Abstract
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene have been identified as an important cause of late-onset, autosomal dominant familial Parkinson disease and contribute to sporadic Parkinson disease. LRRK2 is a large complex protein with multiple functional domains, including a Roc-GTPase, protein kinase, and multiple protein-protein interaction domains. Previous studies have suggested an important role for kinase activity in LRRK2-induced neuronal toxicity and inclusion body formation. Disease-associated mutations in LRRK2 also tend to increase kinase activity. Thus, enhanced kinase activity may therefore underlie LRRK2-linked disease. Similar to the closely related mixed-lineage kinases, LRRK2 can undergo autophosphorylation in vitro. Three putative autophosphorylation sites (Thr-2031, Ser-2032, and Thr-2035) have been identified within the activation segment of the LRRK2 kinase domain based on sequence homology to mixed-lineage kinases. Phosphorylation at one or more of these sites is critical for the kinase activity of LRRK2. Sensitive phopho-specific antibodies to each of these three sites have been developed and validated by ELISA, dot-blot, and Western blot analysis. Using these antibodies, we have found that all three putative sites are phosphorylated in LRRK2, and Ser-2032 and Thr-2035 are the two important sites that regulate LRRK2 kinase activity.
Keywords: Antibodies, Cell Death, Immunochemistry, Neurological Diseases, Phosphotyrosine, LRRK2, Leucine-rich Repeat Kinase 2, Parkinson Disease, Cytotoxicity, Phospho-specific Antibodies
Introduction
Leucine-rich repeat kinase 2 (LRRK2)3 is a large multidomain-containing protein that plays a prominent role in Parkinson disease (PD) (1, 2). Mutations in LRRK2 account for 1–40% of all PD depending upon the ethnic background (3–8). Patients with penetrated mutations have clinical and neuropathologic features that are indistinguishable from idiopathic PD (9, 10). LRRK2 contains repeat sequences beginning at the N terminus as well as leucine-rich repeats. In addition, it contains a kinase effector domain and a guanosine triphosphatase (GTPase) domain that is spanned by a C-terminal Ras domain. There is a WD40 domain at its C terminus that is thought to mediate protein-protein interactions (7). Localization studies indicate that LRKK2 is localized primarily to cytosolic membranous structures (11, 12). The exact physiologic function of LRRK2 is not known, but recent studies suggest that it may regulate neuronal polarity (13, 14).
LRRK2 is regulated by the ubiquitin proteasome system where its levels are controlled by the C terminus of HSP70-interacting proteins (CHIP). CHIP regulates the levels of LRRK2 through an HSP90 chaperone-containing complex. LRRK2 toxicity can be reduced by increasing CHIP E3 ligase activity and reducing HSP90 chaperone activity (15). Familial mutations in LRRK2 are thought to increase LRRK2 kinase activity, although there may be some PD-associated mutations that have minimal detectable effects on kinase activity (16–18). LRRK2 overexpression leads to cytotoxicity that is kinase- and GTP binding-dependent (16, 19, 20). Because LRRK2 toxicity requires kinase activity, there is tremendous interest in understanding the molecular underpinnings of LRRK2 kinase activity as well as developing methods to monitor LRRK2 kinase activity and function. LRRK2 exists as a dimer, and it is autophosphorylated through intramolecular interactions (20, 21). Structurally. LRRK2 is most closely related to mixed lineage kinases. LRRK2 contains a conserved activation segment that is classically defined as a region between two conserved tripeptide motifs (DF/YG) and APE. Within this activation segment, LRRK2 contains three potential autophosphorylation sites, including amino acids Thr-2031, Ser-2032, and Thr-2035. Ser-2032 and Thr-2035 regulate LRRK2 autophosphorylation activity, but do not seem to be involved in LRRK2 dimerization (22). Mass spectrometry identified other amino acids in LRRK2 that are autophosphorylated, and these studies have not shown that these potential phosphorylation sites within the activation loop are indeed phosphorylated where they can regulate the activity of LRRK2 (16). To begin to address this question we developed phophospecific antibodies to Thr-2031, Ser-2032, and Thr-2035 and now show that all of the three possible sites, Thr-2031, Ser-2032, and Thr-2035, are autophosphorylated by LRRK2 and that Ser-2032 and Thr-2035 regulate LRRK2 kinase activity.
EXPERIMENTAL PROCEDURES
Construct Generation
Functional mutations were introduced into LRRK2 cDNA by PCR-mediated, site-directed mutagenesis, using the QuikChange kit (Stratagene), followed by the sequencing to confirm their correct incorporation. Plasmid DNA was prepared using a Maxi-prep column (Qiagen), and concentration of plasmid DNA was normalized by transient transfection assays with subsequent Western blot analysis.
