Abstract
The 26S proteasome is the end point of the ubiquitin-proteasome pathway and degrades ubiquitylated substrates. It is composed of the 20S core particle (CP), where degradation occurs, and the 19S regulatory particle (RP), which ensures substrate specificity of degradation. Whereas the CP is resolved to atomic resolution, the architecture of the RP is largely unknown. We provide a comprehensive analysis of the current structural knowledge on the RP, including structures of the RP subunits, physical protein-protein interactions, and cryoelectron microscopy data. These data allowed us to compute an atomic model for the CP-AAA-ATPase subcomplex. In addition to this atomic model, further subunits can be mapped approximately, which lets us hypothesize on the substrate path during its degradation.
The ubiquitin-proteasome pathway is the major route used by eukaryotic cells for the disposal of misfolded or damaged proteins and for controlling the lifespan of proteins (1–3). As a consequence, the ubiquitin-proteasome pathway regulates a plethora of fundamental cellular processes, such as protein quality control, DNA repair, and signal transduction (4). The 26S proteasome is a molecular machine of ∼2.5 MDa that targets polyubiquitylated proteins. It comprises two subcomplexes, the 20S core particle (CP)1 and one or two copies of asymmetric 19S regulatory particles (RPs), which bind to the end(s) of the barrel-shaped CP.
The active sites of the proteasome are located in the CP cavity where proteolytic cleavage of substrates takes place. Electron microscopy (EM) and x-ray crystallography have revealed that the CP is a cylinder consisting of four concentrically stacked rings (5–7): two identical “α”-rings, each assembled of seven homologous proteins, form the outer rings, and two identical “β”-rings, also assembled of seven homologs, form the two inner rings. Proteolysis is confined to the cavity formed by the β-rings, a nanocompartment sequestered from the cytosol.
The RPs regulate substrate degradation by (i) binding polyubiquitylated substrates, (ii) subsequently deubiquitylating them, (iii) substrate unfolding, and (iv) opening the “gate” to the CP (8). The RPs consists of six AAA-ATPase subunits and at least 13 non-ATPase subunits. In contrast to the CP, the architecture of the RP subunits remains largely unknown. The problems that hamper structural characterization of the RP are manifold. It has proven difficult to obtain homogeneous, concentrated preparations of 26S proteasomes or RPs because the RP tends to disassociate into heterogeneous subcomplexes during purification and concentration. Moreover, many of the RP subunits likely exhibit a significant degree of structural variability. As a consequence, x-ray crystallographic analysis of the entire RP has not been accomplished to date, and only a few subunit fragments have been amenable to high resolution structure determination.
For cryo-EM and protein-protein interaction experiments, the requirements for sample homogeneity are less stringent. Recently, the Drosophila melanogaster 26S proteasome was resolved to ∼20 Å (9). Various proteomics approaches have led to proposals for topological maps of the RP (10–13). The resolution of protein-protein interaction data typically corresponds to the diameters of the proteins or domains found to interact, which are typically far beyond 20 Å. Because of the limited resolution, neither cryo-EM maps nor protein-protein interaction networks are by themselves sufficient to determine the RP architecture (i.e. the localization of the RP subunits in the complex).
The integration of atomic models, cryo-EM maps, and protein-protein interaction data is currently the most promising approach to resolve the architecture of the 26S proteasome (14–18). Here, we provide a comprehensive analysis of the current structural knowledge on the RP, including structures of RP subunits, physical protein-protein interactions, and cryo-EM data. Based on these data, we provide a model for the CP-AAA-ATPase subcomplex. Finally, we outline a path toward resolving the architecture of the 26S proteasome by an integrative structure determination approach, which in turn will provide a basis for a mechanistic understanding.
SUBUNITS OF THE REGULATORY PARTICLE
The RP comprises at least 19 different subunits. In addition to six AAA-ATPases (regulatory particle triple A-ATPases Rpt1–6), the proteasome contains 13 integral regulatory particle non-ATPases, Rpn1–15 (19); the proteins Rpn4 and Rpn14 were erroneously notated as integral subunits but later turned out to be a proteasomal transcription factor and an assembly chaperone, respectively (20–22). All integral subunits are conserved from Saccharomyces cerevisiae to mammals. In addition, two ubiquitin C-terminal hydrolases (UCHs), Ubp6 and UCH37, are commonly found in preparations of purified 26S proteasomes (23, 24).
