Summary
The psychiatric illness risk gene Disrupted in Schizophrenia-1 (DISC1) plays an important role in brain development, however, it is unclear how DISC1 is regulated during cortical development. Here, we report that DISC1 is regulated during embryonic neural progenitor proliferation and neuronal migration through an interaction with DIX domain containing-1 (Dixdc1), the third mammalian gene discovered to contain a Disheveled-Axin (DIX) domain. We determined that Dixdc1 functionally interacts with DISC1 to regulate neural progenitor proliferation by co-modulating Wnt-GSK3β/β-catenin signaling. However, DISC1 and Dixdc1 do not regulate migration via this pathway. During neuronal migration, we discovered that phosphorylation of Dixdc1 by cyclin-dependent kinase 5 (Cdk5) facilitates its interaction with the DISC1-binding partner Ndel1. Furthermore, Dixdc1 phosphorylation and its interaction with DISC1/Ndel1 in vivo is required for neuronal migration. Together, these data reveal that Dixdc1 integrates DISC1 into Wnt-GSK3β/β-catenin-dependent and -independent signaling pathways during cortical development, and further delineate how DISC1 contributes to neuropsychiatric disorders.
Introduction
Psychiatric diseases, including schizophrenia and bipolar disease, are highly prevalent and cause lifelong disability for many people. Despite efforts to better identify disease etiology, our understanding of the pathophysiology of these diseases is still very limited. One of the first risk genes to be identified is Disrupted in schizophrenia-1 (DISC1), which was discovered as a balanced chromosomal translocation t(1;11) (q42.1;q14.3) in a large Scottish pedigree that displayed various psychiatric disorders (Blackwood et al., 2001). A number of studies have examined the biology of DISC1 to determine the underlying mechanisms by which it contributes to risk for psychiatric disorders. Studies have demonstrated that DISC1 regulates embryonic neurogenesis, neuronal migration, axon differentiation and synapse formation, while in the adult brain, DISC1 modulates the genesis and circuit integration of new neurons (Bradshaw et al., 2008; Duan et al., 2007; Enomoto et al., 2009; Hayashi-Takagi et al.,; Kamiya et al., 2005; Kim et al., 2009; Mackie et al., 2007; Niwa et al.,; Pletnikov et al., 2007). Furthermore, the many DISC1 mouse models that have been generated collectively demonstrate that mice with disrupted DISC1 function, particularly during neurodevelopment, display behavioral phenotypes that are consistent with psychiatric disorders such as diminished working memory, increased anxiety/hyperactivity, and increased brain ventricle size (Clapcote et al., 2007; Hayashi-Takagi et al., 2010; Hikida et al., 2007; Ishizuka et al., 2007; Koike et al., 2006; Kvajo et al., 2008; Li et al., 2007; Niwa et al., 2010; Pletnikov et al., 2008). These data support the hypothesis that one of the mechanisms by which psychiatric risk genes function is to disrupt neural development.
Equally interesting is the number of interacting molecules through which DISC1 regulates these events. The more well-known interacting genes include GSK3β, Ndel1, Rac1, the PDE4 family, and Girdin/KIAA1212, which are thought to regulate progenitor proliferation, neuronal migration, synapse formation, cyclic adenosine monophosphate (cAMP) signaling, and adult neuron generation, respectively (Duan et al., 2007; Enomoto et al., 2009; Kamiya et al., 2006; Kim et al., 2009; Mao et al., 2009; Millar et al., 2005; Murdoch et al., 2007; Ozeki et al., 2003; Pletnikov et al., 2007). Although these studies are beginning to shed light on DISC1-mediated signaling pathways, the molecular mechanisms by which DISC1 is regulated during different neurodevelopmental events remains unknown.
We recently identified DISC1 as an essential regulator of neural progenitor proliferation by directly binding to and inhibiting GSK3β to modulate canonical Wnt signaling (Mao et al., 2009). This is interesting given that one of the actions of lithium, the most common mood disorder drug, is to inhibit GSK3β (Beaulieu et al., 2008; Harwood, 2005) and activate TCF/LEF-dependent gene transcription (Stambolic et al., 1996). Furthermore, other schizophrenia risk genes such as Akt or a phosphatidylinositol 4-kinase (PIK4CA), also lie upstream of GSK3β signaling and can therefore potentially impact Wnt signaling. For example, Akt inhibits GSK3β activation, and its protein levels have been shown to be reduced in the brains of schizophrenia patients (Emamian et al., 2004). In addition, the Akt-GSK3β pathway is thought to mediate some of the actions of lithium and antipsychotic drugs in mouse behavioral and biochemical studies (Beaulieu et al., 2009). PIK4CA is a receptor in the chromosome 22q11 deletion region that significantly increases the risk for schizophrenia (Jungerius et al., 2008; Karayiorgou et al., 1995; Vorstman et al., 2009). Interestingly, this family of receptors mediates phosphatidylinositol activity that lies upstream of Akt activation (Stephens et al., 2005). Together, these studies suggest that GSK3β and Wnt signaling may represent one underlying pathogenic pathway in psychiatric disorders. Importantly, these data warrant the need to further examine how risk genes interact with Wnt signaling during brain development to better understand the disease etiology.
In this regard, we investigated how DISC1 is regulated during cortical development by searching for novel DISC1-interacting proteins. We found a novel interaction between DISC1 and DIX domain containing-1 (Dixdc1), a homolog of the Wnt signaling genes Disheveled and Axin. Dixdc1 is the mammalian homologue of Ccd1, which was identified in zebrafish as a positive regulator of Wnt-TCF/LEF signaling (Shiomi et al., 2003). Here we present evidence that Dixdc1 integrates DISC1 within Wnt-GSK3β/β-catenin-dependent and -independent signaling pathways that are essential for embryonic neural progenitor proliferation and neuronal migration, respectively. Together, these results delineate the requirement of canonical Wnt signaling in neural progenitor proliferation but not neuronal migration, and further sheds light on the mechanisms by which DISC1 may contribute to psychiatric disease.
Results
Dixdc1 is a DISC1-interacting protein
To determine if Dixdc1 interacts with DISC1 during embryonic development, we first performed biochemical experiments using embryonic day 14 (E14) brain tissue. We found that Dixdc1 and DISC1 co-immunoprecipitated at this embryonic time point, when neural progenitor proliferation is highly prevalent (Figure 1A), demonstrating that they form a complex in vivo. We then mapped the binding domains between Dixdc1 and DISC1 by creating and expressing full-length GFP-tagged Dixdc1 with FLAG-tagged DISC1 fragments in HEK293 cells. These experiments revealed that Dixdc1 binds most strongly to the C-terminus of DISC1 (fragment 4), and weakly to the middle region of DISC1 (fragment 3; Figure 1B,C). Conversely, we mapped the region(s) of Dixdc1 which associate with DISC1. We generated and expressed FLAG-tagged Dixdc1 fragments with full-length GFP-DISC1 in HEK293 cells. We determined that DISC1 strongly binds to the N-terminal Dixdc1 region (fragment 2) that lies between the calpain homology and coiled-coil domains (Figure 1D,E). Taken together, these data demonstrate that Dixdc1 binds DISC1 in vivo during early brain development.
Figure 1. DISC1 interacts with Dixdc1.
(A) DISC1 interacts with endogenous Dixdc1. E14 brain lysate was subjected to co-immunoprecipitation with anti-HA (negative control), anti-DISC1 or anti-Dixdc1 antibodies and immunoblotted with anti-DISC1 or Dixdc1 antibodies. (B) Schematic showing the flag-tagged DISC1 fragments used for domain mapping (C) Dixdc1 preferentially binds to the C-terminus of DISC1. Western blot showing lysates from HEK293 cells transfected with GFP-tagged Dixdc1 and each of the flag-tagged DISC1 domains, immunoprecipitated with a flag antibody and immunoblotted with a GFP antibody. (D) Schematic of the different flag-tagged Dixdc1 fragments used for domain mapping. (E) DISC1 preferentially binds to the N-terminus of Dixdc1, between residues 181–370 (Dixdc1 Frag2). Western blot showing HEK293 cells transfected with full length GFP- tagged DISC1 together with flag-tagged Dixdc1 fragments, immunoprecipitated with a flag antibody and immunoblotted for GFP. See also Figure S1.
