Abstract
Although biochemotherapy appears to be a promising treatment for metastatic melanoma, its impact remains unpredictable. Microsatellite markers for loss of heterozygosity (LOH) appear to have prognostic significance when identified in primary tumors and serum and/or plasma from cancer patients. However, their association with response to systemic therapy has yet to be assessed. To determine whether microsatellite markers are associated with response to therapy, serum from 41 patients with metastatic melanoma, drawn before the initiation of biochemotherapy, was analyzed for LOH with nine microsatellite markers. During a median follow-up of 13 months, the overall response rate for these 41 patients was 56%, including 13 (32%) complete responses and 10 (24%) partial responses. LOH was detected in sera from 12 (29%) of the 41 patients. The response rate of these 12 patients was 17% (95% confidence interval [CI] = 5% to 45%), whereas that of the 29 patients without LOH was 72% (95% CI = 54% to 85%) (P = .001). All statistical tests were two-sided. The presence of LOH was statistically significant and independently associated with disease progression (multivariable analysis, P = .003). Circulating tumor DNA markers may be useful in assessing prognosis for advanced melanoma patients and their response to biochemotherapy.
Melanoma is characterized by increased genetic instability with advancing stage (1–3). One of the most frequent genetic alterations in melanoma is loss of heterozygosity (LOH) that occurs nonrandomly at certain chromosome loci, suggesting the involvement of putative tumor suppressor and regulatory genes (4–10). Acquisition of genetic alterations is an ongoing phenomenon that occurs throughout disease progression (11–13). Consequently, identification of molecular biomarkers associated with LOH may aid in treatment planning and provide more specific prognostic information.
Melanoma patients with systemic metastasis have an average median survival of 6–9 months (14–17). Such patients treated with biochemotherapy regimens have increased response rates compared with those for patients treated with chemotherapy and biologic response modifiers alone (18–22), which require inpatient hospitalization and are associated with considerable toxicity. We found (23) that use of a concurrent biochemotherapy regimen modified by administering interleukin 2 in a decrescendo fashion had a response rate equivalent to that of chemotherapy and biologic response modifiers alone but with less toxicity. However, the impact on overall survival was modest, and individual patient responses were unpredictable (24,25). Current staging criteria from the American Joint Committee on Cancer (AJCC) (26) do not predict treatment outcomes. To date, there are no established prognostic factors to assess or predict treatment response in patients with advanced-stage melanoma.
Circulating nucleic acids in the serum and/or plasma of cancer patients have allelic losses similar to those found in the primary tumor, suggesting the utility of such nucleic acids as potential markers for disease surveillance (27–32). The purpose of this study is to determine whether microsatellite markers for LOH detected in serum from patients with metastatic melanoma receiving a modified concurrent biochemotherapy regimen were associated with a patient’s response to treatment.
Blood (10 mL) was collected from 41 patients with American Joint Committee on Cancer (AJCC) stage IV melanoma at enrollment into our program of modified concurrent biochemotherapy between July 1995 and September 1997 at the John Wayne Cancer Institute after providing written informed consent (23). The collection, storage, and processing of serum and lymphocyte DNA were as previously described (33). Pre-treatment evaluation, protocol criteria, biochemotherapy (consisting of dacarbazine, cisplatin, vinblastine, decrescendo interleukin 2, interferon alfa-2b, and tamoxifen) dosing and schedule, and patient monitoring and follow-up have been described previously (23).
Treatment response was evaluated only in patients completing at least two cycles of biochemotherapy, as described elsewhere (23). Patients were categorized as having a complete or partial response or stable or progressive disease, as defined previously (23). The duration of response was measured from the first day of treatment to the first evidence of progressive disease, last follow-up date, or death from any cause. Survival was measured from the first day of treatment to death or the date of the last follow-up.
LOH analysis was performed in a blinded fashion. The following nine microsatellite markers, on seven different chromosomes, that show frequent LOH in melanoma tumors were selected: D1S228 at 1p36, D3S1293 at 3p– 3p24.2, D6S264 at 6q25–27, D9S157 and D9S161 at 9p21–23, D10S212 at 10q24–26, D11S2000 at 11q23–24, and D14S51 and D14S62 at 14q31–32. Primer set acquisition, labeling, polymerase chain reaction amplification, and the electrophoresis and analysis of products are as previously described (33). LOH was scored when the intensity of the band for an allele from serum was more than 50% lower than that of the corresponding band from match-paired lymphocytes.