AAA-ATPases
AAA-ATPases possess a 250–300-residue ATP-binding domain (AAA domain) and usually assemble into hexameric rings (Fig. 1) (25). Thus, the proteasomal AAA-ATPases Rpt1–6 are expected to form a heterohexamer. The crystal structures of the major domains of the proteasome-activating nucleotidase (PAN), a homohexameric ortholog of the 26S proteasomal AAA-ATPases found in some archaea (26), have led to insights into the structure and mechanism of the AAA-ATPase modules: each PAN monomer consists of coiled coils protruding from an oligonucleotide binding (OB) fold (PAN-N) and an AAA fold, which have been crystallized separately (27, 28). The PAN-N fragments as well as the AAA folds assemble into hexameric rings (N ring and AAA ring, respectively), but the exact spatial relationship between both rings is not yet known (Fig. 2A).
Fig. 1.
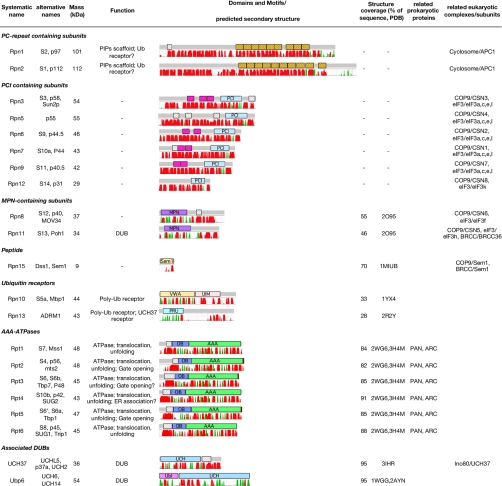
Overview of RP subunits. PC repeat (orange), coiled coils (white, stripes), TPR-like repeats (magenta), PCI module, MPN domain, Sem1 fold, von Willebrand domain (VWA), UIM, PRU, OB fold, AAA-ATPase domain, UCH domain, and ubiquitin-like domain (Ubl) are shown. Secondary structures (red, α-helical; green, β-strand) were predicted using the MPI bioinformatics toolkit (109) as a front end to PSIPRED (110). Putative coiled coils with probabilities >0.5 according to PCOILS (111) are indicated. TPR motifs were assigned using TPRpred (80). ER, endoplasmic reticulum; PIP, proteasome interacting protein.
Fig. 2.
Structures or probable folds of RP subunits. A, AAA-ATPases. Crystal structures of PAN-N (orange) and the PAN AAA fold (red-orange) were determined separately (Protein Data Bank codes 2WG6 and 3H4M). B, Rpn1 and Rpn2. The PC repeats of Rpn1 and Rpn2 probably adopt a solenoid fold similar to protein phosphatase PP2A (residues 282–584 of Protein Data Bank code 1B3U; sequence identity, 15%). C, PCI-module containing subunits. The PCI module of the COP9 subunit CSN7 (Protein Data Bank code 3CHM) consists of a C-terminal winged helix domain (light blue) and a helical bundle (blue). In CSN7, as probably also in most proteasomal PCI subunits, the PCI module is preceded by one or more bihelical domains (light red). D, Rpn8 and Rpn11. The MPN domain of Rpn8 (Protein Data Bank code 2O95) is shown. E, ubiquitin receptors. The Rpn10 UIM (blue) consists of three helices that lack a defined tertiary structure (Protein Data Bank code 1YX5). Ubiquitin (red) binds to the UIM helices. In contrast, ubiquitin binds to loops of the globular PRU domain of Rpn13 (blue). F, associated DUBs. UCH37 exhibits a globular UCH fold and a C-terminal domain, which contains a coiled coil motif. In Ubp6, a UCH fold (Protein Data Bank code 2AYN) is preceded by an N-terminal Ub-like domain (Protein Data Bank code 1WGG) both of which have been solved separately.
