Abstract
Gephyrin and collybistin are key components of GABAA receptor (GABAAR) clustering. Nonetheless, resolving the molecular interactions between the plethora of GABAAR subunits and these clustering proteins is a significant challenge. We report a direct interaction of GABAAR α2 and α3 subunit intracellular M3–M4 domain (but not α1, α4, α5, α6, β1–3, or γ1–3) with gephyrin. Curiously, GABAAR α2, but not α3, binds to both gephyrin and collybistin using overlapping sites. The reciprocal binding sites on gephyrin for collybistin and GABAAR α2 also overlap at the start of the gephyrin E domain. This suggests that although GABAAR α3 interacts with gephyrin, GABAAR α2, collybistin, and gephyrin form a trimeric complex. In support of this proposal, tri-hybrid interactions between GABAAR α2 and collybistin or GABAAR α2 and gephyrin are strengthened in the presence of gephyrin or collybistin, respectively. Collybistin and gephyrin also compete for binding to GABAAR α2 in co-immunoprecipitation experiments and co-localize in transfected cells in both intracellular and submembrane aggregates. Interestingly, GABAAR α2 is capable of “activating ” collybistin isoforms harboring the regulatory SH3 domain, enabling targeting of gephyrin to the submembrane aggregates. The GABAAR α2-collybistin interaction was disrupted by a pathogenic mutation in the collybistin SH3 domain (p.G55A) that causes X-linked intellectual disability and seizures by disrupting GABAAR and gephyrin clustering. Because immunohistochemistry in retina revealed a preferential co-localization of collybistin with α2 subunit containing GABAARs, but not GlyRs or other GABAAR subtypes, we propose that the collybistin-gephyrin complex has an intimate role in the clustering of GABAARs containing the α2 subunit.
Keywords: GABA Receptors, Genetic Diseases, Neuroscience, Protein Targeting, Synapses, Collybistin, GABAAR α2, Gephyrin, Synaptic Clustering, Yeast Tri-hybrid Interactions
Introduction
The clustering proteins gephyrin (1) and collybistin (2, 3) are thought to represent key players in the synaptic clustering of both glycine receptors (GlyRs)4 and GABAARs. Studies using gephyrin knock-out mice or mRNA knockdown (4–7) have shown a loss of postsynaptic clustering of GlyRs and GABAARs containing α2 and γ2 subunits. However, certain GABAAR subtypes still appear to cluster in neurons lacking gephyrin (6–9). Hence, although the majority of GlyRs are likely to be clustered by gephyrin (10), exactly which GABAAR subtypes are subject to gephyrin-dependent clustering remains unclear. What is certain is that the subcellular localization of gephyrin is dependent on certain GABAAR subtypes. For example, targeted deletion of the GABAAR α1, α3, and γ2 subunit genes results in a loss of synaptic gephyrin and GABAAR clusters (11–15), with cytoplasmic gephyrin aggregates indicating disrupted synaptic targeting.
Although the interaction of the GlyR β subunit with the gephyrin E domain has been well characterized (3, 16, 17), the same cannot be said for GABAARs. Different mechanisms have been proposed to explain the complex interactions of GABAARs with gephyrin, including alternative splicing (18), intermediate accessory proteins (19, 20), post-translational modifications (21), or even binding sites formed at intracellular subunit-subunit interfaces (22, 23). However, a recent report suggesting a direct interaction of the GABAAR α2 subunit with gephyrin (24) prompted us to examine interactions of GABAAR α, β, and γ subunits with gephyrin. We also explored the potential role of the RhoGEF collybistin because collybistin knock-out mice show defects in the clustering of selected GABAAR subtypes in the hippocampus and the basolateral amygdala, but unexpectedly do not show a comparable loss of synaptic GlyRs (25–27).
Using a range of molecular, cell biology, and immunohistochemical techniques, we show that both GABAAR α2 and α3 subunits interact directly with gephyrin and define the molecular basis for the α2-gephyrin interaction by mapping the reciprocal binding sites. We also demonstrate that GABAAR α2 can “activate ” collybistin-mediated gephyrin clustering in recombinant systems via interactions with a distinct non-PXXP motif in the α2 subunit M3–M4 domain. This interaction is disrupted by a pathogenic mutation in collybistin (p.G55A) linked to X-linked intellectual disability and seizures (3). We confirm the intimate association of collybistin with the GABAAR α2 subtype in vivo using a novel collybistin antibody. Taken together, these data suggest that GABAAR α2 can trigger the synaptic clustering machinery via dual interactions with collybistin and gephyrin.
EXPERIMENTAL PROCEDURES
Expression Constructs and Site-directed Mutagenesis
GABAAR α2 subunit and gephyrin and collybistin cDNAs were amplified from rat spinal cord or human whole brain first-strand cDNA (Clontech) using Pfx DNA polymerase (Invitrogen) and were cloned into the yeast two- or three-hybrid vectors (pYTH15, pYTH16, or pACT2) (ref. 45) or the mammalian expression vectors pRK5myc, pRK5FLAG, pHcRed1-C1, or pEGFP-C1 (Clontech). Cloning resulted in an in-frame fusion of the GAL4 DNA binding domain (vector pYTH16), GAL4 activation domain (vector pACT2), HcRed, EGFP, myc, or FLAG epitope tags to the N termini of all expressed proteins with the exception of pRK5-GABAAR α2-FLAG, where a C-terminal FLAG tag was engineered. Mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene), and all constructs were verified by DNA sequencing.
Yeast Two-hybrid Assays
The yeast strain Y190 was co-transformed with pYTH16-GABAAR α, β, and γ or pYTH16-GlyR β subunit intracellular M3–M4 regions or pYTH16-gephyrin bait constructs together with pACT2-collybistin or pACT2-gephyrin prey constructs. pYTH16-GlyR β, pYTH16-gephyrin, pACT2-gephyrin deletions, alanine block mutants, and pACT2-collybistin constructs were described previously (3, 36). Additional pACT2-gephyrin mutants, pYTH16-GABAAR baits, pYTH16-GABAAR α2 deletion mutants, and pYTH16-GABAAR α2/α1 chimeras were generated during this study. Transformations were plated on selective dropout media (either −LWH +30 mm 3-amino-1,2,4-triazole or −LW). After incubation at 30 °C for 3–6 days, LacZ reporter gene assays were performed as described (3).