Chi-square (for categorical variables) and t (for numerical variables) tests were used to compare characteristics of patients who did and did not respond to biochemotherapy and characteristics of patients with and without LOH. A logistic regression model was developed to investigate whether the biochemotherapy response was associated with the following covariates: sex, age, Eastern Cooperative Oncology Group (ECOG) performance status, site of metastasis, number of metastatic sites, level of lactate dehydrogenase, prior therapy, and LOH status. A stepwise procedure was used to identify statistically significant covariates. The survival distribution was estimated with the Kaplan–Meier method. The log-rank test was used to compare the distributions of progression-free survival and overall survival between patients with and without LOH. The Cox proportional hazards model was used to study the association of LOH with survival, adjusting for the other clinical factors. The covariates listed above were included in the original model, and the stepwise procedure was also applied for final model selection. Residual plots (Cox–Snell, Martingale) were used for checking the assumption of the Cox proportional hazards model. Statistical analysis was performed with SAS software (SAS Institute, Cary, NC). All statistical tests were two-sided.
LOH was identified in serum from 12 (29%) of the 41 patients: eight (20%) patients for one marker, one (2%) for two markers, and three (7%) for three markers. LOH was not detected in serum samples for 29 (71%) patients or for 40 healthy donors. LOH was most common at microsatellite marker D9S157, occurring in four (12%) of 33 patients’ serum samples informative for the tested marker, followed by D3S1293 (11%), D14S51 (9%), D6S264 (8%), D11S2000 (5%), D14S62 (3%), D1S228 (3%), D9S161 (3%), and D10S212 (0%).
Patients were separated into two groups (23 responders and 18 nonresponders) by their clinical and radiographic responses to biochemotherapy. Responders had a complete or partial response, and nonresponders had stable or progressive disease (Table 1). The overall response rate was 56%, including complete responses in 13 (32%) of the 41 patients and partial responses in 10 (24%) of the 41 patients. Responders completed a mean of six cycles of biochemotherapy (range = 4–8 cycles); nonresponders, those patients demonstrating stable or progressive disease during treatment, were discontinued from the study after completing a mean of three cycles (range = 2–6 cycles). At the start of biochemotherapy, LOH for any one marker was identified in serum from only two (9%) of 23 responders: one with LOH for D3S1293 and the other with LOH for D6S264. Both patients had a complete response to biochemotherapy. In contrast, LOH was detected in sera from 10 (56%) of 18 nonresponders: six (33%) had LOH for one marker, one (6%) had LOH for two markers, and three (17%) had LOH for three markers. LOH was most frequently detected in informative nonresponders for D9S157 (27%), followed by D14S51 (25%), D3S1293 (17%), D6S264 (13%), D11S2000 (12%), D9S161 (6%), D14S62 (6%), and D1S228 (6%).
Table 1.
Patient demographics*
| Factor | Nonresponders (n = 18) |
Responders (n = 23) |
P value† |
|---|---|---|---|
| Sex, No. | |||
| Female | 6 | 4 | .289 |
| Male | 12 | 19 | |
| Age, y‡ | 41.6 ± 10.6 | 47.0 ± 12.1 | .140 |
| Age group, No. | |||
| ≤50 y | 15 | 14 | .117 |
| >50 y | 3 | 9 | |
| LOH group, No. | |||
| Absent | 8 | 21 | .001 |
| Present | 10 | 2 | |
| ECOG performance status, No. | |||
| 0–1 | 5 | 14 | .035 |
| 2 | 13 | 9 | |
| Metastasis to soft tissue/lymph nodes/lung only, No. | |||
| No | 16 | 14 | .075 |
| Yes | 2 | 9 | |
| Lactate dehydrogenase >190 U/L, No. | |||
| No | 5 | 13 | .065§ |
| Yes | 12 | 9 | |
| Prior chemotherapy, No. | |||
| No | 15 | 23 | .077 |
| Yes | 3 | 0 | |
| No. of disease sites | |||
| 1–2 | 9 | 15 | .326 |
| ≥3 | 9 | 8 |
LOH = loss of heterozygosity; ECOG = Eastern Cooperative Oncology Group.
Two-sided chi-square test was used.
Data are the mean ± standard deviation.
Data are missing for two patients.
