Abstract
Talin is an adaptor protein that couples integrins to F-actin. Structural studies show that the N-terminal talin head contains an atypical FERM domain, whereas the N- and C-terminal parts of the talin rod include a series of α-helical bundles. However, determining the structure of the central part of the rod has proved problematic. Residues 1359–1659 are homologous to the MESDc1 gene product, and we therefore expressed this region of talin in Escherichia coli. The crystal structure shows a unique fold comprised of a 5- and 4-helix bundle. The 5-helix bundle is composed of nonsequential helices due to insertion of the 4-helix bundle into the loop at the C terminus of helix α3. The linker connecting the bundles forms a two-stranded anti-parallel β-sheet likely limiting the relative movement of the two bundles. Because the 5-helix bundle contains the N and C termini of this module, we propose that it is linked by short loops to adjacent bundles, whereas the 4-helix bundle protrudes from the rod. This suggests the 4-helix bundle has a unique role, and its pI (7.8) is higher than other rod domains. Both helical bundles contain vinculin-binding sites but that in the isolated 5-helix bundle is cryptic, whereas that in the isolated 4-helix bundle is constitutively active. In contrast, both bundles are required for actin binding. Finally, we show that the MESDc1 protein, which is predicted to have a similar fold, is a novel actin-binding protein.
Keywords: Actin, Cell Adhesion, Crystal Structure, Crystallography, NMR, MESDc1, Integrins, Synemin, Talin, Vinculin
Introduction
Talin is a large (2541 amino acids) dimeric cytoskeletal protein that provides a direct link between the integrin family of cell adhesion molecules and the actin cytoskeleton (1–3), and it has a pivotal role in integrin activation (4, 5) and clustering (6). Talin depletion blocks cell spreading and the assembly of cell-matrix junctions (focal adhesions; FA)2 in vitro (7–9), whereas knock-out of the gene encoding the talin1 isoform is embryonic lethal in mice at gastrulation (10). Use of a conditional talin1 allele confirms that talin1 is essential to integrin activation in platelets (11, 12) and the stability of the membrane cytoskeletal interface in megakaryocytes (13), and deletion of both the talin1 and talin2 isoforms in skeletal muscle leads to disruption of the myotendinous junction and inhibition of myoblast fusion (14, 15).
The N-terminal talin head (47 kDa) (Fig. 1) contains a FERM domain composed of F1, F2, and F3 domains, although it is atypical in that F1 contains a large unstructured insert and is preceded by a previously unrecognized domain, F0 (16, 17). The F3 domain has a phosphotyrosine-binding domain-like fold and binds to both the membrane proximal NPXY motif in β3-integrin tails and the membrane proximal helix (18–20). This is thought to disrupt the inter-subunit interactions between the α- and β-integrin cytoplasmic tails and also their trans-membrane helices (21) leading to integrin activation, although it is now clear that integrin activation also requires cooperation between talin and the kindlin family of FERM domain proteins (22).
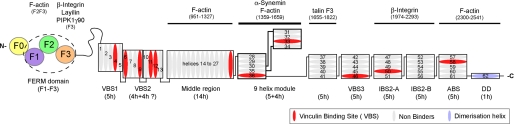
FIGURE 1.
Domain structure and binding partners of talin. Schematic diagram of the talin molecule indicating the regions involved in binding to various ligands. The talin head (residues 1–400) contains a FERM domain (comprising F1, F2, and F3 subdomains) preceded by a domain region referred to as F0 (16). The rod domain contains 62 predicted α-helices (ovals) organized into a series of amphipathic helical bundles. Domain boundaries based on structural determination are indicated by solid lines. Dashed lines indicate boundaries that are tentative. The ∼11 VBS are shown in red. The last α-helix (blue) contains the dimerization domain (DD) (27).
The talin rod (220 kDa) is composed of 62 α-helices organized into a series of amphipathic helical bundles (Fig. 1). It contains a second integrin-binding site (IBS2) (23, 24), at least two actin-binding sites (25), the best characterized of which is at the C terminus (26, 27), and numerous vinculin-binding sites (VBSs) (28). Using limited proteolysis and a large series of recombinant polypeptides, we have begun to identify some of the domains that make up the talin rod and to determine their structures (19, 20, 23, 27, 29–33) and their mode of interaction with integrins, vinculin, and F-actin (2). The rod starts with a 5-helix bundle (residues 482–655) that packs against a 4-helix bundle (residues 656–786) via an extensive hydrophobic interface (32). The C-terminal region of the rod (residues 1655–2482) is composed of five 5-helix bundles coupled by short flexible linkers and ends with a single helix that is responsible for talin dimer formation (27). An intramolecular interaction between the F3 FERM domain and one of the C-terminal rod domains (residues 1655–1822) masks the integrin-binding site in F3 (31, 34) and is thought to contribute to the regulation of talin activity and to the compact form of the molecule seen in electron microscopy (35, 36).
However, the domain organization of the central region of the rod has been difficult to resolve. Interestingly, residues 1359–1659 show significant homology to the gene referred to as mesoderm development candidate 1 (Mesdc1) (37), which is predicted to encode a 364-amino acid protein of unknown function. We therefore expressed this region of talin in Escherichia coli and found that the protein was soluble, stable, and easy to crystallize. We now report the structure of this region of talin and show that it is made up of a 5- and a 4-helix bundle with unique topology. It contains an actin-binding site, two VBSs, and a binding site for the intermediate filament protein α-synemin, which colocalizes with talin in the costameres of skeletal muscle (38). Moreover, we demonstrate that the MESDc1 protein is a novel actin-binding protein.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The regions encoding murine talin residues 1359–1659, 1359–1659 Δ1454–1586, 1458–1584, and 1461–1580 were synthesized by PCR using mouse Talin1 cDNA as template and cloned into the expression vector pET-151/D-TOPO (Invitrogen). Because MESDc1 is encoded by one exon, full-length murine MESDc1 (residues 1–362) was amplified from genomic DNA using PCR and cloned into the expression vector pET-151/D-TOPO. Constructs were expressed in E. coli BL21 Star (DE3), cultured either in LB or, for preparation of 15N-labeled samples for NMR, in minimal media containing 1 g of [15N]ammonium chloride/liter. For selenomethionine incorporation, recombinant His-tagged talin(1359–1659) was expressed in E. coli B834 and cultured in appropriate minimal media. Recombinant His-tagged talin polypeptides were purified by nickel-affinity chromatography following standard procedures. The His tag was removed by cleavage with an enhanced form of tobacco etch virus (TEV) protease (Invitrogen), and the protein was further purified by anion-exchange chromatography.
Recombinant His-tagged chicken vinculin domain 1 (Vd1; residues 1–258) was expressed using a pET-15b expression plasmid and purified as described previously (32). The concentration of the purified proteins was determined using extinction coefficients at 280 nm.
X-ray Crystallography
Crystals of talin residues 1359–1659 were obtained at 6 °C by vapor diffusion equilibration against 18% (w/v) PEG 8000, 100 mm Tris, 180 mm sodium phosphate dibasic at pH 8.5. Protein at 15 mg/ml in 0.2 m NaCl, 2 mm DTT, and 20 mm Tris-HCl, pH 8.0, was mixed with an equal volume of precipitant. The crystals adopted the space group P212121 with 1 molecule per asymmetric unit (see Table 1) and a solvent content of 66.8%.
TABLE 1.