The order of the AAA-ATPase subunits Rpt1–6 in the heterohexamer was suggested to be Rpt1/2/6/4/5/3 based on cross-linking experiments (29). However, the PAN-N structure challenges this model: only Rpt2, Rpt3, and Rpt5 possess an invariant proline (corresponding to Pro-62 of PAN-N) that is required to adopt a cis conformation for coiled coil formation (27). The PAN-N structure suggests strongly that Rpt2, Rpt3, and Rpt5 each pair with an Rpt subunit lacking the cis-proline. Thus, direct adjacencies of Rpt2, Rpt3, or Rpt5, which occur in the previously suggested order (29), are not consistent with the PAN structures. Upon analysis of all available structural and protein-protein interaction data, we concluded that the subunit order in the AAA-ATPase module is most likely Rpt1/Rpt2/Rpt6/Rpt3/Rpt4/Rpt5 (see below) (30).
PAN and the 26S AAA-ATPases promote unfolding of substrates prior to translocation through the pore formed in the hexamer center (31). The C-terminal residues of the AAA-ATPases are responsible for binding of the RP to the CP (32, 33). The C termini probably bind to groves at the interfaces of the CP subunits in a fashion similar to that of the 11 S activator (34) as demonstrated for the C termini of PAN (35). The C termini of Rpt2, Rpt3, and Rpt5 share a common 3-residue HbYX motif of a hydrophobic residue (Hb) and a tyrosine followed by a residue of any type (Fig. 1) responsible for gate opening of the CP in PAN (32). Of all RP AAA-ATPase subunits, only the C termini of Rpt2 and Rpt5 were validated to induce CP gate opening (32).
Proteasome Cyclosome Repeat-containing Non-ATPases
The non-ATPases Rpn1 and Rpn2 are by far the largest proteasome subunits (Fig. 1). Rpn1 and Rpn2 share a significant degree of sequence similarity (30–45% identity). Characteristic for Rpn1 and Rpn2 is a 400–500-residue stretch containing repetitive elements, which can also be found in subunits of the cyclosome complex, the proteasome cyclosome (PC) repeats (36). Structure prediction algorithms suggest that the PC repeats adopt an α-solenoid fold (37). Indeed, circular dichroism spectra indicate that the repeat-containing fragments of Rpn1 and Rpn2 consist almost exclusively of α-helices, whereas the full-length proteins also contain substantial amounts of β-sheet in addition to helical elements (38), which is in agreement with secondary structure prediction (Fig. 1).
The PC repeats likely form two-helix armadillo (ARM)/HEAT units that assemble into a superhelical quaternary structure typical of α-solenoids (Fig. 2B). Regular, α-helical repeat structures are common scaffolds in large protein assemblies, such as the nuclear pore complex (NPC) (39), where they can form highly diverse interfaces (40). The observed structural diversity of solenoids makes prediction of their structure, in particular the superhelical twist, difficult. Recent cryo-EM analysis suggests that the PC repeat segment of Rpn2 adopts a horseshoe-like shape (38). In the RP, the probably twisted Rpn1 and Rpn2 function as scaffolds for the assembly of the ATPase subunits (21, 22, 41, 42).
PCI Module-containing Non-ATPases and Rpn15
The primary structures of Rpn3, Rpn5, Rpn6, Rpn7, Rpn9, and Rpn12 contain a common module that is not only observed in subunits of the 26S proteasome but also in subunits of the COP9 signalosome, and the translation initiation factor eIF3 (i.e. the PCI module (43)). Structures of the PCI modules of the eIF subunit eIFk (44) and the COP9 subunit CSN7 (45) have been solved by x-ray crystallography. The PCI module is composed of two subdomains: an N-terminal helical bundle fold and a C-terminal winged helix fold (45) (Fig. 2C). The Rpn12 PCI domain is evolutionarily distant from the other PCI-containing subunits and substantially shorter (43).
In the full-length RP subunits, the PCI modules are mostly preceded by repetitive bihelical blocks, reminiscent of tetratricopeptide repeats (TPRs), which may form continuous α-solenoids together with the PCI helical bundle folds (43). The TPRs upstream of the PCI domain are occasionally referred to as PCI-associated modules (46). In addition to α-helical repeats, our bioinformatics analysis suggests that some PCI-containing subunits contain coiled coils upstream of the PCI domains (Fig. 1). In summary, fold prediction suggests that PCI-containing subunits consist of α-helical elements, most likely TPRs and coiled coils, besides the PCI modules. Protein-protein interaction data suggest that the PCI-containing subunits are highly interlinked (see Fig. 4 and Table I) through interactions between their PCI modules (45).
Fig. 4.