Dixdc1 is essential for neural progenitor proliferation
DISC1 is required for the regulation of neural progenitor proliferation and neuronal migration during embryonic cortical development (Kamiya et al., 2005; Mao et al., 2009) Given our data that Dixdc1 binds DISC1, we first examined the developmental expression profile of Dixdc1 to determine if it might have functions similar to DISC1. Western blot analysis of brain lysates from different ages revealed that the long and short isoforms of Dixdc1 (l-Dixdc1 and s-Dixdc1) are highly expressed as early as embryonic day 10 (E10) and persist throughout the neurogenic period (E11–17; Figure S1A). However, following E18, the expression of s-Dixdc1, is downregulated to nearly undetectable levels, while l-Dixdc1 expression persists into adulthood. We then performed immunostaining on embryonic brains and found that Dixdc1 co-localizes with Nestin-positive radial glial cells, which give rise to neurons, in the E12 and E15 cortex (Figure S1B). We further determined that Dixdc1 also co-localizes with the neuronal marker β-III tubulin (Tuj1) in the E15 and E17 cortex, demonstrating that Dixdc1 is expressed in both neural progenitor cells (radial glia) and postmitotic neurons (Figure S1C). This staining pattern is in good agreement with a previous study that examined the expressed pattern of Dixdc1 using in situ hybridization (Shiomi et al. 2003). Together, the early expression pattern of Dixdc1 suggests that it may play a role in progenitor proliferation and neuronal differentiation.
To determine whether Dixdc1 regulates neural progenitor proliferation, we knocked down the expression of Dixdc1 using shRNA. We tested different shRNA constructs for their ability to reduce exogenous and endogenous Dixdc1 expression, and selected two that produced the most robust knockdown (Figure S2A–C). We utilized in utero electroporation to introduce Dixdc1 shRNA constructs together with a GFP-encoding construct into neural progenitor cells of the developing cortex in E13 mouse embryos, and analyzed brains at E16. We found that the knockdown of Dixdc1 resulted in a considerable change in cell distribution compared to scrambled shRNA controls. There was a significant loss of GFP-positive cells from the ventricular/subventricular zones (VZ/SVZ), a phenotype that is similar to that seen with DISC1 knockdown (Figure 2A) (Mao et al., 2009). This effect was also accompanied by a reduction of GFP-positive cells in the cortical plate (CP), suggesting an early migration defect. This resulted in Dixdc1 shRNA-treated brains having the majority of the GFP-positive cells accumulating in the intermediate zone (IZ) compared to control brains, which had an approximately equal percentage of GFP-positive cells in each of the three zones (Figure 2A).
Figure 2. Dixdc1 regulates neural progenitor cell proliferation in vivo.
(A) Knockdown of Dixdc1 in utero causes cell positioning defects. Control or Dixdc1 shRNA constructs were electroporated into E13 embryonic mouse brains which were analyzed at E16. Left, confocal images of electroporated E16 brains from each condition. The scale bar represents 50 μm. Right, graph showing the percentage of GFP cells in each region (n=5, ***p<0.0005, mean ± SEM). CP, cortical plate; IZ, intermediate zone; VZ/SVZ, ventricular zone/sub-ventricular zone. (B) Reduced BrdU incorporation in Dixdc1 knockdown cells in utero. E13 brains were electroporated and Brdu (100 mg/kg) was administered after 48 hours, followed by analysis of brains at 72 hours. Left, confocal images of sections stained for Brdu and GFP. Arrow heads indicate cells double positive for GFP and BrdU. Right, graph showing the percentage of double-labeled GFP-BrdU cells relative to total GFP cell number (n=5, ***p<0.0005, mean ± SEM). The scale bar represent 25 μm. (C) Knockdown of Dixdc1 leads to increased premature cell cycle exit. E13 mouse brains were electroporated with control or Dixdc1 shRNA, BrdU (100 mg/kg) was administered at E15 and analyzed 24 hours later at E16. Left, confocal images of brain sections stained for GFP, Ki67 and BrdU. Arrowheads indicate GFP+BrdU+Ki67+ cells while arrows indicate GFP+BrdU+Ki67−cells. Right, graph showing quantification of the cell cycle exit index (n=5, ***p<0.0005, **p<0.005, mean±SEM). The scale bar represents 25 μm. (D) Increased neuronal differentiation of Dixdc1 knockdown cells. Top left, confocal images of E16 brain sections stained for Tuj1 after electroporation of control or Dixdc1 shRNA constructs into E13 brain. Top right, quantification of the percentage of GFP-Tuj1 double positive cells relative to total GFP positive cells is shown as a bar graph. Bottom left, confocal images of E16 brains stained for GFP after E13 brains were electroporated with control or Dixdc1 shRNA together with constructs expressing mCherry and the neuronal-specific pNeuroD-GFP. Bottom right, graph represents the percentage of mCherry positive cells that are double positive for GFP (n=4 for each experiment, **p<0.005, *p<0.01, mean±SEM). The scale bar represent 50 μm. See also Figure S2.
To examine if the loss of GFP-positive cells from the VZ/SVZ was due to reduced neural progenitor proliferation, we injected BrdU into pregnant dams 24 hours prior to brain analysis at E16. Knockdown of Dixdc1 resulted in a significant decrease in BrdU incorporation into GFP-positive cells (Figure 2B), suggesting that Dixdc1 is required for neural progenitor proliferation. These results, together with the observed reduction in the number of GFP-positive cells in the VZ/SVZ, imply that the loss of Dixdc1 function leads to premature cell cycle exit and neuronal differentiation. To test this directly, we first performed an analysis of cell cycle exit by electroporating embryonic brains at E13, pulse labeling with BrdU at E15, and collecting brains at E16, followed by immunocytochemistry for GFP, BrdU and Ki67. Using this analysis, we found that Dixdc1 shRNA caused a significant increase in the number of cells exiting the cell cycle (Figure 2C; see Methods for calculation). To examine if this result directly leads to increased neuronal differentiation, we stained electroporated brains with a neuronal βIII-tubulin (Tuj1) antibody. Quantification of GFP-Tuj1 double-positive cells revealed that the knockdown of Dixdc1 led to a significant increase in the percentage of GFP-labeled cells that were also positive for Tuj1, demonstrating that the loss of Dixdc1 resulted in an increase in neuronal differentiation (Figure 2D). Since there is high expression of Tuj1 in the axons located in the SVZ, this can potentially lead to an overestimation of the percentage of GFP-Tuj1 double-positive cells. To circumvent this, we co-electroporated control or Dixdc1 shRNA plasmids together with an mCherry construct and an expression plasmid encoding GFP under the neuron-specific pNeuroD promoter (Yokota et al., 2007). We quantified the percentage of mCherry-positive cells that were positive for pNeuroD-GFP, and found that the loss of Dixdc1 expression led to a significant increase in neuronal differentiation (Figure 2D), in accordance with our Tuj1 immunochemistry results.
Dixdc1 has also been shown to bind actin directly through its calpain homology domain (Wang et al., 2006). It is therefore possible that the loss of Dixdc1 expression may produce progenitor proliferation defects by disrupting the apical-basal polarity of radial glial cells. To examine this possibility, we performed immunolabeling for Nestin, to label radial glia, and phalloidin staining for F-actin, which is highly concentrated in the end feet of radial glial cells at the apical surface. We found that the staining pattern of these markers was not disrupted after Dixdc1 knockdown (Figure S2E–G). Taken together, these experiments demonstrate that the downregulation of Dixdc1 expression leads to reduced progenitor proliferation, which temporarily leads to premature cell cycle exit and increased neuronal differentiation.
