Abstract
Human Papillomavirus 16 (HPV-16) has been identified as the causative agent of 50% of cervical cancers and many other HPV-associated tumors. The transforming potential/tumor maintenance capacity of this high risk HPV is mediated by two viral oncoproteins, E6 and E7, making them attractive targets for therapeutic vaccines. Of 21 E6 and E7 peptides computed to bind HLA-A*0201, 10 were confirmed through TAP-deficient T2 cell HLA stabilization assay. Those scoring positive were investigated to ascertain which were naturally processed and presented by surface HLA molecules for CTL recognition. Because IFNγ ELISpot frequencies from healthy HPV-exposed blood donors against HLA-A*0201-binding peptides were unable to identify specificities for tumor targeting, their physical presence among peptides eluted from HPV-16-transformed epithelial tumor HLA-A*0201 immunoprecipitates was analyzed by MS3 Poisson detection mass spectrometry. Only one epitope (E711–19) highly conserved among HPV-16 strains was detected. This 9-mer serves to direct cytolysis by T cell lines, whereas a related 10-mer (E711–20), previously used as a vaccine candidate, was neither detected by MS3 on HPV-transformed tumor cells nor effectively recognized by 9-mer specific CTL. These data underscore the importance of precisely defining CTL epitopes on tumor cells and offer a paradigm for T cell-based vaccine design.
Keywords: Cancer Therapy, Mass Spectrometry (MS), Papilloma Viruses, Peptides, Viral Immunology, CTL, MHC-1, MS<SUP>3</SUP>, Human Papillomavirus HPV-16
Introduction
The transforming potential of human Papillomavirus (HPV),4 first suspected in the 1970s, has now been firmly established both biologically and epidemiologically (1–3). The single most important variable linked to malignant transformation is persistent infection with one of the high-risk HPV types. The E6 and E7 proteins encoded by high-risk HPVs have transforming activities and functionally inactivate the p53 and retinoblastoma (Rb) tumor suppressor proteins, respectively (3, 4). HPV-16 is the most abundant high risk HPV and has been detected in >50% of cervical cancer cases and in most other HPV-induced tumors, such as carcinomas of the vagina, anus, vulva, penis, and oropharynx (3, 5, 6). Worldwide, high risk HPVs are thought to be responsible for >500,000 malignancies per year, representing more than 5% of human cancers (7).
A major breakthrough in combating HPV-induced disease was the development of prophylactic vaccines to prevent HPV infection in previously unexposed individuals. These vaccines are based on virus-like particles consisting of the L1 capsid protein (8, 9). Virus-like particles resemble natural virions and are able to induce high titers of L1-neutralizing antibodies. Two vaccines, one against HPV-16, -18, -6, and -11 and another against HPV-16 and -18, were approved for clinical use in 2006 (10–12). Although the impact of prophylactic HPV vaccination on the incidence of vaccine type HPV-associated disease and cancer is unquestionable over time, these vaccines have no therapeutic efficacy for established HPV infections. Antibodies neutralize virus particles only before infection. Moreover, as HPV capsid proteins are exclusively expressed late in the viral replication cycle within the upper layers of the epithelium, immune responses against capsid proteins do not affect persistently infected basal cells and, thus, fail to clear the infection (13). Moreover, high risk HPV-associated cancers generally represent nonproductive infections, and the capsid proteins are not expressed (5, 6). For these reasons, viral capsid based strategies are not useful in the development of therapeutic HPV vaccines.
HPV-16 infection is widespread in the sexually active population, but >95% of infections are either transient and/or cleared by the immune system (13). Regression of lesions has been shown to be dependent on strong localized cell-mediated immune responses. In particular, antigen-specific T cell-mediated immunity is required for the clearance of persistent high-risk HPV infections (14). Hence, the immune system is capable of terminating high risk HPV-associated lesions and tumors. Therapeutic vaccines aimed to induce targeted T cell-mediated immune responses against dysplastic and neoplastic cells, therefore, seem a logical extension for achieving beneficial clinical results. Given that E6 and E7 are consistently expressed in HPV-associated cancers, these proteins themselves represent promising targets for vaccine design. Although most tumor-specific antigens are derived from normal or mutated endogenous self-proteins (15) (TANTIGEN: Tumor T cell Antigen Database), E6 and E7 are foreign viral antigens. These two proteins are required for the induction and maintenance of the malignant phenotype of high-risk HPV-associated cancer cells (5, 6), and because HPV uses the cellular DNA replication machinery for genome synthesis, the mutation rate of HPV proteins is low. Thus, it is unlikely that HPV will evade immune attack through loss or mutation of the E6 and/or E7 gene products (16).
Studies on therapeutic vaccines, therefore, have mostly focused on E6 and E7 as target antigens. To date these targets have been delivered as naked DNA vaccines, with recombinant viral or bacterial vectors, as protein or peptide vaccines, and as fusion constructs with toll-like receptor agonists or proteins that enhance antigen delivery or presentation (for review, see Refs. 13, 17, and 18). Most clinical studies have been performed using DNA vaccines. These include a DNA HPV-16 E7 vaccine that has been tested with various fusion partners to enhance antigen presentation and with another DNA vaccine encoding E6 and E7 peptides from HPV-16 and -18 (for review, see Ref. 19). Live viral vectors also have been tested in the clinic, with vaccinia virus constructs coding for either bovine Papillomavirus E2 (20, 21) or HPV-16 and -18 E6 and E7 (22). As for protein vaccines, a fusion protein of heat shock protein 65 with HPV-16 E7 has been tested in three phase II trials (23–25). In addition, a fusion protein of HPV-16 E6, E7, and L2 was also in a phase II trial (26), and a HPV-16 E7 fusion protein with a Haemophilus influenza protein or HPV-16 E6 and E7 were applied in phase I trials in various adjuvants (13, 27). Such studies, however, have yielded disappointing clinical responses.
For induction of HPV-specific T lymphocytes in a focused manner, vaccination against defined epitopes is an attractive option. Indeed, various MHC class I-restricted CTL epitopes of HPV-16 E6 and E7 have been tested in early phase clinical studies (28–34). Nonetheless, little or no benefit over historic controls has been observed. Recently, multiple long synthetic peptide fragments of E6 and E7 have been used to create a polyepitope vaccine, which when tested in patients with HPV-16-positive vulvar intraepithelial neoplasia, exhibited promising clinical efficacy (35). This type of vaccination-induced clinical response has been the most efficacious to date and argues that a robust outcome can be engendered by peptides in conventional adjuvants.
One of the key practical challenges to specific epitope-based vaccines to stimulate cytotoxic T lymphocytes stems from the fundamental nature of T cell receptor (TCR)-based recognition. TCR recognition is referred to as MHC-restricted as, unlike antibody-based recognition, a TCR physiologically interacts with a peptide in complex with an MHC molecule (pMHC) (for review, see Refs. 36 and 37). Further complexity to TCR-based recognition is that a given peptide binds to some but not all MHC molecules. Each human being expresses 3–6 MHC class I molecules (so-called HLA molecules) and at least as many MHC class II molecules. More than 3000 variants of human MHC class I and 1000 variants of MHC class II have been characterized throughout the world to date (38). Cytotoxic T cell recognition of foreign protein antigens occurs via short (generally 9–10 amino acids long) peptides produced through proteolytic cleavage in the cytoplasmic proteasome complex. These are subsequently transported into the endoplasmic reticulum, bound to MHC class I molecules and ultimately displayed on the cell surface as a pMHC. The viral pMHC serves as a flag to target an infected or transformed cell for destruction by a CTL.
Bioinformatic approaches are important tools for peptide-based vaccines and immunotherapy. Computational methods now offer accuracies that are useful in reducing the number of potential candidate peptides that must be tested experimentally for binding to a given MHC allele (39–41). In silico methods cannot predict, however, which MHC class I-binding peptides are actually processed and displayed on a cell surface. We have developed an MS3 Poisson detection mass spectrometry approach to directly assess the physical presence of predicted CTL target epitopes on tumors and infected cells. Our “predict/detect” method achieves sensitivities comparable with that of a T cell with a dynamic range of one peptide among 100,000 pMHCs displayed per cell.
Here for the first time we have interrogated the MHC class I peptide array of several HLA-A*0201 HPV-16-transformed epithelial tumor cells for the presence of any and all predicted HLA-A*0201-binding E6- and E7-derived peptides. Among E6 and E7 proteins, only a single 9-mer epitope was found on all HPV-16 transformants tested. This conserved peptide, termed E711–19, is predicted to have the capacity to bind to the vast majority of globally distributed A2 alleles (100 of 116 HLA-A2 alleles). We suggest that the lack of prior clinical effectiveness of targeted CTL epitope vaccination (32) is a consequence of misidentification of peptides displayed on tumor cells because of the use of indirect immunological surrogates (killing, proliferation, cytokine production, etc.) to judge T cell epitope expression. Our results offer a direct path to select allele-specific targets that should afford tumor protection to a broad population of patients.
EXPERIMENTAL PROCEDURES
Cell Lines
Three HLA-A*0201-positive, HPV-16-positive human cervical carcinoma cell lines and two cell lines transfected with HPV-16 E6 and E7 were used in this study (Table 1). CaSki (ATCC CRL-1550, HPV-16 genome integrated) was grown in DMEM (Sigma) supplemented with 10% FCS, 1% l-glutamine, and 1% penicillin/streptomycin. Cell lines C66-3 (HPV-16 genome integrated) and C66-7 (HPV-16 genome episomal) were kindly supplied by John H. Lee (Dept. of Otolaryngology, University of Iowa) and grown in E media consisting of 3:1 DMEM:Ham's F-12 supplemented with 10% FCS, 1% penicillin/streptomycin, 10 μg/ml epidermal growth factor, 10 mg/ml insulin, 25 mg/ml transferrin, 25 mg/ml hydrocortisone, 200 μg/ml tri-iodo-thyronine, and 250 μg/ml cholera toxin (42). The two transfected cell lines, N/E6E7 and OKF6/E6E7, were kindly supplied by James G. Rheinwald (Dept. of Dermatology and Harvard Skin Disease Research Center, Brigham and Women's Hospital and Harvard Medical School) and grown in K-SFM (Invitrogen).
TABLE 1.
