Abstract
Mesenchymal stem cells (MSCs) are bone marrow stromal cells that can differentiate into multiple lineages. We previously demonstrated that BMP9 is one of the most potent BMPs to induce osteogenic differentiation of MSCs. BMP9 is one of the least studied BMPs. Whereas ALK1, ALK5, and/or endoglin have recently been reported as potential BMP9 type I receptors in endothelial cells, little is known about type I receptor involvement in BMP9-induced osteogenic differentiation in MSCs. Here, we conduct a comprehensive analysis of the functional role of seven type I receptors in BMP9-induced osteogenic signaling in MSCs. We have found that most of the seven type I receptors are expressed in MSCs. However, using dominant-negative mutants for the seven type I receptors, we demonstrate that only ALK1 and ALK2 mutants effectively inhibit BMP9-induced osteogenic differentiation in vitro and ectopic ossification in MSC implantation assays. Protein fragment complementation assays demonstrate that ALK1 and ALK2 directly interact with BMP9. Likewise, RNAi silencing of ALK1 and ALK2 expression inhibits BMP9-induced BMPR-Smad activity and osteogenic differentiation in MSCs both in vitro and in vivo. Therefore, our results strongly suggest that ALK1 and ALK2 may play an important role in mediating BMP9-induced osteogenic differentiation. These findings should further aid us in understanding the molecular mechanism through which BMP9 regulates osteogenic differentiation of MSCs.
Keywords: Bone, Bone Morphogenetic Protein (BMP), Cell Differentiation, Protein-Protein Interactions, Signal Transduction, ALk Receptors, BMP-9, TGFb Type I Receptors, Mesenchymal Stem Cells, Osteogenic Differentiation
Introduction
Mesenchymal stem cells (MSCs),2 representing a very small fraction of the total population of nucleated cells in bone marrow are adherent marrow stromal cells that can self-renew and differentiate into osteogenic, chondrogenic, adipogenic, and myogenic lineages (1–4). Bone morphogenetic proteins (BMPs), members of the TGFβ superfamily, play an important role in stem cell biology (5, 6) and function to regulate cell proliferation and differentiation during development (7, 8). Several BMPs have been shown to regulate osteoblast differentiation and subsequent bone formation (3, 4, 7–9) and genetic disruptions of these factors have resulted in various skeletal and extraskeletal abnormalities during development (9, 10). We have conducted a comprehensive analysis of the osteogenic activity of 14 human BMPs and demonstrated that BMP9 is one of the most potent BMPs in promoting osteogenic differentiation of MSCs (3, 11, 12). We also demonstrated that osteogenic BMP9 regulates a distinct set of downstream targets in MSCs (13–16).
BMP9 (a.k.a., GDF2) was originally identified from fetal mouse liver cDNA libraries, and is a relatively uncharacterized member of the BMP family (17). BMP9 is highly expressed in the developing mouse liver, and recombinant human BMP9 stimulates hepatocyte proliferation (17, 18). It has been reported that BMP9 may play role in regulating glucose and iron homeostasis in liver (19, 20). BMP9 has been shown to be a potent synergistic factor for hematopoietic progenitor generation and colony formation (21) and may play a role in the induction and maintenance of the neuronal cholinergic phenotype in the central nervous system (22). Interestingly, the recombinant human BMP9 protein was shown to exert negligible osteoinductive activity in vivo (17), while we and others have demonstrated that exogenously expressed BMP9 is highly capable of inducing osteogenic differentiation (3, 4, 11, 12, 23). Nonetheless, the signaling mechanism through which BMP9 regulates osteogenic differentiation of mesenchymal stem cells remains to be fully elucidated.
Members of the TGFβ/BMP superfamily initiate their signaling events through an interaction of their type I and type II receptors, both of which are transmembrane serine/threonine kinases (24–26). Seven type I receptors (a.k.a. activin receptor-like receptors; ALK1 to ALK7) and four type II receptors have been identified in mice and humans (25, 26). In the TGFβ paradigm, the type II receptor binds with high affinity and is responsible for cooperative recruitment and transphosphorylation of its low-affinity type I pair (24). However, many BMP receptors have mixed affinities for their ligands. For example, ActRII has moderate affinity for BMP-7 and interacts weakly with BMP-2, whereas BMPRIA (e.g. ALK3) binds with high affinity to BMP-2 but binds weakly to BMP-7 (25, 26). As one of the least characterized BMPs, BMP9 has been recently shown that ALK1, ALK5 and/or endoglin may act as BMP9 type I receptors in endothelial cells (27–30). However, it remains unclear which type I receptor(s) play an essential role in BMP9-induced osteogenic differentiation of MSCs. Furthermore, it is conceivable that distinct type I receptor(s) may play a major role at different stages of MSC differentiation.
We have conducted a comprehensive analysis of the seven type I receptors for their role in BMP9 osteogenic signaling and demonstrated that ALK1 and ALK2 play an important role in mediating BMP9-induced osteogenic differentiation. Using dominant-negative mutants of seven type I receptors, we have demonstrated that ALK1 and ALK2 mutants effectively inhibit BMP9-induced early osteogenic differentiation in vitro and ectopic ossification in vivo. RNAi silencing of ALK1 and ALK2 inhibits the BMPR-Smad mediated transcription activity, the early osteogenic marker ALP activity, and the ectopic ossification of MSCs stimulated with BMP9. Taken together, these results reveal an important functional role of ALK1 and ALK2 in BMP9-induced osteogenic differentiation of MSCs.
EXPERIMENTAL PROCEDURES
Cell Culture and Chemicals
HEK-293, C2C12, C3H10T1/2, and HCT116 lines were obtained from the ATCC (Manassas, VA). HEK-293 and C2C12 cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM). C3H10T1/2 cells were maintained in complete Basal Medium Eagle. HCT116 cells were cultured in complete McCoy's 5A medium. Unless otherwise indicated, all chemicals were purchased from Sigma or Fisher Scientific.
Isolation of MEFs and Bone Marrow Stromal Cells (BMSCs)
MEFs were isolated from postcoitus day 13.5 mice, as previously described (31). Embryos were dissected into 10 ml of sterile PBS, voided of internal organs, and sheared through an 18-gauge syringe in the presence of 1 ml of 0.25% trypsin and 1 mm EDTA. After 15min incubation with gentle shaking at 37 °C, DMEM with 10% FCS was added to inactivate trypsin. Cells were plated on 100-mm dishes and incubated for 24 h at 37 °C. The adherent cells were used as MEF cells. Aliquots were kept in liquid nitrogen tanks. All MEFs used in this study were less than 5 passages.
BMSCs were harvested from young adult C57 mice by flushing marrow from the femurs and tibias with DMEM supplemented with 10% FCS, 1% penicillin/streptomycin, and 1% l-glutamine. Cells were passed through a 21-gauge needle syringe several times and washed by centrifugation in DMEM. Cells were then seeded into T-75 flasks at 37 °C in 5% CO2 and allowed to adhere to the flasks without disturbance for 3–5 days. All non-adherent cells were then removed, and the medium was changed every 3 days thereafter. The adherent cells (representing BMSCs) were trypsinized and passaged weekly. All BMSCs used in this study were within 3 passages.