Summary of crystallographic analysis and refinement statistics for talin residues 1359–1659
Rsym = Σ|I − 〈I〉|ΣI, where I is the observed intensity and 〈I〉 is the average intensity of the multiple observations of symmetry-related reflections. R = Σ‖Fo| − |Fc|/|Σ|Fo|; Rfree is calculated for a randomly selected 5% of the reflections; Rfactor is calculated for the remaining 95% of the reflections used in refinement. Values in parentheses represent the outer resolution shell. r.m.s.d. is root mean square deviation.
| Selenomethionine | Native | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | a = 38.1 Å | a = 38.0 Å |
| b = 67.2 Å | b = 66.8 Å | |
| c = 185.6 Å | c = 185.7 Å | |
| α, β, γ = 90° | α, β, γ = 90° | |
| No. of molecules in arbitrary units | 1 | 1 |
| Dataset | Peak | High resolution |
| Wavelength | 0.97905 Å | 0.933 Å |
| Resolution | 20-2.15 Å | 50-2.0 Å |
| Measured reflections | 191,345 | 118,234 |
| Unique reflections | 26,864 | 33121 |
| Completeness (%) | 99.9 (99.9) | 99.7 (98.4) |
| Rsym | 8.0 (33.4) | 3.9 (33.5) |
| I/σI | 15.5 (4.6) | 19.3 (3.7) |
| Refinement statistics | ||
| Resolution range | 46.4 to 2.0 Å | |
| Unique reflections (free) | 31,178 (1641) | |
| Rwork | 21.4% (24.3%) | |
| Rfree | 26.4% (30.0%) | |
| No. of residues/atoms | 308/2455 | |
| No. of solvent molecules | 184 | |
| Average B value | 37.7 Å2 | |
| r.m.s.d. bond length | 0.029 Å2 | |
| r.m.s.d. bond angles | 2.17 Å2 | |
Diffraction data were collected from native crystals at ESRF beamline ID14-2 and from selenomethionine crystals at beamline ID23-1, recorded on ADSC Q315R CCD detectors. Data were processed with XDS (39). Phases were determined from the anomalous data collected at the selenium absorption peak (λ = 0.97905) from selenomethionine crystals. 7 of the 10 possible selenium atoms were located using SHELX (40), and a map was constructed using these preliminary phases at 2.15 Å resolution. An initial atomic model was built using the ARP/wARP helical recognition approach (41), and the model was rebuilt manually with Coot (42) and refined using maximum likelihood refinement in Refmac5 (43). Subsequently, the structure was refined against a new 2.0 Å data set collected from native crystals.
The final model converged to an Rwork of 21.4% for all data between 50.0 and 2.0 Å and an Rfree of 26.4%. The final Ramachandran plot shows 93.5% of residues in favored regions, 4.2% in additional favored regions, and 2.3% in generously allowed regions, as defined by PROCHECK (44). The structure has been submitted to the Protein Data Bank with the accession number 2x0c. Figures were generated with CCP4mg (45).
Vinculin Binding
Analytical gel filtration chromatography was used to determine the ability of recombinant talin polypeptides (50 μm) to bind to the vinculin Vd1 domain (50 μm) using a Superdex-75 (10/300) GL column (Amersham Biosciences). Polypeptides were incubated at various temperatures for 30 min prior to loading onto the column, which was pre-equilibrated and eluted with 20 mm Tris, pH 8.0, 150 mm NaCl, and 2 mm DTT at a flow rate of 0.8 ml/min at room temperature. The fractions were analyzed using SDS-PAGE.
Actin Cosedimentation Assays
G-actin was purified from rabbit skeletal muscle (46) and polymerized in 10 mm Tris, 50 mm NaCl, 100 μm ATP, 1 mm DTT, 1 mm MgCl2, pH 7.0. Assays were performed using 4 μm talin and 10 μm F-actin. The mixture was incubated for 60 min at room temperature and centrifuged at 100,000 rpm for 30 min at 22 °C using a Beckman OptimaTM ultracentrifuge. Supernatants and pellets were analyzed on SDS-polyacrylamide gels and stained using Coomassie Blue.
CD Spectroscopy
CD spectra were recorded using a Jasco J-715 spectropolarimeter equipped with a Jasco PTC-348WI temperature control unit. Far-UV CD spectra were recorded at 20 °C over the wavelength range 200–250 nm in a quartz cell of 0.1 cm path length (scan rate 50 nm·min−1). Proteins were dissolved in 20 mm sodium phosphate, pH 6.5, 50 mm NaCl at a concentration of 10 μm. For denaturation studies, the unfolding of α-helices was followed at 222 nm.
Cell Culture and Transfection
NIH3T3 cells were cultured on uncoated plastic dishes in DMEM with 5% FCS and penicillin/streptomycin (Invitrogen) at 37 °C in 5% CO2. Subconfluent cells (6 × 106 cells/ml) were trypsinized and washed in PBS, and 1 × 105 cells were electroporated (using a NEON Microporator from Invitrogen; 1300 mV/20 ms/2 pulses) with 10 μg of DNA per transfection and plated on uncoated glass coverslips (Raymond A. Lamb, East Sussex, UK). 48 h after transfection, cells were washed once with PBS, permeabilized for 4 min with 0.5% Triton X-100 in 20 mm Tris, pH 7.6, 4 mm MgCl2, 300 mm sucrose, 1 mm PMSF, and fixed for 10 min using 3.7% paraformaldehyde in PBS (47). F-actin was stained with phalloidin-TRITC (Molecular Probes) for 20 min, and the coverslips were sealed on glass slides with Mowiol solution (Sigma). Epifluorescence images were taken on an inverted Nikon TE300 microscope equipped with a Hamamatsu ORCA-ER digital camera and an X-cite 120 fluorescence illumination system controlled by Openlab software (Improvision). Images were analyzed using Irfanview and Adobe Photoshop.
RESULTS
Talin Residues 1359–1659 Are Homologous to MESDc1
To identify talin rod domains for structural studies, we initially aligned the talin sequence with other proteins in the database, an approach previously used to identify the C-terminal actin-binding domain in talin that is homologous to that of the HIP1R family of proteins (48). The alignment showed (supplemental Fig. S1A) that the central region of the talin rod (residues 1359–1659) was homologous to a protein of unknown function encoded by a gene called mesoderm development candidate 1 (Mesdc1) (37). This region of talin is conserved across species (supplemental Fig. S2A), and secondary structure predictions on both talin(1359–1659) and MESDc1 identified nine α-helices with almost identical boundaries suggesting that both proteins have a similar tertiary structure.
Crystal Structure of Talin Residues 1359–1659 Reveals a Novel Protein Fold
We expressed recombinant talin(1359–1659) in E. coli, and the purified polypeptide was soluble and stable. Removal of helices from either the N or C terminus resulted in an insoluble polypeptide, suggesting that the nine helices form a discrete domain. The purified polypeptide yielded crystals that diffracted x-rays, and we determined initial phases using anomalous scattering from a selenomethionine derivative (see “Experimental Procedures”) at 2.15 Å resolution, and subsequently we collected a native diffraction set at 2.0 Å resolution for refinement (Table 1). The final high resolution model includes one copy of talin residues 1359–1659 within the asymmetric unit. As predicted, this region of talin contains nine α-helices forming a 5- and a 4-helix bundle with an unusual domain linkage (Fig. 2A). Both the bundles are made up of amphipathic helices and have a hydrophobic core.