Semiquantitative and Quantitative Yeast Tri-hybrid Assays
pYTH15-collybistin and pYTH15-GephE constructs express CB3SH3+ and the gephyrin E domain (amino acids 305–706) under the control of the methionine promoter after integration into SDY191 (45). Integrated yeast strains were co-transformed with pYTH16-GABAAR α2, pYTH16-GlyR β, or pYTH16-gephyrin bait constructs with either pACT2-CB3SH3+ or pACT2-gephyrin P1 prey constructs. Transformations were plated on selective dropout media −LWM to induce expression of CB3SH3+ or gephyrin E domain, or supplemented with 2 mm l-methionine to suppress expression of these proteins.
Quantitative Yeast Assays
Cell pellets were resuspended in Z-buffer containing 40 mm β-mercaptoethanol followed by lysis in 0.1% (w/v) SDS and 0.1% (v/v) chloroform (46). All protein interactions were assayed in four to six independent experiments in triplicate. After addition of chlorophenol red-β-d-galactopyranoside the color change was recorded at 540 nm, and readings were adjusted for turbidity of the yeast suspension at 620 nm. The background signal (co-transfected pACT2 negative control) was subtracted from each co-transformation reading. Established yeast two-hybrid interactions (gephyrin-collybistin and gephyrin-gephyrin) were set as 100%. Statistically significant differences in the interaction strength of two proteins in the presence or absence of a third protein were determined using a Student's two-tailed t test.
Cell Culture, Immunocytochemistry, Confocal Microscopy, and Image Analysis
HEK293 cells (ATCC CRL1573) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin G, and 100 μg/ml streptomycin at 37 °C in 95% air and 5% CO2. Exponentially growing cells were transfected with constructs encoding epitope-tagged gephyrin, collybistin, and GABAAR α subunit constructs using Lipofectamine LTX reagent (Invitrogen). After 24 h cells were fixed in 4% (w/v) paraformaldehyde and stained with antibodies recognizing myc (Sigma) and FLAG (Sigma) and secondary anti-mouse and anti-rabbit antibodies conjugated to Alexa Fluor 488, Alexa Fluor 546, or Cy5 fluorochromes (Molecular Probes). Confocal microscopy was performed using a Zeiss LSM 510 META. All images were taken with a 63× objective. Fluorescence excited by the 488 nm, 543 nm, and 633 nm laser lines of argon and helium/neon lasers was detected separately using only one laser at the time (multitrack function) and a combination of band pass filters (BP 505–530, BP 560–615), long pass (LP 560) filters, and meta function (649–798) dependent on the combination of fluorochromes.
Co-immunoprecipitation and Western Blotting
HEK293 cells were transfected as above and harvested 48 h post-transfection, lysed in a solution containing 150 mm NaCl, 50 mm Tris (pH 7.5), 5 mm EDTA (pH 8), 0.25% (v/v) Nonidet P-40/complete protease inhibitor mixture (Roche Applied Science) and homogenized using a tissue grinder. Following centrifugation (4 °C, 15 min, 16,000 × g), 1 ml of cell lysate containing ∼700 μg of protein was added to 40 μl of anti-FLAG M2 affinity gel (Sigma) and incubated overnight at 4 °C on a turning disk. The affinity gel was subjected to centrifugation (4 °C, 3 min, 100 × g) followed by two washes in 50 mm NaCl, 50 mm Tris, 0.1% (v/v) Triton X-100, two washes in 150 mm NaCl, PBS, 0.1% (v/v) Triton X-100, and two washes in PBS, 0.1% (v/v) Triton X-100. The FLAG fusion proteins were eluted with 150 ng of 3 × FLAG peptide (Sigma) for 30 min at room temperature. The eluates were analyzed by SDS-PAGE and immunoblotting. Approximately 10 μg of protein was loaded into 4–12% (w/v) BisTris precast gels (Invitrogen). Proteins were transferred to polyvinylidine fluoride (PVDF) membranes (Millipore) and nonspecific binding blocked with 5% (w/v) skimmed milk in PBS plus 0.1% (v/v) Tween 20 or with 20% (v/v) horse serum in PBS. Anti-myc antibody (Sigma) was used at a 1:2,000 dilution, and anti-FLAG antibody (Sigma) was used at a 1:3,000 dilution at 4 °C overnight. For detection, a HRP-conjugated anti-rabbit secondary antibody (Santa Cruz Biotechnology) was used at a 1:2,000 dilution together with the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Immunohistochemistry, Microscopy, and Image Analysis
Polyclonal antisera against collybistin were generated using the peptide sequence CFWQNFSRLTPFKK by Multiple Peptide Systems (NeoMPS, San Diego, CA). This peptide was purified to 97% purity by HPLC, coupled to keyhole limpet hemocyanin via an N-terminal cysteine and used to immunize three New Zealand White rabbits using standard procedures. This generated two high affinity collybistin antibodies, as assessed by ELISA results against the source peptide (B2652 pre-bleed <50, 3rd bleed 103,400; B2653 pre-bleed <50; 136,200). One of these antibodies (B2653) was affinity-purified (pABCB3; 0.98 mg/ml, titer 123,800). Immunohistochemistry on rat retina was performed using primary antibodies recognizing GABAAR α subunits (a generous gift from Prof. Jean-Marc Fritschy), GlyRs (mAb4a), collybistin, and gephyrin (mAb7a) as previously described (47, 48). Briefly, adult rats were anesthetized with intraperitoneal ketamine-xylazine 1:1 (0.1 ml/kg) and decapitated. The posterior eyecups were fixed for 30 min in formaldehyde (4% (w/v) in 0.1 m phosphate buffer (pH 7.4)). The eyecups were then rinsed in phosphate buffer and cryoprotected in sucrose (10, 20, and 30% (w/v) in phosphate buffer). The retina was dissected and sectioned vertically with a cryostat. Details about the double immunofluorescence procedure are provided in Ref. 49). The sections were analyzed with a confocal microscope (Zeiss LSM5 Pascal), using a sequential acquisition system (multitrack mode). For analysis of co-localization, confocal images were segmented manually and processed with the co-localization module of the software Imaris (Bitplane, Zurich, Switzerland). Single- and double-labeled clusters were then quantified by a particle detection algorithm using Image J (National Institutes of Health).