Dixdc1 and DISC1 co-regulate Wnt-GSK3β/β-catenin signaling
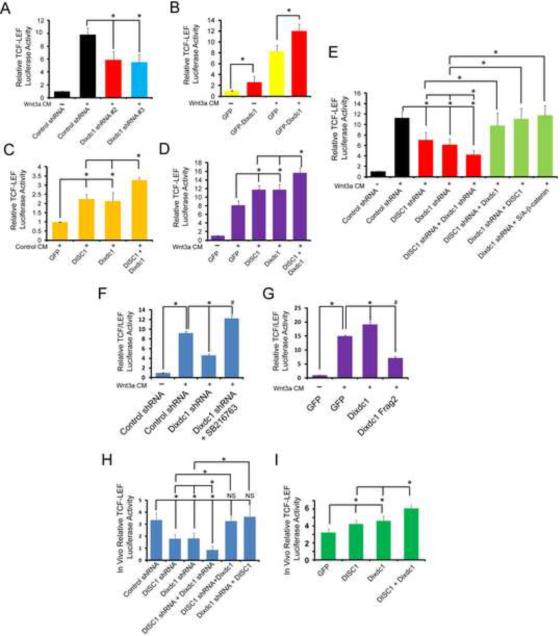
Given our results that DISC1 and Dixdc1 bind one another in vivo, and that DISC1 modifies the Wnt pathway in progenitor cells (Mao et al., 2009), we hypothesized that Dixdc1 and DISC1 may functionally interact to modulate Wnt-mediated GSK3β/β-catenin signaling and thereby regulate neural progenitor proliferation. We first investigated the function of Dixdc1 in Wnt signaling, since Dixdc1 has been reported to stimulate TCF/LEF activity (Shiomi et al., 2003). To measure Wnt-mediated GSK3β/β-catenin activity, we utilized a luciferase reporter construct containing eight copies of the TCF/LEF binding site (8XSuperTOPFLASH), since β-catenin stimulates transcription of genes containing TCF/LEF binding sites (Molenaar et al., 1996; van de Wetering et al., 1997). We knocked-down Dixdc1 expression, and determined that the Wnt3a-stimulated TCF/LEF-reporter activity was reduced by approximately 50% compared to shRNA controls (Figure 3A). We also found that overexpression of Dixdc1, compared to GFP alone, in cells that were stimulated with either control conditioned media (Control CM) or Wnt3a conditioned media (Wnt3a CM), resulted in a significant increase in TCF/LEF-reporter activity (Figure 3B). These findings are similar to our previously reported results for DISC1 (Mao et al., 2009), and suggest that DISC1 and Dixdc1 may co-modulate the Wnt pathway in neural progenitor cells.
Figure 3. Dixdc1 and DISC1 co- regulate Wnt-GSK3β/β-catenin signaling.
(A) Dixdc1 regulates canonical Wnt-GSK3β/β-catenin signaling in vitro. Left, P19 cells were transfected with the TOPFLASH reporter and control or Dixdc1 shRNA for 30 hours, followed by a 16-hour stimulation with control (CM) or Wnt3a conditioned media (Wnt3a CM) and subjected to a luciferase assay. Graph shows the relative TCF/LEF luciferase activity (n=5, *p<0.05, mean±SEM). (B) Dixdc1 can stimulate TCF/LEF activity. HEK293 cells were transfected with GFP or GFP-Dixdc1 and TOPFLASH for 24 hours, stimulated with control conditioned media (CM) or Wnt3a CM or an additional 16 hours followed by luciferase assay. Graph shows relative TCF/LEF luciferase activity. (C and D) Dixdc1 and DISC1 synergistically activate canonical Wnt-GSK3β/β-catenin signaling. HEK293 cells were transfected with TOPFLASH and the indicated constructs, and then treated as in (B) (n=5, *p<0.05, mean±SEM). (E) Dixdc1 and DISC1 are both required for TCF/LEF signaling. P19 cells were transfected with the indicated constructs together with TOPFLASH and then treated as in (A). (n=6, *p<0.05, mean±SEM). (F) The interaction between DISC1 and Dixdc1 is required for TCF/LEF signaling. HEK293 cells were transfected with TOPFLASH and the indicated constructs and then treated as in (B) with 10μM SB216763. (n=3, *p<0.05, mean±SEM). (G) Dixdc1 regulates β-catenin through GSK3β. P19 cells were transfected with TOPFLASH and the indicated constructs and then treated as in (A). (n=3, *p<0.05, mean±SEM). (H) Dixdc1 and DISC1 are both required for TCF/LEF signaling in vivo. E13 brains were electroporated with TOPFLASH together with the indicated constructs followed by dissection of the transfected cortical area and a luciferase assay at E15. The graph shows the relative TCF/LEF activity of the transfected area of E15 brains (n=4, *p<0.05, NS=not significant, mean±SEM). (I) Dixdc1 and DISC1 synergistically regulate TCF/LEF signaling in vivo. E13 brains were treated as in (F) but electroporated with Dixdc1, DISC1 or both plasmids together. (n=5, *p<0.05, mean±SEM).
To further elucidate the concomitant roles of these proteins, we examined whether DISC1 and Dixdc1 functioned together to regulate Wnt-TCF/LEF signaling. We overexpressed Dixdc1 and DISC1 together, and observed that when cells were stimulated with either Control CM or Wnt3a CM, the expression of the two genes together additively activated TCF/LEF-reporter activity compared to either gene alone (Figure 3C,D). We then tested the relationship between Dixdc1 and DISC1 in Wnt signaling. First, we found that treatment with both Dixdc1 and DISC1 shRNA together resulted in significantly reduced TCF/LEF-reporter activity compared to knockdown of either gene alone, demonstrating the interaction between these genes strongly regulate Wnt signaling (Figure 3E). Interestingly, we also found that the decreased TCF/LEF-reporter activity due to Dixdc1 knockdown could be completely rescued by the overexpression of DISC1 (Figure 3E). Conversely, the DISC1 shRNA-mediated decrease in TCF/LEF-reporter activity could be rescued by overexpression of Dixdc1 (Figure 3E), suggesting that Dixdc1 and DISC1 can functionally compensate for each other's TCF/LEF activity when the reciprocal gene is overexpressed. Interestingly, we also found that overexpressing Dixdc1 Fragment 2, which inhibits interaction between DISC1 and Dixdc1 (Figure S7), also significantly reduced TCF/LEF-reporter activity, demonstrating the interaction between DISC1and Dixdc1 is required for Wnt signaling (Figure 3F). Importantly, overexpression of a degradation-resistant β-catenin, which stabilizes its expression levels, also rescues the Dixdc1 shRNA-mediated decrease in reporter activity (Figure 3E), demonstrating that β-catenin signals downstream of Dixdc1, similar to DISC1 (Mao et al., 2009). Finally, we determined that the Dixdc1 shRNA-mediated reduction in TCF/LEF-reporter activity could be completely rescued by application of GSK3β inhibitor (SB216763), suggesting that Dixdc1 regulates β-catenin through GSK3β, similar to DISC1 (Figure 3G).
Since these experiments were performed in vitro, we asked if Dixdc1 and DISC1 co-regulated Wnt-TCF/LEF signaling in vivo. Using in utero electroporation, we found that both DISC1 and Dixdc1 shRNA significantly reduced TCF/LEF-reporter activity in neural progenitor cells to the same degree (Figure 3H). Similar to the in vitro results, we found there was a further significant decrease in TCF/LEF-reporter activity when expression of both DISC1 and Dixdc1 were reduced in vivo. In addition, we found the DISC1 or Dixdc1 shRNA reduced TCF/LEF-reporter activity could be completely rescued by overexpression of the reciprocal gene (Figure 3H). Lastly, we performed gain of function experiments in vivo, and demonstrated that overexpression of either Dixdc1 or DISC1 alone significantly increases TCF/LEF-reporter activity, while overexpression of both genes together additively enhances TCF/LEF signaling (Figure 3I). Taken together, these results demonstrate both Dixdc1 and DISC1 are necessary for Wnt signaling, and functionally interact in an additive manner to regulate Wnt-GSK3β/β-catenin signaling in neural progenitor cells.
Dixdc1 and DISC1 functionally interact to modulate neural progenitor proliferation
Our results demonstrating that Dixdc1 and DISC1 are both required for Wnt-GSK3β/β-catenin signaling suggested that these factors functionally interact to regulate neural progenitor proliferation in vivo. To test this, we knocked down expression of Dixdc1, DISC1 or both genes together in vivo, and observed that the double gene knockdown led to significantly fewer GFP-positive cells remaining in the VZ/SVZ compared to knocking down expression of either gene alone (Figure 4A). In addition, we found the double gene knockdown also led to significantly reduced Brdu incorporation, increased cell cycle exit and increased neuronal differentiation compared to reducing expression of either gene alone (Figure 4B–D, Figure S3). These data suggest that Dixdc1 is a major partner of DISC1 in the regulation of neural progenitor proliferation.