Construction of Adenoviral Vectors Expressing BMP9 and Dominant-negative ALK (dnALKs)
Recombinant adenovirus expressing BMP9 were generated using the AdEasy technology as previously described (11, 12, 32–34). For generating recombinant adenoviruses expressing dnALKs, the coding regions containing extracellular and transmembrane domains of ALK1 to ALK7 were PCR amplified using the primers listed in supplemental Table S1, and subcloned into pAdTrace-TO4 and subsequently used to generate adenoviral recombinants. Recombinant adenoviruses (i.e. Ad-dnALKs) were produced and amplified in packaging HEK293 cells as described (11, 12, 32–34). The Ad-dnALKs also co-express RFP. An analogous adenovirus expressing only GFP or RFP (AdGFP or AdRFP) was used as a control (32–34). All PCR-amplified fragments and cloning junctions were verified by DNA sequencing. Details about the vector construction are available upon request.
Construction of siRNA-expressing Vectors Targeting Mouse ALK1 and ALK2
We used our recently developed pSOS system (35) to select and validate efficacious siRNA target sites of mouse ALK1 and ALK2, and designed three pairs of oligonucleotides containing siRNA target sites for the coding region of ALK1 or ALK2 using Dharmacon's siDESIGN program (supplemental Table S1). Oligo pairs were tested for their silencing efficiencies of mouse ALK1 and ALK2 in the pSOS vector. Meanwhile, the oligo pairs were annealed and subcloned into the Sfi I site of pSES. The shuttle vectors were used to generate adenoviral recombinant plasmids, which were pooled to produce adenovirus Ad-simALK1 and Ad-simALK2 using the AdEasy system (32–34). The resultant adenoviral vectors also express monomeric RFP. Knockdown efficiency was assessed by qPCR analysis. Authenticity of the oligonucleotide cassettes were verified by DNA sequencing. Cloning and construction details are available upon request. For adenovirus infections, the optimal MOIs (multiplicity of infection; usually 10–15 infectious virus units per cell) were determined for each batch adenovirus preparation in each cell line.
Preparation of BMP9-conditioned Medium
BMP9-conditioned medium (BMP9-CM) was prepared as described (31, 36). Briefly, subconfluent HCT116 cells (in 75 cm2 flaks) were infected with an optimal titer of AdBMP9, or AdGFP control. At 15 h post-infection, the culture medium was changed to serum-free DMEM. Conditioned medium was collected at 48 h postinfection and used immediately.
Total RNA Isolation, RT-PCR, and Quantitative Real-time PCR (qPCR) Analysis
Subconfluent cells were seeded in 75-cm2 cell culture flasks in a medium supplemented with 0.5% FCS with or without adenovirus infection. Total RNA was isolated using TRIzol Reagents (Invitrogen) according to the manufacturer's instructions. RT-PCR and qPCR were carried out as described (14–16, 31, 37). Ten micrograms of total RNA were used to generate cDNA templates by reverse transcription with hexamer and Superscript II reverse transcriptase (Invitrogen). The first strand cDNA products were further diluted and used as qPCR templates. The qPCR primers (supplemental Table S1) were 18-mers, designed using the Primer3 program to amplify the 3′-end (∼120 bp) of the gene of interest. SYBR Green-based qPCR analysis was carried out using the Opticon II DNA Engine (M J Research). The specificity of each qPCR reaction was verified by melting curve analysis and by resolving the PCR products on 1.5% agarose gels. 5-fold serially diluted pUC19 was used as a standard. Duplicate reactions were carried out for each sample. All samples were normalized by the expression level of GAPDH.
Transfection and Luciferase Reporter Assay
Exponentially growing cells were seeded in 25 cm2 cell culture flasks and transfected with 2 μg per flask of BMP Smad-responsive luciferase reporter, p12×SBE-Luc using Lipofectamine (Invitrogen). At 16 h after transfection, cells were replated to 24-well plates and infected with Ad-dnALKs or AdRFP at 4 h after replating. At 24 h after infection, cells were stimulated with BMP9 (or RFP) conditioned medium. At indicated time points, cells were lysed and cell lysates were collected for luciferase assays using Promega's Luciferase Assay kit. Each assay condition was performed in triplicate. Reporter activity was expressed as mean ± S.D.
Measurement of Alkaline Phosphatase (ALP) Activity
ALP was assessed by the modified SEAP chemiluminant assay (BD Clontech) and/or histochemical staining assay (using a mixture of 0.1 mg/ml napthol AS-MX phosphate and 0.6 mg/ml Fast Blue BB salt) as previously described (11, 12, 14–16, 31, 37).
Matrix Mineralization Assay
Exponentially growing C3H10T1/2 cells and MEFs were seeded in 24-well cell culture plates and were infected with Ad-dnALKs or AdRFP. Infected cells were cultured in the presence of BMP9-CM, ascorbic acid (50 μg/ml) and β-glycerophosphate (10 mm). At 14 and 21 days after infection, mineralized matrix nodules were stained for calcium precipitation by means of Alizarin Red S staining, as described previously (11, 12, 14–16, 31, 37). Cells were fixed with 0.05% (v/v) glutaraldehyde at room temperature for 10 min. After being washed with distilled water, fixed cells were incubated with 0.4% Alizarin Red S (Sigma-Aldrich) for 5 min, followed by extensive washing with distilled water. The staining of calcium mineral deposits was recorded under bright field microscopy.
Protein Fragment Complementation Assay (PCA)
Gaussia luciferase (GLuc) has 185 amino acids, in which the first 16 amino residues serve as signal peptide. According to Remy and Michnick (38), GLuc can be divided into two functionally complemented fragments, GLuc1-(17–93) and GLuc2-(94–185). The extracellular domains of ALK1 and ALK2 were fused to GLuc1, while the full-length BMP9 was fused with GLuc2. These fragments were cloned into expression vectors pBGLuc1 and pBGLuc2, resulting in ALK1-GLuc1, ALK2-GLuc1, and BMP9-GLuc2. The amplified fragments and cloning junctions were verified by DNA sequencing.
For PCA Gaussia luciferase assays, subconfluent 293 cells were seeded in 12-well plates and transfected with ALK1-GLuc1, ALK2-GLuc1, and/or BMP9-GLuc2 (0.5 μg/well). At 24 and 48 h after transfection, Gaussia luciferase activity was measured by using the Gaussia Luciferase Assay kit (New England Biolabs) according to the manufacturer's instructions. Each assay condition was done in triplicate.
Stem Cell Implantation and Ectopic Ossification
The use and care of animals were approved by the Institutional Animal Care and Use Committee. Subconfluent C3H10T1/2 cells were co-infected with Ad-dnALKs, Ad-simALKs or AdRFP and AdBMP9 or AdGFP for 15 h, and collected for subcutaneous injection (5 × 106 cells per injection) into the flanks of athymic nude (nu/nu) mice (four injections per group, 4–6-week-old, male, Harlan Sprague-Dawley). At 6 weeks after implantation, animals were sacrificed for microCT imaging and the implantation sites were retrieved for histologic evaluation.
MicroCT Imaging Analysis
Animals were sacrificed at 6 weeks and subjected to a high performance microCT imager that has a spatial resolution of 10 μ to 50 μ and a high contrast resolution as previously described (31). This unit provides quantitative measurements regarding the number and volume of each mass in each animal. MicroCT data were acquired and reconstructed into a three-dimensional image, and bone mass was quantified. To calculate the volume of each mass, the Image J program was used to determine the surface area of each planar slice of the microCT, the surface areas were summed, and volume was calculated as ((sum of the surface area of each slice) × 0.0543). These volumes were averaged by dividing by the number of samples for each respective injection condition (n = 4).