RESULTS
Selective GABAAR α Subunit Interactions with Gephyrin and Collybistin
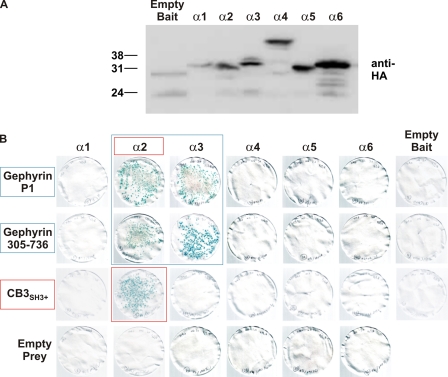
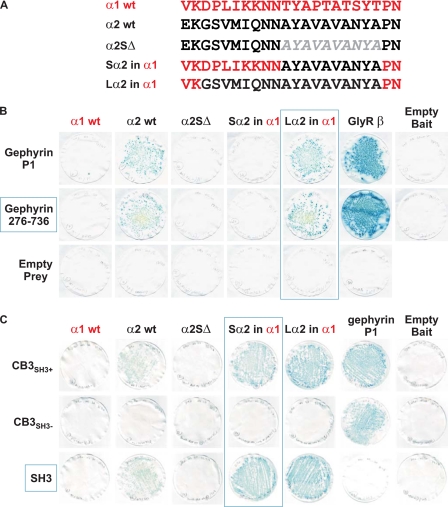
We utilized the yeast two-hybrid system to test for direct interactions between M3–M4 intracellular loops of the GABAAR α1–6, β1–3, γ1–3 subunits with gephyrin and collybistin. Although all bait proteins were expressed in yeast (Fig. 1A and supplemental Fig. 1), only the GABAAR α2 and α3 baits interacted with full-length gephyrin as assessed by LacZ freeze-fracture assays (Fig. 1B and supplemental Fig. 1). A fragment of gephyrin containing part of the C domain (collybistin binding motif) plus the entire E domain (Geph305–736) was sufficient for the interaction with GABAAR α2 or α3 baits (Fig. 1B). Curiously, the GABAAR α2 bait (but not the α3 bait) also interacted with a collybistin prey (CB3SH3+; Fig. 1B). Using deletion and domain swap mutations (Fig. 2A), we mapped the binding sites for gephyrin and collybistin in the GABAAR α2 M3–M4 loop. Deletion of a 10-amino acid motif in this region (α2SΔ, lacking AYAVAVANYA) abolished the interaction of the GABAAR α2 subunit with both gephyrin and collybistin (Fig. 2, B and C). However, inserting this motif into the GABAAR α1 bait (Sα2 in α1) allowed interaction with collybistin but not gephyrin (Fig. 2). To confer both collybistin and gephyrin binding to the GABAAR α1 bait, insertion of an 18-amino acid motif (Lα2 in α1; GSVMIQNNAYAVAVANYA) was necessary (Fig. 2, B and C). Hence, the binding sites for gephyrin and collybistin on GABAAR α2 are distinct but overlapping. It is also noteworthy that GABAAR α2-gephyrin and α3-gephyrin interactions are substantially weaker in LacZ freeze-fracture assays than the well characterized GlyR β-gephyrin interaction (Fig. 2B).
FIGURE 1.
GABAAR subunit interactions with gephyrin and collybistin. A, Western blot showing expression of GAL4 DNA binding domain-tagged GABAAR α subunit baits in the yeast strain Y190 detected using an anti-HA antibody. wt, wild type. B, GABAAR α subunit baits tested for interactions with full-length gephyrin, N-terminally truncated gephyrin (Geph(305–736)), and collybistin (CB3SH3+). LacZ freeze-fracture assays demonstrate that GABAAR α2 and α3 baits interact with gephyrin preys and that gephyrin residues 305–736 are sufficient for this interaction. Note that collybistin only interacts with the GABAAR α2 bait. See also supplemental Fig. 1 for experiments with GABAAR β and γ subunit baits.
FIGURE 2.
Overlapping binding sites for gephyrin and collybistin on GABAAR α2. A, alignment showing the position of an artificial GABAAR α2 subunit deletion and exchange chimeras. wt, wild type. B, GABAAR α2-gephyrin interactions abolished by deletion of a 10-amino acid sequence (AYAVAVANYA) in the GABAAR α2 M3–M4 region (α2SΔ). This sequence is not sufficient to mediate interactions between the Sα2 in α1 chimera and gephyrin. However, exchange of a longer GABAAR α2 motif (GSVMIQNNAYAVAVANYA) into the GABAAR α1 bait is sufficient to mediate interaction between the Lα2 in α1 chimera and gephyrin. C, GABAAR α2-collybistin interaction abolished by deletion of the AYAVAVANYA motif. This sequence is sufficient to mediate interaction between the Sα2 in α1 chimera and collybistin. The N-terminal collybistin SH3 domain is also necessary and sufficient for the GABAAR α2-collybistin interaction.
Mapping Binding Sites for GABAAR α2 on Gephyrin and Collybistin
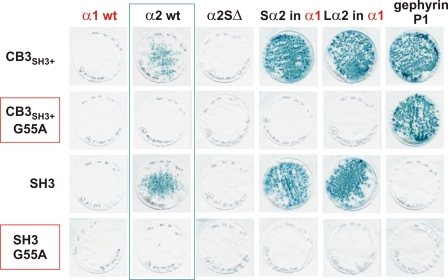
Interestingly, the GABAAR α2 bait did not interact with a prey for an alternatively spliced form of collybistin that lacks the N-terminal SH3 domain (CB3SH3−; Fig. 2C), although this prey did interact with gephyrin. We subsequently verified that a collybistin SH3 domain prey was capable of interacting with GABAAR α2, Sα2 in α1, and Lα2 in α1 baits, but not the α2 construct lacking the collybistin binding motif, α2SΔ (Fig. 3). These interactions were also disrupted by inserting a pathogenic mutation (p.G55A) (3) into the CB3SH3+ or SH3 domain preys (Fig. 3). To confirm that this interaction also occurs in mammalian cells, we expressed HcRed1-tagged GABAAR α1 and α2 M3–M4 domain fusion proteins with myc-tagged collybistin isoforms (myc-CB3SH3− and myc-CB3SH3+) in HEK293 cells. As previously observed for GlyR α1 and α3 intracellular loops (28) the HcRed-GABAAR α1, α2, and α3 fusion proteins show nuclear localization, due to cryptic nuclear localization sequences in the M3–M4 domain. This localization was supported by PSORTII analysis that predicts that the isolated GABAAR α1–3 M3–M4 domains should localize to the nucleus (calculated probabilities: α1, 74%; α2, 70%; α3, 61%). Despite this unusual subcellular compartmentalization, co-expression leads to redistribution of myc-CB3SH3+ (but not myc-CB3SH3−) to nuclear aggregates of HcRed-GABAAR α2 (Fig. 4, E–L). This effect was not observed for the equivalent HcRed-GABAAR α1 or α3 fusion proteins. Taken together, these results suggest that GABAAR α2 binds to the collybistin SH3 domain via a non-PXXP motif (AYAVAVANYA) and that this interaction is disrupted by a known pathogenic mutation in ARHGEF9.