Figure 4. Dixdc1 and DISC1 functionally interact to regulate neural progenitor proliferation.
(A) Cell positioning defects induced by Dixdc1/DISC1 shRNA are rescued by reciprocal gene overexpression. Left, confocal images of GFP-positive cells. Right, graph showing quantification of GFP positive cell positioning in each zone (n=6, **p<0.005, NS=not significant, mean±SEM). The scale bar represent 50 μm. (B) Reduced BrdU incorporation due to DISC1 or Dixdc1 shRNA is rescued by reciprocal overexpression. Left, confocal images of brain sections stained for BrdU and GFP. Arrowheads indicate double positive cells for GFP and BrdU. Right, graph showing quantification of the percentage of double positive cells divided by the total GFP positive cells (n=5, *p<0.05, **p<0.005, NS=not significant, mean±SEM). The scale bar represents 25 μm. (C) Dixdc1/DISC1 shRNA-induced cell cycle exit defects are rescued by reciprocal overexpression. Left, confocal images of brain sections stained for GFP, BrdU and Ki67. Arrowhead, GFP+BrdU+Ki67−cells; Arrow, GFP+BrdU+Ki67+ cells. Right, graph showing the quantification of the cell cycle index for each condition (see Methods for calculation) (n=5, *p<0.05, NS=not significant, mean±SEM). The scale bar represent 25 μm. (D) Defects in neuronal differentiation due to DISC1/Dixdc1 shRNA are rescued by reciprocal overexpression. E13 brains were electroporated with the indicated constructs together with mCherry and pNeuroD-GFP. Left, confocal images of brain sections stained for GFP. Right, graph showing the quantification of all double-positive GFP+mCherry+ cells divided by the total mCherry cell number (n=5, *p<0.05, NS=not significant, mean±SEM). The scale bar represents 50 μm. See also Figure S3.
Since our previous results demonstrated that loss of Dixdc1 or DISC1 leads to reduced Wnt signaling which can be restored by overexpression of the reciprocal gene, we hypothesized that overexpression of either gene would also rescue the other's loss-of-function cellular phenotype in the developing cortex. For these experiments, we first co-expressed full length DISC1 together with Dixdc1 shRNA in the developing mouse cortex. Remarkably, we found that DISC1 overexpression could completely rescue the Dixdc1 shRNA-mediated loss of GFP-positive cells from the VZ/SVZ (Figure 4A). Further analysis revealed that the reduction in BrdU incorporation, increase in cell cycle exit and increased neuronal differentiation due to Dixdc1 shRNA were also completely rescued (Figure 4A–D, Figure S3). These experiments demonstrate that the overexpression of DISC1, which stimulates TCF/LEF activation, is sufficient to restore normal progenitor proliferation after Dixdc1 knockdown.
To test whether directly increasing TCF/LEF signaling rescues the Dixdc1 shRNA-mediated progenitor proliferation defects, we co-expressed Dixdc1 shRNA together with a degradation-resistant β-catenin construct. Importantly, we found that the Dixdc1 shRNA-mediated defects in cortical VZ/SVZ cell positioning, decrease in BrdU incorporation, increase in cell cycle exit and increased neuronal differentiation were all completely restored to control shRNA levels when β-catenin levels were stabilized (Figure 5A–C; Figure S4). However, the loss of cells in the cortical plate after Dixdc1 knockdown was not rescued after stabilizing β-catenin levels, suggesting β-catenin signaling plays no role in neuronal migration. This demonstrates that β-catenin mediates TCF/LEF signaling and neural progenitor proliferation downstream of Dixdc1, similar to our previously reported results regarding DISC1 (Mao et al., 2009).
Figure 5. Dixdc1 and DISC1 regulate progenitor proliferation, but not neuronal migration, via β-catenin signaling.
(A) Cell positioning defects induced by Dixdc1 shRNA are rescued by overexpression of a degradation-resistant β-catenin (stabilized β-catenin). Left, confocal images of GFP-positive cells. Right, graph showing quantification of GFP positive cell positioning in each zone (n=3, *p<0.05, NS=not significant, mean±SEM). The scale bar represents 50 μm. (B) Reduced BrdU incorporation due to Dixdc1 shRNA is rescued by stabilized β-catenin. Left, confocal images of brain sections stained for BrdU and GFP. Arrowheads indicate double positive cells for GFP and BrdU. Right, graph showing quantification of the percentage of double positive cells divided by the total GFP positive cells (n=3, *p<0.05,**p<0.005, NS=not significant, mean±SEM). The scale bar represents 25 μm. (C) Dixdc1 shRNA-induced cell cycle exit defects are rescued by stabilized β-catenin. Left, confocal images of brain sections stained for GFP, BrdU and Ki67. Arrowhead, GFP+BrdU+Ki67−cells; Arrow, GFP+BrdU+Ki67+ cells. Right, graph showing the quantification of the cell cycle index for each condition (n=3, *p<0.05, NS=not significant, mean±SEM). The scale bar represents 25 μm. (D) Increasing Wnt-β-catenin signaling does not rescue neuronal migration deficits due to knockdown of Dixdc1 or DISC1. Left, confocal images of E15 brains that were electroporated with a GFP-encoding plasmid together with either control shRNA, Dixdc1 shRNA with or without DISC1 or pNeuroD-S/A-β-catenin, and brains were analyzed at E19. Right, graphs showing quantification of the percentage of cells in each cortical zone. (n=4, ***p<0.0005, NS=not significant, mean±SEM). The scale bar represents 50 μm. (E) Increasing Wnt-β-catenin signaling does not rescue neuronal migration deficits due to knockdown of DISC1. Left, confocal images of E15 brains that were treated as in (D) but electroporated with either control shRNA, DISC1 shRNA with or without Dixdc1 or pNeuroD-S/A-β-catenin. Right, graph showing quantification of GFP-positive cells in the subdivisions (n=5,***p<0.0005, mean±SEM). The scale bar represents 50 μm. See also Figure S4 and S5.
We also examined whether overexpression of Dixdc1, which stimulates Wnt-TCF/LEF activity, could rescue the neural progenitor proliferation deficits induced by DISC1 shRNA. We co-electroporated embryonic brains with full-length Dixdc1 and DISC1 shRNA, and determined that the reduced neural progenitor proliferation, increased cell cycle exit and increased neuronal differentiation were completely rescued compared to the DISC1 shRNA conditions (Figure 4B–D, Figure S3). Together, these studies demonstrate that the loss-of-function of one gene in vivo leads to the cellular phenotypes observed due to reduced downstream Wnt-TCF/LEF signaling in neural progenitor cells. Furthermore, these deficits can be rescued by overexpression of the reciprocal gene. These data suggest that progenitor proliferation is tightly regulated downstream of the DISC1-Dixdc1 interaction by the levels of β-catenin, which modulate TCF/LEF signaling.
Dixdc1 regulates neuronal migration in the developing cortex
Initial studies of DISC1 reported that it regulates the radial migration of newborn neurons during the later, but not early, stages of cortical development (Kamiya et al., 2005; Mao et al., 2009). To analyze whether Dixdc1 regulates migration, we performed in utero electroporation with Dixdc1 shRNA at a later time point (E15) and analyzed brains at E19, a period of time when neuronal migration is prevalent. We found that knocking down the expression of Dixdc1 resulted in a profound migration defect, where the majority of GFP-positive cells were arrested in the IZ compared to control conditions, where the majority of GFP-positive migrated to the upper cortical plate (Figure S5A). Since knocking down expression of Dixdc1 led to progenitor proliferation defects (Figure 2), we confirmed that the inhibition of migration was not due to disrupted neural differentiation by designing a Dixdc1 shRNA-encoding plasmid under the control of the pNeuroD promoter to restrict Dixdc1 knockdown specifically to neurons (Figure S2D). Using this approach, we confirmed that the knockdown of Dixdc1 in neurons also inhibited neuronal migration (Figure S5A). To determine if the inhibition of neuronal migration was a temporary effect, we electroporated E15 embryonic brains with Dixdc1 shRNA, and analyzed the brains at postnatal day 6 (P6). Interestingly, the downregulation of Dixdc1 expression caused a persistent neuronal migration deficit (Figure S5B), where GFP-positive cells were still arrested in the IZ which, at this time point, is populated with white matter projections. The inhibited radial migration could also be due to a change in cell fate, where Dixdc1 shRNA-expressing cells may acquire the identity characteristic of neurons present in the deeper cortical layers. Therefore, we stained the cortex with different layer specific markers. First, we found that GFP-positive cells lacking Dixdc1 still expressed the mature neuronal marker NeuN, demonstrating that these cells are postmitotic neurons (Figure S5C). Second, we observed no difference in the proportion of GFP-positive cells immunoreactive for the upper layer markers Cux1 and Tbr1, or the deeper layer marker Foxp2, compared to control conditions (Figure S5D–F). Collectively these data strongly argue that Dixdc1 is required for neuronal migration during cortical development.