RP topology based on publicly available protein-protein interaction data (Table I). The protein-protein interactions were determined by two-hybrid assays (red), in vivo pulldown experiments (orange), chemical cross-linking (dashed black), in vitro binding (magenta), and co-expression (green). Dotted lines indicate interactions involving more than two proteins.
Table I. Protein interactions in RP involving six or fewer subunits.
Protein-protein interactions were compiled from yeast two-hybrid screens (Y2h), chemical cross-linking (x-link), in vitro binding assays (IVB), different in vivo pulldown experiments (IVPD), and co-expression (co-expr). The data were obtained from 26S proteasomes of H. sapiens (h), S. cerevisiae (y), Schizomycetes pombe (sp), Sus barbatus (p), D. melanogaster (dm), and Caenorhabditis elegans (w).
| Subcomplex | Technique | Refs. | Species |
|---|---|---|---|
| Rpt1, Rpt2 | IVB, x-link, co-expr | 29, 76, 77, 92 | y, h |
| Rpt1, Rpt3 | x-link | 29 | h |
| Rpt1, Rpt2, Rpt3, Rpt6 | IVB | 76 | h |
| Rpt1, Rpt2, Rpt5, Rpn1 | IVPD | 79 | y |
| Rpt1, Rpn1 | Filter, IVB | 20, 77 | h |
| Rpt1, Rpt2, Rpn1 | Filter, IVB | 41, 77 | h |
| Rpt2, Rpt6 | x-link | 29 | h |
| Rpt2, Rpn1 | IVPD, y2h, IVB | 10, 77, 88, 93 | h |
| Rpt2, α4 | IVPD, y2h | 87, 88 | h |
| Rpt2, Rpn12, Rpn2, Rpt1, Rpn13, Rpn8 | IVPD | 94 | y |
| Rpt3, Rpt4 | Y2h | 88, 95, 96 | y |
| Rpt3, Rpt5 | Y2h, x-link | 13, 29, 95–97 | y, h |
| Rpt3, Rpt6 | Y2h, IVB, IVPD | 13, 22, 76, 88, 95 | y, h |
| Rpt3, Rpn11, Rpt6, Rpn8 | IVPD | 98 | y |
| Rpt4, Rpt5 | Y2h, IVB, x-link, IVPD | 10, 13, 29, 41, 76, 97 | w, y, h |
| Rpt4, Rpt6 | Y2h, x-link | 29, 88, 99 | y, h |
| Rpt4, α4 | Y2h | 10, 88 | y, w |
| Rpt4, α6 | x-link | 29 | h |
| Rpt6, α2 | x-link | 100 | p |
| Rpt6, Rpn1 | IVB | 101 | y |
| Rpt6, Rpn2 | x-link | 29 | h |
| Rpn1, Rpn10 | IVB | 20, 102 | y |
| Rpn1, Ubp6 | IVB | 23 | sp |
| Rpn2, Rpn13 | Y2h, IVPD | 97, 103, 104 | dm, y, sp |
| Rpn3, Rpn5 | x-link | 12 | y |
| Rpn3, Rpn5, Rpn8 | x-link | 12 | y |
| Rpn3, Rpn5, Rpn8, Rpn9 | x-link | 12 | y |
| Rpn3, Rpn5, Rpn7, Rpn9, Rpn11 | x-link | 12 | y |
| Rpn3, Rpn7 | Y2h | 10, 13, 97 | y, w, h |
| Rpn3, Rpn12 | Y2h | 13, 88, 105 | y |
| Rpn3, Rpn15 | x-link | 12 | y |
| Rpn5, Rpn6 | Y2h | 88, 97, 106 | y |
| Rpn5, Rpn6, Rpn8 | MS/MS | 12 | y |
| Rpn5, Rpn6, Rpn8, Rpn9 | MS/MS | 12 | y |
| Rpn5, Rpn8, Rpn9 | MS/MS | 12 | y |
| Rpn5, Rpn8 | IVPD | 107 | h |
| Rpn5, Rpn9 | Y2h | 13, 103 | y, dm |
| Rpn6, Rpn8, Rpn9 | MS/MS | 12 | y |
| Rpn7, Rpn15 | x-link | 12 | y |
| Rpn8, Rpn9 | Y2h | 10, 88 | y |
| Rpn8, Rpn11 | Y2h | 10, 13, 95, 103 | y, dm |
| Rpn9, Rpn11 | Y2h | 10, 13 | y |
| Rpn10, UCH37 | IVPD | 23 | sp |
| Rpn12, UCH37 | IVB, y2h | 108 | h |
| Rpn13, UCH37 | IVPD, IVB | 104, 107 | sp |
Our functional understanding of the PCI-containing subunits is still modest. Winged helix domains are known to be important for the interaction with nucleic acids (47), suggesting that the PCI-containing subunits are important for the role of the proteasome in DNA repair. Indeed, the peptide Rpn15, which associates with the PCI-containing proteins (see Fig. 4), has been shown to mediate proteasome association to DNA double strand breaks (48). The formation of minicomplexes of Rpn15 and the PCI-containing subunits Rpn3 and Rpn7 may be pivotal for regulation of proteins involved in DNA repair (49).