HPV-16 transformed and E6/E7 transduced cell lines
The HPV status, names, and origins of cell lines used in the current study are provided. CaSki was obtained from ATCC (CRL-1550TM) and described in Pattillo (94). C66-3 and C66-7 were gifts of J. H. Lee (42), whereas N/E6E7 and OKF6/E6E7 were gifts of J. G. Rheinwald (unpublished data).
| Cell lines | HPV status | Origin |
|---|---|---|
| CaSki | HPV-16-transformed genome-integrated | Cervical epidermoid carcinoma, small intestinal metastasis |
| C66-3 | HPV-16-transformed genome-integrated | Cervical keratinocytes |
| C66-7 | HPV-16-transformed genome-episomal | Cervical keratinocytes |
| N/E6E7 | HPV-16 E6/E7-immortalized stable human skin keratinocyte cell line | Normal foreskin keratinocytes transfected with the LXSN-16E6E7 retroviral vector |
| OKF6/E6E7 | HPV-16 E6/E7-immortalized stable human mucosal keratinocyte cell line | Normal oral keratinocytes transfected with the LXSN-16E6E7 retroviral vector |
In Silico Prediction of Potential T Cell Epitopes of HPV-16 Proteins E6 and E7
Predictions of HLA-A*0201-binding peptides (both 9-mers and 10-mers) were calculated by the three best predictive servers as described previously (43), namely the Immune Epitope Data base and Analysis Resource server, the NetMHC 3.0 server, and the NetMHCpan 2.2 server. The average predicted IC50 was calculated, and peptides were ranked accordingly. The 21 peptides considered in this study were synthesized by SYNBIOSCI (Livermore, CA). HPLC analysis showed that the purity of the synthesized peptides was >95%. All peptides had expected masses as confirmed by mass spectrometry. Peptides were reconstituted in DMSO at 100 μm each.
HLA-A*0201 Binding Assay
HLA-A*0201-positive, TAP-deficient T2 hybridoma cells (ATCC) were plated at a density of 106 cells/ml in 24-well plates. Cells were pulsed with 10 μm HLA-A*0201-restricted HPV-16 peptides or with 10 μm HLA-A*0201-restricted HTLV-TAX11–19 (LLFGYPVYV)-positive control peptide and 5 μg/ml β2-microglobulin (BD Biosciences) for 6 h at 37 °C in serum-free AIM V media (Invitrogen). HLA-A*0201 expression was determined by flow cytometry (FACSAria) using FITC-conjugated BB7.2 mAb (BD Biosciences). Mean cell fluorescence intensities (MFI) were normalized to the HTLV-TAX-positive control peptide using the formula (MFIsample − MFIcontrol)/(MFIHTLV-TAX − MFIcontrol).
Interferon γ (IFNγ) ELISpot Assay
CD8+ T-cell responses to the 10 HPV-16 peptides that were found to be binders in the HLA-A*0201 binding assay were quantified by IFNγ ELISpot assay. Peripheral blood mononuclear cells (PBMC) isolated from six HLA-A*0201-positive healthy donors under Institutional Review Board approval were plated at 200,000 per well with peptides at a final concentration of 10 μm in anti-IFNγ mAb 1-D1K (Mabtech, Cincinnati, OH)-coated polyvinylidene 96-well plates (Millipore, Billerica, MA). For each individual peptide, the assay was run in duplicate. A HLA-A*0201-restricted HIV-1 peptide (LTFGWCFKL-HIV/Nef137–145) was used as a negative control, and a CMV/EBV/influenza peptide mix (CEF Peptide Pool Classic, Cellular Technology Ltd., Cleveland, OH) and phytohemagglutinin as positive controls. Secreted IFNγ was detected by biotin-labeled anti-IFNγ mAb 7B6–1, and the reaction was developed with streptavidin-ALP and the color reagent nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Sigma). The number of specific IFNγ-secreting T cells was determined with an automated ELISpot reader, calculated by subtracting the average negative control value, and expressed as the number of spot-forming units (SFU) per 106 input cells. A response was considered positive if the activity was at least three times as great as the mean background activity. Of note, three of these six donors tested for HPV-16 antibody scored positive (data not shown).
Nanoscale Immune-affinity Purification by Immunoprecipitation
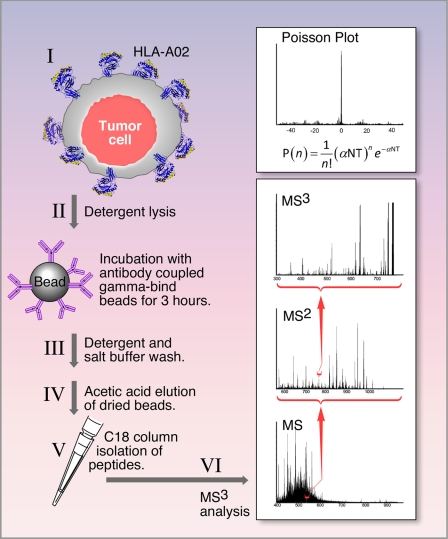
For each immunoprecipitation, 10 μg of anti-HLA-A02 BB7.2 mAb (BD Biosciences) was non-covalently coupled to 20 μl of Gamma Bind beads (GE Biosciences) for 1 h at room temperature. Tumor cells were harvested during the log growth phase and washed with PBS. Cells were pelleted, and the washed and dried cell pellet was lysed using 1.5 ml of lysis buffer consisting of 20 mm Tris, pH 8.0, 1 mm EDTA, 100 mm NaCl, 1% Triton X-100, and 60 mm n-octylglucoside (protease inhibitor tablet, Roche Applied Science, and phenylmethylsulfonyl fluoride) for 10 min on ice. Cell debris was removed using centrifugation for 30 min at maximum speed (13,000 rpm) at 4 °C. Cleared supernatant was incubated with 20 μl of antibody coupled Gamma Bind Plus beads for 2–3 h at 4 °C. Beads were washed 4 times using lysis buffer without Triton X-100 and protease inhibitors. Beads were further washed 4 more times with 10 mm Tris pH v8.0. Dried bead-antibody-HLA pellets were stored at −80 °C for a brief period before MS analysis. Peptides were recovered with 10% acetic acid followed by C18 reverse phase extraction and analyzed on MS.
MS3 Poisson Detection Mass Spectrometry
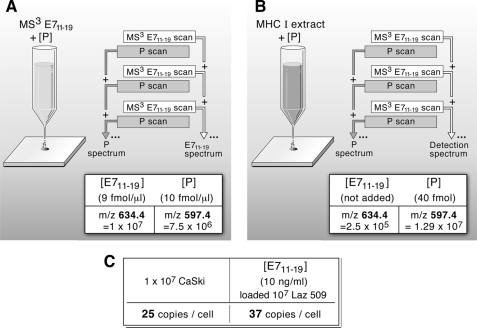
Mathematical details, Poisson scoring, confidence estimation, numerical sampling, and other general principles of MS3 detection are described in a separate manuscript with an analytical focus.5 Nanospray MS3 detection uses a hybrid quadrupole filter, collision cell, and a linear ion trap mass spectrometer (MDS Sciex QTrap 4000). In nanospray MS3 detection a complex mixture is analyzed for a limited number of molecular targets. In place of chromatographic separations, a combination of selective isolations and dissociations filters out a fraction of the ion current highly enriched in fragments specific to a target molecule. This fraction is identified against a background of other ion fragments by using a probabilistic measure. MS3 X/Y spectra are generated by first selectively transmitting a narrow m/z window centered at X into a cell where it dissociates by collision activation. The fragments collect in a linear ion trap downstream of the collision cell. After a collection period, an m/z window centered at Y is isolated in the linear ion trap, and the ion fragments at m/z Y are again dissociated by collision activation. These fragments of a fragment are scanned out and measured to create an MS3 spectrum. For targeted detection, MS3 spectra of synthetic versions are first studied for optimal conditions and MS2 fragment choices, and then reference MS3 spectra are acquired for each of the chosen MS2 fragments. Sample MS3 spectra with corresponding m/z windows and dissociation conditions are acquired, and these spectra are compared against the set of reference spectra using a Poisson probability metric to quantify the likelihood that the experimental spectra contain fragment intensities consistent with the relative arrival rates given by the reference spectra (44).5 In contrast to chromatographic separations, the different ionizable components are simultaneously present in the ion beam. This means molecular abundance of a target can be measured by use of an added calibrant molecule at known concentration. This is done in two steps. In the first step one measures a solution with known target and calibrant concentrations to relate the MS3 ion flux of the target to the calibrant in the detection spectra. For example, to quantitate E711–19, known amounts of this and a control peptide P (KSPWFTTK) are added to a mock MHC I workup using 1.2 pmol of β-galactosidase digest as a carrier. After C18 trapping and elution into the electrospray buffer, the detection MS3 spectra of both peptides is taken in an alternating series to compensate for time variation in the nanospray ion flux. The MS3 signal amplitudes of P (base peak at m/z 597.4) and E711–19 (base peak at m/z 634.4), corrected for relative amounts, are recorded. As noted in Fig. 8A, after a 2-min MS3 collection for E711–19 at 9 fmol/μl in the β-galactosidase sample, one measures a signal amplitude of 107 (arbitrary units) for m/z 634, whereas P at 10 fmol/μl gives 7.5 × 106 for m/z 597. This gives the relative molar MS3 signal response of P to E711–19 at about 0.68. In the second step a known amount of the calibrant peptide is added to the sample being quantitatively analyzed for target. For E711–19 quantitation in 10 million CaSki cells, 40 fmol of KSPWFTTK is added to the sample at the beginning of the acid elution step. The detection MS3 spectra of peptide P and E711–19 is again taken in alternating sequence. The measured ratio of MS3 ion flux for E711–19 and P in the CaSki sample is 2 × 105/1.3 × 107 (m/z 634.4/m/z 597.4). Corrected for the molar response (0.68) as determined in the first step, one has the relative molar amounts of E711–19 to P in the sample as 0.0105. As 40 fmol of P was added, this gives the amount of E711–19 as 422 amol or 254 million molecules from 10 million CaSki cells. As the nanospray ion source often shows significant intensity variations with time and MS3 spectra may be collected for long periods, quantitation data are always collected in a series where a single scan of the target alternates with a single scan of the calibrant and the respective scans are then summed.
FIGURE 8.
Quantitation of the number of E711–19 epitopes per cell on HPV epithelial transformants and peptide-pulsed lymphoid cells. To quantitate the amount of E711–19 recovered from HLA-A2 extracts, the MS3 signal abundance relative to an added control peptide is measured. A, the MS3 signals of the target E711–19 and control P (KSPWFTTK) peptides were measured at known concentrations in a mock MHC I workup. B, a known amount of the control peptide P was added to the HLA-A2 sample being analyzed, and MS3 spectra of peptide P and E711–19 were again taken. C, combining the MS3 signal ratios measured in A and B with the amount of control peptide added provided the amount of target peptide (see “Experimental Procedures”). Knowing the number of cells lysed, the target copies per cell were calculated assuming full recovery up to the point where the control peptide was added (Step IV, Fig. 4).
