Abstract
Mutually exclusive alternative splicing produces transcripts for 12 serpin-1 isoforms in Manduca sexta that differ only in the region encoding the carboxyl-terminal 36–40-amino acid residues. This variable region includes the reactive center loop, which determines the inhibitory selectivity of the serpin. We investigated mRNA levels of individual serpin-1 isoforms by quantitative PCR. The 12 isoforms were expressed at similar levels in hemocytes, but in fat body isoform B mRNA was present at significantly higher levels than isoforms C, D, E, F, G, J, K, and Z. To investigate the presence of individual serpin-1 isoforms in plasma we used immunoaffinity purification of serpin-1 isoforms from M. sexta plasma, followed by two-dimensional PAGE and identification of protein spots by digestion with a series of proteinases and analysis of the resulting peptides by MALDI-TOF/TOF. We identified nine of the 12 serpin-1 isoforms and, through analysis of putative serpin-1-proteinase complexes, identified three endogenous M. sexta proteinase targets of serpin-1. Our results suggest that M. sexta serpin-1 isoforms A, E, and J can inhibit hemolymph proteinase 8, which activates the cytokine spätzle. At least one isoform of serpin-1 can inhibit hemocyte proteinase 1, another M. sexta blood proteinase. In addition, a complex of serpin-1K in a complex with M. sexta midgut chymotrypsin was identified, suggesting serpin-1 isoforms may also function to protect insect tissues from digestive proteinases that may leak into the hemocoel.
Keywords: Innate Immunity, Insect, Mass Spectrometry (MS), Serine Protease, Serpin, Hemolymph
Introduction
Serpins are a superfamily of proteins that are named for being serine proteinase inhibitors, a function that many serpins perform. Serpins contain an exposed reactive center loop, formed from a region near the carboxyl-terminal end of the protein. A proteinase that begins to cleave a serpin in the reactive center loop typically becomes covalently bound to the serpin and inactivated. The proteinase active site serine forms an ester bond with the acyl group of the serpin P1 residue at the amino-terminal side of the scissile bond, but cannot complete the hydrolysis because the serpin reactive center loop rapidly inserts into the A β-sheet, moving the proteinase ∼70 Å and deforming the proteinase catalytic site (1, 2).
Extracelluar serpins exert control over serine proteinase cascades in vertebrate blood, including complement activation, blood clotting, and fibrinolysis (3, 4). In insects, serine proteinase cascades initiated by microbial infection elicit antimicrobial peptide production and lead to activation of prophenoloxidase, which catalyzes the melanization response (5–13). Serpins inhibit proteinases required for prophenoloxidase activation in Manduca sexta (14–17), Drosophila melanogaster (18–21), Tenebrio molitor (22), and Anopheles gambiae (23). Serpins also regulate the Toll pathway in immune responses of D. melanogaster and T. molitor (22, 24) and in D. melanogaster dorsal-ventral patterning (25, 26).
In insects, serpin genes have evolved alternative exon splicing, which produces variation in the sequence of much of the reactive center loop, producing multiple functional serpins from a single gene. This was first described in M. sexta serpin-1, which has 12 different copies of exon 9 that undergo mutually exclusive alternative splicing to produce 12 putative protein isoforms. These isoforms differ in their carboxyl-terminal 39–46 residues, including the P1 residue, and inhibit serine proteinases with different specificities (Fig. 1) (27–31). Similar alternative splicing occurs in some serpin genes from other insect species, with 3–15 alternative exons encoding the reactive center loop present in genes studied so far (32–34). M. sexta serpin-1 is expressed in fat body and, less strongly, in hemocytes (36, 37). Serpin-1 is secreted into the hemolymph and reaches concentrations of 0.4 mg/ml. However, the presence and amount of the different serpin-1 isoforms in hemolymph has not previously been analyzed. It has been unclear whether both tissues express all 12 isoforms and whether any of the isoforms are preferentially expressed. Analysis of cDNA clones from hemocyte and fat body libraries showed that the hemocyte clones were well distributed over the different isoforms, but 19 of the 21 fat body clones were serpin-1F, which led to the speculation that the fat body preferentially expresses isoform F (30).
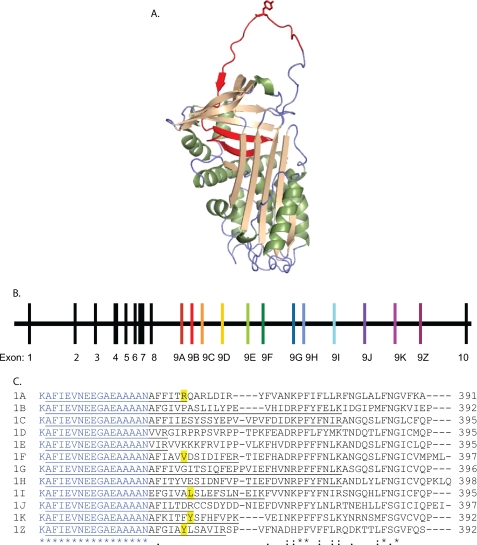
FIGURE 1.
Mutually exclusive splicing of the serpin-1 gene to include different versions of exon 9 produces serpin isoforms with different reactive center loop sequences. A, protein structure of serpin-1K (PDB code 1SEK). The alternatively spliced region is shown in red, and the P1 residue, Tyr359, is shown in the “stick” format. B, structure of the M. sexta serpin-1 gene. The first 8 exons encode the constant region of the protein, which is followed by the carboxyl-terminal variable region, encoded by an exon 9. Each exon 9 includes a stop codon to end the open reading frame. Exon 10 includes the 3′ untranslated region. C, an alignment of the carboxyl termini of the 12 serpin-1 isoforms starting with Lys336. The region encoded by the 3′ end of exon 8 is in blue; exon 9 starts after Asn352. Known P1 residues are highlighted in yellow. The first isoform-specific tryptic peptide for each isoform is underlined.
Little is known about the biological functions of M. sexta serpin-1. Only two of the 12 serpin-1 isoforms have been found to form complexes with M. sexta serine proteases. Serpin-1J can inhibit activation of the phenoloxidase pathway and form a complex with prophenoloxidase activating proteinase-3 in vitro (27, 38), whereas serpin-1I can complex with HP143 in vitro (39). 27 hemolymph proteinases are known in M. sexta (40, 41) and some of these are likely endogenous proteinase targets of serpin-1 isoforms.
In this paper we investigate individual serpin-1 isoform expression at the mRNA level and examine the individual serpin-1 isoform proteins in plasma. We also analyzed putative complexes between serpin-1 and proteinases in plasma samples. Identification of serpin-1 proteinase complexes occurring naturally in hemolymph provides insight into some of the endogenous M. sexta proteinases that serpin-1 inhibits, bringing closer a goal of understanding the function of serpins and proteinases in hemolymph of M. sexta and other insects.
EXPERIMENTAL PROCEDURES
Insects
We originally obtained eggs for the M. sexta colony maintained in our laboratory from Carolina Biological Supply. The insects were reared on an artificial diet as described previously (42).
