1c10·ADP) relative to the bF1·DCCD in the direction of the hydrolysis (Fig. 5D and supplemental Fig. S6D). As a consequence, the presence of the c10-ring and the crystal packing have no significant influence on the γ-subunit twist. The δ-subunit interacts extensively with the γ- and ϵ-subunits along with the c-ring (supplemental Tables S2 and S3). In the δ-subunit, the second helix of the C-terminal domain folds back between the first helix and the N-terminal β-sandwich. The N-terminal helix of the small ϵ-subunit is inserted between the γ-subunit and the two domains of the δ-subunit. Comparison with bF1·DCCD shows that the γ- and δ-subunits have r.m.s.d. values of 2.60 Å and 2.16 Å, respectively. The overall r.m.s.d. of the δ-subunit decreases to 0.8 Å only if the β-sandwich is considered, whereas the helical domain is more variable in sequence and conformation.
Interface between the F0 Rotor Ring and Central Stalk
The foot sole of the central stalk is roughly flat and interacts with the upper surface of the membrane rotor, which adopts a complementary shape with the four Cα atoms of the residues c39–42 lying in a plane slightly tilted down the ring axis. The membrane rotor axis deviates from the pseudo 3-fold axis of F1 by 8.5 ° with a minimum displacement of 2.8 Å (Fig. 2). The c-subunits that interact the most with the δ- or γ-subunit have well defined electron densities and the lowest temperature factors (130 Å2), as reported previously in yF1c10 (2). Only three c-subunits interact with the δ-subunit, four with the γ-subunit, two with both subunits, and one c-subunit does not interact. The buried area with the δ-subunit (660 Å2) is more than twice the area with the γ-subunit (200 Å2) (supplemental Table S3), and the interface area represents only a quarter of the accessible surface of the top side of the rotor ring.
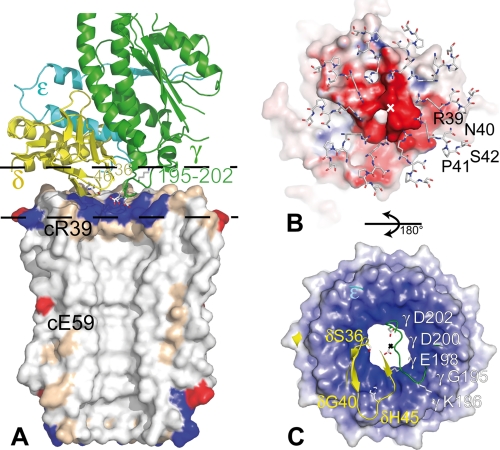
In the γ-subunit, the loop γ195–202 linking the globular domain to the C-terminal helix is almost planar and orthogonal to the rotor axis (Fig. 6A). This loop goes down and crosses over the ring pore with γGly195 and γLys196 on top of the inner ring and γAsp200 nearly in line with the rotor axis (Fig. 6C). The loop contains three acidic residues (γGlu198, γAsp200, and γAsp202, the first being highly conserved Glu/Asp), so the negatively charged stalk surface could interact with the positive rotor ring surface (Fig. 6, B and C).
FIGURE 6.
Views of the F1-F0 rotor interface. A, lateral view of the interface. The γ-, δ-, and ϵ-subunits are shown as green, yellow, and cyan ribbons, respectively. The molecular surface of half a c-ring is colored in white (hydrophobic), beige (polar), red (acid), and blue (basic). The side chains of γGlu198, γAsp200, and γAsp202 nestled in the upper part of the pore are surrounded by the 10 cArg39. B, the loops c39–42 of the c10-ring in interaction with the F1 central stalk surface are viewed from the F0. C, the amino acid residues of the central stalk section defined by the two dashed lines on A in interaction with the F0 rotor surface are viewed from F1. On A and B, the split solvent accessible surfaces are colored in blue (positive), white (neutral), and red (negative). A–C are at the same scale. The x marks the c-ring axis. The side chains of residues located at the interface are drawn using a ball-and-stick diagram.
In the δ-subunit, the region δ36–46 containing the fourth strand and the loop connecting the two sheets of the β-sandwich is again almost planar and orthogonal to the rotor axis (Fig. 6A). It interacts tightly with three c-subunits (with δSer36, δGly40, and δHis45 and cArg39, cAsn40, and cSer42) (Fig. 6, B and C). The last helix of the helical domain runs far from, but quite parallel to, the c-ring surface.