Dixdc1 and DISC1 do not regulate neuronal migration through GSK3β/β-catenin-mediated signaling
Our data suggest that Dixdc1 and DISC1 interact to regulate neural progenitor proliferation via the modulation of Wnt-GSK3β/β-catenin signaling. Since both Dixdc1 and DISC1 loss-of-function result in inhibited neuronal migration, we hypothesized that they also regulate migration together via Wnt signaling. To assess this, we electroporated E15 embryos with either Dixdc1 or DISC1 shRNA, and asked whether stimulation of TCF/LEF signaling by overexpression of the reciprocal gene would rescue the migration deficit. We found that, in both experimental conditions, overexpression of the reciprocal gene did not restore neuronal migration (Figure 5D,E). We then directly examined whether increasing β-catenin levels would rescue the migration deficits specifically in neurons after DISC1 or Dixdc1 knockdown. We co-electroporated Dixdc1 or DISC1 shRNA together with degradation-resistant β-catenin expressed under the neuronal pNeuroD promoter to restrict its expression to neurons. Using this assay, we did not observe a rescue of radial migration after Dixdc1 or DISC1 downregulation (Figure 5D,E). Together, these experiments suggest that Dixdc1 and DISC1 regulate migration via a pathway independent of Wnt-GSK3β/β-catenin signaling, which is in contrast to their regulation of progenitor proliferation via the Wnt-pathway.
Dixdc1 interacts with the DISC1-binding partner Ndel1
We next examined whether an alternative pathway mediates DISC1/Dixdc1-dependent radial migration. Since DISC1 has been previously reported to bind Ndel1 to regulate neuronal migration (Kamiya et al., 2005), we hypothesized that Dixdc1 might also bind Ndel1 in vivo, and regulate neuronal migration via a tripartite Dixdc1/DISC1/Ndel1 complex. In a tandem-affinity purification screen using HA and Flag epitope-tagged Ndel1 as bait, we discovered that Dixdc1 is indeed a Ndel1 binding partner (Figure 6A). We confirmed this interaction in HEK293 cells, finding that overexpressed GFP-tagged Dixdc1 interacted with endogenous Ndel1 (Figure 6B). Furthermore, we determined that Dixdc1 and Ndel1 could endogenously co-immunoprecipitate one another in E17 brain lysate together with Lis1, a known Ndel1 interacting partner that has important roles in radial migration (Figure 6C). To test if DISC1 was also part of the Dixdc1/Ndel1 complex, we performed co-immunoprecipitation experiments in brain tissue at E18. We determined that Dixdc1 can be co-immunoprecipitated in a complex containing DISC1 and Ndel1 (Figure 6D), suggesting that this tripartite complex exists at later embryonic time points. We then mapped the Ndel1-binding site(s) on Dixdc1 using Flag-tagged Dixdc1 fragments expressed together with GFP-tagged Ndel1 in HEK293 cells. Surprisingly, we found that Ndel1 binds to the same Dixdc1 N-terminal region as DISC1, the Dixdc1 fragment 2 (Figure 6E). This suggests that Dixdc1 may facilitate the interaction between DISC1 and Ndel1.
Figure 6. The DISC1/Dixdc1 complex interacts with Ndel1 in a developmental manner and is regulated by Cdk5-mediated phosphorylation of Dixdc1.
(A) Tandem affinity purification for Ndel1 from mouse E16 brain lysate. The labeled bands are proteins that co-immonoprecipitated with Ndel1. (B) Dixdc1 binds Ndel1. Western blot of overexpressed GFP-tagged Dixdc1 in HEK 293 cells immunoprecipitated for GFP and immunoblotted with Ndel1 and GFP antibodies. (C) Endogenous Dixdc1 binds Ndel1. Western blot showing lysates of E17 brain lysate immunoprecipitated with Ndel1 or Dixdc1 antibodies, and immunoblotted with Dixdc1, Ndel1 or Lis1 antibodies. (D) Dixdc1 binds in a complex containing DISC1 and Ndel1. Western blots demonstrating E18 brain lysates immunoprecipitated and immunoblotted with DISC1, Ndel1 and Dixdc1 antibodies. (E) Left, Schematic showing the flag-tagged Dixdc1 fragments used for domain mapping with GFP-Ndel1. Right, Ndel1 preferentially binds to the N-terminus of Dixdc1, within residues 181–370 (Dixdc1 Frag2). Western blot showing lysates from HEK293 cells transfected with GFP-tagged Ndel1 and each of the flag-tagged Dixdc1 domains, immunoprecipitated with a flag antibody and immunoblotted with a GFP antibody. (F) Cdk5 phosphorylates Dixdc1 at residue serine 250. Top, in vitro kinase assay showing GST-Dixdc1 fragments 2, 3, 4 and Dixdc1 Fragment 2 containing a Ser250Ala mutation were incubated with purified Cdk5/p25 proteins, and phosphorylation was detected by autoradiography. Bottom, graph showing quantification of the autoradiogram demonstrating mutation of Dixdc1 at serine 250 significantly abolishes Cdk5 phosphorylation (n=5, *p<0.05, mean±SEM). (G) Dixdc1 Ser250Ala displays reduced binding to Ndel1 but not DISC1. Left, Western blot showing lysates of HEK 293 cells expressing GFP-tagged DISC1 with flag-tagged Dixdc1 or flag-tagged Dixdc1 Ser250Ala, immunoprecipitated with a flag antibody, and immunoblotted with GFP and Ndel1 antibodies. Right, Graph showing quantification of band intensities for binding of flag-Dixdc1 or flag-Dixdc1 Ser250Ala to GFP-DISC1 or Ndel1. (n=3, *p<0.05, mean±SEM). See also Figure S6.
Cdk5 phosphorylates Dixdc1 to regulate binding to Ndel1
Given that both DISC1 and Ndel1 bind to the same N-terminal region of Dixdc1, the interaction of this complex could be regulated by the modification of Dixdc1 in this region. We hypothesized that Dixdc1 may be phosphorylated, and therefore screened different kinases to determine the potential phosphorylation of the N-terminal region. We discovered that Dixdc1 is phosphorylated by cyclin-dependent kinase 5 (Cdk5) at serine 250, which falls within the region of Dixdc1 to which both DISC1 and Ndel1 bind (Figure 6F). Cdk5 has been previously demonstrated by our lab and others to be important for neuronal migration via its regulation of cortical layering through phosphorylation of a number of substrates (Niethammer et al., 2000; Sasaki et al., 2000; Tanaka et al., 2004; Xie et al., 2006; Xie et al., 2003). Based on our discovery that the serine 250 residue of Dixdc1 is phosphorylated by Cdk5, we hypothesized that this phosphorylation event regulates the formation of the Dixdc1/DISC1/Ndel1 complex. To examine this, we asked whether inhibiting phosphorylation at residue 250 on Dixdc1, by changing serine to alanine (Ser250Ala), altered the binding of Dixdc1 to either DISC1 or Ndel1. We overexpressed flag-tagged Dixdc1 or a Dixdc1 Ser250Ala mutant together with GFP-tagged DISC1 or GFP-tagged Ndel1. Interestingly, we found significantly reduced binding of Dixdc1 Ser250Ala to Ndel1, but not DISC1, compared to wild-type Dixdc1 (Figure 6G). This suggests Cdk5-phosphorylation of Dixdc1 does not affect its binding to DISC1 and therefore does not affect its function in Wnt signaling or neural progenitor proliferation. To confirm this, we overexpressed Dixdc1 Ser250Ala and observed that it significantly potentiated Wnt-TCF/LEF reporter activity and increased neural progenitor proliferation similarly to WT-Dixdc1 (Figure S6A,B). Furthermore, we found that knocking down expression of Ndel1 has no effect on Wnt signaling (Figure S6C), suggesting Ndel1 itself is not in the canonical Wnt pathway. Taken together, these data demonstrate that the binding between Dixdc1 and Ndel1 is regulated by Cdk5-mediated phosphorylation of Dixdc1 at serine 250, which may regulate neuronal migration, but not neurogenesis or Wnt signaling.