RNA Preparation, Primer Design, and PCR
An RNeasy Midi Kit (Qiagen) was used to extract RNA from hemocytes or fat body of fifth instar larvae from both naive insects and insects 24 h after injection of 100 μl of a 1 mg/ml of suspension of Micrococcus luteus (Sigma). Hemolymph from eight insects was pooled for each hemocyte sample, and fat body from five insects was used for each fat body sample. RNA was treated with Turbo DNA-free (Ambion) to remove any contaminating genomic DNA. cDNA was synthesized in 20-μl reactions with the SuperScript III kit using an oligo(dT) primer (Invitrogen) from 5.36 μg of RNA (fat body samples), 1.18 μg of RNA (naive hemocytes), and 2.06 μg of RNA (induced hemocytes).
Primers for serpin-1 isoforms and ribosomal protein S3 (rpS3) (supplemental Table S1) were designed using the primer 3 program (Invitrogen). Semi-quantitative reverse transcriptase (RT) PCR was performed using 0.5 μl of midgut, naive fat body or induced fat body cDNA, 1 μl of naive hemocyte cDNA, or 0.6 μl of induced hemocyte cDNA with 0.5 μl of forward primer (10 μm), 0.5 μl of reverse primer (10 μm), and 22.5 μl of Platinum PCR Supermix (Invitrogen) in a total volume of 25 μl. PCR were run for 30 or 35 cycles (30 s at 94 °C, 30 s at 50 °C, and 25 s at 72 °C). The PCR products were analyzed by electrophoresis on a 1% agarose gel and stained with ethidium bromide. The PCR products from the 35 cycle naive hemocyte reactions were excised from the gel, purified (Qiagen kit), and sequenced (Iowa State University DNA Sequencing Facility) using sequence-specific primers. In an experiment to examine sequences of putative allelic variants of serpin-1E, additional PCR was performed with serpin-1E-specific primers using 0.75 μl of naive fat body cDNA in a total volume of 50 μl with 5 cycles of amplification at 54 °C, 5 cycles at 52 °C, and 20 cycles at 50 °C. The PCR product was gel purified and cloned into the pCR4-TOPO vector using a TOPO TA cloning kit (Invitrogen) and sequenced at the Kansas State University Gene Expression facility.
For quantitative RT-PCR, RNA was prepared from fifth instar day 2 larvae 24 h after injection with water as a control (50 μl/larva) or with M. luteus (Sigma, 50 μl/larva, 10 μg/μl). Body fat and hemocyte samples were collected individually from three larvae for each treatment. Total RNA samples were prepared using TRIzol® Reagent (Invitrogen). First-strand cDNA was synthesized following the instructions of BD SprintTM PowerSriptTM PrePrimed Single Shots Kit (Clontech). Each 25-μl reaction for quantitative real time RT-PCR contained 12.5 μl of 2× buffer mixture (Bio-Rad), 1 μl of forward primer (5 μm), 1 μl of reverse primer (5 μm), and 11.5 μl of diluted cDNA. The thermal cycling conditions were 95 °C for 5 min and 40 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 40 s. Amplification was monitored on an iCycler (Bio-Rad) by means of SYBR Green (Bio-Rad) dye. Thresholds were individually calculated for each target gene. Transcript abundance values (ΔCT = CTSerpin-1 isoform − CTrpS3) for each isoform were used to calculate relative mRNA levels using the formula 2−ΔCT. Analysis of variance and the Tukey test was used to compare relative mRNA levels of the isoforms within a single tissue (fat body or hemocytes) and treatment (naive tissue or immune-induced). To compare the expression of individual isoforms within a single tissue under control and immune-induced conditions we performed unpaired t tests with the Welch correction for variance. Statistical analysis was performed using Prism 5 (GraphPad Software).
Protein Concentration Assay
Protein concentrations were determined with the Coomassie Plus Protein Assay Reagent (Pierce) using bovine serum albumin as the standard.
Antisera
Recombinant His6-tagged serpin-1I (27) expressed in Escherichia coli and purified by nickel-nitrilotriacetic acid chromatography and SDS-PAGE was used as antigen for preparation of a rabbit antiserum (Cocalico, Reamstown, PA). Preliminary tests by immunoblot analysis of M. sexta plasma samples indicated that this antiserum was specific for serpin-1 (data not shown). Rabbit antisera to HP6, HP8, and PAP-3 was kindly provided by Dr. Haobo Jiang, Oklahoma State University. Rabbit antisera to peptides representing the reactive center loop region of serpin-1B (GIVPASLILYPEVHI) and serpin-1F (IAVVDSIDIFERTIEFHA) were generated using synthetic peptides conjugated to keyhole limpet hemocyanin (43).
Immunoaffinity Purification of Serpin-1 Isoforms
Rabbit antiserum generated to recombinant serpin-1I, described above, was used to prepare serpin-1-antibody columns for immunoaffinity purification of plasma serpins. Immunoglobulins in 2 ml of serpin-1 antiserum were cross-linked to 2 ml of protein-A beads (Sigma P9424) as described previously (16).
Hemolymph from day 1 and 2 fifth instar larvae was collected from a proleg into a sterile tube containing 2.5 mg of diethylthiocarbamate per larva to inhibit melanization. Hemolymph was centrifuged at 10,000 × g for 15 min to remove hemocytes, and the supernatant (plasma) was either used immediately or frozen in aliquots at −80 °C. Untreated naive plasma was used for immunoaffinity purification except for one experiment, where 4 ml of plasma pooled from 16 naive day 1 fifth instar larvae was incubated with either 50 μl of 40 mg/ml of M. luteus (Sigma M3770) or 100 μl of 10 mg/ml of LPS (Sigma L2630) for 40 min. The LPS-treated plasma was used for immunoaffinity purification of serpin-1 followed by one-dimensional PAGE.
Serpin-1 antibody beads were mixed with 4 ml of plasma and 8 ml of phosphate-buffered saline for 2 h. The beads were centrifuged at 100 × g, washed three times with 12 ml of PBS, and then transferred to a 10-ml disposable chromatography column (Bio-Rad). Serpin-1 was eluted with 10 1-ml applications of elution buffer (100 mm glycine, 10% ethylene glycol, pH 3) collected into 100-μl aliquots of 1 m sodium phosphate, pH 8, for neutralization. Samples were analyzed by reducing SDS-PAGE and, for immunoblotting, transferred to nitrocellulose, and detected with antiserum to serpin-1.
Affinity purified serpin-1 elution fractions with protein concentrations of 50 μg/ml of protein or less were concentrated using a Centriplus YM-3 centrifugal filter device (Amicon) at 3,000 × g, 4 °C, until the volume was reduced from 9 to 2 ml and the concentration of the sample was ∼110 μg/ml. These samples were further concentrated using Microcon YM3 centrifugal filter units (Amicon) at 13,000 × g, 4 °C. Elution fractions that had higher initial protein concentrations (60–130 μg/ml) were also individually concentrated in Microcon YM3 centrifugal filter units. The retentates were pooled and used for two-dimensional SDS-PAGE.