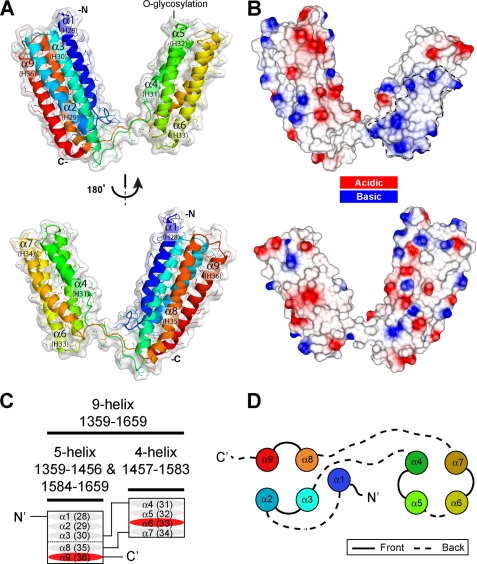
FIGURE 2.
Crystal structure of talin rod residues 1359–1659. A, schematic representation of the talin(1359–1659) crystal structure. This region of talin encodes nine α-helices forming a 5-helix bundle and a 4-helix bundle with an unusual domain linkage. The helix numbers shown in brackets are for full-length talin. B, surface electrostatic potential of the molecule shown in the same orientation as A. There is no evidence of hydrophobic or electrostatic interactions between the two domains. C, this talin module contains nine α-helices (ovals) organized into two different amphipathic helical bundles with an unusual topology. The first three α-helices and the last two helices (α1, α2, α3, α8, and α9) form a 5-helix bundle, whereas helix 4–7 form a 4-helix bundle (α4, α5, α6, and α7). The two vinculin-binding sites (VBS) are shown in red. D, diagram showing the organization of the helices into 5-helix (left) and 4-helix (right) bundles. Solid and dashed lines represent connecting loops on opposite ends of the helices. The coloring is as in A.
A search for similar structures using the DALI server finds 5- and 4-helix bundles, but no other protein in the database has a similar fold to that observed for talin(1359–1659). This novel fold shows a 4-helix bundle inserted into one of the loops in the 5-helix bundle (between helices α3 and α4). This makes the 5-helix bundle unique in that it is composed of nonsequential helices, i.e. helix α1, α2, α3, α8, and α9 (equivalent to helices 28, 29, 30, 35, and 36 of the talin rod). Such an insertion has not previously been identified as judged by DomIns.
There is no interaction between the two bundles in the crystal structure, and none of the contacts in the crystal lattice seem biologically relevant. The topology of the helices and nature of the arrangement are very different from that of the 5- and 4-helix bundles at the N terminus of the talin rod (residues 482–789) (32). In the latter case, the two bundles have a staggered arrangement, with the 4-helix bundle packing against the 5-helix bundle, stabilized by extensive hydrophobic contacts. In the new structure, no hydrophobic patches are present on the surface of either bundle (Fig. 2B). The structure is also different from the IBS2 region (residues 1973–2293) where two 5-helix bundles (IBS2-A and IBS2-B) stack end to end in the crystal structure (23).
The 5-helix bundle in the new structure has the same topology as observed previously in the talin rod, comprising five anti-parallel α-helices (ranging from 25 to 33 residues in length) arranged with right-handed crossover connectivity. A long 10-residue linker connects the first and second helices (α1-α2); otherwise, the helices are connected by short loops (Fig. 2, C and D). This bundle topology has only previously been seen (DALI (49)) in the talin rod 5-helix bundles (23, 31, 32), and this new 5-helix bundle can be superimposed with these domains with a relatively low root mean square difference for backbone atoms (root mean square deviation: 1.9 Å talin(482–655); 2.4 Å talin(1655–1822); 2.3 Å talin(1974–2140), and 2.2 Å talin(2137–2294)).
The 4-helix bundle has a more common right-handed up-down-up-down fold composed of sequential helices α4 to α7 (equivalent to talin rod helices 31 to 34) (Fig. 2, C and D). Helix α4 is the shortest with only 18 amino acids, whereas the other three have a similar length (26–28 residues) and are connected by short loops. Helix α7 is kinked in the middle at Pro-1564 (Fig. 2A), and the bottom of the helix orients toward the gap left by the shorter helix α4 stabilizing this region by interacting with the hydrophobic core (Fig. 3A). Moreover, the bottom of the bundle is further stabilized by a cluster of four phenylalanines (Phe-1467, Phe-1525, Phe-1575, and Phe-1581) with Phe-1581 from the C-terminal end capping the bundle (Fig. 3A). Also two residues from the N-terminal end, Leu-1461 and Val-1462, interact with the hydrophobic core to compensate for the shorter helix α4.
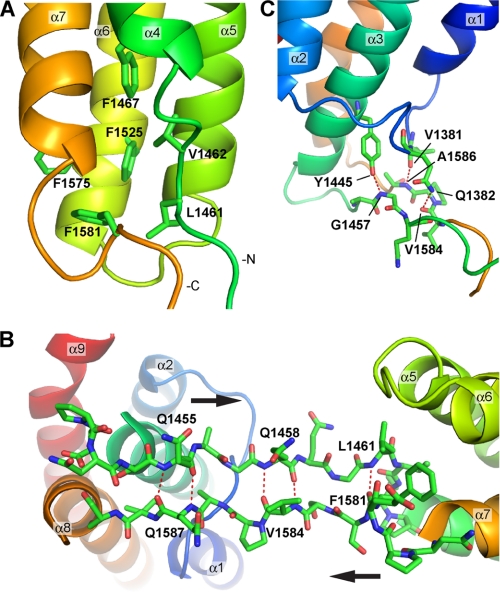
FIGURE 3.
Structural details of the linker region between the two bundles. A, view of the hydrophobic interactions at the bottom of the 4-helix bundle. The short helix α4 leaves a gap that is filled with a cluster of four phenylalanines and two hydrophobic residues from the linker region at the N-terminal end of helix α4, i.e. Leu-1461 and Val-1462. B, view of the linker region that forms a two-stranded anti-parallel β-sheet-like structure. The complementary backbone hydrogen bonds are highlighted by the dashed red lines. C, hydrogen bond networks observed at the bottom of the 5-helix bundle and the linker region.
The linker regions between the two bundles, residues 1451–1464 and 1578–1589, form a two-stranded anti-parallel β-sheet-like structure with complementary backbone hydrogen bonding (Gln-1455 bonds with Gln-1587, Gln-1458, with Val-1584, and Leu-1461 with Phe-1581) (Fig. 3B). This arrangement keeps the two linker regions together limiting their flexibility, and this is supported by the relatively low B-factor values observed for the linker region in the crystal structure. Furthermore, hydrogen bonds are observed between side chains from the bottom of the 5-helix bundle and backbone residues from the linker region, e.g. side chain Gln-1382 with backbone Val-1584, side chain Tyr-1445 with backbone Gly-1457, and backbone Val-1381 with backbone Ala-1586 (Fig. 3C). These interactions are likely to fix the relative orientation between the 5-helix bundle and the linker region. However, such interactions are more limited between the 4-helix bundle and the linker region, and we only observe a single hydrogen bond between the side chain of Arg-1515 with backbone Gln-1459. This would suggest a greater freedom of movement of the 4-helix bundle relative to the linker region, and the B-factor values are higher than average in the vicinity of Gln-1459 and Ser-1583, which are on opposite sides of the sheet just before the 4-helix bundle.