MPN Domain-containing Non-ATPases
Subunits of 26S proteasome, COP9, and eIF3 share another conserved module. The so-called MPN domain was initially found in Mpr1 and Pad1 in the N terminus and is also present in the RP subunits Rpn8 and Rpn11. The MPN domain of Rpn11 contains a zinc-binding JAMM (JAB1/MPN/Mov34 metalloenzyme) motif, which is responsible for its function as a deubiquitylating enzyme (DUB) (50, 51). Rpn11 is required for degradation of ubiquitylated substrates (i.e. it controls substrate degradation positively). Because the deubiquitylating activity of Rpn11 is ATP-dependent, Rpn11 is only active in the assembled RP. Recently, the DUB BRCC36, which is homologous to Rpn11, was identified in the BRCA1/BRCA2-interacting complex (BRCC), a ubiquitin E3 ligase complex that is involved in DNA repair (52, 53). The MPN domain of Rpn8 does not contain the JAMM motif, and its function is not known.
The structure of the Rpn8 MPN domain has been solved by x-ray crystallography (54) (Fig. 2D). Interestingly, the Rpn8 MPN domain forms a dimer in solution. The physical interaction of Rpn8 and Rpn11 is well established (see Fig. 4 and Table I), and it is possible that these two proteins dimerize via their MPN domains, similarly to the Rpn8 crystal structure (54).
Ubiquitin Receptors
Rpn10 was the first proteasome subunit that was shown to function as a ubiquitin receptor (55). Proteomics data indicate that Rpn10 is involved in recognition of ∼25% of all proteasomal substrates (56). Rpn10 consists of a von Willebrand fold and a flexible C-terminal ubiquitin interaction motif (UIM) (Fig. 1). Ubiquitin binding is achieved through helices that are connected by flexible linkers and do not possess a distinct tertiary structure (57) (Fig. 2E). Rpn10 seems to be somewhat loosely associated with the RP (58, 59); as a consequence, Rpn10 is detected substoichiometrically in purified proteasomes (9).
Recently, Rpn13 has been shown to be a second integral Ub receptor (60). Rpn13 binds ubiquitin through a pleckstrin-like receptor of ubiquitin (PRU) domain: in contrast to previously structurally characterized UIMs, ubiquitin binds to loops rather than secondary structural elements in PRU (61) (Fig. 2E). In almost all eukaryotes other than S. cerevisiae, Rpn13 possesses a C-terminal extension (Fig. 1), which serves as an anchor of UCH37 (see below). It will now be interesting to reveal which substrates utilize Rpn13 for degradation.
In addition to integral Ub receptors, several “shuttling” Ub receptors, most notably Dsk2 and Rad23, are transiently associated with the proteasome (62). Furthermore, the RP subunits Rpn1, Rpn2, and Rpt5 are candidates for integral proteasomal Ub receptors that await further confirmation (8): Rpn1 has been shown to interact with the ubiquitin-like domains of Rad23 and Dsk2 (63), suggesting that Rpn1 as well as the related Rpn2 might act as Ub receptors. In cross-linking experiments, ubiquitin bound to Rpt5 (64), indicating that Ub binds either directly to Rpt5 or to a subunit in close proximity (see Chemical Cross-linking below).
Associated Deubiquitylating Enzymes
Two additional DUBs are more loosely associated with the RP and are typically found substoichiometrically in preparations of purified 26S proteasomes. The UCHs Uch37 and Usp14/Ubp6 can dissect ubiquitin chains prior to substrate degradation. In contrast to Rpn11, the UCHs control substrate degradation negatively; i.e. deubiquitylation by UCH can save substrate particles from degradation (65). In addition to its deubiquitylating function, Ubp6 was recently found to induce CP gate opening, i.e. to act as a sensor for substrates to be degraded (66).