Generation of HPV-16-specific T Cell Lines
For use as antigen-presenting cells, dendritic cells were differentiated from adherent donor monocytes in DMEM medium supplemented with 10% human serum, 1% l-glutamine, 1% penicillin/streptomycin, 100 ng/ml granulocyte-macrophage colony-stimulating factor, and 50 ng/ml IL-4 (PeproTech, Rocky Hill, NJ) for 1 week. Differentiated dendritic cells were matured with 10 ng/ml TNF-α, 10 ng/ml IL-6, 10 ng/ml IL-1β, 1 μg/ml prostaglandin E2, and 1 μg/ml peptidoglycan (Sigma) overnight. Cells were pulsed with 10 μm HPV-16 peptides for 3 h and irradiated (3000 radians). Fresh donor PBMC were prepared by Ficoll-Paque (Amersham Biosciences) centrifugation. Donor PBMC were plated at a density of 107cells/ml together with 5 × 104/ml HPV-16 peptide-loaded irradiated antigen-presenting cells in 24-well culture plates in DMEM medium supplemented with 10% human serum, 1% l-glutamine, 1% penicillin/streptomycin, 0.05 mm 2-mercaptoethanol, and 10 ng/ml IL-7. Cultures were fed with 20 IU/ml IL-2 (BD Biosciences) 5 days after stimulation and re-stimulated with peptide-loaded-irradiated donor dendritic cells every week for a total of 4 weeks.
T Cell Proliferation Assay
5 × 105 T cells from the E711–19-specific T cell line were stimulated with 1 × 105 donor B cells loaded with 10 μg/ml E711–19 on 96-well plates. Donor B cells were obtained through stimulation of donor PBMC with 3T3-CD40L cells as described previously (45). CD40-activated B cells were irradiated at 3200 rads before plating. Cells were plated in DMEM medium supplemented with 10% human serum, 1% l-glutamine, 1% penicillin/streptomycin, and 0.05 mm 2-mercaptoethanol. Plates were incubated in a 37 °C tissue culture incubator for 3 days and pulsed with 1 μCi of [3H]thymidine for 16 h. Plates were harvested, and [3H]thymidine incorporation was detected with a liquid scintillation mixture (PerkinElmer Life Sciences Beta Plate Scint) in a luminescence counter (PerkinElmer Life Sciences 1450 LSC).
Analysis of IFNγ Secretion Associated with HPV-16-specific Proliferative Responses
IFNγ quantitation was performed using cytometric bead arrays (BD Biosciences) according to the manufacturer's instructions. Cut-off values were based on the standard curve for IFNγ (100 pg/ml). Antigen-specific cytokine production was defined as a cytokine concentration above cut-off level and >2× the concentration of the medium control.
Characterization of T Cell Lines by IFNγ ELISpot
5 × 104 T cells from the E711–19-specific T cell lines were incubated with 1 × 104 T2 cells loaded with 10 μg E711–19 peptide overnight on precoated IFNγ ELISpot plates. The assay was processed and developed as described above.
Cytotoxicity Assay
Donor B cells from an EBV-immortalized B cell line (Laz 509) were pulsed with 10 μg or 10 ng of the respective HLA-A*0201-restricted HPV-16 peptides at 37 °C overnight. The cells were washed twice with DMEM and pulsed with 100 μCi/ml 51Cr for 90 min at 37 °C. Target cells were washed three times with serum-free Opti-MEM media (Invitrogen) to remove excess 51Cr and plated with peptide-specific CD8+ T cells at 30:1, 10:1, 3:1, and 1:1 ratios. After 4 h of incubation, 50 μl of culture supernatant were mixed with liquid scintillation mixture (PerkinElmer Life Sciences Optiphase Supermix) and analyzed for 51Cr release using a luminescence counter (PerkinElmer Life Sciences 1450 LSC). Percent specific chromium release was calculated using the formula (experimental release − spontaneous release)/(maximum release in 5% Triton X-100 − spontaneous release) × 100.
RESULTS
In Silico Prediction of Potential T Cell Epitopes of HPV-16 Proteins E6 and E7
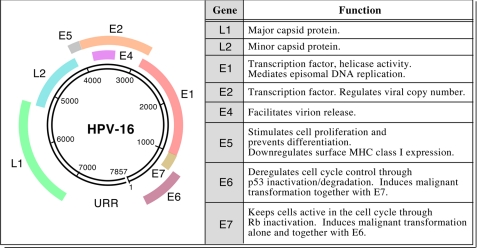
HPV is a small non-encapsulated DNA virus containing ∼8000 bp encoding two major sets of genes (E, early region genes; L, late region genes) that infect stratified squamous epithelium (Fig. 1). As the E6 and E7 proteins bind host regulators of keratinocyte cell division and thereby degrade and/or perturb the cell cycle inhibitors p53 and Rb, respectively, those viral proteins are of keen target interest for immunotherapeutic purposes. Human cell lines transformed by HPV-16 (CaSki, C66-3, and C66-7) or transduced with HPV-16 E6 and E7 containing retroviruses (N/E6E7 and OKF6/E6E7) are listed in Table 1.
FIGURE 1.
HPV-16 genome and transforming activity of E6 and E7. The left panel shows the ∼8000-bp map of this oncogenic DNA virus and its genes. In the right panel, the key functions of the early and late genes are listed (5, 85–93).
Potential HLA-A*0201-binding peptides derived from the HPV-16 E6 and E7 proteins were determined using three prediction servers (Immune Epitope Data base and Analysis Resource, netMHC, and netMHCPAN) previously found to provide the most accurate HLA-A*0201 results (43). Predicted peptides were ranked by average predicted IC50 values as reported in Table 2. When available, experimentally determined IC50 measurements were also included (46). Any peptide containing more than one cysteine residue was excluded from the list to minimize complexities resulting from intramolecular or intermolecular disulfide bond formation. As shown, predictions correlated quite well with published experimental data as all top ranked peptides have been previously defined as binders, aside from peptide E712–20 (46). Within the top group, peptide E711–19 was predicted to be the best binder, whereas peptide E786–93 was experimentally determined to bind most strongly to HLA-A*0201 (46).
TABLE 2.
Bioinformatic predictions of HPV-16 E6- and E7-derived peptides binding to HLA-A*0201
IEDB, Immune Epitope Data base and Analysis Resource server.
| Peptide position | Sequence | Predicted IC50 |
Measured IC50a | |||
|---|---|---|---|---|---|---|
| IEDB | NetMHC | NetMHCpan | Average | |||
| nm | nm | |||||
| E711–19 | YMLDLQPET | 9.6 | 10 | 10.88 | 10.16 | 49 |
| E711–20 | YMLDLQPETT | 22.7 | 27 | 28.85 | 26.18 | 46 |
| E77–15 | TLHEYMLDL | 48.9 | 30 | 64.43 | 47.78 | 188 |
| E782–90 | LLMGTLGIV | 30.9 | 99 | 22.85 | 50.92 | 82 |
| E618–26 | KLPQLCTEL | 115.2 | 123 | 62.64 | 100.28 | 328 |
| E652–60 | FAFRDLCIV | 92.0 | 182 | 153.66 | 142.55 | 130 |
| E629–38 | TIHDIILECV | 386.0 | 44 | 67.12 | 165.70 | 494 |
| E786–93 | TLGIVCPI | 84.4 | 541 | 318.97 | 314.79 | 7 |
| E785–93 | GTLGIVCPI | 286.0 | 535 | 263.26 | 361.42 | 193 |
| E778–87 | TLEDLLMGTL | 485.0 | 587 | 439.66 | 503.89 | |
| E621–30 | QLCTELQTTI | 796.8 | 755 | 569.89 | 707.23 | |
| E777–86 | RTLEDLLMGT | 429.7 | 1381 | 901.31 | 904.00 | |
| E659–68 | IVYRDGNPYA | 1003.4 | 1494 | 761.84 | 1086.41 | |
| E778–86 | TLEDLLMGT | 3079.2 | 422 | 373.13 | 1291.44 | |
| E766–74 | RLCVQSTHV | 952.5 | 4014 | 514.67 | 1827.06 | |
| E644–53 | LLRREVYDFA | 2592.1 | 1894 | 1462.12 | 1982.74 | |
| E782–91 | LLMGTLGIVC | 590.1 | 4206 | 1668.83 | 2154.98 | |
| E689–97 | SLYGTTLEQ | 3836.3 | 923 | 2746.42 | 2501.91 | |
| E712–20 | MLDLQPETT | 3813.3 | 3103 | 1973.12 | 2963.14 | 462 |
| E781–90 | DLLMGTLGIV | 398.6 | 6154 | 6013.88 | 4188.83 | |
| E686–95 | YCYSLYGTTL | 1184.1 | 4394 | 7122.60 | 4233.57 | |
a See Ref. 46.
HLA-A*0201-binding Assay
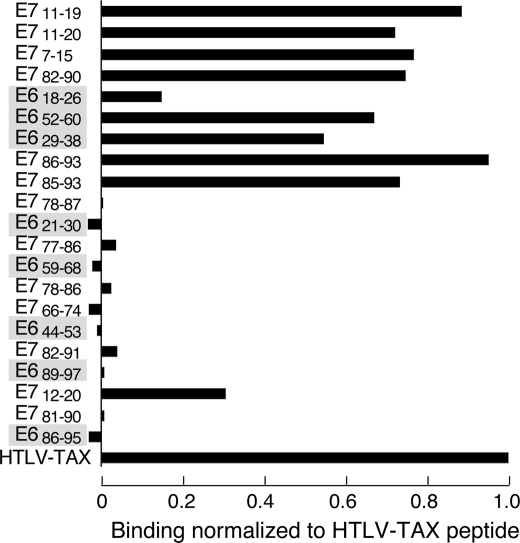
To measure the binding capabilities of predicted peptides, an HLA-A*0201 binding assay was performed using the T2 cell line (47, 48). This cell line is TAP1/2-deficient, displaying low levels of HLA-A*0201 on its surface. After exogenous addition of peptides capable of binding to HLA-A*0201, this HLA complex is stabilized on the surface with a concomitant increase in the number of HLA-A*0201 molecules, as determined by mean fluorescence intensity staining using a fluorochrome-labeled anti-HLA-A2 antibody and flow cytometry. Binding was calculated relative to a known strong HLA-A*0201 binder, the TAX11–19 peptide from the human T cell leukemia virus-1 (HTLV-1). As shown in Fig. 2, peptide E786–93 was found to be the best binder in this assay followed by E711–19. Altogether, the top-ranked predicted peptides were the strongest binders in the T2 assay so that with the exception of E712–20, the peptides with a predicted average IC50 of >500 nm were not found to bind to HLA-A*0201 in the T2 assay. The T2 assay results correlated well with the previously published data (46). Those 10 peptides that were determined to bind experimentally were included in further assays.
FIGURE 2.
HLA-A*0201 T2-based peptide binding assay. HLA-A*0201-positive, TAP-deficient T2 cells were pulsed with 10 μm of the respective peptides given on the graph ordinate for 6 h at 37 °C. Binding was determined with the anti-HLA-A2 antibody BB7.2 by flow cytometry and calculated relative to a known strong binder, the TAX11–19 peptide from the HTLV on the abscissa. The TAX11–19 binding was set at 1.0. The order of peptides is from predicted strongest to weakest binders, top to bottom, respectively. Shaded entries are E6-derived peptides, whereas unshaded entries are E7-derived.