Two-dimensional Gel Electrophoresis
Concentrated affinity purified serpin-1 (38 μg) was prepared for isoelectric focusing (IEF) using the ReadyPrep 2-D Cleanup Kit (Bio-Rad). 11-cm IPG ReadyStrips (4.7–5.9 pH range, Bio-Rad) were rehydrated with 190 μl of affinity purified serpin-1 in strongly chaotropic buffer for 16 h prior to running IEF on a Bio-Rad Protean IEF Cell.
After completion of IEF, the strips were equilibrated with 2% dithiothreitol and then 2.5% iodoacetamide for 12 min each, then electrophoresed in a 4–12% BisTris Criterion gel (Bio-Rad), all performed according Bio-Rad instructions. Immunoblotting was performed with antisera to M. sexta serpin-1, HP1, HP6, HP8, prophenoloxidase-activating proteinase-3, and PPO1 and -2 at 1:2,000 dilution. Coomassie Blue staining was performed with BioSafe Coomassie G-250 stain (Bio-Rad).
Protein Digestion and Identification by Mass Spectrometry
The two-dimensional spots of interest were excised and then analyzed by mass spectrometry at the Nevada Proteomics Center (University of Nevada, Reno). Selected protein spots were digested in bicarbonate buffer with trypsin or a double digest of proteinases lysine-C and aspartic-N (Lys-C/AspN). Other spots were digested by the proteinase glutamic-C (Glu-C) in either bicarbonate or phosphate buffer.
MALDI data analysis peak lists were created using the 4000 Series Explorer software version 3.6 Peaks to the Mascot feature from ABI. MS peak filtering settings included a mass range of 700–4000 Da, minimum S/N filter 0, and peak density filter of 65 peaks/200 Da with a maximum number of peaks set to 200. MS/MS peak filtering included mass ranges of 60 to 20 Da below each precursor mass, minimum S/N filter 0, peak density filter of 65 peaks/200 Da, and cluster area filter with maximum number of peaks 200. The filtered data were searched by Mascot version 2.1.03 (Matrix Science) using the NCBInr data base (NCBInr 20070908), containing 5,454,477 sequences. Searches were performed without restriction to protein species, Mr, or pI and with variable oxidation of methionine residues and carbamidomethylation of cysteines. Maximum missed cleavage was set to 1 and limited to appropriate digestive enzyme and buffer cleavage sites. Precursor mass tolerance and fragment mass tolerance were set to 150 ppm (or 20 ppm, for internally calibrated trypsin digests) and ±0.2 Da. These files were then analyzed using Proteome Software's Scaffold software.
RESULTS
Differential Expression of Serpin-1 Isoform mRNAs
Previous studies of serpin-1 have examined mRNA and protein levels of all the isoforms in aggregate. Northern blot results showed serpin-1 mRNA is present at high levels in fat body and lower levels in hemocytes (specifically granular cells) (36). Whether there may be some preferred splice sites or regulation of isoform production in response to different conditions or during development is unknown. To begin to address these questions, we designed isoform-specific forward primers to the 12 alternatively spliced versions of exon 9 and a common reverse primer in exon 10, which encodes the 3′ untranslated region for all the isoforms. PCR with cDNA from fat body and hemocytes of naive larvae and larvae injected with bacteria (induced) gave products of the expected size for all 12 isoforms (data not shown). Each PCR product from naive hemocytes was gel-purified and directly sequenced using the common exon 10 primer and found to have the sequence of the expected isoform (results not shown), demonstrating the specificity of the exon 9 primers and confirming that all 12 isoforms were expressed.
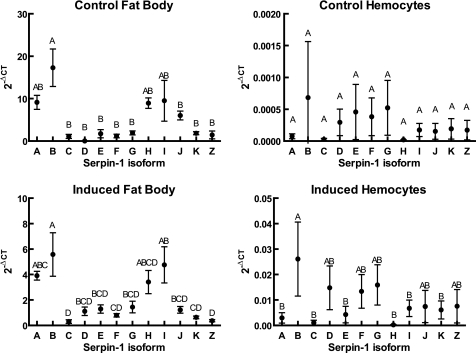
We used quantitative real time PCR to analyze the expression levels of the 12 isoforms in naive fat body and hemocytes and after an immune challenge (Fig. 2). We observed that, in the fat body samples, isoform B was significantly more highly expressed than isoforms C, D, E, F, G, J, K, and Z, whereas isoforms A, H, and I had an intermediate level of expression. In hemocytes, all of the serpin-1 isoforms were expressed at much lower levels relative to rpS3 than in fat body, and there was no significant difference between any of the isoforms in hemocytes from control insects. We compared expression of individual isoforms within a single tissue between naive and immune-challenged conditions and only saw significant differences for two isoforms in fat body, where isoforms H and J both decreased after immune challenge (supplemental Fig. S1).
FIGURE 2.
Relative expression levels of serpin-1 isoform mRNA in fat body and hemocytes from naive and immune-induced insects. RNA isolated from fat body or hemocytes of larvae 24 h after injection with M. luteus or with water as a control was used for analysis by quantitative RT-PCR as described under “Experimental Procedures.” Symbols represent mean ± S.D. (n = 3). Points labeled with the same letter are not significantly different (analysis of variance and Tukey's studentized range test, p < 0.05).
A Proteomic Approach for Identification of Serpin-1 Isoforms
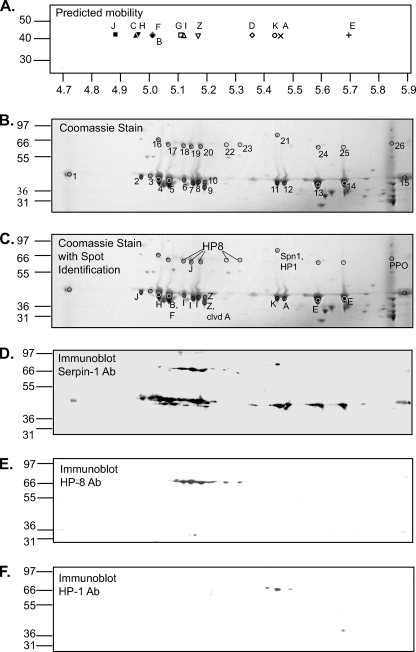
Because the 12 splicing isoforms of serpin-1 are nearly the same size, have mostly the same sequence, and all are recognized by the polyclonal antisera that have been available for serpin-1, little is known about the presence of individual isoforms in hemolymph. To assess the possible separation of serpin-1 isoforms by two-dimensional gel electrophoresis, we plotted predicted isoelectric points of the serpin-1 isoforms and predicted that a reasonable separation would likely be achieved using a narrow pH range isoelectric focusing strip (Fig. 3A). Preliminary separations using larval plasma samples revealed that a series of 11 spots at ∼45 kDa could be detected by immunoblotting after two-dimensional PAGE (supplemental Fig. S2). We confirmed by tryptic peptide mass fingerprinting that all of the spots contained serpin-1, but were not able to identify isoform types for many of the spots (44). To enrich the samples from hemolymph plasma for serpin-1 and increase the amount of serpin-1 isoforms that could be separated in each experiment, we carried out immunoaffinity purification of serpin-1 from plasma samples and used these preparations for further analysis by two-dimensional PAGE. This purification increased the intensity of each putative serpin-1 spot and revealed additional higher molecular weight spots that reacted with serpin-1 antiserum, which were not detected by analysis of crude plasma (Fig. 3D).