FIGURE 3.
Pathogenic p.G55A mutation in collybistin disrupts the GABAAR α2-collybistin interaction. Introduction of the G55A amino acid substitution into preys encoding full-length collybistin (CB3SH3+) or the isolated SH3 domain disrupts the interaction with GABAAR α2. Note that the collybistin SH3 domain alone does not mediate interactions with gephyrin. wt, wild type.
FIGURE 4.
Collybistin and GABAAR α2 co-localize in transfected cells. Confocal microscopy shows co-expression of myc-tagged collybistin isoforms with and without the N-terminal SH3 domain and HcRed1-tagged GABAAR α1-α3 M3–M4 fusion proteins in transfected HEK293 cells. A–D, M–P, HcRed-GABAAR α1 and α3 fusion proteins are localized in the nucleus, whereas myc-tagged collybistin (myc-CB3SH3+) localizes to the cytoplasm. E–H, co-expression of collybistin isoform lacking the SH3 domain (myc-CB3SH3−) has no apparent effect on HcRed-GABAAR α2 distribution. I–L, co-expression of HcRed-GABAAR α2 with SH3-containing collybistin (myc-CB3SH3+) results in redistribution of collybistin and HcRed-GABAAR α2 to nuclear aggregates. Scale bars: 10 μm.
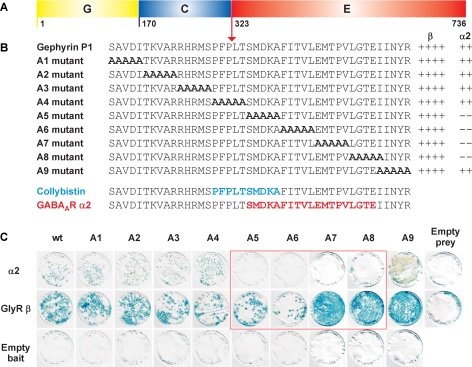
Mapping the GABAAR α2 binding site on gephyrin revealed that although the GABAAR α2 bait interacted with the Geph305–736 prey, shorter deletion constructs (Geph323–736, Geph336–736, Geph305–643, Geph305–674, and Geph305–704) did not mediate this interaction. Because similar findings were previously reported for collybistin binding to gephyrin (3) we used alanine scanning mutagenesis to locate the GABAAR α2 binding site on gephyrin to a 19-amino acid stretch in the N-terminal part of the gephyrin E domain (SMDKAFITVLEMPTVLGTE; Fig. 5). Although 5 alanine block mutants (A5–A8) disrupted GABAAR α2-gephyrin interactions, they did not alter GlyR β-gephyrin interactions (Fig. 5). This newly identified GABAAR α2 binding site overlaps the previously determined collybistin binding site (PFPLTSMDKA) (3) by 5 amino acids.
FIGURE 5.
Mapping the GABAAR α2 binding site on gephyrin. Intracellular GABAAR α2 and GlyR β subunit M3–M4 domain baits were tested for interactions with full-length gephyrin P1 containing the G, C, and E domains (A) and alanine substitution mutants (B) created in this prey. B and C, LacZ freeze-fracture assays demonstrate that the GABAAR α2 bait does not interact with gephyrin mutants A5–A8 whereas the interaction with GlyR β is retained. The GABAAR α2 binding site on gephyrin (amino acids 325–343) is shown together with the previously determined collybistin binding motif (amino acids 320–329) on gephyrin (3). LacZ freeze-fracture assay rankings: ++++, strong interaction; ++, moderate interaction; –, no interaction. wt, wild type.
Synergistic Interactions among GABAAR α2, Collybistin, and Gephyrin
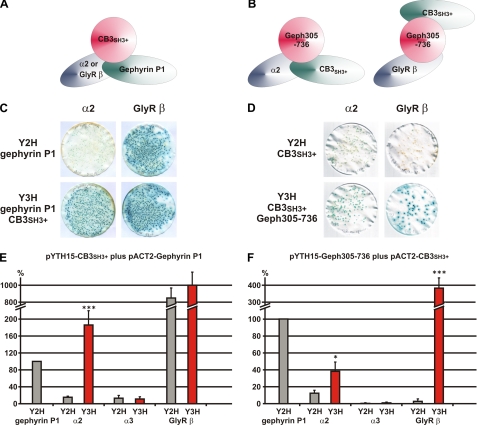
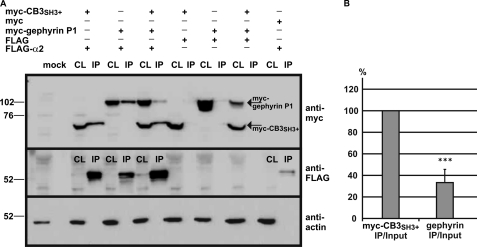
To assess the possibility that GABAAR α2, collybistin, and gephyrin form a multimeric complex, we used the yeast tri-hybrid system to measure changes in apparent interaction strength between interacting proteins in the presence of a third polypeptide expressed under the control of an inducible methionine promoter (Fig. 6, A and B). For example, although the GABAAR α2-gephyrin P1 bait-prey interaction is weaker in yeast strain SDY151 in LacZ freeze-fracture assays (Fig. 6C), this interaction is dramatically strengthened by inducing expression of collybistin (CB3SH3+). Quantitative assays demonstrated that this effect was statistically significant and not observed for either GABAAR α3 or GlyR β baits (Fig. 6, C and E). Similarly, induction of the gephyrin fragment Geph305–736 strengthened collybistin-GABAAR α2 but not collybistin-GABAAR α3 interactions (Fig. 6, D and F). Note that the GlyR β-collybistin interaction mediated by induction of Geph305–736 (Fig. 6D) is also a genuine tri-hybrid interaction because these two proteins occupy separate binding sites on gephyrin (3, 29). We also confirmed the interaction of GABAAR α2 with gephyrin P1 and collybistin by co-immunoprecipitation of a full-length FLAG-tagged GABAAR α2 subunit with myc-CB3SH3+ and/or myc-gephyrin from HEK293 cells (Fig. 7A). Interestingly, GABAAR α2 preferentially co-immunoprecipitates myc-CB3SH3+ rather than myc-gephyrin when all three proteins are co-expressed (Fig. 7B), although this effect may reflect the ready availability of cytoplasmic myc-tagged collybistin versus myc-gephyrin, which typically occurs in large cytoplasmic aggregates when expressed in HEK293 cells (2, 3).