Hematoxylin & Eosin, Trichrome, and Alcian Blue Staining
Retrieved tissues were fixed in 10% formalin overnight and embedded in paraffin. Serial sections of the embedded specimens were stained with hematoxylin and eosin (H & E). Masson's Trichrome and Alcian Blue staining was carried out as described (12, 16, 31, 39).
RESULTS
Endogenous Expression of the Seven Type I Receptors in MSCs and Pre-osteoblast Progenitor Cells
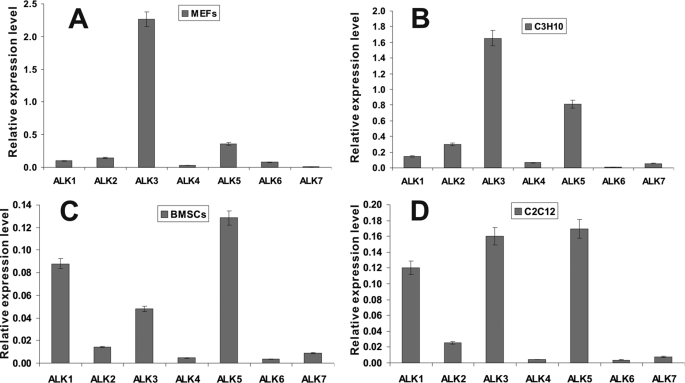
We previously identified BMP9 as one of the most potent BMPs in promoting osteogenic differentiation of MSCs (3, 4, 11, 12). However, the signaling mechanisms and functions of BMP9 largely remain undefined. Here, we sought to determine the obligate type I receptor(s) for BMP9-mediated osteogenic signaling in MSCs. We first examined the endogenous expression of all seven type I TGFβ/BMP receptors in MSCs and osteoblast progenitor cells. Four types of cells were chosen. C3H10T1/2 and MEFs may present early stages of MSCs, while BMSCs may represent a pool of MSCs containing osteoblast progenitor cells at differentiation stages (3, 4). C2C12 cells are myoblastic cells that can be trans-differentiated into the osteoblastic lineage upon osteogenic BMP stimulation (3, 4). The relative expression levels of the seven type I receptors are shown in Fig. 1. ALK3 and ALK5 were in general highly expressed in all four cell types. On the contrary, expression of ALK4, ALK6, and ALK7 was low or undetectable. Expression of ALK2 was modest and remained constant, while ALK1 expression seemed to increase with differentiation status (i.e. lower in C3H10T1/2 and MEFs, and higher in BMSCs and C2C12). Further investigation is required to determine if ALK1 expression plays a role at later stages of MSC differentiation.
FIGURE 1.
Endogenous expression of the seven type I receptors in MSCs and pre-osteoblast progenitor cells. Subconfluent cells were cultured in 1% FCS medium for 24 h. Total RNA was isolated from MEFs (A), C3H10T1/2 (B), BMSCs (C), and C2C12 (D) cells, and was subjected to reverse transcription and qPCR analysis. All samples were normalized for GAPDH expression. Reactions were done in triplicate. Relative expression levels of the type I receptors were expressed as mean ± S.D.
Dominant-negative Mutants of ALK1 and ALK2 Inhibit BMP9-induced Osteogenic Marker ALP Activity in Pre-osteoblast Progenitor Cells
To delineate possible obligate type I receptor(s) for BMP9-induced osteogenic signaling, we constructed dominant-negative mutants for seven type I receptors (i.e. dnALK1 to dnALK7), which contain extracellular and transmembrane domains but lack cytoplasmic domains (Fig. 2A). To effectively transduce these mutants into MSCs, we generated recombinant adenoviral vectors expressing these mutants (i.e. Ad-dnALKs) using the AdEasy technology (32–34). Ad-dnALK viral vectors also express monomeric RFP. As shown in Fig. 2B, the Ad-dnALK and control Ad-RFP vectors effectively transduced C3H10T1/2 cells. We further determined the transgene expression mediated by the Ad-dnALKs. We chose to use semi-quantitative RT-PCR analysis because antibodies are not available for all seven type I receptors and/or different antibodies for the same protein may exhibit different affinities/reactivities. The level of Ad-dnALK-mediated expression was comparable among dnALKs when RT-PCR primers specific for extracellular regions were used (Fig. 2C).
FIGURE 2.
Construction and characterization of adenoviral vectors expressing dominant-negative ALKs (Ad-dnALKs). A, schematic representation of the seven dnALKs. The dotted lines denote the truncated cytoplasmic regions of the type I receptors. B, adenoviral vectors expressing dnALKs effectively transduced MSCs. Subconfluent C3H10T1/2 cells were infected the same titer of Ad-dnALKs (i.e. MOI = 10). Ad-RFP virus was used as a control vector. RFP signal was recorded under a fluorescence microscope at 24 h after infection. C, adenovirus-mediated expression of dnALKs. Subconfluent C3H10T1/2 cells were infected with a comparable titer of Ad-dnALKs or Ad-RFP. At 36 h postinfection, total RNA was isolated from the infected cells and subjected to RT-cDNA reactions with (“+” lanes) or without (“−” lanes) reverse transcriptase. The cDNA products were used for PCR amplification using dnALK-specific primers. The PCR products were resolved on 1.2% agarose gel. M, 1kb+ DNA size ladder (Invitrogen). See text for details.
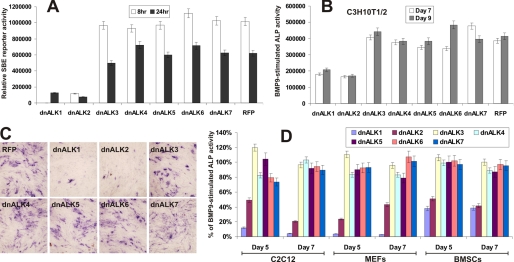
We next determined the function of dnALKs in BMP9 signaling pathway using the BMPR Smad-responsive reporter (40). Experimentally, C3H10T1/2 cells were transfected with 12xSBE-Luc reporter and infected with Ad-dnALKs or Ad-RFP for 24 h, and then stimulated with BMP9-CM. Luciferase activity was measured at 8 and 24 h after BMP9-CM stimulation. Expression of dnALK1 and dnALK2 were shown to effectively inhibit BMPR-Smad reporter activity induced by BMP9, while the other five dnALKs exhibited no significant inhibitory effects (Fig. 3A). When the effect of dnALKs on the BMP9-induced early osteogenic marker ALP activity was examined in C3H10T1/2 cells, both dnALK1 and dnALK2 were shown to inhibit ALP activity quantitatively (Fig. 3B) and qualitatively (Fig. 3C). Accordingly, we obtained similar results using different osteoblast progenitor cells, and found that dnALK1 and dnALK2 exerted inhibitory effect on BMP9-induced ALP activity in MEFs, BMSCs, and C2C12 cells (Fig. 3D). Thus, our results suggest that ALK1 and ALK2 may play an important role in transducing BMP9-initiated osteogenic signaling.
FIGURE 3.