Whereas Ubp6 is the only proteasome-associated DUB in S. cerevisiae, almost all other eukaryotes possess UCH2/UCH37 as an additional UCH (24). Rpn1 recruits Ubp6 to the RP (23). UCH37 interacts physically with the ubiquitin receptors Rpn10 (23) and Rpn13 (67); the binding to Rpn13 seems to be more specific than the binding to Rpn10 (67). Rpn13 possesses a C-terminal domain that seems to have evolved for UCH37 binding (Fig. 1) (67). UCH37 is recruited to Rpn13 via its C-terminal domain (67), which contains a coiled coil domain (Fig. 2F). Inactivated UCH37 is also part of the nuclear Ino80 complex where Rpn13 can activate it in a hit-and-run fashion (68). It will be interesting to study a possible functional interplay between the RP and the Ino80 complex (69).
Subcomplexes of the RP
The RP easily disassociates into two subcomplexes, a distal “lid” consisting of the non-ATPases Rpn3–12 and the “base” comprising the remaining non-ATPases and the AAA-ATPases (70). Recently, the discovery of numerous smaller subcomplexes has provided significant insights into the assembly process of the RP, such as the scaffold function of Rpn1 and Rpn2 for AAA-ATPase assembly (71). The assembly process of the RP and binding of the RP to the CP is facilitated by the assembly chaperones Nas2, Nas6, Hsm3, and Rpn14 in yeast (p27, p28, S5b, and Rpn14 in human) (21, 22, 41, 42).
CRYO-EM ANALYSIS OF THE 26S PROTEASOME
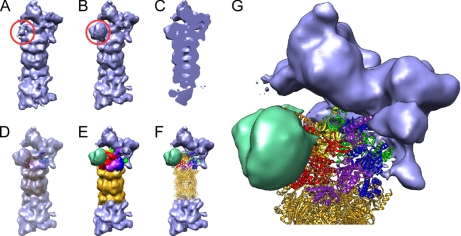
Because of the aforementioned difficulties to obtain homogeneous, concentrated 26S proteasome preparation, most information on the structure of the complete RP to date has been revealed by single particle EM analysis. Reconstructions of air-dried and negatively stained 26S proteasomes from D. melanogaster (72) and Homo sapiens (73) revealed that the double-capped proteasome measured ∼45 nm in length and that the C2 symmetry of the CP is approximately conserved in the two RPs (Fig. 3, A and B).
Fig. 3.
Cryo-EM density map of D. melanogaster 26S proteasome. A, image classification revealed that ∼50% of all analyzed particles lacked an additional density (red circle). B, the remaining 50% possessed an additional density (red circle). C, side cut of density depicted in B. D, atomic models of the CP (gold) and the AAA-ATPases (Rpt1/Rpt2/Rpt6/Rpt3/Rpt4/Rpt5, red/orange/yellow/green/blue/purple) fitted into map B. E, segmentation of B according to the fitted atomic models. F, hybrid representation of the 26S proteasome by atomic models and the cryo-EM density where interpretation on the atomic level is not possible yet. G, enlarged view of F.
More detailed structural insights were obtained from cryo-EM reconstructions of D. melanogaster 26S proteasomes at ∼20-Å resolution (9). The higher resolution allowed segmentation of the AAA-ATPase hexamer in the 26S proteasome. The (pseudo-) 6-fold symmetry axes of both ATPase hexamers do not coincide with the (pseudo-) 7-fold symmetry of the CP; the ATPase axes are shifted by ∼3 nm. In addition, the axes of the ATPases are tilted by ∼10° compared with the CP axis.
Advanced image classification methods revealed that the RPs possess a substantial degree of structural heterogeneity (9). Most significantly, a density of ∼60 kDa can be found at one of the two RPs in ∼50% of the analyzed particles (Fig. 3, A and B). Quantitative mass spectrometry suggests that Rpn10 is present in only 25% of the RPs, indicating that the variable mass may correspond to Rpn10. However, this hypothesis needs to be corroborated for example by cryo-EM imaging of 26S proteasomes with labeled Rpn10.