Phosphorylation of Dixdc1 and its interaction with DISC1/Ndel1 is essential for radial migration
Our data suggest that both DISC1 and Ndel1 bind to the same N-terminal region of Dixdc1, and that the phosphorylation of Dixdc1 at serine 250 is particularly important for its interaction with Ndel1. Based on these results, we first asked whether this phosphorylation site is essential for neuronal migration by inhibiting the phosphorylation of Dixdc1 at serine 250. To achieve this, we used a 3' UTR Dixdc1 shRNA construct that does not target the coding region, and compared the ability of full length WT-Dixdc1 versus the Dixdc1 Ser250Ala mutant to rescue migration. We observed that, while WT-Dixdc1 was able to significantly restore migration, the Dixdc1 Ser250Ala mutant was not able to rescue migration (Figure 7A). These results demonstrate that the Cdk5-mediated phosphorylation of Dixdc1 at serine 250, which regulates binding to the DISC1-interacting protein Ndel1, is essential for radial migration.
Figure 7. The phosphorylation of Dixdc1 and its interaction with the DISC1/Ndel1 complex is essential for radial migration.
(A) The phosphorylation mutant Dixdc1 Ser250Ala is unable restore Dixdc1 shRNA-induced inhibited migration. E15 brains were electroporated with control shRNA or Dixdc1 3' UTR shRNA together with WT-Dixdc1 or Dixdc1 Ser250Ala mutant and analyzed at E19. Left, confocal images depicting the distribution of GFP-positive cells. Right, graph showing quantification of GFP-positive cells in the subdivisions (n=5, **p<0.005, ***p<0.0005, NS=not significant, mean±SEM). The scale bar represents 50 μm. (B) Overexpression of the Ndel1/DISC1 binding domain of Dixdc1 inhibits migration. E15 brains were electroporated with pCAG or pCAG plasmids encoding Dixdc1 Frag1, Dixdc1 Frag2, Dixdc1 Frag3, Dixdc1 Frag4 or full length Dixdc1 together with a GFP-encoding plasmid and analyzed at E19. Left, confocal images of sections of E19 electroporated brains. Right top, schematic of the different Dixdc1 fragments and where DISC1 and Ndel1 bind on Dixdc1. Right bottom, graph showing quantification of cell positioning of GFP-positive cells in the cortex (n=6, ***p<0.0005, NS=not significant, mean±SEM. See also Figure S7.
We also asked whether the interaction between Dixdc1 and DISC/Ndel1 is required for neuronal migration. We overexpressed HA-tagged Dixdc1 fragment 2 (181–370aa), which inhibited the interaction between Flag-tagged Dixdc1 and GFP-tagged Ndel1 or GFP-tagged DISC1 (Figure S7). We then overexpressed the different peptide fragments of Dixdc1 together with GFP in vivo and observed that the overexpression of Dixdc1 fragment 2 was sufficient to completely inhibit radial migration, producing a phenotype very similar to that seen with the application of Dixdc1 or DISC1 shRNA (Figure 7B). Interestingly, the adjacent Dixdc1 fragment 1, which includes the actin binding domain, also produced a neuronal migration phenotype, suggesting that Dixdc1 may also regulate actin dynamics. However, Dixdc1 fragments 3 and 4 did not disrupt migration, demonstrating that the C-terminus of Dixdc1 is not involved in migration. Taken together, these data strongly suggest that the interaction between Dixdc1 and DISC1/Ndel1 in vivo, which is regulated by Cdk5-mediated phosphorylation of Dixdc1, is essential for the radial migration of neurons during cortical development.
Finally, to determine how Cdk5-mediated phosphorylation of Dixdc1 regulates migration, we analyzed the morphology of GFP-positive cells in vivo. Interestingly, we found that GFP-positive cells either lacking Dixdc1 or expressing Dixdc1 Ser250Ala failed to adopt the bipolar morphology that control shRNA or WT-Dixdc1-expressing cells acquired. Instead, these cells were arrested in the intermediate zone and displayed multiple thin and dystrophic processes (Figure 8A). To further determine if this morphological observation was due to a disrupted cytoskeletal network, we cultured GFP-positive cells from electroporated brains. Here we found that GFP-positive cells lacking Dixdc1 or expressing Dixdc1 Ser250Ala had overall diminished F-actin and α-tubulin staining in the neurites (Figure 8B). Taken together, this data suggests Dixdc1, and its phosphorylation by Cdk5, mediates neuronal migration by regulating cellular morphology through modulation of the actin and microtubule cytoskeleton.
Figure 8. Phosphorylation of Dixdc1 regulates the morphology and structural integrity of neurons.
(A) GFP-positive cells lacking Dixdc1 or expressing Dixdc1 Ser250Ala display abnormal morphology in vivo. Left, images of GFP-positive cells taken from the intermediate zone of electroporated brains in the various conditions indicated. Note that either GFP-positive cells lacking Dixdc1 or expressing the Dixdc1 Ser250Ala mutant show a multipolar morphology, while in control or WT-Dixdc1 conditions, cells have adopted the bipolar morphology that is required for migration into the cortex. Right, graph showing quantification of the percentage of GFP cells that are either multipolar or unipolar/bipolar for the various conditions. (n=250 cells from 4 brains for each condition, **p<0.005, NS=not significant, mean±SEM). The scale bar represents 20 μm. (B) Phosphorylation of Dixdc1 regulates the actin and microtubule cytoskeletal network in neurons. Images of GFP-positive cells cultured from electroporated brains for 72 hours and stained with the indicated markers (phalloidin to label filamentous actin and tubulin for microtubules). Arrows indicate the decreased staining of both markers in GFP-cells lacking Dixdc1 or expressing Dixdc1 Ser250Ala compared to non-electroporated cells. The scale bar represents 20 μm.
Discussion
The mechanisms by which the psychiatric illness gene DISC1 is regulated during cortical development remain unknown. We report here that Dixdc1, a homologue of the Wnt signaling genes Disheveled and Axin, regulates DISC1 during embryonic neural progenitor proliferation and radial migration. We provide evidence that DISC1 and Dixdc1 interact to regulate neural progenitor proliferation by modulating Wnt-GSK3β/β-catenin signaling. We further demonstrate that similar to DISC1, Dixdc1 is essential for neuronal migration. Unexpectedly, we discovered that the Wnt-GSK3β/β-catenin pathway plays no role in mediating Dixdc1/DISC1-dependent migration. Instead, Dixdc1 directly binds to the DISC1-interacting partner Ndel1, and this interaction is regulated by phosphorylation of Dixdc1 by the multifunctional enzyme Cdk5. Lastly, our results demonstrate that the phosphorylation of Dixdc1 and its interaction with Ndel1/DISC1 in vivo are required for the appropriate morphology and migration of newborn neurons. These data strongly demonstrate that Dixdc1 is a key binding partner of DISC1 and integrates it into Wnt-GSK3β/β-catenin-dependent and -independent signaling pathways during embryonic brain development (Figure 9).
Figure 9. Model for Dixdc1 function during cortical development.
Model for Dixdc1 function during cortical development. Top, During early cortical development, Dixdc1 binds DISC1 and together they synergistically regulate neural progenitor proliferation via modulation of Wnt-GSK3β/β-catenin signaling. Bottom, During later embryonic development, Dixdc1 integrates DISC1 into a pathway independent of Wnt-GSK3β/β-catenin signaling. In this neuronal migration pathway, Cdk5 phosphorylates Dixdc1, which regulate the formation of the DISC1/Dixdc1/Ndel1 complex that is essential for neuronal migration.