Dominant-negative mutants of ALK1 and ALk2 inhibit BMP9 induced ALP activity in pre-osteoblast progenitor cells. A, dnALK1 and dnALK2 inhibit BMP R-Smad reporter activity induced by BMP9. Subconfluent C3H10T1/2 cells were transfected with 12xSBE-Luc reporter and infected with Ad-dnALKs or Ad-RFP. At 24 h post-transfection/infection, cells were stimulated with BMP9-conditioned medium. Luciferase activity was measured at the indicated time points. Each assay condition was done in triplicate. B and C, inhibition of BMP9 induced ALP activity by dnALK1 and dnALK2. Subconfluent C3H10T1/2 cells were infected with Ad-dnALKs or Ad-RFP. At 24-h postinfection, cells were stimulated with BMP9-conditioned medium. ALP activity was measured at the indicated time points (B) and was stained histochemically (C) at day 7. Each assay condition was done in triplicate. D, dnALK1 and dnALK2-mediated inhibition of BMP9 induced ALP activity in pre-osteoblast progenitor cells. Subconfluent C2C12 cells, MEFs and BMSCs were infected with Ad-dnALKs or Ad-RFP. At 24 h postinfection, cells were stimulated with BMP9-conditioned medium. ALP activity was measured at the indicated time points. Each assay condition was done in triplicate.
Dominant-negative Mutants of ALK1 and ALK2 Inhibit BMP9-induced ALP Activity in a Dose-dependent Manner
The relative expression levels of dnALK1 and dnALK2 over their endogenous wild-type counterparts may affect the dominant-negative inhibitory activities of dnALK1 and dnALK2. Thus, we sought to determine whether BMP9-induced ALP activity could be inhibited by dnALK1 and dnALK2 in a dose-dependent manner. As shown in Fig. 4, A and B, an increase in dnALK1 or dnALK2 mutant in C3H10T1/2 cells led to a significantly higher reduction in BMP9-induced ALP activity. At the highest dose of either dnALK1 or dnALK2, the ALP activity decreased to ∼30 and 40% of the positive control for dnALK1 and dnALK2, respectively. When medium doses of both dnALK1 and dnALK2 were used, there was a slight but detectable synergistic inhibition on BMP9-induced ALP activity (Fig. 4A, far right panel). The dose-dependent inhibition by dnALK1 or dnALK2 on BMP9-induced early osteogenic marker ALP was more pronounced in MEFs. As shown in Fig. 4C, the medium and high doses of dnALK1 almost completely abolished BMP9-induced ALP activity at day 5 and day 7. Accordingly, the high dose of dnALK2 inhibited over 90% of the BMP9-induced ALP activity at day 7 (Fig. 4D). Interestingly, our results also indicate that dnALK1 was seemingly more effective on inhibiting BMP9-induced ALP activity, especially in MEFs.
FIGURE 4.
Dominant-negative mutants of ALK1 and ALk2 inhibit BMP9 induced ALP activity in a dose-dependent manner. A and B, dnALK1 and dnALK2 inhibit BMP9 induced ALP activity in a dose-dependent fashion. Subconfluent C3H10T1/2 cells were infected with Ad-dnALK1, Ad-dnALK2 and/or Ad-RFP at three escalating titers, each of which had a 50% increase increment. At 24 h postinfection, cells were stimulated with BMP9-conditioned medium. ALP activity was measured at the indicated time points (A) and was stained histochemically (B) at day 7. Each assay condition was done in triplicate. C, dnALK1 inhibits BMP9 induced ALP activity in MEFs. Subconfluent MEFs were infected with Ad-dnALK1 and/or Ad-RFP at three escalating titers (50% increment). At 24 h postinfection, cells were stimulated with BMP9-conditioned medium. ALP activity was measured at the indicated time points. Each assay condition was done in triplicate. D, dnALK2 inhibits BMP9 induced ALP activity in MEFs. Subconfluent MEFs were infected with Ad-dnALK2 and/or Ad-RFP at three escalating titers (50% increment). At 24 h post-infection, cells were stimulated with BMP9-conditioned medium. ALP activity was measured at the indicated time points. Each assay condition was done in triplicate.
Dominant-negative Mutants of ALK1 and ALk2 Inhibit BMP9-induced Expression of Inhibitory Smads and in Vitro Matrix Mineralization in MSCs
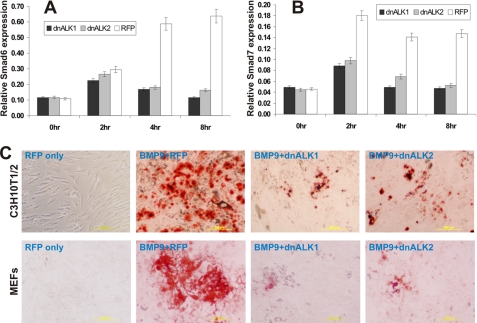
We further examined the inhibitory effects of dnALK1 and dnALK2 on BMP9-activated Smad signaling and late stage BMP9-induced osteogenic differentiation. Both inhibitory Smad6 and Smad7 are known early targets of BMP signaling (41–43). Upon BMP9 stimulation, Smad6 was effectively induced at 4h (Fig. 5A), while Smad7 induction was evident as early as 1-h poststimulation (Fig. 5B). Nonetheless, BMP9-induced expression of both Smads was effectively inhibited by dnALK1 and dnALK2 (Fig. 5, A and B). These results suggest that ALK1 and ALK2 may play an important role in transducing the early signaling events solicited by BMP9 in MSCs.
FIGURE 5.
Dominant-negative mutants of ALK1 and ALk2 inhibit BMP9 induced inhibitory Smad expression and matrix mineralization in MSCs. A and B, BMP9 induced Smad6 and Smad7 expression is inhibited by dnALK1 and dnALK2. Subconfluent C3H10T1/2 cells were infected with Ad-dnALK1, Ad-dnALK2, or Ad-RFP for 24 h, and were stimulated with BMP9-conditioned medium. Total RNA was collected at the indicated time points and subjected to reverse transcriptions and qPCR analysis. The qPCR analysis was done in triplicate. C, BMP9-induced mineralization is inhibited by dnALK1 and dnALK2. Subconfluent C3H10T1/2 cells and MEFs were infected with Ad-dnALK1, Ad-dnALK2, or Ad-RFP for 24 h, and were stimulated with BMP9-conditioned medium. Mineralization was assessed by using Alizarin Red S staining at day 20. Experiments were carried out in duplicate. Representative staining is shown.
We further tested if dnALK1 and dnALK2 would not only affect BMP9-induced ALP activity but also inhibit the BMP9-induced late stage of osteogenic differentiation. Subconfluent C3H10T1/2 cells and primary MEFs were first infected with Ad-dnALK1, Ad-dnALK2, or Ad-RFP for 24 h, and were stimulated with BMP9-conditioned medium. Matrix mineralization was assessed by using Alizarin Red S staining at day 20. As shown in Fig. 5C, BMP9-induced mineralized nodule formation was remarkably inhibited by dnALK1 and dnALK2 in C3H10T1/2 cells and MEFs.