INTRAPROTEASOME INTERACTOME
Measuring Protein-Protein Interactions in the RP
A variety of molecular biology techniques have revealed physical interactions among the RP subunits. Here, we summarize the methods that have been applied to the RP along with the information they provide.
Two-hybrid Assay
One candidate protein (“bait”) is fused to a transcription factor DNA binding domain, and another is fused to the activation domain (“prey”), typically using yeast as the host organism. Activation of a reporter gene implies a physical interaction between the two proteins. However, the assay is prone to false negatives as well as false positives (74, 75). To decrease the number of false-positive two-hybrid interactions in the RP interactome, we considered only those interactions that were reported in two or more independent publications (Table I).
In Vivo Pulldown
Co-immunoprecipitation or tandem affinity purification can be used to purify complexes that include a bait protein. Subsequent analysis, traditionally by Western blotting or more recently by MS, reveals the identity of affinity-purified complexes. Variation of elution buffers often allows purification of different subcomplexes for the same bait. The experiment indicates physical interactions of the detected subunits, but physical contacts between pairs of proteins typically cannot be directly deduced from the data because subcomplexes often comprise more than two proteins.
In Vitro Binding Assays
A bait protein is recombinantly expressed and attached to GST beads. The beads are incubated in cell lysate, again separated from the lysates, and analyzed, typically by Western blotting. Alternatively, the in vitro expressed bait protein may be attached to a nitrocellulose matrix, which is then exposed to cell lysate, washed, and analyzed (76, 77). In vitro binding assays reveal binary physical interactions.
Chemical Cross-linking
The protein complex is chemically cross-linked, typically using relatively extended cross-linking agents, such as bis(sulfosuccinimidyl) suberate, that cross-link specific residues (e.g. lysines) in spatial proximity (for a review, see Leitner et al. (112) in this issue). The cross-linked samples are then separated in a denaturing two-dimensional gel and subsequently analyzed by Western blotting or mass spectrometry. The experiment reveals spatial proximity of two or more proteins. Direct physical interactions cannot be inferred with certainty as the typical cross-linking agents are more than 12 Å long.
Co-expression
Expression of two or more proteins in a heterologous expression system or an in vitro translation system and subsequent purification of a complex of these proteins implies a stable subcomplex of the respective protein.
Mass Spectrometry of Whole Complexes
MS of whole, intact complexes isolated from cells reveals the mass of specific complexes (78). When the subunits are identified by conventional shotgun MS, the stoichiometry of the complex under scrutiny can be determined. In addition, subunits that are peripherally located in the complex can be identified by collision of the complex in the gas phase.
Interactome Topology
The proteasomal interaction map clearly reveals two distinct clusters (Fig. 4). The first consists of the PCI- and MPN-containing non-ATPases as well as Rpn15. The second cluster consists of the AAA-ATPases, the PC repeat-containing proteins Rpn1 and Rpn2, the Ub receptors Rpn10 and Rpn13, and the UCHs. This clustering corresponds to the lid and base subcomplex, which can be purified independently (70).
Base
Numerous protein-protein interactions have been reported among the AAA-ATPases, strongly supporting the notion that the AAA-ATPases form a hexameric subcomplex. The PC repeat-containing subunits Rpn1 and Rpn2 interact with many of the AAA-ATPases, which is in agreement with their likely function as scaffolding proteins for base assembly (79). The Ub receptors Rpn10 and Rpn13 bind to the PC subunits Rpn1 and Rpn2, respectively. Because UCH37 is probably associated with the Ub receptors in substoichiometric amounts, it cannot be excluded that the UCH37 binds to multiple sites on the RP. Thus, Rpn10 and Rpn13 are not necessarily in spatial proximity.
Lid
The PCI subunits form an extensively connected network where only the evolutionary distant Rpn12 is somewhat peripherally bound (12). The peptide Rpn15 binds to Rpn3 and Rpn7. The dimer of the MPN subunits Rpn8 and Rpn11 is integrated into the lid via Rpn5 and Rpn9. The MPN subunits directly interact with the base complex via the AAA-ATPase dimer Rpt3/Rpt6.
INTEGRATIVE STRUCTURE DETERMINATION OF THE RP
Integrative methods for structure determination combine input data of different kinds to obtain models of assemblies with a substantially improved accuracy and completeness compared with the individual input data (also see the review by Lasker et al. (113) in this issue). For example, a comprehensive model of the NPC has been obtained based on protein-protein interaction data, the NPC envelope derived by EM, and ultracentrifugation data about the shapes of the individual proteins (17).