Regulation of the Wnt-GSK3β/β-catenin pathway by DISC1 and Dixdc1
Our studies suggest that both Dixdc1 and DISC1 interact to regulate Wnt signaling during neural progenitor proliferation, since loss-of-function of each gene leads to inhibited progenitor proliferation. Is one protein, however, more important than the other for this process? It is likely that both genes are equally important, since loss of function of either genes leads to similar deficits in Wnt-reporter activity and neural progenitor proliferation. Furthermore, overexpression of each gene can rescue the loss of the other to the same degree, and one gene does not rescue proliferation better than the other, suggesting there is no epistatic interaction between the genes. Although our studies suggest Dixdc1 regulates β-catenin through modulation of GSK3β, future studies are required to determine whether this is a direct inhibition of GSK3β, or indirectly through DISC1. Given that both Dixdc1 and DISC1 loss of function phenotypes can be rescued by stabilizing β-catenin, this also suggests they interact together to determine the state of neural progenitor proliferation by regulating the overall abundance of β-catenin. Indeed, earlier studies have demonstrated that β-catenin levels in neural progenitor cells dictate whether the cells are to undergo further proliferation or neural differentiation in various populations of progenitor cells (Chenn and Walsh, 2002, 2003; Gulacsi and Anderson, 2008).
Given the interplay between DISC1 and Dixdc1, it is tempting to speculate that their functional interaction in the Wnt pathway may also shed light on the incomplete genotype-phenotype penetrance observed in the Scottish DISC1 family. For example, not all members of the Scottish family that carry the translocation manifest a psychiatric disease (Blackwood et al., 2001). One potential answer to this dilemma may be that an individual with a DISC1 mutation might not display symptoms due to a potential compensatory action of Dixdc1. Since Wnt signaling is a crucial regulator of brain growth, aberrant DISC1 polymorphisms that disrupt Wnt signaling might not be tolerated, and thus upregulation of Dixdc1 might compensate for this. This implies additional disease-associated DISC1 polymorphisms might be difficult to detect due to potential compensatory SNPs in Dixdc1. Therefore, based on our data, one avenue to solve this problem might be to analyze both DISC1 and Dixdc1 to determine if SNPs in Dixdc1 segregate with psychiatric disease and DISC1 SNPs, which has been shown to occur for DISC1 and Ndel1 (Burdick et al., 2008).
The regulation of neuronal migration during development
Our data suggests that Dixdc1 is essential for regulating neuronal migration throughout cortical development. However, the loss of DISC1 function only inhibited migration at later, not earlier, stages of migration. What can account for such a mechanism? One explanation might be that, during early stages, because somal translocation is the major mode of migration for newborn neurons (Nadarajah and Parnavelas, 2002), DISC1, but not Dixdc1 might be dispensable for this process. Furthermore, it is possible that, during early brain development, two different pools of Dixdc1 might exist; one pool could function with DISC1 to regulate progenitor proliferation through Wnt-GSK3β/β-catenin signaling, while another pool of Dixdc1 mediates migration through Ndel1. However, during later stages of migration, Cdk5-mediated phosphorylation of Dixdc1 could function as a switch from neurogenesis to neuronal migration by facilitating the interaction between the DISC1/Dixdc1 complex and Ndel1. This possibility is supported by the expression pattern of the Cdk5 activator, p35, which is primarily expressed within the embryonic cortex in postmitotic newborn neurons as they leave the VZ/SVZ (Delalle et al., 1997). Since the microtubule and actin networks are essential for proper neuronal migration (Jaglin and Chelly, 2009), it is likely that Cdk5-mediated regulation of the Dixdc1/DISC1/Ndel1 complex would promote neuronal migration by influencing downstream pathways that impinge upon the cytoskeleton.
It is also equally possible that modifications of DISC1, such as phosphorylation, may regulate the transition from neurogenesis to radial migration. Our findings demonstrate that both DISC1 and Ndel1 bind to the same region of Dixdc1, a region which is a target of Cdk5 phosphorylation. Since Ndel1 itself is a target of Cdk5 (Niethammer et al., 2000; Sasaki et al., 2000), this suggests Cdk5 might phosphorylate DISC1 directly. Although speculative, this suggests Cdk5 could be a downstream component of Wnt signaling, and that activation of Cdk5 might regulate the DISC1/Dixdc1/Ndel1 complex by influencing its activity and/or localization in newborn migrating neurons.
The implications of DISC1 and Dixdc1 in Wnt signaling and psychiatric disease
As GSK3β and Wnt signaling have been implicated in human psychiatric disease both by human genetic studies, as well as by the mechanisms underlying current psychotropic drugs (Beaulieu et al., 2009; Jope and Roh, 2006), it is a pathway that warrants further investigation. Although our study provides a link between DISC1 and Dixdc1 via the Wnt pathway, future studies need to determine whether other candidate psychiatric risk genes also interact with Wnt signaling. This approach will begin to construct a framework from which we may understand the pathways that are disrupted in psychiatric disease.
In conclusion, our study demonstrates that a novel Wnt signaling pathway gene, Dixdc1, is a critical regulator of DISC1 function during cortical development. This discovery suggests that Dixdc1 and DISC1 are involved in Wnt signaling at many levels in the nervous system, and that mutations in DISC1 likely contribute to disease pathology by disrupting Wnt signaling during neural development and in the adult brain.
Experimental Procedures
Reagents (Antibodies and Plasmids)
All antibodies and plasmids generated and used in this study are described in supplementary experimental procedures.
Cell Culture
Primary cortical neurons were isolated from E17 mouse embryonic brains after electroporation at E15 and cultured as described previously (Sanada and Tsai, 2005). 293T, P19 and HT22 cells were cultured in DMEM medium containing 10 % FBS, L-glutamine, and penicillin/streptomycin.
Animals and In utero electroporation
Swiss Webster pregnant female mice were purchased from Taconic for in utero electroporation experiments as described previously (Sanada and Tsai, 2005). Briefly, E13 or E15 embryonic brains were injected with either control or Dixdc1 shRNA constructs (final concentration 2.4 μg/μl) together with an enhanced GFP (EGFP)-expressing plasmid (pCAGIG-enhanced YFP; final concentration 0.8 μg/μl). For rescue experiments where shRNA constructs were co-injected with either full length Dixdc1 or DISC1 constructs (pCAGIG-Dixdc1 or pCAGIG-DISC1), the concentration of these plasmids was 2 fold higher than the final shRNA plasmid concentration. For all cell cycle exit experiments, E13 electroporated mice were intraperitoneally (i.p.) injected with BrdU (100 mg/kg) at E15, sacrificed and processed 24 hr later.
Immunocytochemistry
Embryonic or postnatal brains were fixed in 4% paraformaldehyde and cryoprotected using 30% sucrose overnight. Brains were cryosectioned on a Leica cryostat. Brain sections were rehydrated in PBS and blocked for 1 hour in PBS (containing 10% Donkey serum with 0.3% Triton-X), followed by incubation with the appropriate antibody overnight at 4 degrees. The next day, slides were washed 3 times with PBS, and incubated with the appropriate secondary antibody for 2 hours at room temperature, washed an additional 3 times in PBS and mounted using Prolong Gold antifade (Invitrogen). For brains injected with Brdu, slides were treated with 4N HCl for 2 hours prior to blocking. In brain sections stained with FoxP2, Cux1 or Tbr1 antibodies, citrate antigen retrieval (Biogenex) was performed prior to the blocking step.
Luciferase Assays
Luciferase assays were performed as described in (Mao et al., 2009) using P19 or HEK293 cells for in vitro assays while in utero electroporation of E13 embryonic brains were used for in vivo assays. See supplementary information for further details.
Biochemistry, Kinase assays, and Tandem Affinity Purification Screen
Immunoprecipation and western blot experiments were performed as described in (Mao et al., 2009). The tandem affinity purification screen was previously described in (Shim et al., 2008). For the Cdk5/Dixdc1 in vitro kinase assay, 1 mM of purified Cdk5 protein was incubated with 5 ng/ml GST-Dixdc1 fragments in kinase buffer (2 mM MOPS [pH 7.4], 0.05 mM ethylenediaminetetraacetic acid [EDTA], 2.5 mM MgAcetate, 100 mM ATP, 10 mCi 32P-ATP, 10 mM MgCl2) for 30 min at 30 degrees °C. Kinase activity was measured with radiography.