IFNγ ELISpot Assay
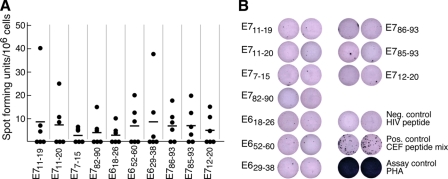
Current epidemiological data suggest that virtually all individuals among the sexually active population have been exposed to HPV infection (for review, see Refs. 13 and 49 and references therein). We, therefore, examined the ability of T cells from fresh peripheral blood mononuclear cells of six HLA-A*0201-positive healthy donors to recognize these HPV candidate peptides in IFNγ ELIspot assay. As shown in Fig. 3A, SFUs per 106 cells were low or undetected in these individuals. However, when SFUs were observed, their size was substantial (Fig. 3B). The low numbers of IFNγ-producing cells reflect the paucity of HPV-specific memory T cells in peripheral blood. These findings are also consistent with previous studies (50) reporting low HPV-specific SFUs and distinct from the robust memory recall SFU response to CEF (a mix of cytomegalovirus, Epstein-Barr virus, and influenza A virus) peptides or the phytohemagglutinin (PHA) assay control (Fig. 3B). The only HPV peptides eliciting an SFU number 4–5-fold over background in one donor each were E711–19 and E629–38 (Fig. 3A). As a consequence of these equivocal responses, we pursued mass spectrometry analysis to identify which viral peptides are physically displayed on HPV-16-transformed, HLA-A*0201-positive cells.
FIGURE 3.
Low or undetectable memory T cells in blood of healthy individuals. Immune recognition of the 10 HLA-A*0201-binding peptides was tested in an IFNγ ELISpot assay. PBMC isolated from 6 HLA-A*0201-positive healthy donors were stimulated with 10 μm respective peptide overnight. In panel A, spots are graphed and presented as SFUs per million PBMC. SFUs of single donors are represented as dots, with a horizontal line corresponding to the mean of six donor samples. Highest SFUs are not from the same donor. In panel B, ELISpot well images from representative plates (2 × 105 cells/well) taken from several donors are shown. CEF, cytomegalovirus/Epstein-Barr-Virus/Influenza positive peptide mix. PHA, phytohemagglutinin, a mitogenic plant lectin used as another positive control.
HLA-A*0201 Immunoprecipitation and MS3 Analysis of Eluted Peptides
For the investigation of HPV-16 antigen presentation by MS analysis, HLA-A*0201+ HPV-16-transformed tumor cell lines as well as E6/E7 expressing human epithelial cell lines enumerated in Table 1 were used. The analytic approach employed is schematically shown in Fig. 4. In brief, tumor cells (∼20–60 × 106) were solubilized using detergent buffers, and then HLA-A*0201 molecules were immunoprecipitated with the BB7.2 anti-HLA-A2-specific mAb coupled to Gamma Bind Plus beads. Peptides were recovered from pMHC complexes by acid elution and analyzed on a hybrid quadrupole-linear ion trap mass spectrometer using MS3 and Poisson statistics as described under “Experimental Procedures.” Peptides from HPV-16 E6 and E7 oncoproteins that were shown by T2 assay to increase surface HLA-A2 expression (Fig. 2) were targeted for detection by mass spectrometry.
FIGURE 4.
Methodology for immunoprecipitation of HLA-A2 molecules, elution of bound peptides, and MS3 analysis of potential CD8 T cell epitopes. MS3 analysis isolates a selected m/z window containing a target ion (e.g. m/z 555.3), fragments (by collision activation) all ions in the selected window, isolates from these fragments a second m/z window containing a target fragment (e.g. m/z 764.3), and dissociates ions in the second m/z window to form an MS3 spectrum (denoted MS3 555.3/764.4). A probabilistic measure quantifies the likelihood the MS3 spectrum generated in these steps contains a reference dissociation pattern obtained from the synthetic peptide. Details are provided under “Experimental Procedures.”
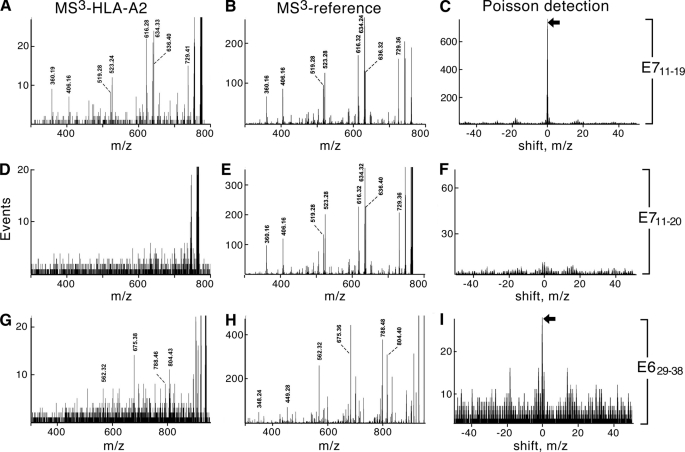
MS3 spectra from immunoaffinity-purified HLA-A*0201 complexes isolated from HPV-16-transformed cell lines (MS3-HLA-A2) were compared against the MS3 patterns of synthetic peptides (MS3-reference) using a probabilistic metric (445 as shown in Fig. 5. Unexpectedly, only one peptide, E711–19, was easily identified, whereas E629–38, the sole other peptide detected, was near the limit of sensitivity. For E711–19, the double-charged molecular ion was selected at m/z 555.3 and dissociated. The proline in E711–19 (YMLDLQPET) tends to direct fragmentation to the amide bond on the amino side of the proline residue. This generates a strong b6 (YMLDLQ-) fragment at m/z 764.4, making an optimal candidate for MS3 detection but suppressing the detection by other fragments (supplemental Fig. S1). The molecular ion is abundant in the peptides recovered from the HPV-16-positive CaSki cervical carcinoma line so that the dissociation pattern of the b6 fragment from the synthetic peptide is immediately recognized in the MS3 555.3/764.3 spectrum (Fig. 5, A and B, and supplemental Fig. S3). The Poisson signature of detection (peak at 0 m/z shift, Fig. 5C) is clear cut but, in this case, largely superfluous. The b8 fragment (YMLDLQPE-) was also detected in the MS3 555.3/990.4 spectrum of recovered peptides (supplemental Fig. S2). In contrast to detecting the b6 fragment of E711–19, detection of E629–38 by MS3 fragmentation of the b8 ion (TIHDIILE-) at m/z 935.5 (MS3 578.3/935.5; Fig. 5, G–I) is not so evident by inspection of the MS3 spectra (Fig. 5, G and H) but does produce the detection signature using the Poisson metric (Fig. 5I). E629–38 detection was further supported by a similar Poisson analysis for the signature of the b7 ion (TIHDIIL-) at m/z 806.5 (data not shown).
FIGURE 5.
MS3 detection analysis of E711–19, E711–20, and E629–38 HPV-16 peptides. Comparison of MS3-HLA-A2 extracts with MS3 reference patterns is shown in the left and middle columns, respectively. Poisson detection signatures are shown in the right columns. A, MS3 555.3/764.4 of HLA-A2-associated peptides extracted from 60 million CaSki cells is shown. B, MS3 555.3/764.4 of synthetic peptide YMLDLQPET corresponding to E711–19 is shown. C, shown is the Poisson detection signature of the synthetic pattern shown in B in the spectrum shown in A. The amplitude at 0 m/z shift relative to nonzero m/z shifts is a probabilistic measure of the uniqueness of the fit and is used as a marker of detection (see “Experimental Procedures” and “Results” sections). D, MS3 605.8/764.4 of HLA-A2 peptides extracted from 60 million CaSki cells is shown. E, MS3 605.8/764.4 of synthetic peptide YMLDLQPETT corresponding to E711–20 is shown. F, the Poisson detection signature of the b6 fragment (YMLDLQ-) is shown. G, MS3 578.3/935.5 of HLA-A2-associated peptides extracted from 20 million CaSki cells is shown. H, a reference spectrum of the b8 fragment (TIHDIILE)- from MS3 578.3/935.5 of the synthetic peptide TIHDIILECV is shown. I, Poisson detection signature of the b8 fragment in MS3 578.3/935.5 spectrum of peptides extracted from CaSki cells is shown.
The longer E711–20 peptide has received considerable attention in the literature as a possible tumor antigen (32), but it was not detected on CaSki (Fig. 5, D–F) nor on any other E7 expressing cell line. MS3 analysis of synthetic E711–19 and E711–20 peptides showed equivalent signal intensity in generating the MS3 spectra used for detection, and both peptides were recovered equivalently from peptide-loaded T2 cells (data not shown). Nonetheless, the difference in surface presentation between E711–19 and E711–20 peptides was substantial. Processing 60 million CaSki cells and MS3 analysis of E711–19 and E711–20 produced a Poisson amplitude of almost 700 events for the E711–19 peptide, whereas the E711–20 detection under identical conditions did not rise above background (10 events).
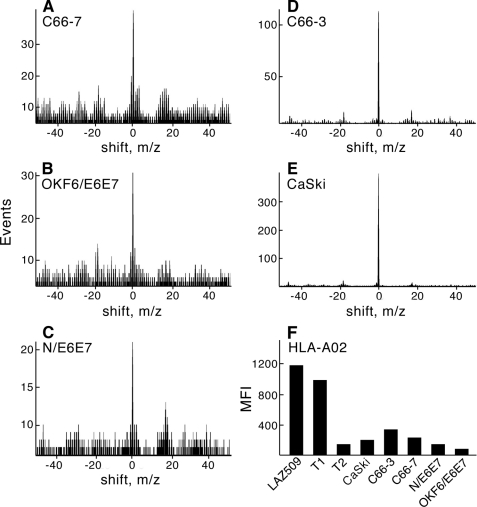
MS3 Detection Analysis Identifies E711–19 on All HPV-16-expressing HLA-A*0201+ Human Epithelial Lines
Poisson detection signatures of E711–19 were obtained from each of the human cell lines described in Table 1. As shown in Fig. 6, from 10–20 × 106 C66-7, OKF6/E6E7, N/E6E7, C66-3, and CaSki cell lines, the HLA-A*0201-bound E711–19 peptide was readily observed by MS3 Poisson detection mass spectrometry (panels A–E, respectively). In contrast, calculating the Poisson signature of the E711–19 b6 fragment against 10 representative MS3 ion backgrounds did not indicate a single instance of positive detection (supplemental Fig. S4). Furthermore, as shown by quantitative flow cytometry analysis of HLA-A2 expression using the anti-HLA-A2 mAb BB7.2, the fraction of surface display due to this epitope is enhanced, as the total HLA-A*0201 copy number is reduced >10-fold on these HPV-16-transformed or -transfected cells relative to the TAP competent T1 cell line or EBV-transformed B cell line Laz 509. Their levels of MHC class I expression are more similar to the TAP-deficient T2 cell line than the TAP-sufficient Laz 509 or T1 cells (Fig. 6F). However, unlike T2 cells, the HPV-16 epithelial cell MS spectra are not dominated by signal peptides of normal proteins (51). In summary, cervical and oral epithelial cells displayed detectable surface HLA-A*0201-bound E711–19 after HPV-16 infection or ectopic expression of HPV-16 gene products. On the other hand, aside from E629–38 in CaSki cells (Fig. 5), MS3 detected no other E6 or E7 epitopes with HLA-A*0201 binding activity as observed in the T2 assay (data not shown).