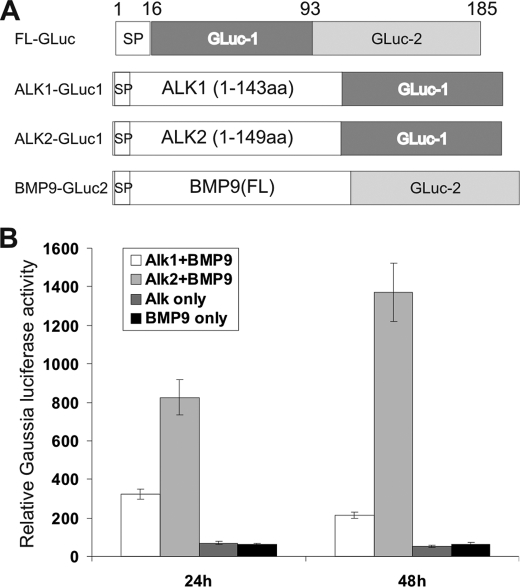
ALK1 and ALK2 Directly Interact with BMP9 as Determined by PCA
PCA has recently been used as a novel technique to detect protein-protein interactions. A recent report has demonstrated that Gaussia luciferase can be split into two functionally complementary fragments for PCA assays (38), GLuc1-(17–93) and GLuc2-(94–185) (Fig. 6A). We constructed ALK1-GLuc1, ALK2-GLuc1, and BMP9-GLuc2 by fusing the extracellular domains of ALK1 and ALK2 to GLuc1, and the full-length BMP9 with GLuc2 (Fig. 6A). We next conducted PCA Gaussia luciferase assay in 293 cells by transfecting ALK1-GLuc1, ALK2-GLuc1, and/or BMP9-GLuc2. Gaussia luciferase activity was measured at 24 and 48 h after transfection. As shown in Fig. 6B, significant increases in Gaussia luciferase activity in the cells co-transfected with ALK1-GLuc1+BMP9-GLuc2 and ALK2-GLuc1+BMP9-GLuc2, while the cells transfected with ALK1-GLuc1, ALK2-GLuc1, or BMP9-GLuc2 only exhibited basal Gaussia luciferase activity. These results suggest that ALK1 and ALK2 may directly interact with BMP9.
FIGURE 6.
Interaction of BMP9 with ALK1 and ALK2 determined by PCA. A, schematic depiction of constructs used for PCA assay. GLuc has 185 amino acids, in which the first 16 amino residues serve as signal peptide. According to Remy and Michnick (38), GLuc can be split into two functionally complemented fragments, GLuc1-(17–93) and GLuc2-(94–185). The extracellular domains of ALK1 and ALK2 were fused to GLuc1, while the full-length BMP9 was fused with GLuc2. FL, full-length; SP, signal peptide. B, subconfluent 293 cells were transfected with ALK1-GLuc1, ALK2-GLuc1, and/or BMP9-Gluc2. Relative Gaussia luciferase activity was determined at 36 h after transfection using the Gaussia luciferase assay kit from New England Biolabs. Each assay condition was done in triplicate.
BMP9-induced Ectopic Bone Formation Is Inhibited by ALK1 and ALK2 Dominant-negative Mutants
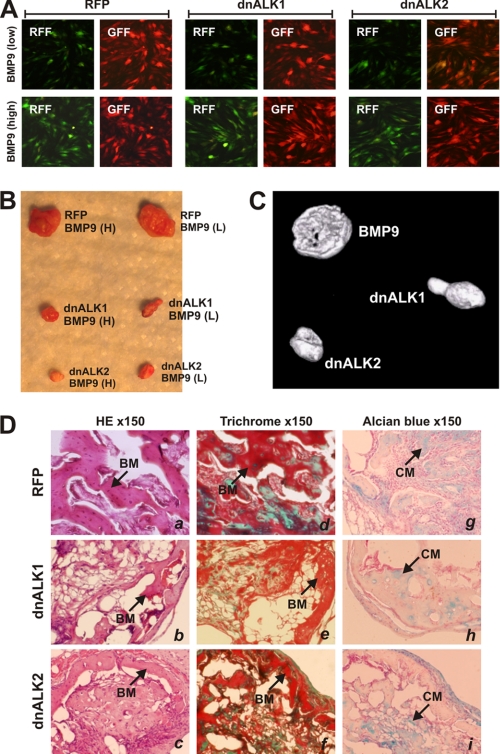
Using the MSC implantation assay, we next determined the effect of ALK1 and ALK2 mutants on BMP9-induced ectopic ossification. Briefly, subconfluent C3H10T1/2 cells were effectively infected with Ad-dnALK1, Ad-dnALK2, AdRFP, and low and high titers of AdBMP9 for 15 h (Fig. 7A). The infected cells were collected and implanted subcutaneously in athymic nude mice. At 6 weeks, animals were sacrificed, and the ectopic bone masses were retrieved. MSCs expressing dnALK1 or dnALK2 formed significantly smaller bony masses (Fig. 7B), which were further confirmed by microCT imaging analysis (Fig. 7C). H & E and Trichrome staining analyses revealed that more immature osteoid matrix and thinner trabeculae were found in bone masses formed by the dnALK1 or adALK2 expressing MSCs (Fig. 7D). However, ALK1 and ALK2 mutants did not seem to affect chondrogenesis (Fig. 7D). Thus, these in vivo results further substantiate the in vitro findings about the important role of ALK1 and ALK2 in BMP9-induced osteogenic differentiation of MSCs.
FIGURE 7.
Inhibition of BMP9-induced ectopic bone formation by dnALK1 and dnALK2. Subconfluent 3H10T1/2 cells were infected with Ad-dnALK1, Ad-dnALK2, AdRFP and low and high titers of AdBMP9 for 15 h (A). The cells were collected and implanted subcutaneously in athymic mice. B, at 6 weeks, animals were sacrificed, and the ectopic bone masses were retrieved. C, retrieved samples were subjected to microCT imaging analysis, and representative three-dimensional reconstructed images are shown. D, histologic analysis of the retrieved samples. The samples were decalcified and paraffin-embedded and sectioned for H & E stain (panels a–c), Trichrome stain (panels d–f), and Alcian blue stain (panels g–i). BM, mineralized bone matrix; CM, chondroid matrix; magnification, ×150.
BMP9 Osteogenic Signaling Is Impaired by RNAi-mediated Silencing of Mouse ALK1 and ALK2 Expression
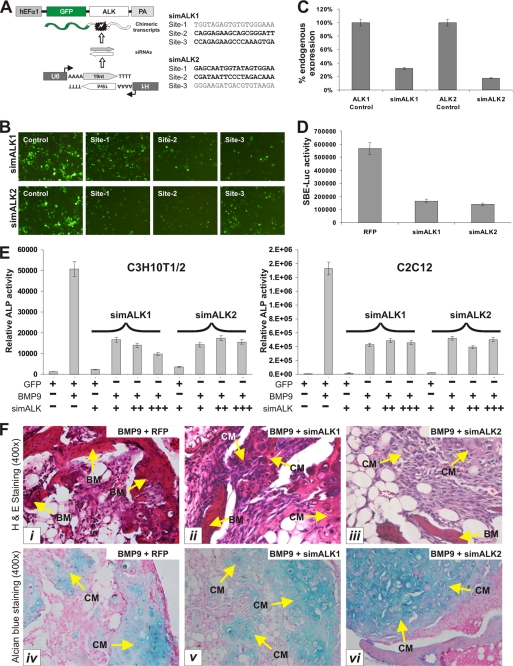
Using the dominant-negative mutants of ALK1 and ALK2 we have demonstrated that both may play an important role in regulating BMP9 osteogenic signaling pathway. One concern over the use of dominant-negative mutants is that the mutants may squelch other factors and result in a nonspecific effect. To further verify the important role of ALK1 and ALK2 in BMP9-mediated osteogenic signaling, we constructed siRNA vectors that target the expression of mouse ALK1 and ALK2 using our recently developed pSOS system (35). We chose three putative siRNA sites for each target gene (Fig. 8A) and tested their knockdown efficiency of the chimeric GFP/ALK1 or GFP/ALK2 in 293 cells. As shown in Fig. 8B, two of the three candidate sites for either ALK1 or ALK2 effectively silenced GFP signal. We subcloned these siRNA sites into a shuttle vector and constructed adenoviral vectors that express siRNAs targeting mouse ALK1 and ALK2 (namely Ad-simALK1 and Ad-simALK2). We were able to show that Ad-simALK1 and Ad-simALK2 effectively knocked down endogenous ALK1 and ALK2 expression in C3H10T1/2 cells by about 70 and 80%, respectively (Fig. 8C). Furthermore, expression of ALK1 and ALK2 siRNAs in C3H10T1/2 cells significantly inhibited BMP9-activated BMPR-Smad reporter activity (Fig. 8D). Collectively, these results demonstrate that the ALK1 and ALK2 siRNA vectors are effective and specific.