DISCUSSION
The S. cerevisiae c-subunit resembles the c-subunit from I. tartaricus (r.m.s.d. 1.2 Å for a monomer, 1.4 Å for a dimer) (16) and Spirulina platensis (r.m.s.d. 1.4 Å for a monomer, 1.6 Å for a dimer) (19) and differs greatly from the solution structure of the E. coli c-monomer (r.m.s.d. ∼4.5 Å) (5). Accordingly, the c-subunit sequence of the mitochondrial H+ F-ATPase (S. cerevisiae) is closer to bacterial Na+ F-ATPase (32% identity with I. tartaricus) than to cyanobacterial (25% with S. platensis), chloroplastic (21% with Spinacea oleracea) and bacterial (18% with E. coli) H+ F-ATPase (supplemental Fig. S3). In addition, the essential residue cGlu59 conserved in all V- and F-ATPase is replaced by cAsp61 in E. coli (supplemental Fig. S3).
In the I. tartaricus Na+ F-ATPase, the Na+ is bound by Glu65, two polar side chains (Gln32 and Ser66), one backbone carbonyl (Val63), and a water molecule (35). In chloroplastic and cyanobacterial H+ F-ATPase, the carboxyl oxygen atoms of the essential Glu61/62 are bound by Gln28/29, Tyr66/67, and the backbone carbonyl Phe59/60. In S. cerevisiae, the polar residues are replaced by hydrophobic ones (Ile26, Ala60, and Phe64) (supplemental Fig. S3 and Fig. 7) and the kink in outer helix H2 disrupts the intrahelical hydrogen bond networks, so cAla56 and cLeu57 backbone carbonyl groups are not hydrogen-bonded. The cLeu57 carbonyl is 5.1 Å apart from the cGlu59 carboxyl oxygen atoms (Fig. 3C); therefore, a nonbonded interaction would require a molecular bridge. As a result, a water molecule could be inserted at a position close to that of the Na+ in I. tartaricus c11-ring and could establish hydrogen bonds with cGlu59 carboxyl and cLeu57 carbonyl groups. The equivalent distance is 4.5 Å in the Na+ binding site (16) and 2.7 Å in the proton binding site with a direct hydrogen bond (16, 19). Hence, the current structure does not rule out the involvement of a hydronium ion in the proton translocation mechanism in yeast. Two types of proton binding sites could exist, with or without hydronium ion requirement. Recently, the observed bell-shaped pH profile for DCCD labeling of the acidic c-ring residues of H+-transporting F-ATP synthase (36) were taken to be the involvement of a hydronium ion in the binding site, as proposed earlier by Boyer (37). A putative hydronium binding scheme for yeast H+ F-ATP synthase could be built on that of the sodium binding of Na+ F-ATPases (38), with a working mode similar to the “push and pull” functional model (39).
FIGURE 7.
Comparison of sodium and proton binding sites. A, proton binding site in S. cerevisiae yF1c10·ADP; B, Na+ binding site in I. tartaricus c11-ring (PDB code 2WGM); C, proton binding site in the c15-ring from S. platensis cyanobacterium (PDB code 2WIE); and D, proton binding site in the c14-ring from S. oleracea chloroplast (PDB code 2W5J) with Gln28 built in the best rotamer conformation.
The outer surface of the c-ring is essentially hydrophobic with the exception of the essential cGlu59 embedded at 24 Å from the two sides of the membrane, if cLys44/cAsp45 and cLys8/carboxyl-terminal groups, at a distance of ∼43 Å, mark the inner and outer surfaces of the mitochondrial membrane, respectively (Fig. 3A). On the matrix side, the pore is closed partly by the cArg39 side chains that could act as a socket to receive the foot of the central stalk. An enlargement to 25 Å near a collar of cAsn35 could indicate the top of a lipid plug. At the pore openings, a few residual density peaks cannot be attributed with certainty to ordered lipids or detergent molecules. However, there is enough space to accommodate, in vivo, two opposite plugs of a few lipids. It has been shown in E. coli that the pore is occupied by phospholipids (40).
The βDP and βTP binding sites contain an Mg·ADP molecule with the “arginine finger” αArg375(373) pointing in toward the site (supplemental Fig. S4). The structure reported here is probably equivalent to the ADP-inhibited state of the enzyme because our crystallization conditions leading to bound Mg·ADP and no free phosphate (41). This form is probably not an intermediate in either ATP hydrolysis or ATP synthesis but rather a pause state from which reactivation occurs upon the presence of protonmotive force in mitochondrial ATP synthase (7, 42). The yF1c10·ADP crystals were grown in the presence of ADP and azide without ATP analogs like various bovine F1-ATPase crystals. Azide is known to enhance Mg·ADP inhibition (43, 44). The bF1·ADP·N3− structure at 1.95-Å resolution revealed how azide enhances the binding of the ADP molecule in the βDP-subunits (45). Afterward, the hypothetical presence of an azide molecule was proposed in bovine structures in the βDP catalytic sites (3, 10, 13). Similarly, it cannot be ruled out that an azide molecule is bound to the βDP and βTP sites of yF1c10·ADP.