FIGURE 6.
Poisson detection of E711–19 on all HPV-16-transformed and -transfected human epithelial cells and their HLA-A2 surface expression. Panels A–E represent Poisson detection signatures of E711–19 from 20 million C66-7 cells (A), 10 million E6/E7 transfected oral OKF6/E6E7 keratinocytes (B), 10 million E6/E7 transfected foreskin keratinocytes (N/E6E7) (C), 20 million C66-3 cells (D), and 20 million CaSki cells (E). Panel F shows mean cell fluorescence intensity values of the indicated cells reacting with the FITC-labeled anti-HLA-A2 mAb BB7.2.
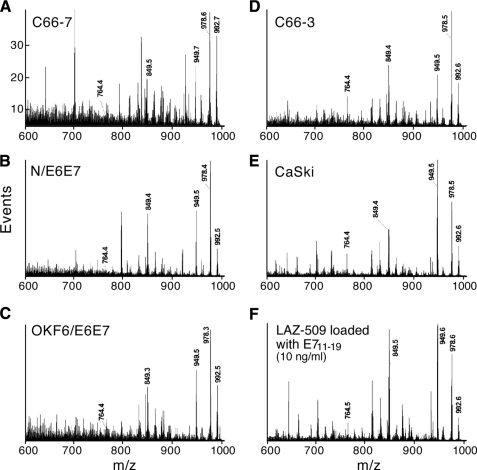
Unexpectedly, the peptide recovery as characterized by mass spectrometry from the C66-7, N/E6E7, and OKF6/E6E7 lines compared with the recovery from the C66-3 and CaSki lines was substantially lower than the amount expected from comparing HLA-A2 expression by flow cytometry. The reason for this is currently under study, but the large variation in overall recovery is best addressed by a direct method of comparing the relative fraction of E711–19 to total peptide among the different cell lines. Such a measure can be obtained from the MS2 555.3 spectra (Fig. 7). These spectra show a number of intense shared peaks at the high m/z end that arise from common terminal losses. Different peptides with molecular masses near 1108.6 Da are co-selected as double-charged ions in the m/z 555.3 window, and if these ions lose a common amino acid residue at the amino or carboxyl terminus, their product ions will appear as a single m/z peak. For example, m/z 992.6, 978.6, 949.5, and 859.5 could be b-type ions that arise from the carboxyl-terminal loss of Val, Leu or Ile, Ala-Ala, and (Leu or Ile)-Glu, respectively. The peak at m/z 978.6 could be a y-type ion arising from the amino-terminal loss of methionine and so on. Because these high m/z peaks reflect the contribution of many peptides, their collective intensity is a qualitative measure of the peptide background. In contrast the peak at m/z 764.4 that appears above the background in the C66-3-, CaSki-, and E711–19-loaded Laz 509 B cell line spectra is shown by MS3 to be predominantly a fragment of the E711–19 peptide. The ratios of m/z 764.4 to the peaks at m/z 992.6, 978.6, 949.5, and 859.5 (Fig. 7) are a relative measure of the E711–19 fraction and show that the low MS3 ion flux for the E711–19 peptide in the C66-7, N/E6E7, and OKF6/E6E7 lines (Fig. 6) is not just a reflection of low peptide recovery overall but that the fraction of E711–19 to total peptide is reduced compared with that fraction in C66-3-, CaSki-, or 10 ng/ml E711–19-loaded Laz 509.
FIGURE 7.
The MS2 555.3 spectra of HLA-A*0201 peptide extracts from different cell lines expressing the E7 oncoprotein and Laz 509 B cells loaded with 10 ng/ml E711–19. The common peaks at the high m/z end are fragments from different peptides sharing amino or carboxyl terminal amino acids and a molecular mass near 1108.6 Da (hence, co-selected in the m/z 555.3 window). Because the intensity of these high m/z peaks is an average of many peptides, their intensity serves as an approximate measure of the peptide background. Their amplitude relative to m/z 764.4 provides in a single spectrum a characterization of the fraction of A2-bound peptide that is E711–19 (see “Results”). The C66-7, N/E6E7, and OKF6/E6E7 samples show not just lower absolute amounts of E711–19 (Fig. 6) but also that E711–19 is a smaller relative fraction of the total peptide population.
To quantitate the number of E711–19 epitopes bound to HLA-A*0201, we developed a method using a calibrant peptide (P) whereby the MS3 spectrum of the calibrant and E711–19 are taken in alternating series as described in Fig. 8 (panels A and B and the legend). Fig. 8C shows that CaSki cells endogenously express 25 copies of E711–19 per cell, whereas exogenous addition of 10 ng/ml synthetic E711–19 to Laz 509 loads 37 copies of this HLA-A*0201 epitope per cell.
Generation and Characterization of T Cell Lines
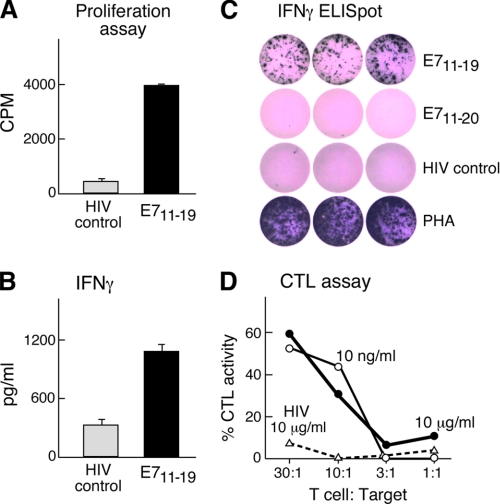
To next test if PBMC from an HLA-A*0201-positive donor could be activated by E711–19-loaded autologous dendritic cells, in vitro stimulation was performed. After 4 weekly stimulations, the resulting T cell lines consisted of up to 70% CD8+ T cells. As shown in Fig. 9A, these CD8+ T cells proliferated when stimulated with E711–19 peptide-loaded autologous CD40-activated B cells but did not proliferate against control HIV-peptide (LTFGWCFKL-HIV/Nef137–145)-loaded CD40-activated B cells as judged by tritiated thymidine incorporation. Upon stimulation with peptide-loaded antigen-presenting cells or T2 target cells, E711–19-specific T cells produced IFNγ as detected by multiplex cytometric bead array or ELISpot assay (Fig. 9, B and C, respectively). Collectively, these data show that the T cell line is specific for E711–19. ELISpot analysis suggested that the frequency of antigen-specific T cells in the cell line was ≥100-fold that detected in fresh autologous PBMC from the same donor (compare Figs. 3 and 9).
FIGURE 9.
Specificity of CD8 T cells elicited against E711–19 was directed against E711–19 but not E711–20. E711–19-specific T cells were generated by four weekly stimulations with autologous dendritic cells and tested for their antigen-specific proliferation by tritiated thymidine incorporation (panel A), IFNγ secretion by cytometric bead assay (panel B), IFNγ production by ELISpot (panel C), and ability to lyse autologous EBV-transformed B cells pulsed with the indicated amounts of E711–19 or an irrelevant Nef137–145 (LTFGWCFKL) HIV peptide (panel D). PHA, phytohemagglutinin.
To assess whether the T cell line manifests cytolytic activity able to lyse target cells displaying a number of E711–19 epitopes comparable with those arrayed on CaSki, we loaded the autologous donor EBV-immortalized B cell line Laz 509 with 10 ng/ml or 10 μg/ml E711–19. As shown in Fig. 9D, E711–19-specific T cells lyse both E711–19 peptide-loaded Laz 509 target cells in a 51Cr release assay. Up to 52% CTL activity was observed against 10 ng/ml E711–19-loaded target cells at a 30:1 E/T ratio. By contrast, no killing was observed with the same Laz 509 cells pulsed with 10 μg/ml HIV-1 peptide (Nef137–145). These same T cells also lysed CaSki, consistent with display of E711–19 at equivalent density. However, given that CaSki and the T cell line differ at class I alleles other than HLA-A*0201, alloreactivity might have been, at least in part, responsible for that cytolysis (data not shown). The use of autologous B cells pulsed with E711–19 excludes alloreactivity as the basis for target lysis. Fig. 9C also indicates that whereas the T cell line raised against E711–19 shows specificity for E711–19, it lacks detectable E711–20 reactivity as judged by ELIspot. In a reciprocal manner, a T cell line raised against E711–20 showed no specificity for B cells pulsed with E711–19 (data not shown).
The E711–19 Epitope Is Highly Conserved among HPV-16 Strains and Binds to the Vast Majority of A2 Alleles
Given the expression of E711–19 on HPV-16 transformed or transfected cell lines, we determined whether known strains of HPV-16 conserve this epitope. Using the Human Papillomavirus T Cell Antigen Database with 791 HPV protein entries, we performed multiple sequence alignment of the 16 HPV-16 E7 sequences. As shown in Fig. 10, E711–19 is fully conserved in 15 of 16 sequences. A single conservative amino acid substitution is found in the remaining sequence. The latter represents a single variant among 35 HPV-16 cervical cancer or cervicitis patients analyzed. Analysis of the complete E7 open reading frame from those 35 patients revealed four nucleotide variations in three (8.5%) patients, whereas the other 32 did not contain any nucleotide changes compared with the prototype (52). Of the identified changes, two were silent nucleotide changes, and two were missense substitutions, resulting in the amino acid changes L15V and S31R. This rare LI5V conservative mutation falls within the E711–19 epitope.
FIGURE 10.
Conservation of E711–19 in all high risk HPV-16 strains. 15 of the 16 HPV-16 E7 sequences in the HPV data base include the E711–19 sequence. The corresponding UniProt accession number of each sequence is shown at the left of each line. Asterisk, this column contains identical amino acid residues in all sequences; colon, this column contains different but highly conserved (very similar) amino acids; no symbol indicates that this column contains dissimilar amino acids or gaps.
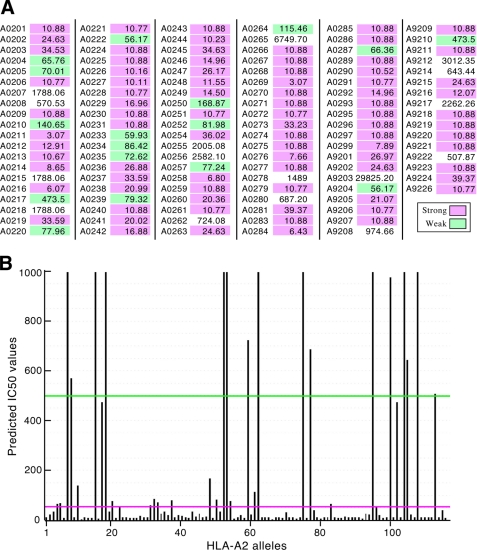
Fig. 11, panel A, lists E711–19 HLA binding predictions on all 116 HLA-A2 alleles using NetMHCpan (53) and their calculated IC50 values (in nm). Significant binding is predicted for 100 of the 116 HLA-A2 alleles. As graphically depicted in panel B, E711–19 is a strong binder to 85 alleles (<50 nm), a weak binder to 15 alleles (50–500 nm), and a non-binder to 16 alleles (>500 nm).