Antibody Generation and Purification
The peptide (sequence see Fig. 1B) was synthesized and used to immunize rabbits. This peptide was conjugated to SulfoLink coupling gel (Pierce) for affinity purification. Briefly, antiserum was incubated with a peptide-conjugated SulfoLink column for 48 h at 4 °C. The column was washed in TBS buffer (50 mm Tris, pH 7.4, 400 mm NaCl) three times with 1 column volume each time. The antibody was eluted with 4 m MgCl2 10 times with 1 ml each time. These 10 1-ml aliquots were collected and subjected to A280 measure. The two fractions with the highest A280 values were pooled together and dialyzed immediately against 50 mm Tris, 50 mm NaCl, and 0.01% sodium azide solution overnight at 4 °C. The dialysis buffer was changed to 50 mm PBS, 20% sucrose, 0.01% NaN3 and dialyzed for another 24 h at 4 °C. The final antibodies were frozen at −80 °C in 100-μl aliquots.
FIGURE 1.
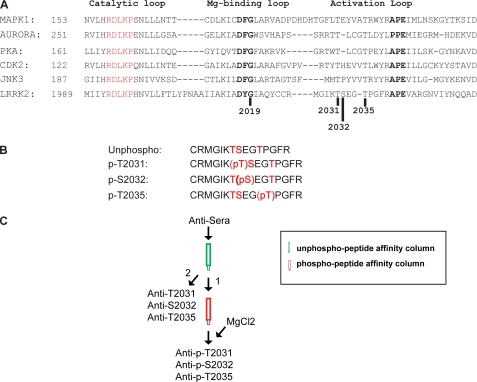
Overview of the three possible phosphorylation sites within LRRK2 kinase activation loop and the peptides used for the production of phospho-LRRK2 specific antibodies. A, sequence alignment of the activation loop of LRRK2 and other homologous MAP kinases. The activation loop is defined as the region between two conserved motifs, DF/YG and APE. Three possible sites for the autophosphorylation of LRRK2, Thr-2031, Ser-2032, and Thr-2035, are denoted by a dark bar. B, peptides used in the production of LRRK2 phospho-specific antibodies. Phosphopeptides pT2031, pS2032, and pT2035 were used for the generation and purification of the antibodies. Nonphosphorylated peptide was used for the purification and characterization of the antibodies. C, schematic diagram depicting the purification procedure of LRRK2 phospho-specific antibodies.
Autophosphorylation Assay
HEK-293FT (Invitrogen) cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). For each experiment, cells were plated in 10-cm dishes, and transfection was accomplished using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. Cells were harvested 48 h after transfection and incubated into lysis buffer containing 0.5% Nonidet P-40, 150 mm NaCl, 50 mm HEPES, pH 7.4, 5 mm EGTA, 1× Complete protease inhibitors (Roche), and 1× phosphatase inhibitor cocktails 1 and 2 (Sigma), for 1 h at 4 °C with rotation. Insoluble material was removed with a 153,000 rpm centrifugation for 15 min. Magnetic Dynabeads (50 ml; Invitrogen) were preincubated with anti-myc antibody (clone 9E10; Roche) for 3 h at 4 °C with rotation and added to the precleared lysate. The bead-lysate mixture was allowed to rotate overnight at 4 °C, and the Dynabeads were subsequently washed five times with lysis buffer supplemented with 500 mm NaCl. Dynabeads were suspended in 50 ml of kinase buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm EGTA, and 20 mm β-glycerol phosphate), and kinase reactions were initiated with the addition of 10 mm ATP (0.5 mCi of [γ-32P]ATP (PerkinElmer Life Sciences)) and 20 mm MgCl2. Reactions were incubated at 30 °C for 30 min with shaking, and the supernatants were removed. Beads were suspended in 2× Laemmli buffer and heated for 10 min at 70 °C. Kinase reactions were resolved on SDS-polyacrylamide gels and transferred onto nitrocellulose membrane. The autophosphorylation signal was estimated using a Storm PhosphorImager and ImageQuant 6.0. Input levels of protein present on the nitrocellulose membrane were determined by Western blotting.
Primary Cortical Neuronal Cultures and Cell Viability
Primary cortical neuron cultures were prepared from gestational day 15–16 fetal mice. Cortices were dissected and the cells dissociated by trituration in modified Eagle's medium, 20% horse serum, 25 mm glucose, and 2 mm l-glutamine, following a 15-min digestion in TrypLE (Invitrogen). The cells were plated on 24-well plates coated with poly-l-ornithine and were maintained in modified Eagle's medium, 10% horse serum, 25 mm glucose, and 2 mm l-glutamine in a 7% CO2 humidified 37 °C incubator. The glial cell growth was inhibited by addition of 5-fluoro-20-deoxyuridine (30 mm; Sigma) to the culture medium on in vitro day 4. For assessment of LRRK2 toxicity, LRRK2 and EGFP constructs were combined in a molar ratio of 10:1, respectively, and mix with Lipofectamine 2000 (Invitrogen) in serum-free medium. DNA·Lipofectamine complexes were added to in vitro day 10 cultures. After a 4-h incubation, the medium was replaced with original neuron culture medium. LRRK2-transfected neurons were subsequently fixed with 4% paraformaldehyde after 48-h transfection. TUNEL staining was performed using the In Situ Cell Death Detection kit (Roche Applied Science) according to the manufacturer's instructions. Fluorescent images were collected on a Zeiss Automatic stage with Axiovision 6.0. Percent viability was calculated by normalizing the surviving green cells with total EGFP-positive cells in each culture. Most of the green transfected cells are LRRK2-transfected neurons as shown previously (23).