The consequence of this unique arrangement of the 5- and 4-helix domains is that it interrupts the otherwise linear organization of the helical bundle domains that make up the talin rod, and it seems likely that the 4-helix bundle will protrude out of the rod (Fig. 1). Interestingly, all of the talin rod domains identified to date have an average calculated isoelectric point (pI) of ∼5.0 (4.4–6.1), and this is also the case for this talin 9-helix module (pI of 5.76). However, the two bundles have very different pI values, 4.6 and 7.8 for the 5- and 4-helix bundles, respectively. The electrostatic surface representation of the molecule (Fig. 2B) shows a basic surface on the 4-helix bundle that points away from the 5-helix bundle, and this may be a functionally important feature of this region of the talin rod. (The talin head domain also has a basic surface, and this interacts with acidic phospholipids (6, 20). However, the 9-helix module does not bind negatively charged phospholipids vesicles in vitro; data not shown.)
Talin 9-Helix Module Is Relatively Compact in Solution
Solution studies support the crystallographic data pointing to a “V-shape” arrangement of the two bundles in the 9-helix module with limited flexibility, as judged by analytical gel filtration, NMR, and small angle x-ray scattering (see supplemental “Results” and supplemental Fig. S3). Gel filtration shows that at a concentration of 50 μm, the polypeptide is monomeric with an apparent molecular mass of 39 kDa, significantly higher than the calculated value (32 kDa) indicative of a nonglobular conformation (large radius of gyration).
Sequence Analysis
The residues in the linker regions between the two bundles (residues 1451–1464 and 1578–1589) are relatively well conserved across species (supplemental Fig. S2A) suggesting that this region has an important structural function; perhaps it allows some flexibility between the two domains. The four phenylalanines (Phe-1467, Phe-1525, Phe-1575, and Phe-1581) that stabilize the 4-helix bundle are also conserved. Phe-1581, which caps the C-terminal end of the bundle (Fig. 3A), is invariant in the talin sequences analyzed and is also conserved in MESDc1 (supplemental Figs. S1A and S2A), implying that bundle capping is important in stabilizing the hydrophobic core. Phe-1575 (replaced by a tyrosine in Caenorhabditis elegans talin) and Phe-1525 (replaced by leucine in Drosophila talin) are both substantially conserved. Phe-1467 is less conserved, being replaced in both C. elegans and Drosophila talin and in MESDc1 by leucine or valine, thus keeping the hydrophobic core intact.
Two O-glycosylation sites have been identified in the talin rod, Thr-1487 and Thr-1890 (50). Thr-1487 is located within the 4-helix bundle in a loop between helix α4 and α5 (Fig. 2A). This threonine is conserved in C. elegans talin and MESDc1 but not in Drosophila melanogaster talin (supplemental Fig. S1A and supplemental Fig. S2A). The positioning of Thr-1487 at the tip of the bundle and its conservation suggest that it may be biologically important, although the role of talin glycosylation has not been investigated.
A number of phosphorylation sites have been mapped within human platelet talin (51), and two of these (Ser-1461 and Ser-1508) are located within residues 1359–1659 and are potentially phosphorylated by protein kinase C. Both of these serine residues are solvent-exposed in the crystal structure; Ser-1641 (located in the middle of helix α9) is fully conserved in all talins as well as in MESDc1 (supplemental Figs. S1A and S2A) suggesting that phosphorylation of this residue might have an important biological role. However, Ser-1508 in helix 5 is not conserved in other talins or in MESDc1.
Vinculin Binding
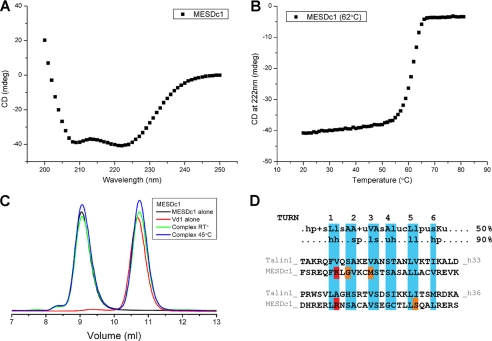
Talin(1359–1659) contains two potential VBS. The VBS in helix α9 contained in the 5-helix bundle (equivalent to talin rod helix 36) showed weak binding in the initial SPOT-peptide analysis, although we were able to crystallize it in complex with vinculin Vd1 (28). In contrast, the VBS in helix α6 and contained in the 4-helix bundle (equivalent to talin rod helix 33) bound Vd1 quite strongly in the SPOT-peptide analysis (28). As with all such VBS in talin, the residues involved in vinculin binding are buried in the hydrophobic core of the bundles (28), and one of the factors that determines the activity of the VBS is the inherent stability of the bundle in which it is embedded (52).
Preincubation of the 9-helix module with vinculin Vd1 (1:1 molar ratio) followed by analytical gel filtration provided clear evidence of complex formation. Interestingly, binding occurred even at room temperature (Table 2 and Fig. 4C), in marked contrast to most other VBS-containing talin rod domains that only bind vinculin at higher temperatures (23, 33, 52).
TABLE 2.
Properties of talin domains
The melting temperature was determined using CD. Vinculin binding was assessed using gel filtration after 30 min of incubation of talin domains with vinculin Vd1 (residues 1–258).
| Domain | Coordinates | Melting | Vinculin binding |
|---|---|---|---|
| °C | |||
| 9-Helix | 1357–1653 | 53.3 | Yes |
| 5-Helix | 1359–1659 Δ1454–1586 | 58.6 | Cryptic |
| 4-Helix | 1458–1584 | 42.3 | Yes |
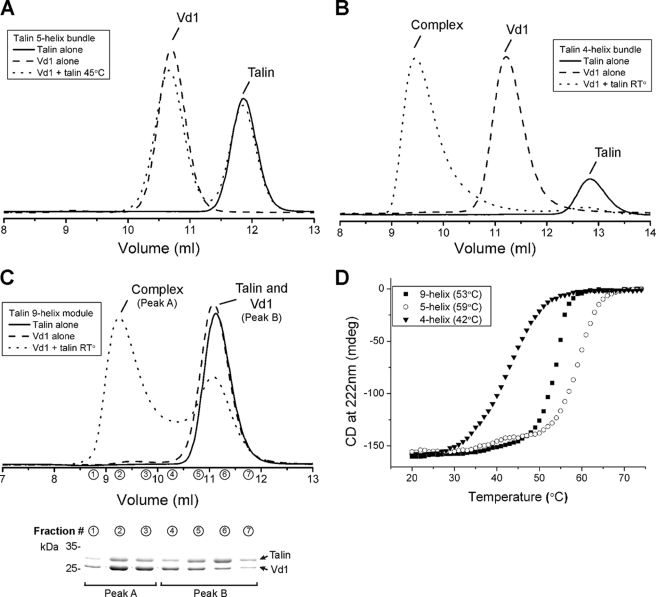
FIGURE 4.
Vinculin Vd1 binding analyzed by gel filtration. Vinculin Vd1 was incubated using a 1:1 ratio with the talin 5-helix (A), the 4-helix (B), or the 9-helix (C) polypeptides at various temperatures, and complex formation was analyzed on a Superdex-75 (10/300) GL gel filtration column at room temperature (RT). Incubation of the 4-helix (B) or 9-helix polypeptides (C) with Vd1 resulted in complex formation. Preincubation of the proteins at 37 °C did not increase significantly the formation of a talin-Vd1 complex (data not shown). No binding was observed with the 5-helix bundle alone. D, denaturation profiles for the talin rod polypeptides were measured by monitoring the change in circular dichroism at 222 nm with increasing temperatures. Profiles are shown for the 9-helix module (squares), the 5-helix bundle (circles), and the 4-helix bundle (triangles). The melting temperature (Tm) for each domain is indicated.