FIGURE 8.
Inhibition of BMP9 signaling by RNAi-mediated knockdown of ALK1 and ALK2 gene expression. A, schematic representation of siRNA selection strategy for mouse ALK1 and ALK2. Detailed information about the pSOS system was previously described (35). B, selection of siRNAs targeting mouse ALK1 and ALK2. The target sites were subcloned and tested using the pSOS system. The resultant vectors were transfected into 293 cells, and knockdown of chimeric GFP/ALK1 or GFP/ALK2 expression was recorded 5 days after transfection. C, verification of ALK1 and ALK2 knockdown in C3H10T1/2 cells. Total RNA was collected from subconfluent transfected cells and subjected to qPCR analysis using primers corresponding to the 3′-UTR of mouse ALK1 and ALK2. All samples were normalized for GAPDH expression. D, inhibition of BMP R-Smad reporter activity by ALK1 and ALK2 knockdown in MSCs. Representative results of three independent experiments are shown. E, effect of silencing ALK1 and ALK2 expression on BMP9-induced ALP activity. Subconfluent C3H10T1/2 and C2C12 cells were co-infected with AdBMP9 and AdGFP or various titers of Ad-simALK1 and Ad-simALK2. ALP activity was measured at day 5 after infection. Each assay condition was done in triplicate. F, effect of silencing ALK1 and ALK2 expression on BMP9-induced ectopic ossification. C3H10T1/2 cells were co-infected with AdBMP9 and AdGFP, Ad-simALK1, or Ad-simALK2 for 15 h, collected, and subjected to subcutaneous injection into flanks of athymic mice. At 6 weeks, animals were sacrificed, and the ectopic bone masses were retrieved and subjected to H & E stain (panels i-iii) and Alcian blue stain (panels iv-vi). BM, mineralized bone matrix; CM, chondroid matrix; magnification, ×150.
We sought to determine if BMP9-initiated osteogenic signaling was affected by silencing ALK1 and ALK2 in MSCs. Silencing ALK1 and ALK2 was shown to effectively inhibit BMP9-induced ALP activities in both C3H10T1/2 and C2C12 cells (Fig. 8E). These results were further confirmed by the in vivo stem cell implantation assay. Co-expression of BMP9 and ALK1 or ALK2 knockdown vectors in MSCs led to a decrease in ectopic ossification and bone matrix mineralization, and to an increase in undifferentiated MSCs (Fig. 8F, top panel). However, there was no significant effect on chondrogenesis (Fig. 8F, bottom panel). Therefore, the results obtained from ALK1 and ALK2 knockdown experiments were consistent with that from the use of ALK1 and ALK2 dominant-negative mutants. Taken together, our findings have collectively demonstrated that ALK1 and ALK2 play an important role in transducing BMP9-initiated osteogenic signaling in mesenchymal stem cells.
DISCUSSION
We have recently identified BMP9 as one of the most potent osteogenic BMPs both in vitro and in vivo (3, 4, 11, 12). Further expression profiling analysis has identified several important signaling mediators (such as Id HLH, CTGF, and Hey1) of BMP9-induced osteogenic differentiation of MSCs (13–16). However, BMP9 remains as one of least characterized BMPs. Little is known about initial signaling events involved in BMP9 signaling. It has been recently reported that ALK1 may function as a potential receptor for BMP9 based on BIAcore assay (27), whereas other studies suggest that ALK1, ALK5, and/or endoglin may act as BMP9 type I receptors in endothelial cells (28–30). However, it remains unclear which type I receptor(s) play essential role in BMP9-induced osteogenic differentiation of MSCs.
In this study, we conducted a comprehensive analysis of the seven type I receptors for their role in BMP9 osteogenic signaling and demonstrated that ALK1 and ALK2 play an important role in mediating BMP9-induced osteogenic differentiation. Using dominant-negative mutants of seven type I receptors, we have demonstrated that ALK1 and ALK2 mutants effectively inhibit BMP9-induced early osteogenic differentiation in vitro and ectopic ossification in vivo. RNAi silencing of ALK1 and ALK2 inhibits the BMPR-Smad mediated transcription activity, the early osteogenic marker ALP activity, and the ectopic ossification of MSCs stimulated with BMP9. Thus, our results reveal an important functional role of ALK1 and ALK2 in BMP9-induced osteogenic differentiation of MSCs.
We have conducted mostly the functional analyses of the involvement of type I receptors in BMP9 osteogenic signaling. We attempted unsuccessfully to pull down the BMP9 and ALK1 or ALK2 protein complex through immunoprecipitation (data not shown). We reason that the ternary complex formed by BMP9 and its type I receptors may be transient and highly sensitive to changes in ionic strength or detergents during cell lysis and/or in vitro manipulations. It is also possible that the affinity of BMP9 binding to its type I receptors is relatively low. This is consistent with our observation that the secreted forms of ALK1 and ALK2 extracellular dominant-negative mutants exerted very limited inhibitory effect on BMP9-induced osteogenic signaling (data not shown). Nonetheless, our functional analysis has demonstrated that ALK1 and ALK2 are important type I receptors for BMP9-induced osteogenic differentiation in MSCs.
Although in endothelial cells ALK1, ALK3, or ALK5 has been reported as BMP9 receptors, it is conceivable that different receptor combinations may play an important role in determining the biological outcomes of BMP9 action. We have found most of the seven type I receptors are expressed in MSCs or osteoblastic progenitor cells. Interestingly, ALK3 and ALK5 are abundantly expressed in MSCs. However, their dominant-negative mutants do not effectively block BMP9 osteogenic signaling activity in vitro and in vivo. These findings suggest that interaction between BMP9 and its type I receptors may be cell type-specific and/or cell context-dependent. However, the detailed molecular mechanism behind BMP9-receptor interaction requires further investigation.
The crystal structure of BMP9 and its differences with the existing crystal structures of other BMPs, both in isolation and in complex with their receptors, has recently been reported (27). Like other TGFβ/BMP ligands, BMP9 is translated as precursors, with pro-regions that after cleavage from the ligand pro-region of BMP9 remains tightly associated after secretion (27). However, it was found that the activities of BMP9 and BMP9 pro-region complex were equivalent (27). Using surface plasmon resonance studies (BIAcore) and cell-based assays to test the ability of soluble ALK1 to block the activity of BMP9 pro-region complex, Brown MA et al. (27) identified ALK1 as a potential receptor for BMP9. Thus, the structural data support that at least ALK1 may function as a cognate type I receptor for BMP9.