FIGURE 3.
Immunoaffinity-purified serpin-1 separated by isoelectric focusing and SDS-PAGE. A, the calculated isolectric points and masses of each serpin-1 isoform were used to predict separation after isoelectric focusing on a 4.7–5.9 pH range strip followed by SDS-PAGE. B, immunoaffinity-purified serpin-1 was separated by two-dimensional PAGE, using separation by IEF with a pH range of 4.7–5.9, followed by SDS-PAGE on a 4–12% acrylamide gradient gel. Coomassie Blue-stained spots corresponding to those from a duplicate gel detected by immunoblot analysis with serpin-1 antiserum (see panel D) were picked for trypsin digestion followed by MALDI-TOF and MALDI-TOF/TOF mass spectrometry analysis. Replicate gels were also analyzed after similar treatment with Glu-C and Lys-C/AspN (supplemental Fig. S3). C, summary of identification of serpin-1 isoforms and other proteins, based on mass spectrometry and immunoblot data. Panels D–F show immunoblot analysis of replicate two-dimensional PAGE samples using antisera to serpin-1 (D), HP8 (E), and HP1 (F).
Detection of multiple protein isoforms by two-dimensional PAGE and mass spectrometry is more challenging than general protein identification (45). Detection of individual serpin-1 isoforms proved to be particularly difficult because only the carboxyl-terminal region of ∼40 amino acid residues, after residue 353, are isoform-specific. Additionally, the end of the common portion of serpin-1 (residues 337–352) contains a long region with no trypsin cleavage sites and multiple glutamic acids, making it a difficult region to detect by proteinase digestion and MS. We used multiple proteinases (46) to improve our detection of specific serpin-1 isoforms, especially for isoforms B, C, G, and H (Table 1) and used enzyme cleavage rules as listed on the ExPASy proteomics server.
TABLE 1.
Serpin-1 isoform-specific peptides expected after cleavage by different proteases
| Enzyme | Number of isoform-specific peptidesa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | Z | |
| Trypsin | 3 | 1 | 1 | 4 | 4 | 3 | 1 | 1 | 3 | 3 | 4 | 3 |
| Glu-C bicarbonate | 0 | 3 | 1 | 2 | 0 | 2 | 2 | 3 | 2 | 3 | 2 | 0 |
| Glu-C phosphate | 2 | 3 | 3 | 3 | 3 | 2 | 2 | 4 | 2 | 3 | 2 | 3 |
| Lys-C/AspN | 3 | 3 | 1 | 3 | 4 | 4 | 1 | 4 | 2 | 2 | 4 | 3 |
a The number of serpin-1 isoform-specific peptides expected assuming 0 missed cleavages and a peptide mass between 700 and 3500 Da. Peptides with possible modifications are only listed once.
Spots at ∼45 kDa (spots 1–15) were excised from three gels and analyzed by digestion with trypsin, Lys-C/AspN, or Glu-C in either bicarbonate buffer or phosphate buffer (Fig. 3B and supplemental Fig. S3). MALDI-TOF/TOF data from proteinase-digested spots was searched against the NCBI data base, and the top Mascot results are listed in supplemental Table S2. We based isoform identification on the presence of at least 1 MS/MS peptide by scaffold (spectra in supplemental Fig. S4) or at least two peptides identified by peptide mass fingerprinting (PMF).
Spots 1 and 15, at the extreme ends of the IEF strip, were found by trypsin digestion to have unresolved serpin-1 isoforms and were not analyzed further. Trypsin digestion followed by MALDI-TOF/TOF analysis was adequate for identification of serpin-1 isoforms in spots 2 (J), 4 (H), 6–8 (I), 9 (A and Z), 10 (Z), 11 (K), 12 (A), and 13 and 14 (E). Multiple isoforms were detected in a few spots. Spot 5 clearly contained both isoforms B and F. Antisera generated to peptides from the variable regions of serpin-1B or -1F recognized spot 5 (supplemental Fig. S5), in agreement with the MS results.
Spot 9 contained three isoform Z peaks, one of which was confirmed by MS/MS, and also a serpin-1A peak corresponding to residues 337–355. The pI of serpin-1A after cleavage between P1-P1′ is 5.1, close to the pI of spot 10, which, along with the slightly lower size of spot 9, raises the possibility that the serpin-1A detected in spot 9 is cleaved serpin-1A.
Serpin-1 F and H have predicted tryptic peptides of very similar size, 1751.83 and 1751.88, respectively. A peptide with the observed mass 1752.029 was present in spot 4, which was inconclusive by peptide mass fingerprinting at the 150 ppm error tolerance appropriate for that spot. However, MS/MS analysis of this peptide led to its identification as serpin-1H. Additionally, three peptides from Lys-C/AspN digestion of spot 4 were identified as serpin-1H peptides, two of which were confirmed by MS/MS. One serpin-1H peptide was also identified in spot 4 by MS/MS after Glu-C digestion in phosphate buffer. All together we are quite confident of the presence of serpin-1H in spot 4.
PMF results from Lys-C/AspN digestion supported the results from trypsin digestion in spots 10 (Z), 11 (K), 13 (E), and 14 (E). Lys-C/AspN digestion also led to MS/MS identification of a serpin-1B peptide in spot 5. The only evidence for the presence of serpin-1 isoform D was in spot 8 by Lys-C/AspN digestion. This location, pH 5.2, is a little more acidic than 5.36, the predicted pI of isoform D.
MALDI-TOF/TOF analysis of peptides from Glu-C cleavage in bicarbonate buffer helped confirm the presence of serpin-1J in spot 2 and serpin-1B in spot 5. The same serpin-1B peptide was also detected by Glu-C cleavage in phosphate buffer. Glu-C cleavage in phosphate buffer also detected two masses that could match isoform C in spot 14p, although this spot is at a much higher pH, 5.7, than would be expected for isoform C (pI 4.94) and spot 14 was definitively shown to contain isoform E in both trypsin and Lys-C/AspN digestions.
The observed positions of all of the identified isoforms after isoelectric focusing was remarkably similar to the expected locations based on predicted pI (Fig. 3, A–C), with the exception of serpin-1C, which was only detected in spot 14, a spot that also contains serpin-1E. Isoform G was not detected in any of the samples.