RESULTS
Generation of Phospho-specific Antibodies
LRRK2 contains three potential amino acids, Thr-2031, Ser-2032, and Thr-2035, within its activation loop that could be involved in autophosphorylation and regulation of LRRK2 (Fig. 1A). Accordingly, we made phospho-mimetic peptides (2025–2039) to Thr-2031, Ser-2032, and Thr-2035 and raised rabbit polyclonal antibodies to the three phosphopeptides (Fig. 1B). These antibodies were subjected to an affinity purification protocol, which first involved incubating the antisera with a nonphosphopeptide affinity column to remove nonphosphopeptide-specific antibodies followed by incubation with a phospho-specific peptide column. Both columns were eluted with 4 m MgCl2, and antibodies from the nonphosphopeptide column were designated anti-T2031, anti-S2032, and anti-T2035, and antibodies from the phosphopeptide column were designated anti-p-T2031, anti-p-S2032, and anti-p-T2035 (Fig. 1C).
Characterization of Novel Phospho-LRRK2 Antibodies
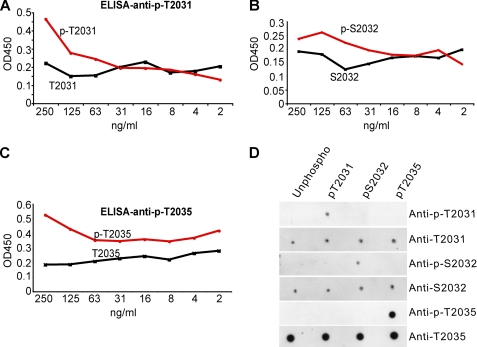
ELISA data using serial dilution of synthetic phospho- and nonphosphorylated peptide amino acids 2025–2039 which contain Thr-2031, Ser-2032, and Thr-2035 reveal that anti-p-T2031, anti-p-S2032, and anti-p-T2035 react strongly with their corresponding phosphorylated peptide, but not with the nonphosphorylated peptide (Fig. 2, A–C). To test the specificity of each phospho-antibody, a dot-blot analysis was performed where 1 ng of each peptide was blotted to a nitrocellulose membrane followed by immunodetection with anti-p-T2031, anti-p-S2032, and anti-p-T2035, and anti-T2031, anti-S2032, and anti-T2035 (Fig. 2D). Each phospho-specific antibody specifically recognizes its corresponding amino acid with little or no overlap with other phosphopeptides (Fig. 2D). Antibody anti-p-T2031 and anti-p-S2032 show slight cross-reactivity to other phosphopeptides at higher concentration (10 ng of peptide) (data not shown).
FIGURE 2.
Characterization of the purified LRRK2 phospho-specific antibodies using specific peptides. A, ELISA using antibody anti-pT2031 and phospho-LRRK2 peptide (pT2031) and nonphosphorylated peptide control. B, ELISA using antibody anti-pS2032 and phospho-LRRK2 peptide (pS2032) and nonphosphorylated peptide control. C, ELISA using antibody anti-pT2035 and phospho-LRRK2 peptide (pT2035) and nonphosphorylated peptide control. D, dot blot using specific phospho- and its corresponding nonphosphoantibody.
To determine whether these phosphoantibodies recognize full-length LRRK2, HEK293 cells were transfected with myc-tagged LRRK2 (Fig. 3). Anti-p-T2031 recognizes wild type LRRK2, and there is minimal reduction in immunoreactivity with the phospho-deficient T2031A mutant, whereas there is increased immunoreactivity against the phospho-mimetic mutants T2031D or T2031E (Fig. 3A). Anti-p-S2032 recognizes wild type LRRK2, and there is a modest reduction in immunoreactivity with the phospho-deficient S2032A mutant, whereas there is increased immunoreactivity against the phospho-mimetic mutants S2032D or S2032E (Fig. 3B). Anti-p-T2035 recognizes wild type LRRK2, and there is a marked reduction in immunoreactivity with the phospho-deficient T2035A mutant, whereas there is increased immunoreactivity against the phospho-mimetic mutants T2035D and T2035E (Fig. 3C).
FIGURE 3.
Characterization of the purified LRRK2 phospho-specific antibodies using overexpressed LRRK2 protein. Immunoprecipitated LRRK2 wild type (WT) protein and various phosphorylation mutant proteins, derived from transfected HEK-293 cells, were analyzed by SDS-PAGE and Western blotting using specific phosphoantibody and anti-myc antibody. Phosphorylation-deficient mutant proteins were T2031A (A), S2032A (B), and T2035A (C); phospho-mimic mutant proteins were T2031D and T2031E (A), S2032D and S2032E (B), and T2035D and T2035E (C).