FIGURE 6.
Yeast tri-hybrid assays reveal synergistic interactions among GABAAR α2, collybistin, and gephyrin. A and B, schematics depicting the principle of yeast tri-hybrid experiments. C, LacZ freeze-fracture assays showing a strengthening of the GABAAR α2-gephyrin interaction upon co-expression of collybistin (CB3SH3+). Note that no obvious effect is observed on the GlyR β-gephyrin interaction upon induction of collybistin. D, LacZ freeze-fracture assays showing a strengthening of the GABAAR α2-collybistin interactions upon co-expression of Geph305–736. Note that although GlyR β and collybistin do not usually interact, a genuine tri-hybrid interaction is observed when Geph305–736 reveals binding sites for both proteins, bringing the GAL4 DNA binding domain and activation domain domains into close proximity. E and F, quantification of the results shown in C and D. Error bars represent the mean ± S.D. n = 6, Student's two-tailed t test: ***, p < 0.001; **, p < 0.01; *, p < 0.05.
FIGURE 7.
Epitope-tagged GABAAR α2 co-immunoprecipitates collybistin and gephyrin in transfected mammalian cells. A, FLAG-tagged full-length GABAAR α2 was co-expressed with myc-tagged collybistin and/or myc-gephyrin in HEK293 cells. Note that myc-collybistin and myc-gephyrin are present in the cell lysates (CL) and FLAG-GABAAR α2 immunoprecipitation (IP) samples purified using FLAG beads, but not in IP samples from cells co-transfected with the FLAG tag alone. Interestingly, FLAG-GABAAR α2 appears to co-immunoprecipitate myc-collybistin preferentially rather than myc-gephyrin when all three proteins are co-transfected. B, this phenomenon was quantified after correcting for the protein input and normalizing bound collybistin to 100%. Error bars represent the mean ± S.D. n = 5, Student's two-tailed t test: ***, p < 0.001.
GABAAR α2 Activates Collybistin-mediated Submembrane Gephyrin Clustering
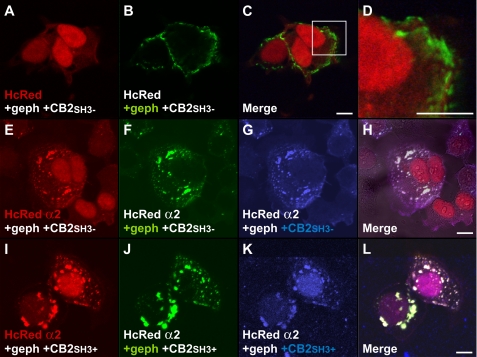
To assess whether GABAAR α2 interactions with the collybistin SH3 domain can activate gephyrin submembrane aggregation, we used a cellular model of clustering (2, 3). As previously described, collybistin isoforms lacking the SH3 domain (myc-CB2SH3− or myc-CB3SH3−) were able to cluster EGFP-gephyrin in submembrane compartments (Fig. 8, A–H). Although in previous experiments HcRed-GABAAR α2 M3-M4 fusion protein targeted to the cell nucleus (Fig. 4, E–L), on co-expression with EGFP-gephyrin and myc-collybistin isoforms lacking the SH3 domain (e.g. myc-CB2SH3−), HcRed-GABAAR α2 redistributed to cytoplasmic and submembrane gephyrin/collybistin aggregates (Fig. 8, E–H). This effect is likely to be mediated by GABAAR α2-gephyrin interactions because it was not seen for HcRed (Fig. 8, A–D). Co-expression of SH3 containing collybistin isoforms (e.g. myc-CB2SH3+) with EGFP-gephyrin does not normally lead to submembrane gephyrin accumulation (2, 3). However, co-expression of HcRed-GABAAR α2 was able to activate collybistin-mediated gephyrin clustering: all three proteins co-localized in both submembrane and intracellular aggregates.
FIGURE 8.
GABAAR α2 activates collybistin-mediated submembrane aggregation of gephyrin. A–D, confocal microscopy showing expression of myc-tagged collybistin, EGFP gephyrin, and HcRed. Co-expression of collybistin without the SH3 domain with EGFP-gephyrin results in submembrane gephyrin aggregates (3); however, HcRed does not co-localize to these aggregates. E–H, HcRed-GABAAR α2 M3-M4 fusion protein localizes to the EGFP-gephyrin/myc-collybistin (CB2SH3− or CB3SH3−) aggregates upon triple expression. Note that this co-localization is likely to be mediated by the GABAAR α2-gephyrin interaction. I–L, co-expression of collybistin containing a SH3 domain (CB3SH3+), EGFP-gephyrin and HcRed-GABAAR α2 leads to co-localization of all three proteins in submembrane aggregates and intracellular aggregates. Note that collybistin isoforms containing the SH3 domain are usually incapable of targeting gephyrin to the submembrane compartment (3, 6). Scale bars: 10 μm.