Whatever the crystallization method, the detergent concentration or the inhibitor used to crystallize the yeast F1F0-ATP synthase, all subunits of the peripheral stalk were lost as indicated by SDS-PAGE analysis of crystals. SDS-PAGE analysis of three-dimensional crystals of the bacterial F1F0-ATP synthase from Chloroflexus aurantiacus obtained by slow detergent removal also indicates the loss of the peripheral stalk during crystallization (46). The crystallization conditions may induce the destabilization of the a-subunit/c-ring and the peripheral stalk/F1 interfaces, leading to the F1-c-ring subcomplex.
In E. coli (47) and I. tartaricus (48), three acidic residues have been identified in the γ-subunit as good candidates for interaction with the c-ring. However, we cannot exclude that the positive charges of conserved cArg39 could be partially neutralized by the polar heads of a phospholipid plug. The loss of the peripheral stalk and the large crystal lattice interface (980 Å2) between the c-ring and the β-barrels domains of the (αβ)3 could disturb the interface of a similar area (890 Å2) between the central stalk and the c-ring (supplemental Table S3), so the densities of cArg39 and γGlu198 side chain residues are very poorly defined.
The winding of the central stalk observed in different conformational states of yeast F1 complexes, with or without the c-ring, appears specific to the yeast F1. The structure of a complex between bovine F1 and a truncated stator containing OSCP-, b-, d-, and F6-subunits reveals the position of the peripheral stalk along the cleft of a noncatalytic interface (49) and suggests a low degree of freedom for the location of the membrane domain of the stator relative to the membrane rotor. The peripheral stalk prevents rotation of the (αβ)3 head but does not prevent its essential conformational changes. The predictions of the transmembrane regions and the secondary structures of the yeast subunit b (subunit 4) suggest that the peripheral stalks are highly similar. In yF1c10·ADP, the stoichiometry of 10 monomers per c-ring and the rotation of ∼40 ° of the central stalk foot with respect to bovine are compatible with a step-by-step rotation of 36 ° of the rotor, without involving any shift of the membrane stator and hence modification of the overall conformation of the peripheral stalk. Finally, when comparing the yF1(I) and yF1c10·ADP structures, it appears that the presence of the c-ring does not modify the position of the γ-subunit and of the foot sole of the central stalk (Fig. 5).
Refinement of the crystal structure of the yeast F1c10 inhibited by AMP-PNP should unveil some unknown features of this complex. High resolution structures of rotor rings of bacterial or mitochondrial H+-transporting F-ATP synthase are now required to clarify the nature of the proton binding site in mitochondrial ATP synthases.
Supplementary Material
Acknowledgments
We thank Bernard Pucci for providing the H-TAC detergent and Lucile Moynié, Ray Cooke, and reviewers who contributed remarks to improve the manuscript. We also thank the beamline staffs of the European Synchrotron Radiation Facility (Grenoble, France) and the synchrotron SOLEIL (Saint-Aubin, France) for help in performing X-ray diffraction.
This work was supported by the Région Aquitaine, the Agence Nationale de la Recherche (ANR-06-PCVI-0016), the European Synchrotron Radiation Facility, and the synchrotron SOLEIL.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S6.
The atomic coordinates and structure factors (code 2WPD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AMP-PNP
- adenylyl imidodiphosphate
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank
- DCCD
- dicyclohexylcarbodiimide
- DCU
- dicyclohexyl-N-acylurea.