FIGURE 11.
E711–19 binds to the vast majority of HLA-A2 alleles. HLA binding predictions of E711–19 were performed using NetMHCpan on the 116 known HLA-A2 alleles. Panel A shows the predicted IC50 values for each allele (in nm) with strong binding (<50 nm) shaded magenta, weak binding (50–500 nm) shaded green, and no binding (>500 nm) unshaded. Panel B gives a bar graph representation of E711–19 IC50 for each allele along the x axis corresponding to those going from the left to right columns in the order defined in panel A.
DISCUSSION
HPV-induced dysplasia and cancer cause significant morbidity worldwide (7). Although prophylactic vaccines are now available, immunization does not reach everyone at risk. Given that HPV-associated cancers develop years and often decades after initial infection, it was predicted that no measurable decline of HPV-associated cancers in women may occur before 2040. This prediction was based upon higher acceptance rates for the vaccines than is currently achieved in the United States (for review, see Ref. 13). Furthermore, the approved prophylactic vaccines have no therapeutic effects (54), leaving HPV-infected individuals in need of treatment options. Fortunately, most HPV infections are cleared naturally by the immune system (14). If the lesions do not regress, surgical treatments are necessary. These procedures are associated with significant morbidity ranging from dysfunction to infertility depending on the site and stage of the lesion. A noninvasive treatment such as a therapeutic vaccine fostering an effective anti-HPV state would be an attractive alternative. A vaccine could be offered to patients who do not clear HPV infection spontaneously during a finite observation period as well as to patients with established lesions.
HPV-related diseases represent an ideal set of clinical disorders to test development of a therapeutic cancer vaccine as the viral oncoproteins E6 and E7 are consistently expressed in HPV-associated cancers (16), and they represent “non-self” cancer antigens. Given their key role in cancerous transformation, a large body of research focusing on E6 and E7 as therapeutic vaccine targets has been conducted. Multiple MHC class I- and class II-restricted epitopes, mostly of HPV-16 and HPV-18, have been reported (19, 30, 46, 55–69). Several HLA-A*0201-restricted HPV-16 E7 epitopes have already been applied in clinical trials, namely E711–20 (32, 33), E786–93 (31–34), and E712–20 (34). Although induction of peptide-specific T cell responses could be demonstrated in these studies, no clinical improvements exceeding the rate of spontaneous tumor regression were observed.
The lack of clinical impact was thought to be a consequence of an advanced stage of disease in the patient groups. However, as there is evidence that HPV infection influences antigen presentation, this lack of success might be caused by a paucity (or even absence) of epitopes presented on HPV-16-transformed cells. In this regard several HPV immune evasion mechanisms have been described (for review, see Refs. 70–72) including down-regulation of components of the antigen-processing machinery and MHC class I molecules (73, 74), resulting in decreased presentation of antigenic peptides. Furthermore, precise and direct identification of T cell epitopes expressed on HPV-transformed cells has been lacking. Instead, determination of relevant epitopes has been inferred by bioinformatic prediction, synthetic peptide HLA binding studies, and peripheral T cell functional activation readouts employing various immunologic assays. However, because the success of a therapeutic vaccine is dependent on accurate identification of HPV epitopes displayed as pMHC on HPV-infected target cells or HPV-transformed tumor cells, it is essential to define HPV-16 E6 and E7 T cell epitopes that are naturally processed and presented on the surface of virally altered cells. Only those HPV peptide/MHC class I complexes are capable of being recognized by cytolytic T lymphocytes to target destruction of transformed cells.
To this end, we have developed a new methodology, nanospray MS3 Poisson detection mass spectrometry. This methodology works by filtering the ion beam through two stages of mass selection and fragmentation (generating MS3 spectra) and detecting a target molecule by a probabilistic measure of the target's known dissociation patterns in the MS3 spectra. The methodology combines instrumental and ionization optimizations in a detection mode format to provide a high dynamic range from limited sample amounts. An instrumental geometry in which a quadrupole filter is placed in front of an ion trap (QTrap 4000) achieves a high duty cycle for MS3 spectra. Static nanospray avoids losses from surface exposure associated with chromatography and in its low (a few nanoliters per minute) flow, an optimal conversion of molecules in the condensed phase into gas phase ions.
Our findings are that none of the clinically targeted A2 peptides employed in epitope-based T cell vaccines to date could be detected on HPV-16-transformed cell lines tested herein. Two examples of this discordance are of particular note. The E786–93 peptide has been previously reported to be by far the best HLA-A*0201-binding peptide derived from HPV-16 (46). It is among the top predicted binders in the present study and the strongest binder in the HLA-A*0201 T2 binding assay. Nonetheless, E786–93 could not be detected by mass spectrometry on any of the HPV-16-transformed tumor cell lines. Furthermore, MS and fragmentation analyses of the peptides recovered from E786–93-loaded T2 cells showed that more than 90% of the E786–93 peptide complexed with HLA-A*0201 was modified with an additional cysteine that could be localized to the backbone cysteine residue and is most likely linked via a disulfide bridge (data not shown). This modification was also checked by MS3 analysis but was not detected on an HPV-16-transformed cell line. Likewise, E711–20 was not present on HPV-16 human tumor cells, although E711–19 was rather abundant. Based on our current observations, we suggest that one reason for the lack of clinical efficacy of epitope-based therapeutic T cell vaccines for HPV-16 and, by extension, other infectious diseases and cancers may be that vaccine-elicited responses were misdirected.
The E711–19 9-mer was detected on all five HLA-A*0201+ HPV-16-expressing cell lines examined (Fig. 6). As shown, the amount of E711–19 displayed relative to other endogenous HLA-A2 bound peptides varies significantly. These differences fit well with our knowledge about integrated versus episomal HPV-16 genome expression (75). The two cell lines with the integrated HPV-16 genome, CaSki and C66-3, which can have no E2-induced repression of E6 and E7 expression, display relatively more E7-derived peptide on their surface. By contrast, the transformed cell line C66-7 with the episomal HPV-16 genome in which E2 is still present displays far less E711–19. The two transfectant cell lines, N/E6E7 and OKF6/E6E7, have comparable amounts to the episomal cell line.
The striking specificity of human CTL for E711–19 is worth underscoring. E711–19-specific T cells recognize and lysed E711–19-loaded but not E711–20-loaded target cells even though both peptides bind well to HLA-A*0201. Alloreactivity as a basis for cytotoxicity in the assay was excluded by using a peptide pulsed autologous B cell line (Laz 509) as a target. Although not shown, E711–20 10-mer-specific T cells lysed autologous E711–20 but not E711–19-pulsed B cells as well. Why should this recognition be so discrete in view of the fact that the two peptides differ from one another by only a single carboxyl-terminal threonine residue? The answer lies in the general nature of peptide binding to MHC class I molecules. One pocket exists for the amino terminus and a second for the carboxylate of the peptide, thereby docking the peptide ends in the HLA groove between α1 and α2 helices. The fixed “ends” mandate that the 10-mer peptide bulges further out of the HLA binding groove than the 9-mer, with greater surface area exposed to the TCR. Both the position of the main chain and side chain positions are altered. Hence, these two peptides will differ in their TCR recognition features (Ref. 76 and references therein). Failure to identify the precise epitope for use in the vaccine elicitation strategy will likely engender a misdirected response even for such similar sequences.
A recent study of 19 women with grade 3 vulvar intraepithelial neoplasia vaccinated with a non-epitope targeted mix of long peptides from HPV-16 viral E6 and E7 oncoproteins in incomplete Freund's adjuvant showed promising clinical responses (35). At 3 months post-vaccination, 5 women had complete regression, and at 12 months, 79% of subjects appeared to show significant clinical responses. These results demonstrate that therapeutic vaccination harnessing cellular immunity can be effective. But vaccinating with any set of long peptides that span the target antigen and incorporate the expected T cell epitopes is unlikely to be optimal. Broad and nonselective immune responses arising from an uncharacterized processing of vaccine components in secondary lymphoid organs coupled with restricted presentation by primary tumor cells would limit the population of responding CTLs at the tumor and therein directly reduce TCR-mediated cytolysis and secondarily dilute requisite inflammatory signals in the microenvironment. Dendritic cell presentation of multiple T cell epitopes, few of which are relevant, combined with well known mechanisms of immunodominance may further result in misguided responses (76). Targeting a response in a precise way in future immunotherapeutic efforts offering appropriate adjuvant and delivery to dendritic cells will focus T cells on relevant protective/therapeutic epitopes. In addition, by precisely selecting T cell epitopes in vaccine formulation, bioinformatics can be used to calculate population protection coverage, ensuring that there is an adequate breadth of epitopes incorporated.
The landmark studies of Benacerraf (77) on immune response genes demonstrated many years ago that immune responses to chemically defined antigens are strictly dependent on MHC alleles. In this regard, the basis for the complete response in only a subset of patients with vulvar intraepithelial neoplasia (35) is almost certainly related to differences in HLA alleles expressed by the subjects. An analysis of the frequencies of HLA-A, -B, and -C alleles in the five major ethnic groups of the United States involving 1296 unrelated subjects from five major outbred groups (African American, Caucasians, Asian, Hispanic, and North American Natives) was performed (78). The frequency of A*0201, which binds strongly to E711–19, is higher than 10% in each group aside from Asians. In the Korean population, A*0201 and A*0206, which are E711–19 binding alleles, compose 85% of the A2 positive population (79). Among the 16 HLA-A2 alleles that are not predicted to bind E711–19, we find that A*0207, A*0278, A*9208 and A*9217 are found in the Chinese population, with the frequency of A*0207 higher than 10% in the southern Chinese Han population (80, 81).
Nine major HLA class I supertypes (A1, A2, A3, A24, B7, B27, B44, B58, and B62) have been identified (82). Each of the four HLA class I supertypes (A2, A3, B7, and B44) allows coverage of about 35–55% of the general population regardless of ethnicity (83). When epitopes from the A2, A3, B7, and B44 supertypes are combined, general population coverage is higher than 90%. Common alleles of the four supertypes are: A2 (A*0201-07, A*6802, and A*6901), A3 (A*0301, A*1101, A*3101, A*3301, and A*6801), B7 (B*0702, B*3501–03, B*5101-02, B*5301, and B*5401) and B4 (B*3701, B*4001, B*4006, B*4402, and B*4403). Within the A2 supertype, predicted binding affinities for the conserved E711–19 epitope to A*0201, A*0202, A*0203, A*0204, A*0205, and A*0206 are excellent (Fig. 11). By selecting for promiscuous peptide binders parsed for conservation of sequence among viruses and whose presentation on target cells is verified by physical methods, a new paradigm for T cell-based vaccines can evolve predicated on T cell epitope specificity and precise HLA restriction. This paradigm should be added to vaccinology in the genomic era (84).