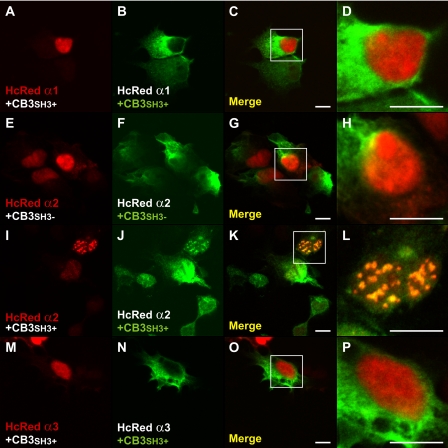
Collybistin Preferentially Co-localizes with GABAAR α2 in Vivo
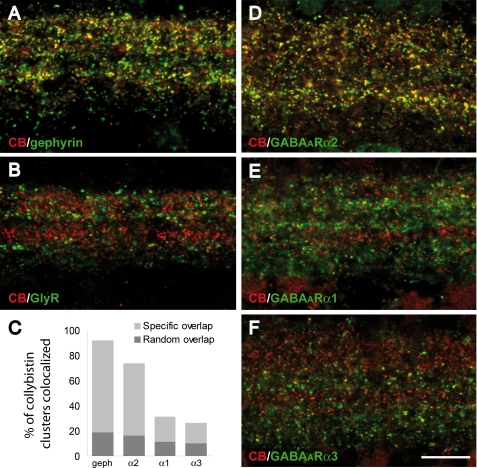
One major barrier to understanding the function of collybistin in vivo has been the lack of collybistin antibodies that function in immunostaining experiments. To overcome this issue, we generated a polyclonal antibody against a peptide epitope at the collybistin CB3 C terminus (FWQNFSRLTPFKK) because this is one of two commonly expressed alternatively splice variants: CB2 and CB3 (3). This antiserum specifically recognizes collybistin CB3 splice variants in Western blotting and immunocytochemistry (supplemental Fig. 2). Immunohistochemistry on rat retina sections with this novel antiserum revealed a punctate staining pattern indicative of synaptic labeling, as shown by extensive co-localization with gephyrin (Fig. 9A). Notably, 60% of gephyrin clusters co-localized with collybistin, whereas 92% of collybistin puncta co-localized with gephyrin, suggesting that our antiserum recognizes the predominant collybistin isoform(s) expressed in the retina. Double labeling with mAb4a (Fig. 9B), which recognizes all GlyR α subunits (10), demonstrated that collybistin is not associated with GlyRs in the retina. This is consistent with previous findings in collybistin knock-out mice, where normal postsynaptic aggregation of gephyrin and GlyRs was observed at glycinergic synapses (25). By contrast, there was a prominent co-localization of collybistin with GABAAR α2 (75%; Fig. 9, C and D) and more limited co-localization with GABAAR α1 (31%; Fig. 9, C and E) and GABAAR α3 (27%; Fig. 9, C and F). These observations support a preferential association of the collybistin-gephyrin complex with GABAARs containing the α2 subunit.
FIGURE 9.
Collybistin preferentially co-localizes with the GABAAR α2 subunit in rat retina. Confocal images of the internal plexiform layer of the rat retina show double labeling with an antiserum against collybistin (red) in combination with antibodies directed against gephyrin (mAb7a; A), GlyRs (mAb4a; B), and GABAAR α2 (D), α1 (E), and α3 (F). Collybistin puncta show extensive co-localization with gephyrin (A) and GABAAR α2 (D), whereas the overlap with glycine receptors is negligible (B). Only limited co-localization is observed with GABAAR α1 (E) and GABAAR α3 (F). The graph in C shows the percentage of collybistin clusters co-localized with gephyrin and GABAAR subunits in correctly aligned images (specific overlap) and randomly superimposed images (random overlap). In all cases there was a statistically significant difference between the specific overlap and the random overlap (Mann-Whitney test, p < 0.05, n = 4–6 confocal fields). The density of collybistin puncta co-localized with GABAAR α2 was significantly higher than the density of puncta co-localized with the other GABAAR α subunits (ordinary ANOVA, post hoc Tukey test, p < 0.001). Scale bar: 10 μm.
DISCUSSION
The aim of this study was to determine the molecular basis of the clustering of GABAARs by gephyrin and collybistin. Using proteomic techniques we found that the GABAAR α2 and α3 subunits harbor binding sites for gephyrin, whereas GABAAR α2 also binds to the RhoGEF collybistin. For GABAAR α2, these interactions are mediated by two overlapping binding sites in the intracellular M3–M4 domain, the collybistin SH3 domain and a motif at the start of the gephyrin E domain that is distinct from the GlyR β binding site (29). This suggests that gephyrin is capable of binding GlyRs and GABAARs simultaneously, providing a molecular explanation for the co-localization of different inhibitory receptors under presynaptic terminals releasing distinct neurotransmitters. Our data also provide for gephyrin-dependent and gephyrin-independent modes of GABAAR clustering. GABAARs containing α2 and α3 subunits are typically postsynaptic receptors, whereas receptors containing the GABAAR α4 and α6 subunits play an important role in tonic inhibition and are located preferentially at extrasynaptic sites (30), consistent with a lack of interaction with gephyrin in our assays. Similarly, the majority of GABAARs containing the α5 subunit do not form clusters that co-localize with gephyrin (6, 31, 32) because their synaptic localization is mediated by activated radixin (33). Although the GABAAR α1 subunit did not interact with gephyrin in our assays, both gephyrin-dependent and gephyrin-independent clustering of GABAAR α1 has been described (7, 9), perhaps hinting at a more complex regulatory mechanism, e.g. subtle post-translational modifications or GABAARs containing more than one α subunit.
One major unexpected finding of our study was the association of GABAAR α2 with the RhoGEF collybistin in multiple in vitro assays and retina sections. GABAAR α2 associates with collybistin via a distinct alanine-rich non-PXXP motif (34, 35) that is not conserved in other GABAAR α subunits (AYAVAVANYA). Interestingly, an extended version of this motif (GSVMIQNNAYAVAVANYA) was required for GABAAR α2 interactions with gephyrin. Tretter et al. (24) previously suggested that the collybistin binding site described here (AYAVAVANYA) was sufficient for the interaction between the GABAAR α2 and gephyrin, but this assumption was based on targeting experiments in primary neuronal cell cultures. Thus, the collybistin-binding AYAVAVANYA motif is likely to be sufficient for targeting GABAAR α2 to postsynaptic sites where both collybistin and gephyrin are present and therefore does not contradict our observations. Consistent with this view, the postsynaptic localization of GABAAR fusion proteins harboring the collybistin binding motif (AYAVAVANYA) was equivalent to that of constructs expressing the complete M3–M4 intracellular GABAAR α2 domain (24). It is also relevant that the collybistin SH3 domain binding site on GABAAR α2 is not a typical PXXP motif like that recently described for neuroligin 2 (36) but is mediated via a non-PXXP motif. This allows simultaneous interactions of both neuroligin 2 and GABAAR α2 with collybistin and is also consistent with recent data suggesting a preferential co-localization of neuroligin 2 with GABAergic synapses (37). Like neuroligin 2, GABAAR α2 was also able to activate submembrane gephyrin clustering by SH3 domain containing collybistin isoforms in cellular assays. By inference, GABAAR α2 could also act as a signal for triggering synaptic accumulation of collybistin and gephyrin and subsequent assembly of inhibitory postsynaptic densities. Although the nature of this activation is speculative at present, perhaps binding of peptide ligand to the SH3 domain relieves autoinhibition of collybistin in terms of RhoGEF domain activation of the small GTPase Cdc42 (38, 39) or enables interactions of the pleckstrin homology domain with trafficking proteins or phosphoinositides (40).