ALK1 is an orphan receptor in the TGF-β family. It has been implicated as an inhibitor of lateral TGF-β/ALK-5 signaling (44), correlated with vasculogenesis and angiogenesis (45). ALK1 may be involved in hereditary hemorrhagic telangiectasia as heterozygotes with mutations in the ALK1 gene develop hereditary hemorrhagic telangiectasia type 2 (HHT2), also known as Osler-Rendu-Weber syndrome, an autosomal dominant disorder characterized by multisystemic vascular dysplasia and recurrent hemorrhage (46). ALK1 homozygous embryos die at midgestation, exhibiting severe vascular abnormalities characterized by excessive fusion of capillary plexes into cavernous vessels and hyperdilation of large vessels (47). The vascular defects are associated with enhanced expression of angiogenic factors and proteases, and are characterized by deficient differentiation and recruitment of vascular smooth muscle cells, which are reminiscent of mice lacking TGFβ1, TGFβ type II receptor, or endoglin (47). This suggests that ALK1 signaling pathway in endothelial cells plays a crucial role in determining vascular endothelial properties during angiogenesis. Nonetheless, the ALK1 role in BMP9-induced osteogenic differentiation of MSCs requires further investigation.
It has been reported that ALK2 can bind both activin and BMPs in conjunction with the activin and BMP type II receptors. In mice, ALK2 is primarily expressed in the extra-embryonic visceral endoderm before gastrulation and later in both embryonic and extra-embryonic cells during gastrulation (48). Homozygous ALK2 mutant embryos were arrested at the early gastrulation stage, displaying abnormal visceral endoderm morphology and severe disruption of mesoderm formation (48). In fact no homozygous mutants were recovered after E9.5 (49). Subsequent studies suggest that signaling through ALK2 is essential in extra-embryonic tissues at the time of gastrulation for normal mesoderm formation and that subsequent ALK2 signaling is essential for normal development after gastrulation (48, 49). Thus, these findings may indirectly support ALK2's role in BMP9-induced mesenchymal stem cell differentiation. Consistent with this possibility, it has been recently reported that ALK2 R206H mutation linked to FOP confers constitutive activity to the BMP Type I receptor and sensitizes mesenchymal cells to BMP-induced osteoblast differentiation and bone formation, suggesting that aberrant activation of BMP9 in soft tissues may cause fibrodysplasia ossificans progressiva (FOP) (50).
In conclusion, we have conducted a comprehensive analysis of the functional role of type I receptors in BMP9 induced osteogenic signaling in MSCs. Using dominant-negative mutants of the seven type I receptors, we have demonstrated that ALK1 and ALK2 are important receptors for BMP9-induced osteogenic differentiation both in vitro and in vivo. These results have been confirmed by using siRNAs to specifically target ALK1 and ALK2. Future studies should be devoted to the elucidation of detailed mechanism behind BMP9 and ALK1 or ALK2 interaction in the context of MSC differentiation.
Supplementary Material
Acknowledgment
We thank Dr. Di Chen of the University of Rochester Medical Center for kindly providing p12xSBE-Luc reporter.
This work was supported, in whole or in part, by Research Grants CA106569, AT004418, AR50142, and AR054381 (to T.-C. H., R. C. H., and H. H. L.) from the National Institutes of Health. This work was also supported by The Brinson Foundation (to T.-C. H.), the Orthopaedic Research and Education Foundation (to R. C. H. and H. H. L.), the 863 Program of Ministry of Science and Technology of China (2007AA2z400, to T.-C. H. and Z.-L. D.), the Natural Science Foundation of China (30901530, to X. L., 30800658, to J. L., and 30772211, to Z.-L. D.), and the Natural Science Foundation Project of Chongqing Science and Technology Commission 2008BB5396 (to L. C. and M. T.) and 2009BB5060 (to J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- MSC
- mesenchymal stem cells
- GLuc
- Gaussia luciferase
- BMP
- bone morphogenetic protein
- BMSC
- bone marrow stromal cells
- ALP
- alkaline phosphatase
- PCA
- protein fragment complementation assay.
REFERENCES
- 1.Prockop D. J. (1997) Science 276, 71–74 [DOI] [PubMed] [Google Scholar]
- 2.Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S., Marshak D. R. (1999) Science 284, 143–147 [DOI] [PubMed] [Google Scholar]
- 3.Luu H. H., Song W. X., Luo X., Manning D., Luo J., Deng Z. L., Sharff K. A., Montag A. G., Haydon R. C., He T. C. (2007) J. Orthop. Res. 25, 665–677 [DOI] [PubMed] [Google Scholar]
- 4.Deng Z. L., Sharff K. A., Tang N., Song W. X., Luo J., Luo X., Chen J., Bennett E., Reid R., Manning D., Xue A., Montag A. G., Luu H. H., Haydon R. C., He T. C. (2008) Front Biosci. 13, 2001–2021 [DOI] [PubMed] [Google Scholar]
- 5.Varga A. C., Wrana J. L. (2005) Oncogene 24, 5713–5721 [DOI] [PubMed] [Google Scholar]
- 6.Zhang J., Li L. (2005) Dev. Biol. 284, 1–11 [DOI] [PubMed] [Google Scholar]
- 7.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 8.Attisano L., Wrana J. L. (2002) Science 296, 1646–1647 [DOI] [PubMed] [Google Scholar]
- 9.Hogan B. L. (1996) Genes Dev. 10, 1580–1594 [DOI] [PubMed] [Google Scholar]
- 10.Zhao G. Q. (2003) Genesis 35, 43–56 [DOI] [PubMed] [Google Scholar]
- 11.Cheng H., Jiang W., Phillips F. M., Haydon R. C., Peng Y., Zhou L., Luu H. H., An N., Breyer B., Vanichakarn P., Szatkowski J. P., Park J. Y., He T. C. (2003) J. Bone Joint Surg. Am. 85-A, 1544–1552 [DOI] [PubMed] [Google Scholar]
- 12.Kang Q., Sun M. H., Cheng H., Peng Y., Montag A. G., Deyrup A. T., Jiang W., Luu H. H., Luo J., Szatkowski J. P., Vanichakarn P., Park J. Y., Li Y., Haydon R. C., He T. C. (2004) Gene Ther. 11, 1312–1320 [DOI] [PubMed] [Google Scholar]
- 13.Peng Y., Kang Q., Cheng H., Li X., Sun M. H., Jiang W., Luu H. H., Park J. Y., Haydon R. C., He T. C. (2003) J. Cell. Biochem. 90, 1149–1165 [DOI] [PubMed] [Google Scholar]
- 14.Peng Y., Kang Q., Luo Q., Jiang W., Si W., Liu B. A., Luu H. H., Park J. K., Li X., Luo J., Montag A. G., Haydon R. C., He T. C. (2004) J. Biol. Chem. 279, 32941–32949 [DOI] [PubMed] [Google Scholar]