Our results strongly imply that the presence or absence of memory responses in the peripheral blood is not a useful surrogate to guide immunity to relevant tumor target antigens. Precursor frequencies may be too low, especially if cells have already trafficked to target organs as effector memory populations. Moreover, as antigen is transported to lymph node and presented to T cells on dendritic cells, a focusing of immune response through cross-presentation or other means may select for immunodominant epitopes not necessarily reflective of relevant peptide array displayed on tumor cells. Immune responses against the latter are what will lead to protection. The MS technology described here offers an approach toward making identification of such a display tractable using a limited number of cells.
Supplementary Material
Acknowledgments
We thank K. Münger (Channing Laboratory, Department of Medicine, Brigham and Women's Hospital/Harvard Medical School) for expert advice and review of this manuscript and J. H. Lee (Department of Otolaryngology, University of Iowa) and James G. Rheinwald (Department of Dermatology and Harvard Skin Disease Research Center, Brigham and Women's Hospital and Harvard Medical School) for kind gifts of the C66 and E6E7 cell lines, respectively.

The on-line version of this article (available at http://www.jbc.org) contains supplemental S1–S4.
B. Reinhold, D. B. Keskin, and E. L. Reinherz, submitted for publication.
- HPV
- human Papillomavirus
- CTL
- cytotoxic T lymphocyte
- PBMC
- peripheral blood mononuclear cells
- pMHC
- a complex of a peptide and an MHC molecule
- TCR
- T cell receptor
- Rb
- retinoblastoma
- HTLV
- human T cell leukemia virus
- SFU
- spot-forming unit.
REFERENCES
- 1.zur Hausen H. (1977) Curr. Top. Microbiol. Immunol. 78, 1–30 [DOI] [PubMed] [Google Scholar]
- 2.Kreimer A. R., Clifford G. M., Boyle P., Franceschi S. (2005) Cancer Epidemiol. Biomarkers Prev. 14, 467–475 [DOI] [PubMed] [Google Scholar]
- 3.Munoz N., Castellsague X., de Gonzalez A. B., Gissmann L. (2006) Vaccine 24, Suppl. 3, 1–10 [DOI] [PubMed] [Google Scholar]
- 4.Moscicki A. B., Schiffman M., Kjaer S., Villa L. L. (2006) Vaccine 24, Suppl S3, 42–51 [DOI] [PubMed] [Google Scholar]
- 5.Longworth M. S., Laimins L. A. (2004) Microbiol. Mol. Biol. Rev. 68, 362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Münger K., Baldwin A., Edwards K. M., Hayakawa H., Nguyen C. L., Owens M., Grace M., Huh K. (2004) J. Virol. 78, 11451–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkin D. M., Bray F. (2006) Vaccine 24, Suppl 3, S3/11–25 [DOI] [PubMed] [Google Scholar]
- 8.Berzofsky J. A., Ahlers J. D., Janik J., Morris J., Oh S., Terabe M., Belyakov I. M. (2004) J. Clin. Invest. 114, 450–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowy D. R., Schiller J. T. (2006) J. Clin. Invest. 116, 1167–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Printz C. (2009) Cancer 115, 5130. [DOI] [PubMed] [Google Scholar]
- 11.Ramqvist T., Andreasson K., Dalianis T. (2007) Expert Opin. Biol. Ther. 7, 997–1007 [DOI] [PubMed] [Google Scholar]
- 12.Vetter K. M., Geller S. E. (2007) J. Womens Health 16, 1258–1268 [DOI] [PubMed] [Google Scholar]
- 13.Frazer I. H. (2004) Nat. Rev. Immunol 4, 46–54 [DOI] [PubMed] [Google Scholar]
- 14.Stanley M. (2006) Vaccine 24, Suppl 1S, 16–22 [Google Scholar]
- 15.Boon T., Coulie P. G., Van den Eynde B. (1997) Immunol Today 18, 267–268 [DOI] [PubMed] [Google Scholar]
- 16.Devaraj K., Gillison M. L., Wu T. C. (2003) Crit. Rev. Oral. Biol. Med. 14, 345–362 [DOI] [PubMed] [Google Scholar]
- 17.Leggatt G. R., Frazer I. H. (2007) Curr. Opin. Immunol. 19, 232–238 [DOI] [PubMed] [Google Scholar]
- 18.Roden R. B., Ling M., Wu T. C. (2004) Hum. Pathol. 35, 971–982 [DOI] [PubMed] [Google Scholar]
- 19.Peng S., Trimble C., Wu L., Pardoll D., Roden R., Hung C. F., Wu T. C. (2007) Clin. Cancer Res. 13, 2479–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Hernández E., González-Sánchez J. L., Andrade-Manzano A., Contreras M. L., Padilla S., Guzmán C. C., Jiménez R., Reyes L., Morosoli G., Verde M. L., Rosales R. (2006) Cancer Gene Ther. 13, 592–597 [DOI] [PubMed] [Google Scholar]
- 21.Albarran Y., Carvajal A., de la Garza A., Cruz Quiroz B. J., Vazquez Zea E., Díaz Estrada I., Mendez Fuentez E., López Contreras M., Andrade-Manzano A., Padilla S., Varela A. R., Rosales R. (2007) BioDrugs 21, 47–59 [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann A. M., Stern P. L., Rankin E. M., Sommer H., Nuessler V., Schneider A., Adams M., Onon T. S., Bauknecht T., Wagner U., Kroon K., Hickling J., Boswell C. M., Stacey S. N., Kitchener H. C., Gillard J., Wanders J., Roberts J. S., Zwierzina H. (2002) Clin. Cancer Res. 8, 3676–3685 [PubMed] [Google Scholar]
- 23.Einstein M. H., Kadish A. S., Burk R. D., Kim M. Y., Wadler S., Streicher H., Goldberg G. L., Runowicz C. D. (2007) Gynecol. Oncol. 106, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palefsky J. M., Berry J. M., Jay N., Krogstad M., Da Costa M., Darragh T. M., Lee J. Y. (2006) Aids 20, 1151–1155 [DOI] [PubMed] [Google Scholar]
- 25.Derkay C. S., Smith R. J., McClay J., van Burik J. A., Wiatrak B. J., Arnold J., Berger B., Neefe J. R. (2005) Ann. Otol. Rhinol. Laryngol. 114, 730–737 [DOI] [PubMed] [Google Scholar]
- 26.Fiander A. N., Tristram A. J., Davidson E. J., Tomlinson A. E., Man S., Baldwin P. J., Sterling J. C., Kitchener H. C. (2006) Int. J. Gynecol Cancer 16, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 27.Hallez S., Simon P., Maudoux F., Doyen J., Noël J. C., Beliard A., Capelle X., Buxant F., Fayt I., Lagrost A. C., Hubert P., Gerday C., Burny A., Boniver J., Foidart J. M., Delvenne P., Jacobs N. (2004) Cancer Immunol. Immunother. 53, 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kast W. M., Brandt R. M., Drijfhout J. W., Melief C. J. (1993) J. Immunother. Emphasis Tumor Immunol. 14, 115–120 [DOI] [PubMed] [Google Scholar]
- 29.Feltkamp M. C., Smits H. L., Vierboom M. P., Minnaar R. P., de Jongh B. M., Drijfhout J. W., ter Schegget J., Melief C. J., Kast W. M. (1993) Eur. J. Immunol. 23, 2242–2249 [DOI] [PubMed] [Google Scholar]
- 30.Ressing M. E., Sette A., Brandt R. M., Ruppert J., Wentworth P. A., Hartman M., Oseroff C., Grey H. M., Melief C. J., Kast W. M. (1995) J. Immunol. 154, 5934–5943 [PubMed] [Google Scholar]
- 31.Steller M. A., Gurski K. J., Murakami M., Daniel R. W., Shah K. V., Celis E., Sette A., Trimble E. L., Park R. C., Marincola F. M. (1998) Clin. Cancer Res. 4, 2103–2109 [PubMed] [Google Scholar]
- 32.van Driel W. J., Ressing M. E., Kenter G. G., Brandt R. M., Krul E. J., van Rossum A. B., Schuuring E., Offringa R., Bauknecht T., Tamm-Hermelink A., van Dam P. A., Fleuren G. J., Kast W. M., Melief C. J., Trimbos J. B. (1999) Eur. J. Cancer 35, 946–952 [DOI] [PubMed] [Google Scholar]
- 33.Ressing M. E., van Driel W. J., Brandt R. M., Kenter G. G., de Jong J. H., Bauknecht T., Fleuren G. J., Hoogerhout P., Offringa R., Sette A., Celis E., Grey H., Trimbos B. J., Kast W. M., Melief C. J. (2000) J. Immunother. 23, 255–266 [DOI] [PubMed] [Google Scholar]
- 34.Muderspach L., Wilczynski S., Roman L., Bade L., Felix J., Small L. A., Kast W. M., Fascio G., Marty V., Weber J. (2000) Clin. Cancer Res. 6, 3406–3416 [PubMed] [Google Scholar]
- 35.Kenter G. G., Welters M. J., Valentijn A. R., Lowik M. J., Berends-van der Meer D. M., Vloon A. P., Essahsah F., Fathers L. M., Offringa R., Drijfhout J. W., Wafelman A. R., Oostendorp J., Fleuren G. J., van der Burg S. H., Melief C. J. (2009) N. Engl. J. Med. 361, 1838–1847 [DOI] [PubMed] [Google Scholar]
- 36.Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 37.Wang J. H., Reinherz E. L. (2002) Mol. Immunol. 38, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 38.Robinson J., Waller M. J., Fail S. C., McWilliam H., Lopez R., Parham P., Marsh S. G. (2009) Nucleic Acids Res. 37, D1013–D1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brusic V., August J. T., Petrovsky N. (2005) Expert Rev. Vaccines 4, 407–417 [DOI] [PubMed] [Google Scholar]
- 40.De Groot A. S., Berzofsky J. A. (2004) Methods 34, 425–428 [DOI] [PubMed] [Google Scholar]
- 41.Purcell A. W., McCluskey J., Rossjohn J. (2007) Nat. Rev. Drug Discov. 6, 404–414 [DOI] [PubMed] [Google Scholar]