The nature and sequence of the gephyrin binding motif on GABAAR α2 is distinct from that found in GlyR β (16, 17), consistent with overlapping collybistin-GABAAR α2 binding sites at the start of the gephyrin E domain and cooperative GABAAR α2-collybistin-gephyrin interactions. The binding site for collybistin on gephyrin (3) was previously mapped to a 10-amino acid stretch (PFPLTSMDKA) that partially overlaps with the GABAAR α2 binding site described here (SMDKAFITVLEMPTVLGTE). The close proximity of the collybistin/GABAAR α2 binding sites on gephyrin and collybistin/gephyrin binding sites on GABAAR α2 suggests an intimate relationship of these three proteins. Yeast tri-hybrid experiments confirmed a significant increase in the interaction strength between GABAAR α2 and gephyrin in the presence of collybistin and GABAAR α2 and collybistin in the presence of gephyrin. In addition, GABAAR α2 exhibits a preference for collybistin rather than gephyrin in co-immunoprecipitation assays, suggesting a dynamic process of competition mediated by the GABAAR α2 M3–M4 domain. Immunohistochemical analysis in retina, chosen because of the substantial separation of GABAergic and glycinergic synapses (10), also support a selective function of collybistin at a subset of GABAergic (but not glycinergic) synapses, predominantly those containing GABAAR α2.
Collybistin knock-out mice suffer from increased anxiety and impaired spatial learning due to changes of GABAergic inhibition, network excitability, and synaptic plasticity (25–27). These results are mirrored in humans, where defects in ARHGEF9, encoding collybistin, give rise to a diverse range of symptoms linked to gephyrin and GABAAR mislocalization, with intellectual disability as a common feature (3, 40, 41). Our proposed link between collybistin and GABAAR α2 may help to explain why in collybistin knock-out mice, such a distinct loss of GABAAR clustering was observed in parts of the hippocampus and basolateral amygdala, but no apparent effect on GlyR clustering was observed (25). Interestingly, the affected brain regions preferentially express the GABAAR α2 subunit transcripts and only relatively small amounts of GABAAR α1 and α3 subunit mRNAs (42). Loss of GABAAR α2 is also consistent with the heightened anxiety observed in collybistin knock-out mice because GABAARs containing the α2 subunit have previously been linked to anxiety (43, 44). The disruption of the GABAAR α2-collybistin interaction by a known mutation affecting the collybistin SH3 domain mutation (p.G55A) also illustrates the pathophysiological importance of our findings. We previously reported that the G55A mutation found in a male patient suffering from hyperekplexia, epilepsy, and intellectual disability led to the loss of GABAARs at postsynaptic sites in primary neuronal cell cultures and dendritic accumulation of gephyrin (3). We now know that the G55A missense mutation results in the loss of both neuroligin 2 and GABAAR α2 interactions, suggesting that this substitution completely perturbs the structure of the collybistin SH3 domain. Additional variable symptoms observed in patients harboring different collybistin mutations that are not easily explained by the loss of GABAAR α2 at postsynaptic sites are likely due to dominant negative effects of expressed but dysfunctional collybistin proteins, interfering with gephyrin-mediated clustering of other GABAAR and GlyR subtypes (3, 40). However, because GABAAR α2 shows such a distinct association and functional interplay with collybistin, it is highly likely that additional RhoGEFs or unknown clustering factors mediate synaptic localization of gephyrin and/or GABAAR and GlyR subtypes.
Supplementary Material
This work was supported by Medical Research Council New Investigator Award G0501258 (to K. H.) and by a Compagnia di San Paolo and Regione Piemonte Ricerca Sanitaria Finalizzata 2008 grant (to M. S.-P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GlyR
- glycine receptor
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- CB
- collybistin
- GABAAR
- GABAA receptor
- SH3
- src homology 3.
REFERENCES
- 1.Prior P., Schmitt B., Grenningloh G., Pribilla I., Multhaup G., Beyreuther K., Maulet Y., Werner P., Langosch D., Kirsch J., Betz H. (1992) Neuron 8, 1161–1170 [DOI] [PubMed] [Google Scholar]
- 2.Kins S., Betz H., Kirsch J. (2000) Nat. Neurosci. 3, 22–29 [DOI] [PubMed] [Google Scholar]
- 3.Harvey K., Duguid I. C., Alldred M. J., Beatty S. E., Ward H., Keep N. H., Lingenfelter S. E., Pearce B. R., Lundgren J., Owen M. J., Smart T. G., Lüscher B., Rees M. I., Harvey R. J. (2004) J. Neurosci. 24, 5816–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng G., Tintrup H., Kirsch J., Nichol M. C., Kuhse J., Betz H., Sanes J. R. (1998) Science 282, 1321–1324 [DOI] [PubMed] [Google Scholar]
- 5.Kneussel M., Brandstätter J. H., Laube B., Stahl S., Müller U., Betz H. (1999) J. Neurosci. 19, 9289–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer F., Kneussel M., Tintrup H., Haverkamp S., Rauen T., Betz H., Wässle H. (2000) J. Comp. Neurol. 427, 634–648 [DOI] [PubMed] [Google Scholar]
- 7.Lévi S., Logan S. M., Tovar K. R., Craig A. M. (2004) J. Neurosci. 24, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu W., Jiang M., Miralles C. P., Li R. W., Chen G., de Blas A. L. (2007) Mol. Cell. Neurosci. 36, 484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kneussel M., Brandstätter J. H., Gasnier B., Feng G., Sanes J. R., Betz H. (2001) Mol. Cell. Neurosci. 17, 973–982 [DOI] [PubMed] [Google Scholar]
- 10.Wässle H., Heinze L., Ivanova E., Majumdar S., Weiss J., Harvey R. J., Haverkamp S. (2009) Front. Mol. Neurosci. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweizer C., Balsiger S., Bluethmann H., Mansuy I. M., Fritschy J. M., Möhler H., Lüscher B. (2003) Mol. Cell. Neurosci. 24, 442–450 [DOI] [PubMed] [Google Scholar]