Univariate analysis detected a statistically significant association between nonresponders and the presence of LOH. The response rate of the 12 patients with LOH was 17% (95% CI = 5% to 45%), whereas that among the 29 patients without LOH was 72% (95% CI = 54% to 85%; P = .001, χ2 test). Failure to respond to biochemotherapy was associated with increased numbers of serum LOH markers (P = .002, Wilcoxon rank sum test). The only other factor associated with biochemotherapy response was ECOG performance status (P = .035, χ2 test). A logistic regression multivariable analysis was performed to investigate whether biochemotherapy response was associated with various covariates. The presence of LOH was the only factor statistically significantly associated with response to biochemotherapy (estimated odds ratio = 0.08, 95% CI = 0.01 to 0.43; P = .003).
After a median follow-up of 13 months, 32 of the 33 patients whose disease progressed had expired; at time of publication, nine remaining patients were still alive, one with stable disease and eight with no clinical evidence of disease after additional biochemotherapy and/or surgery. The median time to progression for nonresponders was 2.3 months and that for responders was 10.3 months (P<.001, log-rank test), and median overall survival was 4.7 months for nonresponders and 41.6 months for responders (P<.001, log-rank test). Patients with LOH in serum had statistically significantly decreased median progression-free survival (3 months versus 9.5 months; P<.001) and overall survival (6.3 months versus 15 months; P = .022) compared with patients without LOH (Fig. 1). Univariate analysis also showed that the site of metastasis, level of lactate dehydrogenase, and history of prior therapy were associated with disease progression and/or recurrence and with overall survival from melanoma. Multivariable analysis showed that an LOH status (risk ratio [RR] =3.87, 95% CI = 1.71 to 8.77; P<.001, Wald test) and a lactate dehydrogenase level of more than 190 U/L (RR = 3.76, 95% CI = 1.56 to 9.02; P<.003, Wald test) were the only statistically significant predictors of disease recurrence. The site of metastasis (soft tissue, lymph nodes, and lung versus other organs; RR = 0.32, 95% CI = 0.12 to 0.85) and a prior history of therapy (RR = 6.71, 95% CI = 1.67 to 27.00) were also statistically significantly associated with the overall survival of patients with advanced-stage melanoma treated with biochemotherapy.
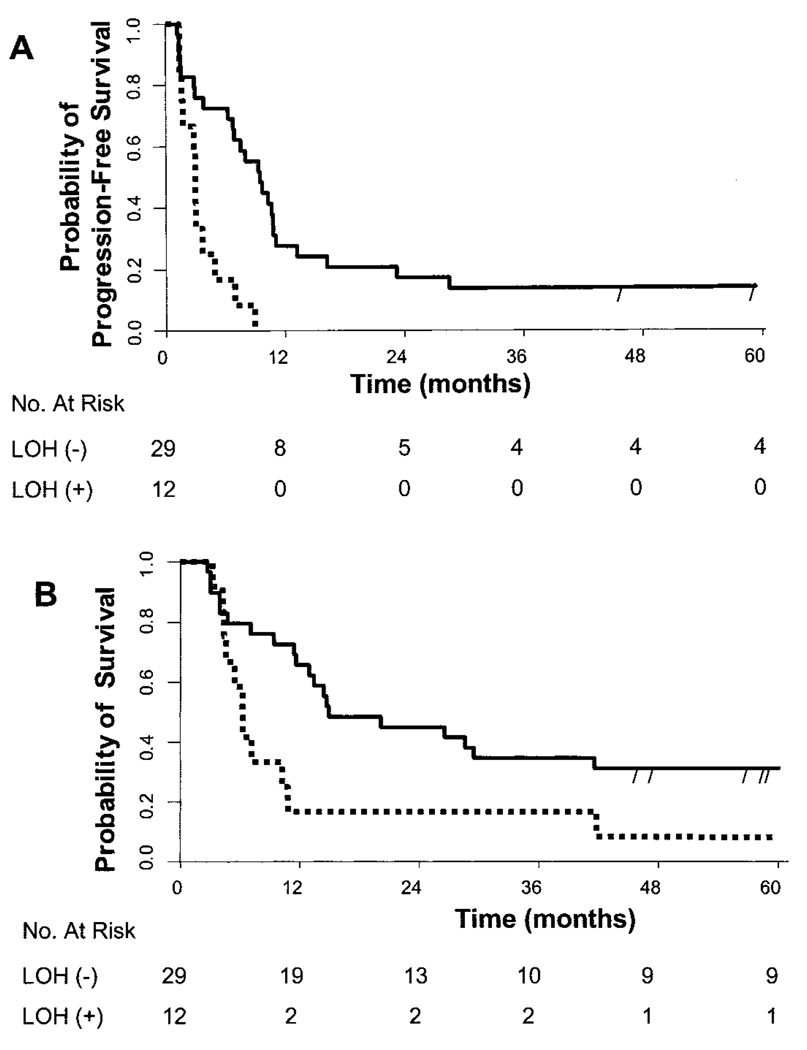
Fig. 1.