One possible explanation for the two serpin-1E spots (13 and 14) is allelic variation involving charged amino acids in the isoform-specific region of serpin-1E. To investigate this we performed PCR with primers that started in exon 8 (forward) or exon 10 (reverse) but extended into the exon 9E region and cloned the products. Of 10 clones sequenced, five matched the serpin-1E sequence in NCBI (accession AAC47335.1), whereas the other five differed by a single difference at nucleotide 10138 (C to T). This polymorphism leads to a change in amino acid 371 from His to Tyr (H371Y). The predicted isoelectric point of this serpin-1E Tyr371 allele is 5.6, which corresponds to spot 13, whereas the His371 variant has a predicted pI of 5.7, matching spot 14. Mass spectrometry results demonstrated that a peptide with the correct mass to include residue His371 (1566.95) is present in spot 14 but absent in spot 13, where a new peak is visible at 1592.95, consistent with Tyr371 in spot 14 (supplemental Fig. S6).
Identification of Serpin-1 Complexes with Hemolymph Proteinases
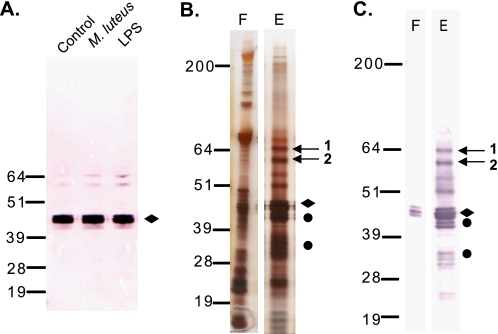
Serpin-1 protein isoforms are expressed in fat body and hemocytes and then secreted into the hemolymph. Immunoblots of plasma showed the presence of putative serpin-1-protease complexes, which were increased after the plasma was treated with LPS or M. luteus (Fig. 4A). To further analyze the putative serpin-1 proteinase complexes, we performed SDS-PAGE of serpin-1 immunoaffinity purified from plasma incubated with LPS (Fig. 4B). We observed two bands at 60–64 kDa, an appropriate size for serpin-protease complexes (15, 16), which were strongly recognized by the serpin-1 antiserum (Fig. 4C). These bands were excised and analyzed by MALDI-TOF/TOF, and both contained serpin-1 (Table 2 and supplemental Fig. S7). Band 1 contained an isoform-specific peptide corresponding to serpin-1J that was confirmed by additional TOF/TOF fragmentation. Band 1 also contained masses corresponding to isoform-specific peptides from serpin-1 isoforms A, E, and K. Band 2 contained two isoform-specific peptides matching serpin-1K and an identifiable proteinase, M. sexta chymotrypsin, identified previously as a midgut digestive proteinase (47). This result is consistent with the conclusion that band 2 contains serpin-1K in a covalent complex with chymotrypsin. Additional searches of the MALDI-TOF/TOF data with catalytic domain sequences of known M. sexta proteinases failed to identify a proteinase in band 1, suggesting that this may represent a complex with a proteinase whose sequence has not yet been determined.
FIGURE 4.
Immunoaffinity purification of serpin-1 and two putative serpin-1 protease complexes from M. sexta plasma. A, M. sexta plasma incubated without elicitor (C, control) or activated for 40 min in the presence of M. luteus or LPS from E. coli was analyzed by immunoblot using serpin-1 antisera. Open arrows indicate putative serpin-1 proteinase complexes. Serpin-1 was immunoaffinity purified from LPS-treated plasma, and elution fractions were analyzed by silver staining (B) or immunoblot (C) with serpin-1 antiserum. F, flow-through; E, elution fraction. Putative serpin-1 proteinase complex bands are marked with arrows. Full-length serpin-1 is indicated with a diamond, cleaved serpin-1 is indicated by a circle.
TABLE 2.
Proteins identified from bands 1 and 2 by Mascot search of the NCBInr database
| Band | Rank | Protein name | Accession No. | Isoform | Protein score | Protein score C.I.% | Peptide count | Serpin-1 isoform-specific peptides |
|
|---|---|---|---|---|---|---|---|---|---|
| Observed mass(es) | Position | ||||||||
| 1 | 1 | Serpin 1 | AAC47340.1 | J | 415 | 100 | 16 | 2422.190a | 337–359 |
| 1 | 2 | Serpin 1 | AAC47342.1 | A | 375 | 100 | 15 | 2341.148 | 337–358 |
| 1 | 3 | Serpin 1 | AAC47334.1 | K | 368 | 100 | 15 | 1951.960 | 337–355 |
| 1 | 4 | Serpin 1 | AAC47335.1 | E | 367 | 100 | 15 | 1974.002 | 337–355 |
| 2 | 1 | Serpin 1 | AAC47334.1 | K | 467 | 100 | 19 | 1951.941 | 337–355 |
| 3169.569 | 326–355 | ||||||||
| 2 | 2 | Serpin 1 | AAC47340.1 | J | 450 | 100 | 18 | 1870.931 | 382–397 |
| 2 | 3 | Serpin 1 | AAC47332.1 | H | 450 | 100 | 18 | 1751.894 | 382–396 |
| 2 | 9 | Chymotrypsinogen | AAA58743.1 | NAb | 73 | 88.9 | 10 | NA | |
a Peptide verified by MS/MS with a confidence interval of greater than 95%.
b NA, not applicable.
We suspected that spots 16–26 from the two-dimensional gels (Fig. 3) were serpin-1-proteinase complexes, based on molecular weight. With data from MALDI-TOF/TOF we identified serpin-1 in spots 16–25 and prophenoloxidase in spot 26, results that are consistent with the pattern of serpin-1 antisera binding to two-dimensional immunoblots, which gave positive results for spots 16–25 but not 26 (Fig. 3D).
To identify the proteinase components in these putative serpin-proteinase complexes we constructed a list of catalytic domain sequences from known M. sexta proteinases (HP1–24) (40) and used Mascot to search those sequences against MALDI-TOF/TOF data from trypsin-cleaved spots. Using this method, at least four HP8 catalytic domain peptides were found in spots 19, 20, 22, and 23 (Table 3), and MS/MS data also confirmed the presence of HP8 in spots 22 and 23. Two-dimensional immunoblots with antiserum against HP8 supported these results, showing a signal for HP8 corresponding to spots 18–20 and 22–23 (Fig. 3E). Also, Scaffold analysis of MS/MS data identified a peptide matching HP8 in spots 19 and 20 digested by Lys-C/AspN, and another HP8 peptide was detected in spot 19 after Glu-C bicarbonate digestion (supplemental Table S2).
TABLE 3.
Protein identification from Mascot search against M. sexta protease catalytic domain sequences
| Trypsin spot | Protein name | Accession No. | Protein score | Protein score C.I.% | Total ion score | Total ion C.I.% | Peptide count | % Sequence coverage |
|---|---|---|---|---|---|---|---|---|
| 19t | HP8 | AAV91006.1 | 19 | 64.927 | 0 | 4 | 21 | |
| 20t | HP8 | AAV91006.1 | 18 | 61.543 | 0 | 4 | 19 | |
| 21t | HP1 | AAB94557.1 | 36 | 99.418 | 18 | 99.992 | 4 | 17 |
| 22t | HP8 | AAV91006.1 | 94 | 100 | 62 | 100 | 6 | 18 |
| 23t | HP8 | AAV91006.1 | 33 | 98.784 | 9 | 99.978 | 5 | 20 |
Spot 21 clearly contained serpin-1, but no isoform-specific peptides were detected. HP1 was detected in spot 21 in the Mascot search against the activated M. sexta proteinase sequences by PMF (Table 3). This identification was also confirmed by immunoblotting with HP1 antibody (Fig. 3F). The expected mass of a serpin-1-HP1 complex is ∼64.6 kDa, quite close to the observed size of the serpin-1-HP1 complex in spot 21 (∼66 kDa). Similar two-dimensional immunoblots of immunoaffinity purified serpin-1 isoforms using antisera to HP6 and prophenoloxidase-activating proteinase-3 had no signal, indicating HP6 and prophenoloxidase-activating proteinase-3 were not present in these samples (data not shown).