- 12.Alldred M. J., Mulder-Rosi J., Lingenfelter S. E., Chen G., Lüscher B. (2005) J. Neurosci. 25, 594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R. W., Yu W., Christie S., Miralles C. P., Bai J., Loturco J. J., De Blas A. L. (2005) J. Neurochem. 95, 756–770 [DOI] [PubMed] [Google Scholar]
- 14.Kralic J. E., Sidler C., Parpan F., Homanics G. E., Morrow A. L., Fritschy J. M. (2006) J. Comp. Neurol. 495, 408–421 [DOI] [PubMed] [Google Scholar]
- 15.Studer R., von Boehmer L., Haenggi T., Schweizer C., Benke D., Rudolph U., Fritschy J. M. (2006) Eur. J. Neurosci. 24, 1307–1315 [DOI] [PubMed] [Google Scholar]
- 16.Meyer G., Kirsch J., Betz H., Langosch D. (1995) Neuron 15, 563–572 [DOI] [PubMed] [Google Scholar]
- 17.Kneussel M., Hermann A., Kirsch J., Betz H. (1999) J. Neurochem. 72, 1323–1326 [DOI] [PubMed] [Google Scholar]
- 18.Meier J., Grantyn R. (2004) J. Physiol. 559, 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., Bedford F. K., Brandon N. J., Moss S. J., Olsen R. W. (1999) Nature 397, 69–72 [DOI] [PubMed] [Google Scholar]
- 20.Kneussel M., Haverkamp S., Fuhrmann J. C., Wang H., Wässle H., Olsen R. W., Betz H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8594–8599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang C., Deng L., Keller C. A., Fukata M., Fukata Y., Chen G., Lüscher B. (2006) J. Neurosci. 26, 12758–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nymann-Andersen J., Sawyer G. W., Olsen R. W. (2002) J. Neurochem. 83, 1164–1171 [DOI] [PubMed] [Google Scholar]
- 23.Fritschy J. M., Harvey R. J., Schwarz G. (2008) Trends Neurosci. 31, 257–264 [DOI] [PubMed] [Google Scholar]
- 24.Tretter V., Jacob T. C., Mukherjee J., Fritschy J. M., Pangalos M. N., Moss S. J. (2008) J. Neurosci. 28, 1356–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadopoulos T., Korte M., Eulenburg V., Kubota H., Retiounskaia M., Harvey R. J., Harvey K., O'Sullivan G. A., Laube B., Hülsmann S., Geiger J. R., Betz H. (2007) EMBO J. 26, 3888–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papadopoulos T., Eulenburg V., Reddy-Alla S., Mansuy I. M., Li Y., Betz H. (2008) Mol. Cell. Neurosci. 39, 161–169 [DOI] [PubMed] [Google Scholar]
- 27.Jedlicka P., Papadopoulos T., Deller T., Betz H., Schwarzacher S. W. (2009) Mol. Cell. Neurosci. 41, 94–100 [DOI] [PubMed] [Google Scholar]
- 28.Melzer N., Villmann C., Becker K., Harvey K., Harvey R. J., Vogel N., Kluck C. J., Kneussel M., Becker C. M. (2010) J. Biol. Chem. 285, 3730–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim E. Y., Schrader N., Smolinsky B., Bedet C., Vannier C., Schwarz G., Schindelin H. (2006) EMBO J. 25, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belelli D., Harrison N. L., Maguire J., Macdonald R. L., Walker M. C., Cope D. W. (2009) J. Neurosci. 29, 12757–12763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crestani F., Keist R., Fritschy J. M., Benke D., Vogt K., Prut L., Blüthmann H., Möhler H., Rudolph U. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8980–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serwanski D. R., Miralles C. P., Christie S. B., Mehta A. K., Li X., De Blas A. L. (2006) J. Comp. Neurol. 499, 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loebrich S., Bähring R., Katsuno T., Tsukita S., Kneussel M. (2006) EMBO J. 25, 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douangamath A., Filipp F. V., Klein A. T., Barnett P., Zou P., Voorn-Brouwer T., Vega M. C., Mayans O. M., Sattler M., Distel B., Wilmanns M. (2002) Mol. Cell 10, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q., Berry D., Nash P., Pawson T., McGlade C. J., Li S. S. (2003) Mol. Cell 11, 471–481 [DOI] [PubMed] [Google Scholar]
- 36.Poulopoulos A., Aramuni G., Meyer G., Soykan T., Hoon M., Papadopoulos T., Zhang M., Paarmann I., Fuchs C., Harvey K., Jedlicka P., Schwarzacher S. W., Betz H., Harvey R. J., Brose N., Zhang W., Varoqueaux F. (2009) Neuron 63, 628–642 [DOI] [PubMed] [Google Scholar]
- 37.Hoon M., Bauer G., Fritschy J. M., Moser T., Falkenburger B. H., Varoqueaux F. (2009) J. Neurosci. 29, 8039–8050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid T., Bathoorn A., Ahmadian M. R., Collard J. G. (1999) J. Biol. Chem. 274, 33587–33593 [DOI] [PubMed] [Google Scholar]
- 39.Xiang S., Kim E. Y., Connelly J. J., Nassar N., Kirsch J., Winking J., Schwarz G., Schindelin H. (2006) J. Mol. Biol. 359, 35–46 [DOI] [PubMed] [Google Scholar]
- 40.Kalscheuer V. M., Musante L., Fang C., Hoffmann K., Fuchs C., Carta E., Deas E., Venkateswarlu K., Menzel C., Ullmann R., Tommerup N., Dalprà L., Tzschach A., Selicorni A., Lüscher B., Ropers H. H., Harvey K., Harvey R. J. (2009) Hum. Mutat. 30, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marco E. J., Abidi F. E., Bristow J., Dean W. B., Cotter P., Jeremy R. J., Schwartz C. E., Sherr E. H. (2008) J. Med. Genet. 45, 100–105 [DOI] [PubMed] [Google Scholar]
- 42.Wisden W., Laurie D. J., Monyer H., Seeburg P. H. (1992) J. Neurosci. 12, 1040–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon C. I., Rosahl T. W., Stephens D. N. (2008) Pharmacol. Biochem. Behav. 90, 1–8 [DOI] [PubMed] [Google Scholar]
- 44.Möhler H. (2009) Biochem. Soc. Trans. 37, 1328–1333 [DOI] [PubMed] [Google Scholar]
- 45.Fuller K. J., Morse M. A., White J. H., Dowell S. J., Sims M. J. (1998) BioTechniques 25, 85–92 [DOI] [PubMed] [Google Scholar]
- 46.Sancho R. M., Law B. M., Harvey K. (2009) Hum. Mol. Genet. 15, 3955–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sassoè-Pognetto M., Wässle H., Grünert U. (1994) J. Neurosci. 14, 5131–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patrizi A., Scelfo B., Viltono L., Briatore F., Fukaya M., Watanabe M., Strata P., Varoqueaux F., Brose N., Fritschy J. M., Sassoè-Pognetto M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13151–13156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider Gasser E. M., Straub C. J., Panzanelli P., Weinmann O., Sassoè-Pognetto M., Fritschy J. M. (2006) Nat. Protocols 1, 1887–1897 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.