REFERENCES
- 1.Lau W. C., Baker L. A., Rubinstein J. L. (2008) J. Mol. Biol. 382, 1256–1264 [DOI] [PubMed] [Google Scholar]
- 2.Stock D., Leslie A. G., Walker J. E. (1999) Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 3.Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 4.Uhlin U., Cox G. B., Guss J. M. (1997) Structure 5, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 5.Girvin M. E., Rastogi V. K., Abildgaard F., Markley J. L., Fillingame R. H. (1998) Biochemistry 37, 8817–8824 [DOI] [PubMed] [Google Scholar]
- 6.Rubinstein J. L., Walker J. E., Henderson R. (2003) EMBO J. 22, 6182–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer P. D. (1993) Biochim. Biophys. Acta 1140, 215–250 [DOI] [PubMed] [Google Scholar]
- 8.Leslie A. G., Walker J. E. (2000) Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menz R. I., Walker J. E., Leslie A. G. (2001) Cell 106, 331–341 [DOI] [PubMed] [Google Scholar]
- 10.Kagawa R., Montgomery M. G., Braig K., Leslie A. G., Walker J. E. (2004) EMBO J. 23, 2734–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao H. Z., Weber J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18478–18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermolin J., Fillingame R. H. (1989) J. Biol. Chem. 264, 3896–3903 [PubMed] [Google Scholar]
- 13.Gibbons C., Montgomery M. G., Leslie A. G., Walker J. E. (2000) Nat. Struct. Biol. 7, 1055–1061 [DOI] [PubMed] [Google Scholar]
- 14.Kabaleeswaran V., Puri N., Walker J. E., Leslie A. G., Mueller D. M. (2006) EMBO J. 25, 5433–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabaleeswaran V., Shen H., Symersky J., Walker J. E., Leslie A. G., Mueller D. M. (2009) J. Biol. Chem. 284, 10546–10551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier T., Polzer P., Diederichs K., Welte W., Dimroth P. (2005) Science 308, 659–662 [DOI] [PubMed] [Google Scholar]
- 17.Murata T., Yamato I., Kakinuma Y., Leslie A. G., Walker J. E. (2005) Science 308, 654–659 [DOI] [PubMed] [Google Scholar]
- 18.Deleted in proof
- 19.Pogoryelov D., Yildiz O., Faraldo-Gómez J. D., Meier T. (2009) Nat. Struct. Mol. Biol. 16, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 20.Vollmar M., Schlieper D., Winn M., Büchner C., Groth G. (2009) J. Biol. Chem. 284, 18228–18235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talbot J. C., Dautant A., Polidori A., Pucci B., Cohen-Bouhacina T., Maali A., Salin B., Brèthes D., Velours J., Giraud M. F. (2009) J. Bioenerg. Biomembr. 41, 349–360 [DOI] [PubMed] [Google Scholar]
- 22.Leslie A. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 23.CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.Karmali A. M., Blundell T. L., Furnham N. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams P. D., Gopal K., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Pai R. K., Read R. J., Romo T. D., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C. (2004) J. Synchrotron Radiat. 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 29.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31.Frishman D., Argos P. (1995) Proteins 23, 566–579 [DOI] [PubMed] [Google Scholar]
- 32.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 33.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier T., Krah A., Bond P. J., Pogoryelov D., Diederichs K., Faraldo-Gómez J. D. (2009) J. Mol. Biol. 391, 498–507 [DOI] [PubMed] [Google Scholar]
- 36.von Ballmoos C. (2007) J. Bioenerg. Biomembr. 39, 441–445 [DOI] [PubMed] [Google Scholar]
- 37.Boyer P. D. (1988) Trends Biochem. Sci. 13, 5–7 [DOI] [PubMed] [Google Scholar]
- 38.Dimroth P., von Ballmoos C., Meier T. (2006) EMBO Rep. 7, 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Ballmoos C., Dimroth P. (2007) Biochemistry 46, 11800–11809 [DOI] [PubMed] [Google Scholar]
- 40.Oberfeld B., Brunner J., Dimroth P. (2006) Biochemistry 45, 1841–1851 [DOI] [PubMed] [Google Scholar]
- 41.Di Pietro A., Fellous G., Godinot C., Gautheron D. C. (1986) Biochim. Biophys. Acta 851, 283–294 [DOI] [PubMed] [Google Scholar]
- 42.Schouppe C., Vaillier J., Venard R., Rigoulet M., Velours J., Haraux F. (1999) J. Bioenerg. Biomembr. 31, 105–117 [DOI] [PubMed] [Google Scholar]
- 43.Hyndman D. J., Milgrom Y. M., Bramhall E. A., Cross R. L. (1994) J. Biol. Chem. 269, 28871–28877 [PubMed] [Google Scholar]
- 44.Milgrom Y. M., Boyer P. D. (1990) Biochim. Biophys. Acta 1020, 43–48 [DOI] [PubMed] [Google Scholar]
- 45.Bowler M. W., Montgomery M. G., Leslie A. G., Walker J. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8646–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiselyova O. I., Shiryaeva G. N., Efremov R. G., Gordeliy V. I., Yaminsky I. V., Yanyushin M. F., Bueldt G., Yaguzhinsky L. S. (2005) J. Cryst. Growth 275, e1447–1452 [Google Scholar]
- 47.Andrews S. H., Peskova Y. B., Polar M. K., Herlihy V. B., Nakamoto R. K. (2001) Biochemistry 40, 10664–10670 [DOI] [PubMed] [Google Scholar]
- 48.Pogoryelov D., Nikolaev Y., Schlattner U., Pervushin K., Dimroth P., Meier T. (2008) FEBS J. 275, 4850–4862 [DOI] [PubMed] [Google Scholar]
- 49.Rees D. M., Leslie A. G., Walker J. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21597–21601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vonck J., von Nidda T. K., Meier T., Matthey U., Mills D. J., Kühlbrandt W., Dimroth P. (2002) J. Mol. Biol. 321, 307–316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.