To establish whether one or both bundles contain an active VBS, we cloned, expressed, and purified the two individual domains, i.e. the 5-helix domain (residues 1359–1659 Δ1454–1586) and the 4-helix domain (residues 1458–1584). The NMR spectrum of the 9-helix module (supplemental Fig. S4A) shows good peak dispersion with the slight broadening expected from an extended domain in solution. The 1H,15N HSQC spectra of the individual 5- and 4-helix bundles (supplemental Fig. S4, B and C) show good peak dispersion with uniform intensity and line width, suggesting that they are correctly folded. Superimposition of the spectra of the individual domains with that of the 9-helix module indicates that the two domains retain the same structural features when expressed on their own.
Gel filtration showed that the purified 5-helix bundle did not bind vinculin Vd1 even after preincubation at 45 °C for 30 min (Fig. 4A). In contrast, the purified 4-helix bundle readily formed a complex with Vd1 even at 20 °C (Fig. 4B). Interestingly, the 4-helix bundle on its own has a rather low melting temperature (Tm 42 °C, see Table 2 and Fig. 4D) when compared with the 5-helix bundle (Tm of 59 °C), and this correlates with its ability to bind Vd1 at room temperature. However, the N- and C-terminal regions of the 4-helix bundle are normally kept together by the preceding 5-helix bundle, and this most likely stabilizes the domain. Indeed, the 9-helix module melts in a single step with a Tm of 53 °C, much higher than that of the isolated 4-helix bundle (Tm 42 °C) and somewhat lower than that of the isolated 5-helix bundle (Tm of 59 °C).
Incubation of a 1:1 mixture of the talin 9-helix module and vinculin Vd1 at room temperature followed by gel filtration and SDS-PAGE showed the presence of both the complex and free talin and vinculin (Fig. 4C), indicative of an interaction that has intermediate affinity in the micromolar range. In contrast, mixing the isolated 4-helix bundle with Vd1 resulted in quantitative complex formation (Fig. 4B) consistent with tight binding in the nanomolar affinity range (53). This suggests that stabilization of the 4-helix bundle by incorporation in the 9-helix module decreases its affinity for vinculin.
Talin 9-Helix Module Binds F-actin Both in Vitro and in Vivo
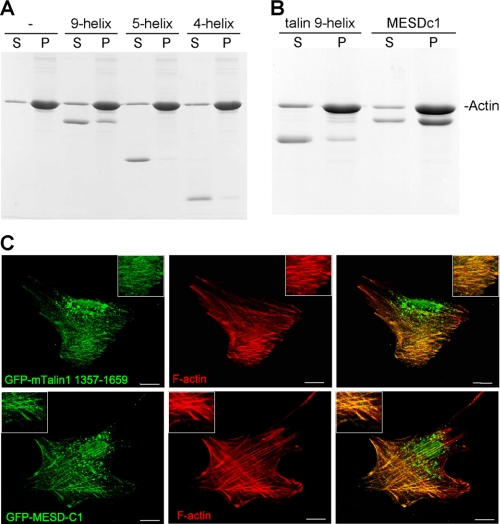
An F-actin-binding site was previously identified in the middle of the talin rod (25), and we therefore tested the ability of the 9-helix bundle to bind F-actin using a cosedimentation assay. As shown in Fig. 5A, the 9-helix polypeptide bound F-actin, albeit rather weakly, and about 30% of the protein cosedimented with F-actin. Both domains were required for binding, because neither the 4- nor 5-helix bundles on their own cosedimented with F-actin, demonstrating that the basic surface observed on the 4-helix bundle (Fig. 2B) is not the sole determinant for F-actin binding. To establish whether the talin 9-helix polypeptide bound F-actin in vivo, we expressed it as a GFP-tagged construct in NIH3T3 cells. Triton X-100 extraction before fixation revealed a clear colocalization of the GFP-talin polypeptide with actin stress fibers (Fig. 5C). However, despite its ability to bind vinculin in vitro, the GFP-tagged polypeptide did not colocalize with vinculin in FAs (data not shown). It has been reported previously that talin domains containing VBSs do not necessarily localize to FAs; this has been shown with the IBS2-A domain (residues 1975–2135) (23) and even a rod fragment spanning residues 434–2197, which consists of multiple domains and at least 10 VBSs (54). Thus, it appears that FA localization of talin polypeptides containing VBSs depends upon additional factors.
FIGURE 5.
Both the talin 9-helix module and MESDc1 bind F-actin. The talin 9-helix, 5-helix, and 4-helix polypeptides were incubated with F-actin, and binding was determined using a cosedimentation assay. After centrifugation, supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE. A, talin 9-helix module binds F-actin, but not the individual subdomains, i.e. the 5- and 4-helix bundles. B, MESDc1 proteins binds F-actin with higher affinity than the talin 9-helix module. C, images of NIH3T3 cells transfected with cDNAs encoding either GFP-talin residues 1359–1659 or GFP-MESDc1 after Triton X-100 extraction. The F-actin cytoskeleton was stained with phalloidin Alexa 594. Scale bar, 10 μm. Both the GFP-tagged talin 9-helix module and MESDc1 colocalize with actin stress fibers.
MESDc1 Is a Novel F-actin-binding Protein
Talin residues 1359–1659 show homology to the protein predicted to be encoded by the MESDc1 gene, which was originally identified as one of five candidate genes on mouse chromosome 7 implicated in mesoderm development (37), although this activity was subsequently assigned to another gene. Inspection of the EST database shows that the MESDc1 mRNA is widely expressed, and MESDc1 protein is predicted to contain 362 amino acids. The N-terminal region (residues 1–43) of MESDc1 has no homologues in the database, and secondary structure predictions suggest that this region forms a 24-amino acid-long helix. The homology with talin is restricted to 301 amino acids at the C terminus of MESDc1, and secondary structure prediction clearly indicates that residues 44–352 of MESDc1 have a similar secondary structure to talin residues 1359–1659 (supplemental Fig. S1A). We generated a three-dimensional model of MESDc1 (supplemental Fig. S5A) using Modeler (55) with the crystal structure of talin as the template. We observed no important clashes, i.e. the hydrophobic residues are kept within the core of the model and charged residues are surface-exposed, suggesting that the model is plausible. One small difference is the insertion of four amino acids in the MESDc1 sequence at the N terminus of helix α10 (talin helix α9, supplemental Fig. S1A) making the loop α9-α10 slightly longer.
The protein encoded by the MESDc1 gene has not been characterized, but recombinant MESDc1 expressed well in E. coli, and the protein was readily purified using standard methods. Circular dichroism measurements showed that it was largely αhelical (Fig. 6A), supporting our three-dimensional model, with a melting temperature of 62 °C (Fig. 6B), suggesting that it was folded correctly. Purified MESDc1 (50 μm) had a rather large apparent molecular mass (74.6 kDa) as determined by gel filtration compared with a calculated value of 38.4 kDa (data not shown). Although the talin 9-helix module has a somewhat higher apparent molecular mass (39 kDa) than the calculated value (32 kDa) due to its extended conformation, the data on MESDc1 suggest that it may form a dimer in solution. MESDc1 contains an extra helix at the N terminus that could mediate dimerization, but its sequence does not show obvious signs of coiled-coil structure unlike the talin C-terminal dimerization helix (27). We expressed and purified the N-terminal 43 amino acids of MESDc1 as thioredoxin fusion protein. Gel filtration shows that it is monomeric with a very similar elution profile to thioredoxin alone (data not shown). Moreover, the 1H,15N HSQC spectrum of this construct shows very sharp resonances with little dispersion for the MESDc1 amino acids, suggesting that this region is unfolded when expressed on its own.