Mass spectrometry detection of the serpin-1 isoform(s) present in proteinase complexes was more difficult than detecting full-length serpin-1 isoforms, because the target proteinase cleaves the serpin at the P1 residue, releasing a carboxyl-terminal that contains much of the variable sequence (see Fig. 1). Only isoforms A, D, E, J, and K have an isoform-specific tryptic peptide after they are cleaved between P1 and P1′. A search of the raw MALDI-TOF peaks from trypsin-treated samples for predicted peptides from Lys336 to the P1 residue for the remaining isoforms in putative complex bands (spots 16–25) failed to reveal any additional isoform-specific information (data not shown). Of the four spots shown to contain HP8 by MS (spots 19, 20, 22, 23), spot 19 contains serpin-1 isoform J based on PMF and MS/MS results. PMF results suggest that spots 19 and 20 contain serpin-1A, whereas spots 22 and 23 contain serpin-1E, but these identifications are more tentative because of the lack of MS/MS corroboration. Altogether, these results suggest that serpin-1 isoforms J, A, and E formed complexes with HP8 in M. sexta plasma and that these three serpin-1 isoforms may inhibit HP8 in vivo.
DISCUSSION
Inhibitory serpins are under positive selection pressure and the reactive center loop evolves rapidly (48, 49). Gene duplications can produce multiple copies of related serpins, which may diverge rapidly and gain new functions. For example, human α1-antitrypsin (SERPINA1) is a single gene but expansions have occurred leading to four genes in guinea pig and rabbit and up to five genes present in some mouse species (50).
Although gene duplication has also expanded the serpin repertoire in insects, another mechanism for increasing serpin diversity involves multiple mutually exclusive alternative exons, which encode the reactive center loop. This was first shown in detail in M. sexta, where 12 different serpin-1 isoforms with different inhibitory specificities are created due to mutually exclusive splicing of 12 exon 9 variants (27, 28).
Homologs of M. sexta serpin-1 are also found in other insects, including the Lepidopterans Bombyx mori, in which there are three alternative exons in serpin-1 and four in serpin-28 (32, 33), Mamestra configurata, which has nine isoforms (51), and Choristoneura fumiferana, which has at least four isoforms (34). Also homologous is serpin-1 from the flea Ctenophalides felis, which consists of 15 isoforms (35).
A. gambiae serpin-10 (four isoforms) and D. melanogaster serpin-4 (four alternatively spliced C-terminal exons) are non-secreted serpins (33, 52, 53). The number of serpin-4 reactive center loop-encoding exons was compared in 12 Drosophila species and the number varied from three (in Drosophila persimilis and Drosophila pseudoobscura) up to nine (in Drosophila grimshawi) (54). Alternative splicing to generate different C termini was also observed in Tribolium castaneum serpins 1, 15, 20, and 28 (33, 55).
Our real time PCR results showed that all 12 M. sexta serpin-1 isoforms are expressed in both fat body and hemocytes The 12 isoforms were expressed at similar levels in hemocytes, but in fat body isoform B mRNA was present at significantly higher levels than isoforms C, D, E, F, G, J, K, and Z. The only significant change in expression observed after injection of larvae with bacteria was a decrease in levels of isoforms H and J in fat body.
Another example of mutually exclusive alternative splicing in insects involving more than two alternative spliced exons is the dscam gene, which contains three positions (exons 4, 6, and 9) with many mutually exclusive exons (12, 48, and 33, respectively, in D. melanogaster). dscam isoforms are differentially expressed in different tissues (whole brain versus eye-antennal imaginal discs), at different stages of development, and within individual cells (56). D. melanogaster muscle myosin also has mutually exclusive alternative splicing at five positions, three of which have 3 or more mutually exclusive exons, and similar patterns are found for myosin in many other insects (57). Isoforms of muscle myosin heavy chain are differentially expressed in different tissues (58).
Alternative splicing is a common strategy for increasing protein diversity across higher eukaryotes and often the identification of different protein isoforms is cited as an advantage of proteomic studies. In practice, isoform identification can be challenging. Techniques for identifying individual protein isoforms by multiple proteinase digestions and MS analysis, as demonstrated in this study, can be helpful in proteomic studies of other systems. To separate and identify the serpin-1 isoform proteins present in plasma we used two-dimensional PAGE followed by MALDI-TOF/TOF analysis. Trypsin digestion led to identification of eight of the 12 isoforms, although serpin-1F was identified by PMF alone. We used three additional digestion conditions (Lys-C/AspN and Glu-C in either phosphate or bicarbonate buffer), which increased peptide coverage for serpin-1 isoform H (spot 4) and allowed identification a ninth isoform, B, in spot 5. Antisera generated from isoform-specific peptides from either serpin-1B or serpin-1F recognized a single spot on two-dimensional PAGE immunoblots, which is consistent with the mass spectrometry results and indicates that peptide-based antisera can be used to identify isoforms from mutually exclusive exons.
A recent proteomic analysis of M. sexta hemolymph identified 11 spots on a 4–7 pH range strip as serine protease inhibitors (spots 18–28) and labeled four of these as serpin-1 (spots 20, 21, 26, and 27) (59). We analyzed the matched peptides given by Furusawa et al. (59) and determined that all 11 identified serine protease inhibitor spots were serpin-1. Isoform-specific peptides were present in eight of the 11 spots, representing isoforms E, I, K, and Z. The locations of the serpin-1 isoforms in the Furusawa paper are largely consistent with our results, including serpin-1E in the two serpin-1 spots with highest pI. This suggests that the two alleles for serpin-1E are found in multiple M. sexta populations.
In addition to identifying serpin-1 isoforms, we were interested in finding endogenous proteinases inhibited by serpin-1. High molecular weight spots on our two-dimensional gels of immunoaffinity purified serpins were putative serpin-protease complexes. There are a few special challenges to identifying serpin-proteinase complexes by mass spectrometry. The first is that a band or two-dimensional spot of a serpin-proteinase complex contains at least two different proteins, which reduces the confidence of the individual identifications by PMF. Many insect hemolymph proteinases contain amino-terminal clip domains that, after activation, are attached to the catalytic domain solely by a disulfide bond and separated by SDS-PAGE under reducing conditions. Identification of a catalytic domain by PMF data base search (such as Mascot) is challenging because the sequence in the data base is the full-length protease zymogen, whereas only the catalytic domain is present in the complex spot. Less coverage leads to a lower score and a higher likelihood of a false negative. Identifying individual serpin-1 isoforms is also more difficult after the serpin complexes with a proteinase. After cleavage of the serpin at the P1 residue by a target proteinase, at most only 1 isoform-specific tryptic peptide remains (in the case of A, D, E, J, K) and, for the other isoforms, there is no remaining isoform-specific tryptic peptide.