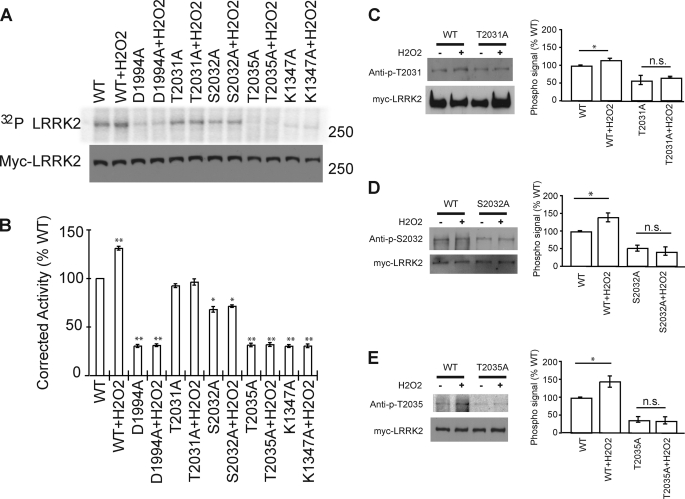
Autophosphorylation Activity of LRRK2 Is Reduced by Mutations at Ser-2032 and Thr-2035
We evaluated the autophosphorylation activity of LRRK2 in the setting of mutations of the potential phosphorylation sites within the activation loop of LRRK2 (Fig. 4). Mutating all three potential phosphoamino acids to alanine (T2031A, S2032A, T2035A) reduces LRRK2 autophosphorylation by 60% (Fig. 4). As previously reported, the disease-causing mutation G2019S increases LRRK2 autophosphorylation by >30% (Fig. 4). Mutating Thr-2031 to the phospho-deficient T2031A mutant or the phospho-mimetic mutants T2031D and T2031E had no significant effect on LRRK2 autophosphoryation (Fig. 4, A and B). Mutating Ser-2032 to the phospho-deficient S2032A reduces LRRK2 autophosphorylation, whereas there are minimal effects on LRRK2 autophosphorylation with phospho-mimetic mutants S2032D or S2032E (Fig. 4, A and B). Mutating Thr-2035 to the phospho-deficient T2035A mutant or the phospho-mimetic mutant T2035D or T2035E reduces LRRK2 autophosphorylation to a level equivalent to the reduction in kinase activity in LRRK2 where the three potential phosphoamino acids are mutated to alanine (T2031A, S2032A, T2035A) (Fig. 4, A and B). Double mutants T2035A/T2031A and T2035A/S2032A showed a decrease of autophosphorylation similar to that of the T2035A single mutant, and T2031A/S2032A had a lower level of autophosphorylation, compared with the T2031A and S2032A single mutants (Fig. 4, C and D).
FIGURE 4.
Regulation of LRRK2 autophosphorylation activity by the three possible phosphorylation sites (Thr-2031, Ser-2032, and Thr-2035). A and C, immunoblot and autoradiogram of autophosphorylated LRRK2, as resolved by SDS-PAGE. LRRK2 protein levels were determined by Western blot analysis, using anti-myc antibody. B and D, normalization of incorporated 32P compared with LRRK2 protein content. Data represent three independent experiments, in arbitrary units, where wild type (WT)-LRRK2 kinase activity is defined as 100%. Control bar represents mean ± S.E. *, p < 0.05; **, p < 0.01 compared with wild type LRRK2 kinase activity, assessed by a two-tailed one-sample Student's t test.
Hydrogen Peroxide (H2O2) Activates LRRK2 Kinase
H2O2 was shown to potentiate LRRK2 toxicity (16), thus we wondered whether H2O2 affected LRRK2 kinase activity. H2O2 activates LRRK2 kinase by ∼30% (Fig. 5, A and B) to a level equivalent to the disease-causing mutation G2019S (see Fig. 4). H2O2 fails to activate a kinase-dead version of LRRK2, which contains a D1944A mutation in the ATP binding pocket as well as the GTP binding-deficient mutant K1347A (Fig. 5, A and B). Autophosphorylation of the phospho-deficient T2031A, S2032A, and T2035A LRRRK2 mutants is not increased by H2O2 (Fig. 5, A and B).
FIGURE 5.
Hydrogen peroxide (H2O2) activates LRRK2 kinase through the phosphorylation of all three phosphorylation sites (Thr-2031, Ser-2032, and Thr-2035). A, immunoblot and autoradiogram of autophosphorylated LRRK2, as resolved by SDS-PAGE. LRRK2 protein levels were determined by Western blot analysis, using anti-myc antibody. B, normalization of incorporated 32P compared with LRRK2 protein content. Data represent three independent experiments, in arbitrary units, where wild type (WT)-LRRK2 kinase activity is defined as 100%. Control bar represent mean ± S.E. *, p < 0.05; **, p < 0.01 compared with wild type LRRK2 kinase activity, assessed by a two-tailed one-sample Student's t test. C–E, Western blot using phospho-specific antibodies to detect the increasing phosphosignal of LRRK2 under the peroxide treatment. HEK-293 cells transfected with wild type or phospho-deficient mutant LRRK2 were treated with peroxide and immunoprecipitated by anti-myc antibody. Data represent five independent experiments, in arbitrary units, where wild type LRRK2 signal is defined as 100%. Control bar represent mean ± S.E. *, p < 0.05 compared with phosphorylation level of WT-LRRK2, assessed by a two-tailed one-sample Student's t test. n.s is nonsignificant, comparing the treated and untreated LRRK2 mutant samples, assessed by two tailed unpaired Students' t test.