Image and Statistical Analysis
All images were acquired using a confocal Zeiss LSM 510 microscope. Images were further analyzed with Adobe Photoshop and ImageJ v1.42q. Statistical analysis was performed with the student's t test. All bar graphs are plotted as mean ±SEM.
Supplementary Material
Acknowledgements
We would like to thank Z. Xie for the S/A-β-catenin and pNeuroD-GFP constructs. We would like to thank Takahiro Soda and Alison Mungenast for critical reading of the manuscript, and all other Tsai lab members. L.-H. T. is an investigator of the Howard Hughes Medical Institute and the director of the neurobiology program at the Stanley Center for Psychiatric Research. KKS is a recipient of the Human Frontiers Science Program Longterm fellowship and an NSERC postdoctoral fellowship. Y.M. is a recipient of the National Alliance for Research on Schizophrenia and Depression Young Investigator Award. This work was partially supported by a National Institutes of Health grant (NS37007) to L.-H.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beaulieu JM, Gainetdinov RR, Caron MG. Akt/GSK3 signaling in the action of psychotropic drugs. Annu Rev Pharmacol Toxicol. 2009;49:327–347. doi: 10.1146/annurev.pharmtox.011008.145634. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Marion S, Rodriguiz RM, Medvedev IO, Sotnikova TD, Ghisi V, Wetsel WC, Lefkowitz RJ, Gainetdinov RR, Caron MG. A beta-arrestin 2 signaling complex mediates lithium action on behavior. Cell. 2008;132:125–136. doi: 10.1016/j.cell.2007.11.041. [DOI] [PubMed] [Google Scholar]
- Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S, Claessens A, Porteous DJ, Millar JK. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem Biophys Res Commun. 2008;377:1091–1096. doi: 10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, Elashvili S, Arai H, Goldman D, Sawa A, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb Cortex. 2003;13:599–606. doi: 10.1093/cercor/13.6.599. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Delalle I, Bhide PG, Caviness VS, Jr., Tsai LH. Temporal and spatial patterns of expression of p35, a regulatory subunit of cyclin-dependent kinase 5, in the nervous system of the mouse. J Neurocytol. 1997;26:283–296. doi: 10.1023/a:1018500617374. [DOI] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Asai N, Namba T, Wang Y, Kato T, Tanaka M, Tatsumi H, Taya S, Tsuboi D, Kuroda K, et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Gulacsi AA, Anderson SA. Beta-catenin-mediated Wnt signaling regulates neurogenesis in the ventral telencephalon. Nat Neurosci. 2008;11:1383–1391. doi: 10.1038/nn.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood AJ. Lithium and bipolar mood disorder: the inositol-depletion hypothesis revisited. Mol Psychiatry. 2005;10:117–126. doi: 10.1038/sj.mp.4001618. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, Kong S, Wu D, Xue R, Andrade M, Tankou S, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A. 2007;104:14501–14506. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K, Chen J, Taya S, Li W, Millar JK, Xu Y, Clapcote SJ, Hookway C, Morita M, Kamiya A, et al. Evidence that many of the DISC1 isoforms in C57BL/6J mice are also expressed in 129S6/SvEv mice. Mol Psychiatry. 2007;12:897–899. doi: 10.1038/sj.mp.4002024. [DOI] [PubMed] [Google Scholar]
- Jaglin XH, Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 2009;25:555–566. doi: 10.1016/j.tig.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Jope RS, Roh MS. Glycogen synthase kinase-3 (GSK3) in psychiatric diseases and therapeutic interventions. Curr Drug Targets. 2006;7:1421–1434. doi: 10.2174/1389450110607011421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungerius BJ, Hoogendoorn ML, Bakker SC, Van't Slot R, Bardoel AF, Ophoff RA, Wijmenga C, Kahn RS, Sinke RJ. An association screen of myelin-related genes implicates the chromosome 22q11 PIK4CA gene in schizophrenia. Mol Psychiatry. 2008;13:1060–1068. doi: 10.1038/sj.mp.4002080. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kamiya A, Tomoda T, Chang J, Takaki M, Zhan C, Morita M, Cascio MB, Elashvili S, Koizumi H, Takanezawa Y, et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum Mol Genet. 2006;15:3313–3323. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, Kang E, Song H, Ming GL. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H, Arguello PA, Kvajo M, Karayiorgou M, Gogos JA. Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc Natl Acad Sci U S A. 2006;103:3693–3697. doi: 10.1073/pnas.0511189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvajo M, McKellar H, Arguello PA, Drew LJ, Moore H, MacDermott AB, Karayiorgou M, Gogos JA. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci U S A. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhou Y, Jentsch JD, Brown RA, Tian X, Ehninger D, Hennah W, Peltonen L, Lonnqvist J, Huttunen MO, et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci U S A. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie S, Millar JK, Porteous DJ. Role of DISC1 in neural development and schizophrenia. Curr Opin Neurobiol. 2007;17:95–102. doi: 10.1016/j.conb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Murdoch H, Mackie S, Collins DM, Hill EV, Bolger GB, Klussmann E, Porteous DJ, Millar JK, Houslay MD. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci. 2007;27:9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nat Rev Neurosci. 2002;3:423–432. doi: 10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Niwa M, Kamiya A, Murai R, Kubo K, Gruber AJ, Tomita K, Lu L, Tomisato S, Jaaro-Peled H, Seshadri S, et al. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 65:480–489. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnikov MV, Ayhan Y, Nikolskaia O, Xu Y, Ovanesov MV, Huang H, Mori S, Moran TH, Ross CA. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–186. 115. doi: 10.1038/sj.mp.4002079. [DOI] [PubMed] [Google Scholar]
- Pletnikov MV, Xu Y, Ovanesov MV, Kamiya A, Sawa A, Ross CA. PC12 cell model of inducible expression of mutant DISC1: new evidence for a dominant-negative mechanism of abnormal neuronal differentiation. Neurosci Res. 2007;58:234–244. doi: 10.1016/j.neures.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Shim SY, Samuels BA, Wang J, Neumayer G, Belzil C, Ayala R, Shi Y, Tsai LH, Nguyen MD. Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J Biol Chem. 2008;283:12232–12240. doi: 10.1074/jbc.M710200200. [DOI] [PubMed] [Google Scholar]
- Shiomi K, Uchida H, Keino-Masu K, Masu M. Ccd1, a novel protein with a DIX domain, is a positive regulator in the Wnt signaling during zebrafish neural patterning. Curr Biol. 2003;13:73–77. doi: 10.1016/s0960-9822(02)01398-2. [DOI] [PubMed] [Google Scholar]
- Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L, Williams R, Hawkins P. Phosphoinositide 3-kinases as drug targets in cancer. Curr Opin Pharmacol. 2005;5:357–365. doi: 10.1016/j.coph.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Tseng HC, Kulkarni AB, Tsai LH, Gleeson JG. Cdk5 phosphorylation of doublecortin ser297 regulates its effect on neuronal migration. Neuron. 2004;41:215–227. doi: 10.1016/s0896-6273(03)00852-3. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Chow EW, Ophoff RA, van Engeland H, Beemer FA, Kahn RS, Sinke RJ, Bassett AS. Association of the PIK4CA schizophrenia-susceptibility gene in adults with the 22q11.2 deletion syndrome. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:430–433. doi: 10.1002/ajmg.b.30827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zheng L, Zeng Z, Zhou G, Chien J, Qian C, Vasmatzis G, Shridhar V, Chen L, Liu W. DIXDC1 isoform, l-DIXDC1, is a novel filamentous actin-binding protein. Biochem Biophys Res Commun. 2006;347:22–30. doi: 10.1016/j.bbrc.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Xie Z, Samuels BA, Tsai LH. Cyclin-dependent kinase 5 permits efficient cytoskeletal remodeling--a hypothesis on neuronal migration. Cereb Cortex. 2006;16(Suppl 1):i64–68. doi: 10.1093/cercor/bhj170. [DOI] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Ring C, Cheung R, Pevny L, Anton ES. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron. 2007;54:429–445. doi: 10.1016/j.neuron.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.