The use of multiple digestive enzymes contributed to the identification of HP8 in putative proteinase complexes. MS/MS analysis of spot 19 led to identification of two different peptides matching HP8, one each from Lys-C/AspN and Glu-C bicarbonate digestions, whereas an isoform-specific peptide for serpin-1J was observed in trypsin-digested spot 19. Lys-C/AspN digestion followed by MS/MS also identified a HP8 peptide in spot 20. Based on single isoform-specific peptides indicated by PMF, we have support for the presence of serpin-1A in spots 19 and 20, and serpin-1E in spots 22–23. The presence of isoform 1J in spot 19 was further supported by MS/MS results. These results and those from immunoblotting indicate that serpin-proteinase complexes form between HP8 and serpin-1 and that the serpin-1 isoforms involved are A, E, and J. In a previous study with recombinant protein, A, E, and J were the only three serpin-1 isoforms observed to inhibit a fungal protease, PR2, which cleaves after Arg residues (27).
HP8 cleaves its substrate, pro-spätzle after an Arg residue and can cleave an artificial peptide substrate with a P1 Arg (6), consistent with the inhibition of HP8 by these serpin-1 variants that have a basic residue at the P1 position of the RCL. In vitro studies confirm serpin-1A, E, and J as possible inhibitors of HP8 and show that serpin-1J is the most effective of the three isoforms (6). Another serpin-proteinase complex was found in spot 21, with unidentified isoform(s) of serpin-1 and the serine proteinase HP1. This proteinase complex was confirmed by immunoblotting with both HP1 and serpin-1 antibodies. HP1 has a predicted trypsin-like specificity and has been found in complexes with serpin-4 and serpin-5 (15). Other proteinases in serine protease cascades have been found to be regulated by multiple serpins (17), which may be related to changes in individual serpin concentrations after infection. HP1 is a candidate for a cleaving serine proteinase homolog-1, and may also function more directly in phenoloxidase activation (60).
We have demonstrated that all 12 variants of serpin-1 are expressed in fat body and hemocytes of M. sexta, and we were able to detect and identify 9 of these isoforms in plasma through proteomic techniques. The functions of the serpin-1 isoforms are not yet well understood. The results presented here provide evidence that serpin-1 can inhibit several known M. sexta proteinases, including HP1 and HP8, which occur in plasma and have immune functions. Serpin-1K was identified as a complex with a digestive chymotrypsin, which may point toward a role for the serpin-1 variants in protection of tissues from endogenous proteinases that escape their normal compartments and enter the hemocoel. The multiple serpin-1 isoforms may function to inhibit specific plasma proteinases and also provide a background level of protection against a broad range of potentially harmful endogenous or exogenous proteinases.
Supplementary Material
Acknowledgments
We thank Rebekah Woolsey from the Nevada Proteomics Center for sample digestion and mass spectrometry. The Nevada Proteomics Center is supported by National Institutes of Health Grant P20 RR-016464 from the INBRE Program of the National Center for Research Resources. We also thank Dr. Maureen Gorman for providing serpin-1I used for antiserum production, Dr. Haobo Jiang for antiserum to HP6, HP8, and prophenoloxidase-activating proteinase-3, Yang Wang for help characterizing isoform-specific antisera, and Dr. Rollie Clem for the use of the BioRad IEF unit.
This work was supported, in whole or in part, by National Institutes of Health Grant GM41247. This is contribution 10-259-J from the Kansas Agricultural Experiment Station.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
- HP
- hemolymph proteinase
- IEF
- isoelectric focusing
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PMF
- peptide mass fingerprinting.
REFERENCES
- 1.Whisstock J. C., Bottomley S. P. (2006) Curr. Opin. Struct. Biol. 16, 761–768 [DOI] [PubMed] [Google Scholar]
- 2.Dementiev A., Dobó J., Gettins P. G. (2006) J. Biol. Chem. 281, 3452–3457 [DOI] [PubMed] [Google Scholar]
- 3.Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., Pemberton P. A., Remold-O'Donnell E., Salvesen G. S., Travis J., Whisstock J. C. (2001) J. Biol. Chem. 276, 33293–33296 [DOI] [PubMed] [Google Scholar]
- 4.Rau J. C., Beaulieu L. M., Huntington J. A., Church F. C. (2007) J. Thromb. Haemost. 5, Suppl. 1, 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An C., Ishibashi J., Ragan E. J., Jiang H., Kanost M. R. (2009) J. Biol. Chem. 284, 19716–19726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An C., Jiang H., Kanost M. R. (2010) FEBS J. 277, 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorman M. J., Wang Y., Jiang H., Kanost M. R. (2007) J. Biol. Chem. 282, 11742–11749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Jiang H. (2007) Insect Biochem. Mol. Biol. 37, 1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang I. H., Chosa N., Kim S. H., Nam H. J., Lemaitre B., Ochiai M., Kambris Z., Brun S., Hashimoto C., Ashida M., Brey P. T., Lee W. J. (2006) Dev. Cell. 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 10.Kambris Z., Brun S., Jang I. H., Nam H. J., Romeo Y., Takahashi K., Lee W. J., Ueda R., Lemaitre B. (2006) Curr. Biol. 16, 808–813 [DOI] [PubMed] [Google Scholar]
- 11.Kim C. H., Kim S. J., Kan H., Kwon H. M., Roh K. B., Jiang R., Yang Y., Park J. W., Lee H. H., Ha N. C., Kang H. J., Nonaka M., Söderhäll K., Lee B. L. (2008) J. Biol. Chem. 283, 7599–7607 [DOI] [PubMed] [Google Scholar]
- 12.Kan H., Kim C. H., Kwon H. M., Park J. W., Roh K. B., Lee H., Park B. J., Zhang R., Zhang J., Söderhäll K., Ha N. C., Lee B. L. (2008) J. Biol. Chem. 283, 25316–25323 [DOI] [PubMed] [Google Scholar]
- 13.Tang H., Kambris Z., Lemaitre B., Hashimoto C. (2006) J. Biol. Chem. 281, 28097–28104 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., Jiang H. (2004) Insect Biochem. Mol. Biol. 34, 387–395 [DOI] [PubMed] [Google Scholar]