To determine whether the three putative autophosphorylation sites in LRRK2 are phosphorylated, LRRK2 phosphorylation was monitored in response to H2O2 with the phospho-specific antibodies (Fig. 5, C–E). All three amino acids (Thr-2031, Ser-2032, and Thr-2035) are slightly phosphorylated in response to H2O2 (Fig. 5, C–E).
All Three Putative Sites Are Autophosphorylated
LRRK2 autophosphorylation can be inhibited by staurosporine, a generic kinase inhibitor, at a low concentration (18, 24). To determine whether any of the three putative sites is phosphorylated in the autophosphorylation reaction, overexpressed wild type LRRK2 was immunoprecipitated and subjected to the standard LRRK2 kinase assay, with or without prior treatment of staurosporine (100 nm). The inhibition of LRRK2 phosphorylation can be monitored by all three specific phospho-LRRK2 antibodies (Fig. 6A), suggesting that all three sites are phosphorylated in LRRK2.
FIGURE 6.
Autophosphorylation of LRRK2 happens on all three putative sites (Thr-2031, Ser-2032 and Thr-2035. A, Western blotting using phospho-specific antibodies to detect the decreasing phosphosignal of LRRK2 under the treatment of kinase inhibitor staurosporine at 100 nm. Overexpressed human wild type LRRK2 protein was immunoprecipitated by anti-myc antibody and subjected to the standard LRRK2 kinase assay, with or without prior treatment of staurosporine (100 nm). B, Western blotting using phospho-specific antibodies to detect the increasing phosphosignal of LRRK2-G2019S mutant. HEK-293 cells transfected with wild type LRRK2 or G2019S mutant were treated with peroxide and immunoprecipitated (IP) by anti-myc antibody. WT+, wild type LRRK2 treated with H2O2; GS, G2019S; GS+, G2019S mutant LRRK2 treated with H2O2; n.s., not significant. Data represent three independent experiments, in arbitrary units, where WT-LRRK2 signal is defined as 100%. Control bar represents mean ± S.E. *, p < 0.05; **, p < 0.01, compared with untreated wild type LRRK2, assessed by a two-tailed one-sample Student's t test.
Similarly, the increasing signal of LRRK2 phosphorylation also can be monitored by these three specific phospho-LRRK2 antibodies (Fig. 6B). Previous studies have shown that LRRK2 mutant G2019S enhanced its kinase activity by 2- or 3-fold (22, 25). The higher phosphorylation signals can be detected in our Western blots using any of these three LRRK2 phosphoantibodies (Fig. 6B), which further proved that all three sites are phosphorylated in LRRK2. H2O2 treatment did not enhance the phosphorylation level of G2019S mutant (Fig. 6B).
LRRK2 Toxicity Is Partially Dependent on LRRK2 Kinase Activity
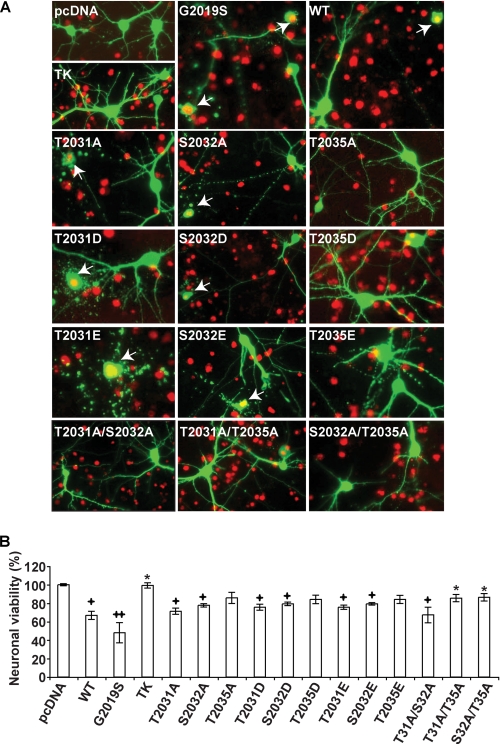
Previously, we and others showed that LRRK2 toxicity is dependent on LRRK2 kinase activity (16, 19). To determine whether mutations of the phosphorylation sites within the activation loop of LRRK2 influence LRRK2 toxicity, cell viability was monitored following overexpression of LRRK2 and the various LRRK2 phosphorylation mutants in mouse cortical neurons (Fig. 7). Toxicity was monitored using TUNEL method. Overexpression of wild type LRRK2 leads to ∼30% cell death, which is slightly less than the cell death induced by G2019S LRRK2 (Fig. 7). Mutating all three phosphoamino acids to alanine (T2031A, S2032A, T2035A) completely attenuates the cytoxicity. The phospho-deficient T2031A mutant or the phospho-mimetic mutants T2031D and T2031E had no effect on LRRK2 cytotoxicity compared with wild type LRRK2, similar to their lack of effect on LRRK2 autophosphorylation (Fig. 7). The phospho-deficient S2032A mutant or the phospho-mimetic mutants S2032D and S2032E also had no effect on LRRK2 cytotoxicity compared with wild type LRRK2, which is in contrast to the effect on LRRK2 autophosphorylation (Fig. 7). The phospho-deficient T2035A mutant or the phospho-mimetic mutants T2035D and T2035E slightly rescued LRRK2 cytotoxicity compared with wild type LRRK2, but still retained modest toxicity compared with control and TK-LRRK2 (Fig. 7).