FIGURE 6.
Biochemical characterization of MESDc1. A, secondary structure analysis of MESDc1 by circular dichroism. The profile suggests that the protein is largely helical. B, denaturation profile for MESDc1 was measured by monitoring the change in circular dichroism at 222 nm with increasing temperatures. The melting temperature (Tm) is indicated. C, vinculin Vd1 (residues 1–258) was incubated with MESDc1 at various temperatures, and complex formation was analyzed on a Superdex-75 (10/300) GL gel filtration column at room temperature (RT). MESDc1 does not bind to Vd1 even after preincubation of the proteins at 45 °C for 30 min. D, VBS peptide sequences were aligned using ClustalW as described in Gingras et al. (28). Residues highlighted in blue align with the buried (>75%) hydrophobic side chains from the VBS1-Vd1 complex crystal structure (32). Residues that clash with the consensus VBS sequence are highlighted in red, i.e. hydrophobic residue substituted by positive residue. The residues that do not fit with the ideal consensus VBS sequence are highlighted in orange. The 50 and 90% VBS consensus sequence is shown at the top as described in Gingras et al. (28). Uppercase letters indicate conserved residues (single-letter amino acid code). Lowercase letters indicate conserved classes of amino acids as follows: h, hydrophobic residues (A, C, F, G, H, I, K, L, M, P, T, V, W, Y); p, polar residues (C, D, E, H, K, N, Q, R, S, T); c, charged resides (D, E, H, K, R); s, small residues (A, C, D, G, N, P, S, T, V); +, positive residues (H, K, R); l, aliphatic residues (I, L, V); and u, tiny (A, G, S, C).
Given the homology between MESDc1 and the talin 9-helix module, we tested its ability to bind F-actin and vinculin. Interestingly, MESDc1 bound to F-actin in a cosedimentation assay better than the equivalent talin domain, and ∼70% of the polypeptide was found in the pellet (Fig. 5B). Analysis of the three-dimensional model of MESDc1 shows two small basic regions on the surface of the 4-helix bundle (supplemental Fig. S5B). In contrast, there is one large basic region on the surface of the talin 9-helix module (Fig. 2B). Thus, surface charge does not explain why MESDc1 binds F-actin better than the equivalent talin domain in the cosedimentation assay. The possibility that MESDc1 is dimeric may be relevant in this regard, the C-terminal actin-binding site in talin only binds F-actin with high affinity as a dimer (27).
MESDc1 did not bind the vinculin Vd1 domain as determined by gel filtration, even after incubation at 45 °C for 30 min (Fig. 6C). Comparison of the sequence of the VBS helices in the talin 9-helix module with the overall consensus for VBSs in the talin rod and the equivalent helices in MESDc1 shows that the latter deviates from the consensus (Fig. 6D). The most important clashes in the MESDc1 sequence are localized in the first helical turn where hydrophobic residues are normally found in talin, whereas basic residues are found in the MESDc1 sequence (Fig. 6D). Consistent with the above findings, GFP-MESDc1 expressed in NIH3T3 cells colocalized with actin stress fibers (Fig. 5C) but not vinculin-containing FAs (data not shown) (52, 54).
DISCUSSION
Here, we report the crystal structure of talin residues 1359–1659 that contains nine α-helices that are organized into a unique fold with two distinct domains. The 5-helix bundle is formed by nonsequential helices α1, α2, and α3 followed by α8 and α9, whereas the 4-helix bundle is composed of consecutive helices and is inserted between helices 3 and 4 of the 5-helix bundle. The two bundles are connected by a linker that forms a two-stranded anti-parallel β-sheet-like structure. This is a completely novel feature of the talin rod that is otherwise composed of a linear arrangement of 5- and 4-helix bundles. Because the 5-helix bundle contains the N and C termini of this module, we propose that it is linked by short loops to the adjacent helical bundles in the talin rod via end-to-end packing, whereas the 4-helix bundle protrudes from the talin rod (Fig. 1) (33).
Another novel feature of the module is that it contains a constitutively active VBS that binds vinculin Vd1 at room temperature. Analysis of the isolated 4-helix bundle shows that it also binds Vd1 at room temperature, whereas the VBS in the 5-helix bundle is cryptic, like most other VBSs in the talin rod (23, 32, 52). To date, the only other region of the talin rod that binds Vd1 at room temperature is the 4-helix bundle spanning residues 755–889 (29); both these 4-helix bundles are unique in that they have a cluster of threonine residues within their hydrophobic core (supplemental Fig. S6), and this likely modulates the stability of the bundle, and hence the availability of the VBS. Indeed, we have shown that replacing the threonine cluster in talin(755–889) with hydrophobic residues markedly suppresses vinculin binding (52). However, within the 9-helix module, the termini of the 4-helix bundle are kept together by the 5-helix bundle, and CD melting experiments show this stabilizes the fold; because unfolding the bundle is required for vinculin binding, this will decrease its affinity for vinculin, and this may explain why Vd1 binding to the 9-helix module has a lower affinity than binding to the isolated 4-helix bundle. How the various VBS behave within the context of full-length talin remains to be explored.
We have previously suggested that activation of the cryptic VBSs in the talin rod might be triggered by force exerted on integrin-talin-actin complexes (52), and experimental evidence in support of the idea of force-induced activation of the VBSs in talin has recently come from elegant in vitro studies using magnetic tweezers (56). Vinculin stabilizes FAs by cross-linking talin to F-actin or to membrane phospholipids (57, 58), and perhaps the fact that two of the VBSs in talin are activated more readily than others allows for a graduated recruitment of vinculin in response to progressive increases in force. However, expression of vinculin Vd1 alone in vinculin null fibroblasts has been shown to lead to larger and more stable FAs even in the absence of actomyosin contraction, an effect that was dependent on the ability of Vd1 to bind talin (59). This suggests that mechanisms that regulate exposure of the talin-binding site in vinculin may also drive the interaction of vinculin with talin, and it will be interesting to establish whether the two domains containing constitutively active VBSs in talin are involved in this response.
The 9-helix module also binds F-actin, although neither the 4- nor 5-helix bundles alone are able to bind. This raises the possibility that vinculin might bind to the VBS in the 4-helix bundle via its N-terminal Vd1 domain leaving the C-terminal vinculin tail to bind to the F-actin bound to the talin 9-helix module. This may stabilize the interaction between the talin 9-helix bundle and F-actin, which on its own is rather weak. Although this remains to be investigated, what is clear is that several regions of talin, including the talin head (60), and at least two regions in the talin rod have the ability to bind F-actin (Fig. 1) (25). Much attention has focused on the C-terminal actin-binding site in talin (26, 27), which is homologous to the Hip1R family of actin-binding proteins (61), and our recent data show that it plays an essential role in the assembly of FAs (65). However, we have shown that the talin C-terminal actin-binding site, which is dimeric, binds along a single actin filament and does not cross-link F-actin (27). Therefore, it seems unlikely that the C-terminal actin-binding site accounts for the ability of talin to cross-link F-actin (62), and actin-binding sites elsewhere in the talin molecule, including that in the 9-helix module, may be relevant in this regard. Similarly, recent studies on filamin show that apart from the well characterized actin-binding site in the N-terminal calponin homology domain, Ig repeats 9–15 in the filamin rod also bind F-actin and increase the overall avidity of filamin for F-actin (63).