- 15.Tong Y., Jiang H., Kanost M. R. (2005) J. Biol. Chem. 280, 14932–14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y., Wang Y., Gorman M. J., Jiang H., Kanost M. R. (2003) J. Biol. Chem. 278, 46556–46564 [DOI] [PubMed] [Google Scholar]
- 17.Zou Z., Jiang H. (2005) J. Biol. Chem. 280, 14341–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Gregorio E., Han S. J., Lee W. J., Baek M. J., Osaki T., Kawabata S., Lee B. L., Iwanaga S., Lemaitre B., Brey P. T. (2002) Dev. Cell. 3, 581–592 [DOI] [PubMed] [Google Scholar]
- 19.Ligoxygakis P., Pelte N., Ji C., Leclerc V., Duvic B., Belvin M., Jiang H., Hoffmann J. A., Reichhart J. M. (2002) EMBO J. 21, 6330–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherfer C., Tang H., Kambris Z., Lhocine N., Hashimoto C., Lemaitre B. (2008) Dev. Biol. 323, 189–196 [DOI] [PubMed] [Google Scholar]
- 21.Tang H., Kambris Z., Lemaitre B., Hashimoto C. (2008) Dev. Cell. 15, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang R., Kim E. H., Gong J. H., Kwon H. M., Kim C. H., Ryu K. H., Park J. W., Kurokawa K., Zhang J., Gubb D., Lee B. L. (2009) J. Biol. Chem. 284, 35652–35658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel K., Suwanchaichinda C., Morlais I., Lambrechts L., Cohuet A., Awono-Ambene P. H., Simard F., Fontenille D., Kanost M. R., Kafatos F. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16858–16863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levashina E. A., Langley E., Green C., Gubb D., Ashburner M., Hoffmann J. A., Reichhart J. M. (1999) Science 285, 1917–1919 [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto C., Kim D. R., Weiss L. A., Miller J. W., Morisato D. (2003) Dev. Cell 5, 945–950 [DOI] [PubMed] [Google Scholar]
- 26.Ligoxygakis P., Roth S., Reichhart J. M. (2003) Curr. Biol. 13, 2097–2102 [DOI] [PubMed] [Google Scholar]
- 27.Jiang H., Kanost M. R. (1997) J. Biol. Chem. 272, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 28.Jiang H., Wang Y., Huang Y., Mulnix A. B., Kadel J., Cole K., Kanost M. R. (1996) J. Biol. Chem. 271, 28017–28023 [DOI] [PubMed] [Google Scholar]
- 29.Jiang H., Mulnix A. B., Kanost M. R. (1995) Insect Biochem. Mol. Biol. 25, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 30.Jiang H., Wang Y., Kanost M. R. (1994) J. Biol. Chem. 269, 55–58 [PubMed] [Google Scholar]
- 31.Li J., Wang Z., Canagarajah B., Jiang H., Kanost M., Goldsmith E. J. (1999) Structure 7, 103–109 [DOI] [PubMed] [Google Scholar]
- 32.Sasaki T. (1991) Eur. J. Biochem. 202, 255–261 [DOI] [PubMed] [Google Scholar]
- 33.Zou Z., Picheng Z., Weng H., Mita K., Jiang H. (2009) Genomics 93, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y. P., He W. Y., Béliveau C., Nisole A., Stewart D., Zheng S. C., Doucet D., Cusson M., Feng Q. L. (2009) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 154, 165–173 [DOI] [PubMed] [Google Scholar]
- 35.Brandt K. S., Silver G. M., Becher A. M., Gaines P. J., Maddux J. D., Jarvis E. E., Wisnewski N. (2004) Arch. Insect Biochem. Physiol. 55, 200–214 [DOI] [PubMed] [Google Scholar]
- 36.Kanost M. R., Prasad S. V., Huang Y., Willott E. (1995) Insect Biochem. Mol. Biol. 25, 285–291 [DOI] [PubMed] [Google Scholar]
- 37.Kanost M. (1990) in Molecular Insect Science (Hagedorn H. H. E. A., ed) pp. 139–146, Plenum Press, New York [Google Scholar]
- 38.Jiang H., Wang Y., Yu X. Q., Zhu Y., Kanost M. (2003) Insect Biochem. Mol. Biol. 33, 1049–1060 [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Jiang H. (2006) J. Biol. Chem. 281, 9271–9278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H., Wang Y., Gu Y., Guo X., Zou Z., Scholz F., Trenczek T. E., Kanost M. R. (2005) Insect Biochem. Mol. Biol. 35, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Zou Z., Jiang H. (2006) Genomics 87, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunn P. E., Drake D. (1983) J. Invertebr. Pathol. 41, 77–85 [Google Scholar]
- 43.Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 44.Ragan E. J. (2008) Immune-related Protein Complexes and Serpin-1 Isoforms in Manduca sexta Plasma. Ph.D. thesis, Kansas State University, Manhattan, KS [Google Scholar]
- 45.John J. P., Chen W. Q., Pollak A., Lubec G. (2007) J. Proteome Res. 6, 2695–2710 [DOI] [PubMed] [Google Scholar]
- 46.Myung J. K., Frischer T., Afjehi-Sadat L., Pollak A., Lubec G. (2008) Amino Acids 35, 485–494 [DOI] [PubMed] [Google Scholar]
- 47.Peterson A. M., Fernando G. J., Wells M. A. (1995) Insect Biochem. Mol. Biol. 25, 765–774 [DOI] [PubMed] [Google Scholar]
- 48.Hill R. E., Hastie N. D. (1987) Nature 326, 96–99 [DOI] [PubMed] [Google Scholar]
- 49.Barbour K. W., Goodwin R. L., Guillonneau F., Wang Y., Baumann H., Berger F. G. (2002) Mol. Biol. Evol. 19, 718–727 [DOI] [PubMed] [Google Scholar]
- 50.Forsyth S., Horvath A., Coughlin P. (2003) Genomics 81, 336–345 [DOI] [PubMed] [Google Scholar]
- 51.Hegedus D. D., Erlandson M., Baldwin D., Hou X., Chamankhah M. (2008) Gene 418, 15–21 [DOI] [PubMed] [Google Scholar]
- 52.Krüger O., Ladewig J., Köster K., Ragg H. (2002) Gene 293, 97–105 [DOI] [PubMed] [Google Scholar]
- 53.Danielli A., Kafatos F. C., Loukeris T. G. (2003) J. Biol. Chem. 278, 4184–4193 [DOI] [PubMed] [Google Scholar]
- 54.Börner S., Ragg H. (2008) Gene 415, 23–31 [DOI] [PubMed] [Google Scholar]
- 55.Zou Z., Evans J. D., Lu Z., Zhao P., Williams M., Sumathipala N., Hetru C., Hultmark D., Jiang H. (2007) Genome Biol. 8, R177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neves G., Zucker J., Daly M., Chess A. (2004) Nat. Genet. 36, 240–246 [DOI] [PubMed] [Google Scholar]
- 57.Odronitz F., Kollmar M. (2008) BMC Mol. Biol. 9, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hodges D., Cripps R. M., O'Connor M. E., Bernstein S. I. (1999) Genetics 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furusawa T., Rakwal R., Nam H. W., Hirano M., Shibato J., Kim Y. S., Ogawa Y., Yoshida Y., Kramer K. J., Kouzuma Y., Agrawal G. K., Yonekura M.(2008) J. Proteome Res. 7, 938–959 [DOI] [PubMed] [Google Scholar]
- 60.Jiang H. (2008) Insect Sci. 15, 53–66 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.