FIGURE 7.
LRRK2-induced neuronal toxicity correlated with its kinase activity. A, mouse cortical neurons stained by TUNEL and an anti-GFP antibody after 48-h transfection (13 total days in vitro) with the indicated LRRK2 (wild type and mutant) constructs in a 10:1 molar ratio with EGFP. White arrows indicate neurons counted as nonviable EGFP- and TUNEL-positive neurons. B, quantitative data analysis of neuronal viability of LRRK2-transfected mouse cortical neurons. Data are representative of three independent experiments. Error bars represent mean ± S.E. +, p < 0.05; ++, p < 0.01 compared with control (EGFP only); *, p < 0.05 compared with wild type LRRK2-transfected neurons, assessed by one-way nonparametric analysis of variance with Dunnett's Multiple Comparison test.
The toxicities of double phospho-deficient mutants were also measured to investigate further the relationship between the phosphorylation of LRRK2 and its pathology. Double mutants, which contain T2035A and either one of the other two deficient mutants (T2031A or S2032A), can rescue the toxicity of LRRK2 to a level similar to TK-LRRK2 (Fig. 7). Double mutant T2031A/S2032A also reduced the toxicity of LRRK2 but to a modest level.
DISCUSSION
In the present study, we investigated LRRK2 autophosphorylation by raising phospho-specific antibodies to the three potential phosphorylation sites within the activation segment of LRRK2. All kinases contain an activation segment flanked by two conserved tripeptide motifs (DF/YG) and APE (26). Within this activation segment it has been suggested that Thr-2031, Ser-2032, and Thr-2035 are potentially autophosphorylated by LRRK2. It is essential to identify potential phosphorylation sites in LRRK2 and ultimately to study the role of autophosphorylation in LRRK2 activity and pathogenesis. Accordingly, we raised polyclonal peptide antibodies recognizing phosphorylated Thr-2031, Ser-2032, and Thr-2035, respectively. ELISA, dot-blot, and immunoblot analysis demonstrate that anti-p-T2031, anti-p-S2032, and anti-p-T2035 are able to detect phosphorylated Thr-2031, Ser-2032, and Thr-2035, respectively.
Most studies to date have investigated LRRK2 phosphorylation by either monitoring constitutive autophosphorylation or phosphorylation of pseudosubstrates (11, 16, 19). No potential activators of LRRK2 kinase have been reported. Previously, we showed that LRRK2 toxicity is enhanced by H2O2, and here we show that H2O2 increases LRRK2 autophosphorylation to a level equivalent to the enhanced phosphorylation observed with the G2019S mutant (16). Thus, H2O2 treatment provides an opportunity to investigate the consequences of perturbation of the phosphorylation state within the activation loop of LRRK2 in response to LRRK2 activation. A number of protein kinases contain a conserved Thr-2035 within their activation segment, where it is thought to serve as an activator of kinase activity. Mutations of Thr-2035 to T2035A abolish LRRK2 autophosphorylation, as previously described, and H2O2 fails to activate LRRK2 (22). A double mutant containing T2035A together with T2031A or S2032A showed similar reduction of LRRK2 autophosphorylation. Substitutions with negatively charged amino acids to act as pseudo-phospho-mimetics T2035D and T2035E also abolished LRRK2 autophosphorylation and activation by H2O2. The anti-p-T2035 antibody shows that Thr-2035 is indeed phosphorylated in LRRK2 protein. Western blotting using this antibody showed decreasing phosphorylation signals of wild type LRRK2 when treated with staurosporine and increasing phosphorylation signals of LRRK2-G2019S mutant compared with its wild type partner, thus suggesting that phosphorylation of this residue is critical for LRRK2 kinase activity, although we cannot exclude the possibility that this residue provides important structural confirmation to LRRK2, as observed previously (22).