Kaplan–Meier analysis of progression-free (A) and overall (B) survival for patients with distant metastatic melanoma according to serum loss of heterozygosity (LOH) status at the initiation of concurrent biochemotherapy. The estimated progression-free survival rate at 6 months for LOH-negative patients [LOH (−)] was 0.72 (95% confidence interval [CI] = 0.56 to 0.87) and for LOH-positive patients [LOH (+)] was 0.17 (95% CI = 0.0 to 0.38). The estimated overall survival rate at 6 and 12 months for LOH-negative patients was 0.79 (95% CI = 0.64 to 0.94) and 0.66 (95% CI = 0.48 to 0.83) and for LOH-positive patients was 0.58 (95% CI = 0.30 to 0.86) and 0.17 (95% CI = 0.0 to 0.38), respectively. Solid line = patients without serum LOH (−); dotted line = patients with serum LOH (+); vertical dashes = censored patients.
Allelic loss on chromosome 9p occurs in lesions of all thicknesses and is one of the most common genetic aberrations identified in melanoma (5,10,34–36). We found that the most frequent serum microsatellite LOH marker in our study corresponded to this chromosomal location. We have previously hypothesized that microsatellite markers in melanoma patients’ plasma had prognostic significance (33); however, the patients analyzed in that study had different disease stages and treatments. Because higher concentrations of genomic DNA can be identified in serum than in plasma (37), LOH may be relatively underestimated, as seen in this study.
Analysis of primary and advanced-stage tumor tissues provides information relative to the time of the procedure and is not practical for serially assessing disease. At present, additional methods are needed to further assess individual prognosis among patients within the same AJCC staging group and to identify potential responders to treatment protocols as early as possible. Molecular markers of metastatic disease obtained in a minimally invasive manner, such as through a blood test, would allow the disease to be followed as it progressed.
The presence of LOH in the serum of patients with advanced metastatic melanoma was associated with a poorer response to induction biochemotherapy and, independently, with patient outcome. To our knowledge, this is the first study to evaluate the association between circulating DNA tumor markers and a patient’s response to systemic therapy for a solid tumor. Genetic alterations found, such as circulating tumor microsatellites, serving as immediate determinants of disease progression and response to treatment will, no doubt, permit a more efficient use of our health care resources.
Acknowledgments
Editor’s note: S. J. O’Day has a research grant from and is a member of the Speaker’s Bureau for Chiron; he also has research grants from Immunex, Berlex, and Schering.
Supported in part by Public Health Service grants P01CA29605 and R21CA100314 (to D. S. B. Hoon) from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, and the Martin H. Weil Fund, Raymond Kathe Fund, and the Roy E. Coats Research Laboratories of the John Wayne Cancer Institute.