Previous studies have shown that the central part of the talin rod (residues 1327–1948) binds the 312-amino acid insert (SNTIII) present within the muscle variant of the intermediate filament protein α-synemin (38). Using the same overlay assay used above, we have shown that both the 4- and 5-helix bundles present in the talin 9-helix module bind SNTIII indicating that there are two α-synemin-binding sites in this region of talin (see supplemental “Results” and supplemental Fig. S7). It is noteworthy that SNTIII also binds the vinculin tail, but this is competitive with talin binding (64).
Talin residues 1359–1659 show homology to residues 44–352 of the protein encoded by the MESDc1 gene, and this region of MESDc1 is predicted to have a similar secondary structure to talin. Indeed we have generated a homology model of MESDc1 using the crystal structure of the talin residues 1359–1659. Proteins with similar folds often have very different functions, but it is striking that both proteins bind to F-actin, and GFP-tagged MESDc1 colocalized with actin stress fibers when expressed in NIH3T3 cells. However, MESDc1 does not bind to vinculin, and analysis of the MESDc1 sequence shows that helices 7 and 10 (equivalent to the VBS helices in talin) diverge from the VBS consensus sequence (28). Interestingly, MESDc1 binds to F-actin better than does the talin 9-helix module. Gel filtration experiments indicate that MESDc1 is dimeric in solution, and the NMR HSQC shows broad signals (data not shown), consistent with this conclusion. The C-terminal actin-binding site in talin is also dimeric, and dimerization is important for high affinity actin binding (26, 27). Thus, the fact that MESDc1 is also dimeric may contribute to its higher affinity for F-actin. However, although the MESDc1 mRNA appears to be widely expressed, there is no literature on the role of this novel actin-binding protein in cells.
In summary, we show that residues 1359–1659 from the central region of the talin rod have a novel structure quite different from that of other regions of the talin rod or any other protein in the Protein Data Bank. It contains binding sites for vinculin, F-actin, and the muscle-specific isoform of the intermediate filament protein α-synemin, and this and its unusual fold suggest that it plays an important role in the function of talin. Moreover, we predict that the previously uncharacterized protein MESDc1 will have a similar structure, and we demonstrate that it also binds F-actin.
Supplementary Material
Acknowledgments
We thank Peter Moody (Leicester, United Kingdom) and Klaus Futterer (Birmingham, United Kingdom) for their help with the crystallography. We thank Emmanuel Debrand for help in analyzing the MESDc1 gene and J. Günter Grossmann (Liverpool, United Kingdom) for help with small angle x-ray scattering data collection and processing. We also thank Ralf Schmid and Imtiaz M. Shafiq (Leicester, United Kingdom) for generating the MESDc1 homology model.
This work was supported, in whole or in part, by National Institutes of Health Grant U54 GM64346 from NIGMS Cell Migration Consortium. This work was also supported by the Wellcome Trust, Cancer Research United Kingdom.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7, “Results,” and additional references.
The atomic coordinates and structure factors (code 2x0c) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FA
- focal adhesion
- VBS
- vinculin-binding site
- TRITC
- tetramethylrhodamine isothiocyanate
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1.Critchley D. R., Gingras A. R. (2008) J. Cell Sci. 121, 1345–1347 [DOI] [PubMed] [Google Scholar]
- 2.Critchley D. R. (2009) Annu. Rev. Biophys. 38, 235–254 [DOI] [PubMed] [Google Scholar]
- 3.Roberts G. C., Critchley D. R. (2009) Biophys. Rev. 1, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderwood D. A. (2004) J. Cell Sci. 117, 657–666 [DOI] [PubMed] [Google Scholar]
- 5.Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., Calderwood D. A. (2003) Science 302, 103–106 [DOI] [PubMed] [Google Scholar]
- 6.Saltel F., Mortier E., Hytönen V. P., Jacquier M. C., Zimmermann P., Vogel V., Liu W., Wehrle-Haller B. (2009) J. Cell Biol. 187, 715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Jiang G., Cai Y., Monkley S. J., Critchley D. R., Sheetz M. P. (2008) Nat. Cell Biol. 10, 1062–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albigès-Rizo C., Frachet P., Block M. R. (1995) J. Cell Sci. 108, 3317–3329 [DOI] [PubMed] [Google Scholar]
- 9.Priddle H., Hemmings L., Monkley S., Woods A., Patel B., Sutton D., Dunn G. A., Zicha D., Critchley D. R. (1998) J. Cell Biol. 142, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monkley S. J., Zhou X. H., Kinston S. J., Giblett S. M., Hemmings L., Priddle H., Brown J. E., Pritchard C. A., Critchley D. R., Fässler R. (2000) Dev. Dyn. 219, 560–574 [DOI] [PubMed] [Google Scholar]
- 11.Petrich B. G., Marchese P., Ruggeri Z. M., Spiess S., Weichert R. A., Ye F., Tiedt R., Skoda R. C., Monkley S. J., Critchley D. R., Ginsberg M. H. (2007) J. Exp. Med. 204, 3103–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieswandt B., Moser M., Pleines I., Varga-Szabo D., Monkley S., Critchley D., Fässler R. (2007) J. Exp. Med. 204, 3113–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Litvinov R. I., Chen X., Bach T. L., Lian L., Petrich B. G., Monkley S. J., Kanaho Y., Critchley D. R., Sasaki T., Birnbaum M. J., Weisel J. W., Hartwig J., Abrams C. S. (2008) J. Clin. Invest. 118, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conti F. J., Felder A., Monkley S., Schwander M., Wood M. R., Lieber R., Critchley D., Müller U. (2008) Development 135, 2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conti F. J., Monkley S. J., Wood M. R., Critchley D. R., Müller U. (2009) Development 136, 3597–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goult B. T., Bouaouina M., Harburger D. S., Bate N., Patel B., Anthis N. J., Campbell I. D., Calderwood D. A., Barsukov I. L., Roberts G. C., Critchley D. R. (2009) J. Mol. Biol. 394, 944–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goult B. T., Bouaouina M., Elliott P. R., Bate N., Patel B., Gingras A. R., Grossmann J. G., Roberts G. C., Calderwood D. A., Critchley D. R., Barsukov I. L. (2010) EMBO J. 29, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., Campbell I. D. (2007) Cell 128, 171–182 [DOI] [PubMed] [Google Scholar]
- 19.García-Alvarez B., de Pereda J. M., Calderwood D. A., Ulmer T. S., Critchley D., Campbell I. D., Ginsberg M. H., Liddington R. C. (2003) Mol. Cell 11, 49–58 [DOI] [PubMed] [Google Scholar]