Substitution of Ser-2032 with S2032A partially reduced constitutive and H2O2 stimulated LRRK2 autophosphorylation. Substitutions with negatively charged amino acids to act as pseudo-phospho-mimetics S2032D or S2032E had no effect on constitutive LRRK2 autophosphorylation, suggesting that this site is also autophosphorylated by LRRK2. Consistent with this notion, anti-p-S2032 antibody indicates that Ser-2032 is autophosphorylated in LRRK2, with higher signals when activated by H2O2 or mutant G2019S, and lower signals when inhibited by staurosporine. Although mutations of Thr-2031 to T2031A, T2031D, or T2031E had no effect on LRRK2 autophosphorylation, anti-p-T2031 indicates that this site is also autophosphorylated in LRRK2. Thus, all three amino acids within the activation loop of LRRK2 are autophosphorylated, with Thr-2035 and Ser-2032 playing a role in the activation state of LRRK2.
LRRK2 toxicity requires kinase activity, and we investigated the role of the phosphorylated amino acids within the activation loop on LRRK2 toxicity in cortical neurons (16). As previously reported, wild type LRRK2 is toxic. Substitution of all three amino acids with alanine (TK-LRRK2) completely attenuates wild type LRRK2 toxicity. The most parsimonious explanation for the reduction in LRRK2 toxicity by TK-LRRK2 is a reduction in kinase activity. However, we cannot exclude the possibility that TK-LRRK2 has functional consequences on the structural confirmation of LRRK2 that impair its kinase activity. Surprisingly, substitution of Thr-2035 to T2035A only partially rescued toxicity, which contrasts with the dramatic effects on autophosphorylation. Substitution of Thr-2031 and Ser-2032 with alanine had essentially no effect on wild type LRRK2 toxicity. Substitutions with negatively charged amino acids to act as pseudo-phospho-mimetics for Thr-2031, Ser-2032, and Thr-2035 also had no effect. Double mutants T2031A/T2035A and S2032A/T2035A can rescue LRRK2-induced toxicity, similar to TK-LRRK2, whereas T2031A/S2032A still retains modest cytotoxicity compared with the pcDNA control. These data suggest that LRRK2 toxicity is not completely dependent on the phosphorylation state of LRRK2 and that phosphorylation of the individual amino acids in the activation loop has as yet unexplained functions and roles in LRRK2 toxicity.
In conclusion, this study reports the development of LRRK2 phosphopeptide antibodies to the phosphopeptides within the activation loop of LRRK2 and shows for the first time that all three amino acids are autophosphorylated. These antibodies can be used to explore LRRK2 activity and the role of autophosphorylation in the pathogenesis of PD.
This work was supported, in whole or in part, by National Institutes of Health Grant NS38377.
- LRRK2
- leucine-rich repeat kinase 2
- CHIP
- C terminus of HSP70-interacting proteins
- EGFP
- enhanced green fluorescent protein
- PD
- Parkinson disease
- TK
- triple kinase mutant of LRRK2 (T2031A, S2032A and T2035A).
REFERENCES
- 1.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R. J., Calne D. B., Stoessl A. J., Pfeiffer R. F., Patenge N., Carbajal I. C., Vieregge P., Asmus F., Müller-Myhsok B., Dickson D. W., Meitinger T., Strom T. M., Wszolek Z. K., Gasser T. (2004) Neuron 44, 601–607 [DOI] [PubMed] [Google Scholar]
- 2.Paisán-Ruíz C., Jain S., Evans E. W., Gilks W. P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A. M., Khan N., Johnson J., Martinez J. R., Nicholl D., Carrera I. M., Pena A. S., de Silva R., Lees A., Martí-Massó J. F., Pérez-Tur J., Wood N. W., Singleton A. B. (2004) Neuron 44, 595–600 [DOI] [PubMed] [Google Scholar]
- 3.Gorostidi A., Ruiz-Martínez J., López de Munain A., Alzualde A., Massó J. F. (2009) Neurogenetics 10, 157–159 [DOI] [PubMed] [Google Scholar]
- 4.Möller J. C., Rissling I., Mylius V., Höft C., Eggert K. M., Oertel W. H. (2008) Eur. J. Neurol. 15, 743–745 [DOI] [PubMed] [Google Scholar]