- 42.Lee J. H., Yi S. M., Anderson M. E., Berger K. L., Welsh M. J., Klingelhutz A. J., Ozbun M. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2094–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin H. H., Ray S., Tongchusak S., Reinherz E. L., Brusic V. (2008) BMC Immunol. 9, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong W., Reche P. A., Lai C. C., Reinhold B., Reinherz E. L. (2003) J. Biol. Chem. 278, 45135–45144 [DOI] [PubMed] [Google Scholar]
- 45.Schultze J. L., Michalak S., Seamon M. J., Dranoff G., Jung K., Daley J., Delgado J. C., Gribben J. G., Nadler L. M. (1997) J. Clin. Invest. 100, 2757–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kast W. M., Brandt R. M., Sidney J., Drijfhout J. W., Kubo R. T., Grey H. M., Melief C. J., Sette A. (1994) J. Immunol. 152, 3904–3912 [PubMed] [Google Scholar]
- 47.Arnold D., Driscoll J., Androlewicz M., Hughes E., Cresswell P., Spies T. (1992) Nature 360, 171–174 [DOI] [PubMed] [Google Scholar]
- 48.Momburg F., Ortiz-Navarrete V., Neefjes J., Goulmy E., van de Wal Y., Spits H., Powis S. J., Butcher G. W., Howard J. C., Walden P., Hammerling G. (1992) Nature 360, 174–177 [DOI] [PubMed] [Google Scholar]
- 49.Muñoz N., Bosch F. X., Castellsagué X., Díaz M., de Sanjose S., Hammouda D., Shah K. V., Meijer C. J. (2004) Int. J. Cancer 111, 278–285 [DOI] [PubMed] [Google Scholar]
- 50.van den Hende M., van Poelgeest M. I., van der Hulst J. M., de Jong J., Drijfhout J. W., Fleuren G. J., Valentijn A. R., Wafelman A. R., Slappendel G. M., Melief C. J., Offringa R., van der Burg S. H., Kenter G. G. (2008) Int. J. Cancer 123, 146–152 [DOI] [PubMed] [Google Scholar]
- 51.Henderson R. A., Michel H., Sakaguchi K., Shabanowitz J., Appella E., Hunt D. F., Engelhard V. H. (1992) Science 255, 1264–1266 [DOI] [PubMed] [Google Scholar]
- 52.Cento V., Ciccozzi M., Ronga L., Perno C. F., Ciotti M. (2009) J. Med. Virol. 81, 1627–1634 [DOI] [PubMed] [Google Scholar]
- 53.Hoof I., Peters B., Sidney J., Pedersen L. E., Sette A., Lund O., Buus S., Nielsen M. (2009) Immunogenetics 61, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiller J. T., Castellsague X., Villa L. L., Hildesheim A. (2008) Vaccine 26, Suppl 10, K53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kadish A. S., Ho G. Y., Burk R. D., Wang Y., Romney S. L., Ledwidge R., Angeletti R. H. (1997) J. Natl. Cancer Inst. 89, 1285–1293 [DOI] [PubMed] [Google Scholar]
- 56.Bourgault Villada I., Bénéton N., Bony C., Connan F., Monsonego J., Bianchi A., Saiag P., Lévy J. P., Guillet J. G., Choppin J. (2000) Eur. J. Immunol. 30, 2281–2289 [DOI] [PubMed] [Google Scholar]
- 57.van der Burg S. H., Ressing M. E., Kwappenberg K. M., de Jong A., Straathof K., de Jong J., Geluk A., van Meijgaarden K. E., Franken K. L., Ottenhoff T. H., Fleuren G. J., Kenter G., Melief C. J., Offringa R. (2001) Int. J. Cancer 91, 612–618 [DOI] [PubMed] [Google Scholar]
- 58.Rudolf M. P., Man S., Melief C. J., Sette A., Kast W. M. (2001) Clin. Cancer Res. 7, 788s–795s [PubMed] [Google Scholar]
- 59.Kather A., Ferrara A., Nonn M., Schinz M., Nieland J., Schneider A., Dürst M., Kaufmann A. M. (2003) Int. J. Cancer 104, 345–353 [DOI] [PubMed] [Google Scholar]
- 60.Bourgault Villada I., Moyal Barracco M., Villada I. B., Barracco M. M., Ziol M., Chaboissier A., Barget N., Berville S., Paniel B., Jullian E., Clerici T., Maillère B., Guillet J. G. (2004) Cancer Res. 64, 8761–8766 [DOI] [PubMed] [Google Scholar]
- 61.Nakagawa M., Kim K. H., Moscicki A. B. (2004) Clin. Diagn. Lab. Immunol. 11, 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warrino D. E., Olson W. C., Knapp W. T., Scarrow M. I., D'Ambrosio-Brennan L. J., Guido R. S., Edwards R. P., Kast W. M., Storkus W. J. (2004) Clin. Cancer Res. 10, 3301–3308 [DOI] [PubMed] [Google Scholar]
- 63.Sarkar A. K., Tortolero-Luna G., Follen M., Sastry K. J. (2005) Gynecol. Oncol. 99, S251–S261 [DOI] [PubMed] [Google Scholar]
- 64.Hara M., Matsueda S., Tamura M., Takedatsu H., Tanaka M., Kawano K., Mochizuki K., Kamura T., Itoh K., Harada M. (2005) Int. J. Oncol. 27, 1371–1379 [PubMed] [Google Scholar]
- 65.Oerke S., Höhn H., Zehbe I., Pilch H., Schicketanz K. H., Hitzler W. E., Neukirch C., Freitag K., Maeurer M. J. (2005) Int. J. Cancer 114, 766–778 [DOI] [PubMed] [Google Scholar]
- 66.Nakagawa M., Kim K. H., Gillam T. M., Moscicki A. B. (2007) J. Virol. 81, 1412–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallagher K. M., Man S. (2007) J. Gen Virol. 88, 1470–1478 [DOI] [PubMed] [Google Scholar]
- 68.Morishima S., Akatsuka Y., Nawa A., Kondo E., Kiyono T., Torikai H., Nakanishi T., Ito Y., Tsujimura K., Iwata K., Ito K., Kodera Y., Morishima Y., Kuzushima K., Takahashi T. (2007) Int. J. Cancer 120, 594–604 [DOI] [PubMed] [Google Scholar]
- 69.Piersma S. J., Welters M. J., van der Hulst J. M., Kloth J. N., Kwappenberg K. M., Trimbos B. J., Melief C. J., Hellebrekers B. W., Fleuren G. J., Kenter G. G., Offringa R., van der Burg S. H. (2008) Int. J. Cancer 122, 486–494 [DOI] [PubMed] [Google Scholar]
- 70.Tindle R. W. (2002) Nat. Rev. Cancer 2, 59–65 [DOI] [PubMed] [Google Scholar]
- 71.Kanodia S., Fahey L. M., Kast W. M. (2007) Curr. Cancer Drug Targets 7, 79–89 [DOI] [PubMed] [Google Scholar]
- 72.Stanley M. A., Pett M. R., Coleman N. (2007) Biochem. Soc. Trans. 35, 1456–1460 [DOI] [PubMed] [Google Scholar]
- 73.Georgopoulos N. T., Proffitt J. L., Blair G. E. (2000) Oncogene 19, 4930–4935 [DOI] [PubMed] [Google Scholar]
- 74.Brady C. S., Bartholomew J. S., Burt D. J., Duggan-Keen M. F., Glenville S., Telford N., Little A. M., Davidson J. A., Jimenez P., Ruiz-Cabello F., Garrido F., Stern P. L. (2000) Tissue Antigens 55, 401–411 [DOI] [PubMed] [Google Scholar]
- 75.Steger G., Corbach S. (1997) J. Virol. 71, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meijers R., Lai C. C., Yang Y., Liu J. H., Zhong W., Wang J. H., Reinherz E. L. (2005) J. Mol. Biol. 345, 1099–1110 [DOI] [PubMed] [Google Scholar]
- 77.Benacerraf B. (1980) Nobel Lecture Physiology or Medicine 1971–1980 (1992) (Lindsten J. ed) pp. 597–623, World Scientific Publishing Co., Singapore [Google Scholar]
- 78.Cao K., Hollenbach J., Shi X., Shi W., Chopek M., Fernández-Viña M. A. (2001) Hum. Immunol. 62, 1009–1030 [DOI] [PubMed] [Google Scholar]
- 79.Lee K. W., Oh D. H., Lee C., Yang S. Y. (2005) Tissue Antigens 65, 437–447 [DOI] [PubMed] [Google Scholar]
- 80.Gao S. Q., Zou H. Y., Cheng L. H., Jing S. Z., Deng Z. H. (2009) Zhonghua Yi Xue Yi Chuan Xue Za Zhi 26, 228–232 [DOI] [PubMed] [Google Scholar]
- 81.Yao Y., Shi L., Shi L., Matsushita M., Yu L., Lin K., Tao Y., Huang X., Yi W., Oka T., Tokunaga K., Chu J. (2009) Tissue Antigens 73, 561–568 [DOI] [PubMed] [Google Scholar]
- 82.Sette A., Sidney J. (1999) Immunogenetics 50, 201–212 [DOI] [PubMed] [Google Scholar]
- 83.Sidney J., Grey H. M., Kubo R. T., Sette A. (1996) Immunol Today 17, 261–266 [DOI] [PubMed] [Google Scholar]
- 84.Rinaudo C. D., Telford J. L., Rappuoli R., Seib K. L. (2009) J. Clin. Invest. 119, 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ashrafi G. H., Haghshenas M., Marchetti B., Campo M. S. (2006) Int. J. Cancer 119, 2105–2112 [DOI] [PubMed] [Google Scholar]
- 86.Davy C. E., Ayub M., Jackson D. J., Das P., McIntosh P., Doorbar J. (2006) Virology 349, 230–244 [DOI] [PubMed] [Google Scholar]
- 87.Davy C. E., Jackson D. J., Raj K., Peh W. L., Southern S. A., Das P., Sorathia R., Laskey P., Middleton K., Nakahara T., Wang Q., Masterson P. J., Lambert P. F., Cuthill S., Millar J. B., Doorbar J. (2005) J. Virol. 79, 3998–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dyson N., Howley P. M., Münger K., Harlow E. (1989) Science 243, 934–937 [DOI] [PubMed] [Google Scholar]
- 89.Kabsch K., Alonso A. (2002) J. Virol. 76, 12162–12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McIntosh P. B., Martin S. R., Jackson D. J., Khan J., Isaacson E. R., Calder L., Raj K., Griffin H. M., Wang Q., Laskey P., Eccleston J. F., Doorbar J. (2008) J. Virol. 82, 8196–8203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O'Brien P. M., Saveria Campo M. (2002) Virus Res. 88, 103–117 [DOI] [PubMed] [Google Scholar]
- 92.Werness B. A., Levine A. J., Howley P. M. (1990) Science 248, 76–79 [DOI] [PubMed] [Google Scholar]
- 93.Zhang B., Li P., Wang E., Brahmi Z., Dunn K. W., Blum J. S., Roman A. (2003) Virology 310, 100–108 [DOI] [PubMed] [Google Scholar]
- 94.Pattillo R. A., Hussa R. O., Story M. T., Ruckert A. C., Shalaby M. R., Mattingly R. F. (1977) Science 196, 1456–1458 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.