REFERENCES
- 1.Parmiter AH, Nowell PC. Cytogenetics of melanocytic tumors. J Invest Dermatol. 1993;100:254S–258S. [PubMed] [Google Scholar]
- 2.Albino AP, Fountain JW. Molecular genetics of human malignant melanoma. Cancer Treat Res. 1993;65:201–255. doi: 10.1007/978-1-4615-3080-0_8. [DOI] [PubMed] [Google Scholar]
- 3.Fountain JW, Bale SJ, Housman DE, Dracopoli NC. Genetics of melanoma. Cancer Surv. 1990;9:645–671. [PubMed] [Google Scholar]
- 4.Goldberg EK, Glendening JM, Karanjawala Z, Sridhar A, Walker GJ, Hayward NK, et al. Localization of multiple melanoma tumor-suppressor genes on chromosome 11 by use of homozygosity mapping-of-deletions analysis. Am J Hum Genet. 2000;67:417–431. doi: 10.1086/302999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Healy E, Rehman I, Angus B, Rees JL. Loss of heterozygosity in sporadic primary cutaneous melanoma. Genes Chromosomes Cancer. 1995;12:152–156. doi: 10.1002/gcc.2870120211. [DOI] [PubMed] [Google Scholar]
- 6.Healy E, Belgaid C, Takata M, Vahlquist A, Rehman I, Rigby H, et al. Allelotypes of primary cutaneous melanoma and benign melanocytic nevi. Cancer Res. 1996;56:589–593. [PubMed] [Google Scholar]
- 7.Gonzalgo ML, Bender CM, You EH, Glendening JM, Flores JF, Walker GJ, et al. Low frequency of p16/CDKN2A methylation in sporadic melanoma: comparative approaches for methylation analysis of primary tumors. Cancer Res. 1997;57:5336–5347. [PubMed] [Google Scholar]
- 8.Flores JF, Walker GJ, Glendening JM, Haluska FG, Castresana JS, Rubio MP, et al. Loss of the p16INK4a and p15INK4b genes, as well as neighboring 9p21 markers, in sporadic melanoma. Cancer Res. 1996;56:5023–5032. [PubMed] [Google Scholar]
- 9.Jimenez P, Canton J, Concha A, Cabrera T, Fernandez M, Real LM, et al. Microsatellite instability analysis in tumors with different mechanisms for total loss of HLA expression. Cancer Immunol Immunother. 2000;48:684–690. doi: 10.1007/s002620050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmieri G, Cossu A, Ascierto PA, Botti G, Strazzullo M, Lissia A, et al. Definition of the role of chromosome 9p21 in sporadic melanoma through genetic analysis of primary tumours and their metastases. The Melanoma Cooperative Group. Br J Cancer. 2000;83:1707–1714. doi: 10.1054/bjoc.2000.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbst RA, Mommert S, Casper U, Podewski EK, Kiehl P, Kapp A, et al. 11q23 allelic loss is associated with regional lymph node metastasis in melanoma. Clin Cancer Res. 2000;6:3222–3227. [PubMed] [Google Scholar]
- 12.Herbst RA, Weiss J, Ehnis A, Cavenee WK, Arden KC. Loss of heterozygosity for 10q22-10qter in malignant melanoma progression. Cancer Res. 1994;54:3111–3114. [PubMed] [Google Scholar]
- 13.Nakayama T, Taback B, Turner R, Morton DL, Hoon DS. Molecular clonality of intransit melanoma metastasis. Am J Pathol. 2001;158:1371–1378. doi: 10.1016/S0002-9440(10)64088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth A, Wanek L, Morton D. Prognostic factors in 1,521 melanoma patients with distant metastases. J Am Coll Surg. 1995;181:193–201. [PubMed] [Google Scholar]
- 15.Balch CM, Soong SJ, Murad TM, Smith JW, Maddox WA, Durant JR. A multifactorial analysis of melanoma. IV. Prognostic factors in 200 melanoma patients with distant metastases (stage III) J Clin Oncol. 1983;1:126–134. doi: 10.1200/JCO.1983.1.2.126. [DOI] [PubMed] [Google Scholar]
- 16.Roses DF, Harris MN, Gumport SL, Michelassi F, Coffey JA, Dubin N. Regional lymph node dissection for malignant melanoma of the extremities. Surgery. 1981;89:654–659. [PubMed] [Google Scholar]
- 17.Roses DF, Karp NS, Oratz R, Dubin N, Harris MN, Speyer J, et al. Survival with regional and distant metastases from cutaneous malignant melanoma. Surg Gynecol Obstet. 1991;172:262–268. [PubMed] [Google Scholar]
- 18.Anderson CM, Buzaid AC, Legha SS. Systemic treatments for advanced cutaneous melanoma. Oncology (Huntingt) 1995;9:1149–1158. [PubMed] [Google Scholar]