- 5.Mata I. F., Hutter C. M., González-Fernández M. C., de Pancorbo M. M., Lezcano E., Huerta C., Blazquez M., Ribacoba R., Guisasola L. M., Salvador C., Gómez-Esteban J. C., Zarranz J. J., Infante J., Jankovic J., Deng H., Edwards K. L., Alvarez V., Zabetian C. P. (2009) Neurogenetics 10, 347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Healy D. G., Falchi M., O'Sullivan S. S., Bonifati V., Durr A., Bressman S., Brice A., Aasly J., Zabetian C. P., Goldwurm S., Ferreira J. J., Tolosa E., Kay D. M., Klein C., Williams D. R., Marras C., Lang A. E., Wszolek Z. K., Berciano J., Schapira A. H., Lynch T., Bhatia K. P., Gasser T., Lees A. J., Wood N. W. (2008) Lancet Neurol. 7, 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandhi P. N., Chen S. G., Wilson-Delfosse A. L. (2009) J. Neurosci. Res. 87, 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Floris G., Cannas A., Solla P., Murru M. R., Tranquilli S., Corongiu D., Rolesu M., Cuccu S., Sardu C., Marrosu F., Marrosu M. G. (2009) Parkinsonism Relat. Disord. 15, 277–280 [DOI] [PubMed] [Google Scholar]
- 9.Haugarvoll K., Rademakers R., Kachergus J. M., Nuytemans K., Ross O. A., Gibson J. M., Tan E. K., Gaig C., Tolosa E., Goldwurm S., Guidi M., Riboldazzi G., Brown L., Walter U., Benecke R., Berg D., Gasser T., Theuns J., Pals P., Cras P., De Deyn P. P., Engelborghs S., Pickut B., Uitti R. J., Foroud T., Nichols W. C., Hagenah J., Klein C., Samii A., Zabetian C. P., Bonifati V., Van Broeckhoven C., Farrer M. J., Wszolek Z. K. (2008) Neurology 70, 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aasly J. O., Toft M., Fernandez-Mata I., Kachergus J., Hulihan M., White L. R., Farrer M. (2005) Ann. Neurol. 57, 762–765 [DOI] [PubMed] [Google Scholar]
- 11.West A. B., Moore D. J., Biskup S., Bugayenko A., Smith W. W., Ross C. A., Dawson V. L., Dawson T. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16842–16847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biskup S., Moore D. J., Celsi F., Higashi S., West A. B., Andrabi S. A., Kurkinen K., Yu S. W., Savitt J. M., Waldvogel H. J., Faull R. L., Emson P. C., Torp R., Ottersen O. P., Dawson T. M., Dawson V. L. (2006) Ann. Neurol. 60, 557–569 [DOI] [PubMed] [Google Scholar]
- 13.Shin N., Jeong H., Kwon J., Heo H. Y., Kwon J. J., Yun H. J., Kim C. H., Han B. S., Tong Y., Shen J., Hatano T., Hattori N., Kim K. S., Chang S., Seol W. (2008) Exp. Cell Res. 314, 2055–2065 [DOI] [PubMed] [Google Scholar]
- 14.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. (2006) Neuron 52, 587–593 [DOI] [PubMed] [Google Scholar]
- 15.Ko H. S., Bailey R., Smith W. W., Liu Z., Shin J. H., Lee Y. I., Zhang Y. J., Jiang H., Ross C. A., Moore D. J., Patterson C., Petrucelli L., Dawson T. M., Dawson V. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2897–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West A. B., Moore D. J., Choi C., Andrabi S. A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K. L., Dawson V. L., Dawson T. M. (2007) Hum. Mol. Genet. 16, 223–232 [DOI] [PubMed] [Google Scholar]
- 17.Gloeckner C. J., Kinkl N., Schumacher A., Braun R. J., O'Neill E., Meitinger T., Kolch W., Prokisch H., Ueffing M. (2006) Hum. Mol. Genet. 15, 223–232 [DOI] [PubMed] [Google Scholar]
- 18.Anand V. S., Reichling L. J., Lipinski K., Stochaj W., Duan W., Kelleher K., Pungaliya P., Brown E. L., Reinhart P. H., Somberg R., Hirst W. D., Riddle S. M., Braithwaite S. P. (2009) FEBS J. 276, 466–478 [DOI] [PubMed] [Google Scholar]
- 19.Smith W. W., Pei Z., Jiang H., Dawson V. L., Dawson T. M., Ross C. A. (2006) Nat. Neurosci. 9, 1231–1233 [DOI] [PubMed] [Google Scholar]
- 20.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M. P., Beilina A., Blackinton J., Thomas K. J., Ahmad R., Miller D. W., Kesavapany S., Singleton A., Lees A., Harvey R. J., Harvey K., Cookson M. R. (2006) Neurobiol. Dis. 23, 329–341 [DOI] [PubMed] [Google Scholar]
- 21.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J. M., Daniëls V., Lewis P., Jain S., Ding J., Syed A., Thomas K. J., Baekelandt V., Cookson M. R. (2008) J. Biol. Chem. 283, 16906–16914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luzón-Toro B., Rubio de la Torre E., Delgado A., Pérez-Tur J., Hilfiker S. (2007) Hum. Mol. Genet. 16, 2031–2039 [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y., Coombes C. E., Kilaru A., Li X., Gitler A. D., Bowers W. J., Dawson V. L., Dawson T. M., Moore D. J.PLoS Genet. 6, e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen S., Webber P. J., West A. B. (2009) J. Biol. Chem. 284, 36346–36356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaleel M., Nichols R. J., Deak M., Campbell D. G., Gillardon F., Knebel A., Alessi D. R. (2007) Biochem. J. 405, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolen B., Taylor S., Ghosh G. (2004) Mol. Cell 15, 661–675 [DOI] [PubMed] [Google Scholar]