- 20.Anthis N. J., Wegener K. L., Ye F., Kim C., Goult B. T., Lowe E. D., Vakonakis I., Bate N., Critchley D. R., Ginsberg M. H., Campbell I. D. (2009) EMBO J. 28, 3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau T. L., Kim C., Ginsberg M. H., Ulmer T. S. (2009) EMBO J. 28, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meves A., Stremmel C., Gottschalk K., Fässler R. (2009) Trends Cell Biol. 19, 504–513 [DOI] [PubMed] [Google Scholar]
- 23.Gingras A. R., Ziegler W. H., Bobkov A. A., Joyce M. G., Fasci D., Himmel M., Rothemund S., Ritter A., Grossmann J. G., Patel B., Bate N., Goult B. T., Emsley J., Barsukov I. L., Roberts G. C., Liddington R. C., Ginsberg M. H., Critchley D. R. (2009) J. Biol. Chem. 284, 8866–8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moes M., Rodius S., Coleman S. J., Monkley S. J., Goormaghtigh E., Tremuth L., Kox C., van der Holst P. P., Critchley D. R., Kieffer N. (2007) J. Biol. Chem. 282, 17280–17288 [DOI] [PubMed] [Google Scholar]
- 25.Hemmings L., Rees D. J., Ohanian V., Bolton S. J., Gilmore A. P., Patel B., Priddle H., Trevithick J. E., Hynes R. O., Critchley D. R. (1996) J. Cell Sci. 109, 2715–2726 [DOI] [PubMed] [Google Scholar]
- 26.Smith S. J., McCann R. O. (2007) Biochemistry 46, 10886–10898 [DOI] [PubMed] [Google Scholar]
- 27.Gingras A. R., Bate N., Goult B. T., Hazelwood L., Canestrelli I., Grossmann J. G., Liu H., Putz N. S., Roberts G. C., Volkmann N., Hanein D., Barsukov I. L., Critchley D. R. (2008) EMBO J. 27, 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gingras A. R., Ziegler W. H., Frank R., Barsukov I. L., Roberts G. C., Critchley D. R., Emsley J. (2005) J. Biol. Chem. 280, 37217–37224 [DOI] [PubMed] [Google Scholar]
- 29.Fillingham I., Gingras A. R., Papagrigoriou E., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2005) Structure 13, 65–74 [DOI] [PubMed] [Google Scholar]
- 30.Gingras A. R., Vogel K. P., Steinhoff H. J., Ziegler W. H., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2006) Biochemistry 45, 1805–1817 [DOI] [PubMed] [Google Scholar]
- 31.Goult B. T., Bate N., Anthis N. J., Wegener K. L., Gingras A. R., Patel B., Barsukov I. L., Campbell I. D., Roberts G. C., Critchley D. R. (2009) J. Biol. Chem. 284, 15097–15106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papagrigoriou E., Gingras A. R., Barsukov I. L., Bate N., Fillingham I. J., Patel B., Frank R., Ziegler W. H., Roberts G. C., Critchley D. R., Emsley J. (2004) EMBO J. 23, 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goult B. T., Gingras A. R., Bate N., Barsukov I. L., Critchley D. R., Roberts G. C. (2010) FEBS Lett. 584, 2237–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goksoy E., Ma Y. Q., Wang X., Kong X., Perera D., Plow E. F., Qin J. (2008) Mol. Cell 31, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler J., Lünsdorf H., Jockusch B. M. (1997) Eur. J. Biochem. 243, 430–436 [DOI] [PubMed] [Google Scholar]
- 36.Molony L., McCaslin D., Abernethy J., Paschal B., Burridge K. (1987) J. Biol. Chem. 262, 7790–7795 [PubMed] [Google Scholar]
- 37.Wines M. E., Lee L., Katari M. S., Zhang L., DeRossi C., Shi Y., Perkins S., Feldman M., McCombie W. R., Holdener B. C. (2001) Genomics 72, 88–98 [DOI] [PubMed] [Google Scholar]
- 38.Sun N., Critchley D. R., Paulin D., Li Z., Robson R. M. (2008) Exp. Cell Res. 314, 1839–1849 [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 40.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 41.Cohen S. X., Ben Jelloul M., Long F., Vagin A., Knipscheer P., Lebbink J., Sixma T. K., Lamzin V. S., Murshudov G. N., Perrakis A. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 43.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 44.Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1992) Proteins 12, 345–364 [DOI] [PubMed] [Google Scholar]
- 45.Potterton E., McNicholas S., Krissinel E., Cowtan K., Noble M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1955–1957 [DOI] [PubMed] [Google Scholar]
- 46.Pardee J. D., Spudich J. A. (1982) Methods Enzymol. 85, 164–181 [DOI] [PubMed] [Google Scholar]
- 47.Vale R. D., Ignatius M. J., Shooter E. M. (1985) J. Neurosci. 5, 2762–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brett T. J., Legendre-Guillemin V., McPherson P. S., Fremont D. H. (2006) Nat. Struct. Mol. Biol. 13, 121–130 [DOI] [PubMed] [Google Scholar]
- 49.Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 50.Hagmann J., Grob M., Burger M. M. (1992) J. Biol. Chem. 267, 14424–14428 [PubMed] [Google Scholar]
- 51.Ratnikov B., Ptak C., Han J., Shabanowitz J., Hunt D. F., Ginsberg M. H. (2005) J. Cell Sci. 118, 4921–4923 [DOI] [PubMed] [Google Scholar]
- 52.Patel B., Gingras A. R., Bobkov A. A., Fujimoto L. M., Zhang M., Liddington R. C., Mazzeo D., Emsley J., Roberts G. C., Barsukov I. L., Critchley D. R. (2006) J. Biol. Chem. 281, 7458–7467 [DOI] [PubMed] [Google Scholar]
- 53.Izard T., Vonrhein C. (2004) J. Biol. Chem. 279, 27667–27678 [DOI] [PubMed] [Google Scholar]
- 54.Himmel M., Ritter A., Rothemund S., Pauling B. V., Rottner K., Gingras A. R., Ziegler W. H. (2009) J. Biol. Chem. 284, 13832–13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) in Current Protocols in Bioinformatics, Supplement 15, 5.6.1–5.6.30, John Wiley & Sons, Inc., New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunders R. M., Holt M. R., Jennings L., Sutton D. H., Barsukov I. L., Bobkov A., Liddington R. C., Adamson E. A., Dunn G. A., Critchley D. R. (2006) Eur. J. Cell Biol. 85, 487–500 [DOI] [PubMed] [Google Scholar]
- 58.Chandrasekar I., Stradal T. E., Holt M. R., Entschladen F., Jockusch B. M., Ziegler W. H. (2005) J. Cell Sci. 118, 1461–1472 [DOI] [PubMed] [Google Scholar]
- 59.Humphries J. D., Wang P., Streuli C., Geiger B., Humphries M. J., Ballestrem C. (2007) J. Cell Biol. 179, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H. S., Bellin R. M., Walker D. L., Patel B., Powers P., Liu H., Garcia-Alvarez B., de Pereda J. M., Liddington R. C., Volkmann N., Hanein D., Critchley D. R., Robson R. M. (2004) J. Mol. Biol. 343, 771–784 [DOI] [PubMed] [Google Scholar]
- 61.Senetar M. A., Foster S. J., McCann R. O. (2004) Biochemistry 43, 15418–15428 [DOI] [PubMed] [Google Scholar]
- 62.Schmidt J. M., Zhang J., Lee H. S., Stromer M. H., Robson R. M. (1999) Arch. Biochem. Biophys. 366, 139–150 [DOI] [PubMed] [Google Scholar]
- 63.Nakamura F., Osborn T. M., Hartemink C. A., Hartwig J. H., Stossel T. P. (2007) J. Cell Biol. 179, 1011–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun N., Critchley D. R., Paulin D., Li Z., Robson R. M. (2008) Biochem. J. 409, 657–667 [DOI] [PubMed] [Google Scholar]
- 65.Kopp P. M., Bate N., Hansen T. M., Brindle N. P., Praekelt U., Debrand E., Coleman S., Mazzeo D., Goult B. T., Gingras A. R., Pritchard C. A., Critchley D. R., Monkley S. J. (2010) Eur. J. Cell Biol. 89(9):661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.