- 19.Lakhani S, Selby P, Bliss JM, Perren TJ, Gore ME, McElwain TJ. Chemotherapy for malignant melanoma: combinations and high doses produce more responses without survival benefit. Br J Cancer. 1990;61:330–334. doi: 10.1038/bjc.1990.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legha SS, Buzaid AC. Role of recombinant interleukin-2 in combination with interferon-alfa and chemotherapy in the treatment of advanced melanoma. Semin Oncol. 1993;20:27–32. [PubMed] [Google Scholar]
- 21.Buzaid AC, Legha SS. Combination of chemotherapy with interleukin-2 and interferon-alfa for the treatment of advanced melanoma. Semin Oncol. 1994;21(6) Suppl 14:23–28. [PubMed] [Google Scholar]
- 22.O’Day SJ, Boasberg PD, Piro L, Kristedja TS, Wang HJ, Martin M, et al. Maintenance biotherapy for metastatic melanoma with interleukin-2 and granulocyte macrophage-colony stimulating factor improves survival for patients responding to induction concurrent biochemotherapy. Clin Cancer Res. 2002;8:2775–2781. [PubMed] [Google Scholar]
- 23.O’Day SJ, Gammon G, Boasberg PD, Martin MA, Kristedja TS, Guo M, et al. Advantages of concurrent biochemotherapy modified by decrescendo interleukin-2, granulocyte colony-stimulating factor, and tamoxifen for patients with metastatic melanoma. J Clin Oncol. 1999;17:2752–2761. doi: 10.1200/JCO.1999.17.9.2752. [DOI] [PubMed] [Google Scholar]
- 24.O’Day SJ, Kim CJ, Reintgen DS. Metastatic melanoma: chemotherapy to biochemotherapy. Cancer Control. 2002;9:31–38. doi: 10.1177/107327480200900105. [DOI] [PubMed] [Google Scholar]
- 25.Buzaid AC, Atkins M. Practical guidelines for the management of biochemotherapyrelated toxicity in melanoma. Clin Cancer Res. 2001;7:2611–2619. [PubMed] [Google Scholar]
- 26.American Joint Committee on Cancer. Cancer staging manual. 6th ed. New York (NY): Springer; 2002. pp. 209–220. [Google Scholar]
- 27.Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- 28.Chen XQ, Stroun M, Magnenat JL, Nicod LP, Kurt AM, Lyautey J, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2:1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- 29.Taback B, Giuliano AE, Hansen NM, Hoon DS. Microsatellite alterations detected in the serum of early stage breast cancer patients. Ann N Y Acad Sci. 2001;945:22–30. doi: 10.1111/j.1749-6632.2001.tb03860.x. [DOI] [PubMed] [Google Scholar]
- 30.Silva JM, Dominguez G, Garcia JM, Gonzalez R, Villanueva MJ, Navarro F, et al. Presence of tumor DNA in plasma of breast cancer patients: clinicopathological correlations. Cancer Res. 1999;59:3251–3256. [PubMed] [Google Scholar]
- 31.Fujiwara Y, Chi DD, Wang H, Keleman P, Morton DL, Turner R, et al. Plasma DNA microsatellites as tumor-specific markers and indicators of tumor progression in melanoma patients. Cancer Res. 1999;59:1567–1571. [PubMed] [Google Scholar]
- 32.Chan AT, Lo YM, Zee B, Chan LY, Ma BB, Leung SF, et al. Plasma Epstein-Barr virus DNA and residual disease after radiotherapy for undifferentiated nasopharyngeal carcinoma. J Natl Cancer Inst. 2002;94:1614–1619. doi: 10.1093/jnci/94.21.1614. [DOI] [PubMed] [Google Scholar]
- 33.Taback B, Fujiwara Y, Wang HJ, Foshag LJ, Morton DL, Hoon DS. Prognostic significance of circulating microsatellite markers in the plasma of melanoma patients. Cancer Res. 2001;61:5723–5726. [PubMed] [Google Scholar]
- 34.Holland EA, Beaton SC, Edwards BG, Kefford RF, Mann GJ. Loss of heterozygosity and homozygosity deletions on 9p21-p22 in melanoma. Oncogene. 1994;9:1361–1365. [PubMed] [Google Scholar]
- 35.Puig S, Ruiz A, Lazaro C, Castel T, Lynch M, Palou J, et al. Chromosome 9p deletions in cutaneous malignant melanoma tumors: the minimal deleted region involves markers outside the p16 (CDKN2) gene. Am J Hum Genet. 1995;57:395–402. [PMC free article] [PubMed] [Google Scholar]
- 36.Pollock PM, Welch J, Hayward NK. Evidence for three tumor suppressor loci on chromosome 9p involved in melanoma development. Cancer Res. 2001;61:1154–1161. [PubMed] [Google Scholar]
- 37.Lee TH, Montalvo L, Chrebtow V, Busch MP. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001;41:276–282. doi: 10.1046/j.1537-2995.2001.41020276.x. [DOI] [PubMed] [Google Scholar]