- 15.Luo Q., Kang Q., Si W., Jiang W., Park J. K., Peng Y., Li X., Luu H. H., Luo J., Montag A. G., Haydon R. C., He T. C. (2004) J. Biol. Chem. 279, 55958–55968 [DOI] [PubMed] [Google Scholar]
- 16.Sharff K. A., Song W. X., Luo X., Tang N., Luo J., Chen J., Bi Y., He B. C., Huang J., Li X., Jiang W., Zhu G. H., Su Y., He Y., Shen J., Wang Y., Chen L., Zuo G. W., Liu B., Pan X., Reid R. R., Luu H. H., Haydon R. C., He T. C. (2009) J. Biol. Chem. 284, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J. J., Celeste A. J., Kong F. M., Jirtle R. L., Rosen V., Thies R. S. (1995) Endocrinology 136, 4293–4297 [DOI] [PubMed] [Google Scholar]
- 18.Miller A. F., Harvey S. A., Thies R. S., Olson M. S. (2000) J. Biol. Chem. 275, 17937–17945 [DOI] [PubMed] [Google Scholar]
- 19.Chen C., Grzegorzewski K. J., Barash S., Zhao Q., Schneider H., Wang Q., Singh M., Pukac L., Bell A. C., Duan R., Coleman T., Duttaroy A., Cheng S., Hirsch J., Zhang L., Lazard Y., Fischer C., Barber M. C., Ma Z. D., Zhang Y. Q., Reavey P., Zhong L., Teng B., Sanyal I., Ruben S. M., Blondel O., Birse C. E. (2003) Nat. Biotechnol. 21, 294–301 [DOI] [PubMed] [Google Scholar]
- 20.Truksa J., Peng H., Lee P., Beutler E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10289–10293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ploemacher R. E., Engels L. J., Mayer A. E., Thies S., Neben S. (1999) Leukemia 13, 428–437 [DOI] [PubMed] [Google Scholar]
- 22.López-Coviella I., Berse B., Krauss R., Thies R. S., Blusztajn J. K. (2000) Science 289, 313–316 [DOI] [PubMed] [Google Scholar]
- 23.Jane J. A., Jr., Dunford B. A., Kron A., Pittman D. D., Sasaki T., Li J. Z., Li H., Alden T. D., Dayoub H., Hankins G. R., Kallmes D. F., Helm G. A. (2002) Mol. Ther. 6, 464–470 [DOI] [PubMed] [Google Scholar]
- 24.Massagué J., Weis-Garcia F. (1996) Cancer Surv. 27, 41–64 [PubMed] [Google Scholar]
- 25.Miyazono K., Maeda S., Imamura T. (2005) Cytokine Growth Factor Rev. 16, 251–263 [DOI] [PubMed] [Google Scholar]
- 26.Sieber C., Kopf J., Hiepen C., Knaus P. (2009) Cytokine Growth Factor Rev. 20, 343–355 [DOI] [PubMed] [Google Scholar]
- 27.Brown M. A., Zhao Q., Baker K. A., Naik C., Chen C., Pukac L., Singh M., Tsareva T., Parice Y., Mahoney A., Roschke V., Sanyal I., Choe S. (2005) J. Biol. Chem. 280, 25111–25118 [DOI] [PubMed] [Google Scholar]
- 28.Scharpfenecker M., van Dinther M., Liu Z., van Bezooijen R. L., Zhao Q., Pukac L., Löwik C. W., ten Dijke P. (2007) J. Cell Sci. 120, 964–972 [DOI] [PubMed] [Google Scholar]
- 29.Upton P. D., Davies R. J., Trembath R. C., Morrell N. W. (2009) J. Biol. Chem. 284, 15794–15804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao E. S., Lin L., Yao Y., Boström K. I. (2009) Blood 114, 2197–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang N., Song W. X., Luo J., Luo X., Chen J., Sharff K. A., Bi Y., He B. C., Huang J. Y., Zhu G. H., Su Y. X., Jiang W., Tang M., He Y., Wang Y., Chen L., Zuo G. W., Shen J., Pan X., Reid R. R., Luu H. H., Haydon R. C., He T. C. (2009) J. Cell Mol. Med 13, 2448–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 33.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., He T. C. (2007) Nat. Protoc 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 35.Luo Q., Kang Q., Song W. X., Luu H. H., Luo X., An N., Luo J., Deng Z. L., Jiang W., Yin H., Chen J., Sharff K. A., Tang N., Bennett E., Haydon R. C., He T. C. (2007) Gene 395, 160–169 [DOI] [PubMed] [Google Scholar]
- 36.Zhou L., An N., Jiang W., Haydon R., Cheng H., Zhou Q., Breyer B., Feng T., He T. C. (2002) BioTechniques 33, 1126–1128, 1130, 1132 [DOI] [PubMed] [Google Scholar]
- 37.Si W., Kang Q., Luu H. H., Park J. K., Luo Q., Song W. X., Jiang W., Luo X., Li X., Yin H., Montag A. G., Haydon R. C., He T. C. (2006) Mol. Cell Biol. 26, 2955–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remy I., Michnick S. W. (2006) Nat. Methods 3, 977–979 [DOI] [PubMed] [Google Scholar]
- 39.Kang Q., Song W. X., Luo Q., Tang N., Luo J., Luo X., Chen J., Bi Y., He B. C., Park J. K., Jiang W., Tang Y., Huang J., Su Y., Zhu G. H., He Y., Yin H., Hu Z., Wang Y., Chen L., Zuo G. W., Pan X., Shen J., Vokes T., Reid R. R., Haydon R. C., Luu H. H., He T. C. (2009) Stem Cells Dev. 18, 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao M., Harris S. E., Horn D., Geng Z., Nishimura R., Mundy G. R., Chen D. (2002) J. Cell Biol. 157, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imamura T., Takase M., Nishihara A., Oeda E., Hanai J., Kawabata M., Miyazono K. (1997) Nature 389, 622–626 [DOI] [PubMed] [Google Scholar]
- 42.Afrakhte M., Morén A., Jossan S., Itoh S., Sampath K., Westermark B., Heldin C. H., Heldin N. E., ten Dijke P. (1998) Biochem. Biophys. Res. Commun. 249, 505–511 [DOI] [PubMed] [Google Scholar]
- 43.Hata A., Lagna G., Massagué J., Hemmati-Brivanlou A. (1998) Genes Dev. 12, 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goumans M. J., Valdimarsdottir G., Itoh S., Lebrin F., Larsson J., Mummery C., Karlsson S., ten Dijke P. (2003) Mol. Cell 12, 817–828 [DOI] [PubMed] [Google Scholar]
- 45.Roelen B. A., van Rooijen M. A., Mummery C. L. (1997) Dev. Dyn. 209, 418–430 [DOI] [PubMed] [Google Scholar]
- 46.Johnson D. W., Berg J. N., Baldwin M. A., Gallione C. J., Marondel I., Yoon S. J., Stenzel T. T., Speer M., Pericak-Vance M. A., Diamond A., Guttmacher A. E., Jackson C. E., Attisano L., Kucherlapati R., Porteous M. E., Marchuk D. A. (1996) Nat. Genet. 13, 189–195 [DOI] [PubMed] [Google Scholar]
- 47.Oh S. P., Seki T., Goss K. A., Imamura T., Yi Y., Donahoe P. K., Li L., Miyazono K., ten Dijke P., Kim S., Li E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2626–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu Z., Reynolds E. M., Song J., Lei H., Feijen A., Yu L., He W., MacLaughlin D. T., van den Eijnden-van Raaij J., Donahoe P. K., Li E. (1999) Development 126, 2551–2561 [DOI] [PubMed] [Google Scholar]
- 49.Mishina Y., Crombie R., Bradley A., Behringer R. R. (1999) Dev. Biol. 213, 314–326 [DOI] [PubMed] [Google Scholar]
- 50.van Dinther M., Visser N., de Gorter D. J., Doorn J., Goumans M. J., de Boer J., Ten Dijke P. (2010) J. Bone Miner. Res. 25, 1208–1215 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.