CP-AAA-ATPase Subcomplex
One option to use protein-protein interaction data for structural research is to discriminate between different candidate models. We have used this strategy to build a model of the subcomplex of the CP and the AAA-ATPases (30). First, we built comparative models of the hexameric N ring and AAA ring based on the PAN crystal structures in all possible subunit orders and fitted the models into the density corresponding to the AAA-ATPases from the 26S density determined by cryo-EM at 20-Å resolution (9). Because the EM maps are not of sufficient resolution to rank the different subunit arrangements, we evaluated the different models based on their agreement with protein-protein interaction data and alternate positioning of Rpt2, Rpt3, and Rpt5. Given these filters, the most likely subunit order is Rpt1/Rpt2/Rpt6/Rpt3/Rpt4/Rpt5. Next, we fitted the CP into the cryo-EM density map and determined the rotation of the AAA-ATPase hexamer around its symmetry axis based on protein-protein interactions between CP subunits and AAA-ATPase subunits. In the resulting model, Rpt6 and Rpt3 are largely buried by the proteasome lid, which is in excellent agreement with recently reported assembly intermediates (81). The coiled coil pair Rpt1/Rpt2 is joined by a distinct density, which in part consists of the variable mass detected by image classification (Fig. 3, E and F). The coiled coils of Rpt4/Rpt5 apparently do not bind to additional subunits.
Taking into account the interactome, additional subunits can be localized approximately using the CP-AAA-ATPase model. The interactome suggests that the density adjacent to Rpt1/Rpt2 corresponds to Rpn1 and Rpn10 (Fig. 3, E and F). Indeed, the volume of the density (∼2 × 105Å3) is in good agreement with the expected volume of the two proteins (∼2.2 × 105Å3). Moreover, Rpn8 and Rpn11 should be in close proximity to the Rpt6/Rpt3 pair. Thus, substrates of the Rpn10 pathway get recognized at the peripherally located Rpn10 and are subsequently deubiquitylated by Rpn11, which is presumably located in the RP cavity near Rpt6/Rpt3.
Solving the Proteasomal Puzzle
To facilitate the approximate localization of all RP subunits (“molecular architecture”) by integration of cryo-EM maps, protein-protein interaction data, and atomic models of subunits, additional data are required. Some subunits can be localized by means of cryo-EM single particle analysis of 26S proteasomes with appropriate genetic tags or deletions of the corresponding genes. Moreover, high throughput data acquisition and image processing will yield higher resolution density maps of the 26S proteasome (14, 82).
An increase in the resolution of protein-protein interaction data will be pivotal for accurate model building. Toward this goal, identification of cross-linked peptides appears to be the most promising technology (Ref. 83; for a review, see Leitner et al. (112) in this issue). Whereas the resolution of the current RP interactome is determined approximately by the diameters of the constituent proteins or, at best, their domains, the resolution of a residue-specific cross-linking restraint is much higher. In addition, the cross-linking/MS technology promises to be less prone to false positives than some established protein-protein interaction methods, such as two-hybrid assays.
Most RP non-ATPase subunits are still structurally poorly characterized (Fig. 1); further structural analysis of these subunits will be required to position them accurately. Given that most structural data available today have only been obtained in the last 5 years, structural coverage can be expected to increase rapidly in the coming years. Structural genomics initiatives may also play a significant role in expanding structural data on the RP subunits as suggested by the recently deposited structure of UCH37 (Protein Data Bank code 3IHR).
Based on our CP-AAA-ATPase model and the RP interactome, we can already hypothesize on the mechanism of the RP in substrate degradation (Fig. 5). Substrates using the Rpn10 pathway are probably recruited to the RP near Rpt1/Rpt2. The interactome suggests that the Ub receptor Rpn13 is located in proximity to the Rpt4/Rpt5 pair. Thus, Rpn10 and Rpn13 substrates may enter the proteasome through different portals (84). The gate to the CP may then be opened differently for both types of substrates: Rpn10 substrates might use Rpt2 as a “gate opener” to the CP, possibly facilitated by Ubp6 (66), whereas Rpt5 might open the CP gate for Rpn13 substrates. Subsequently, substrates are deubiquitylated at Rpn11, which appears to be centrally located in the RP cavity before being unfolded in the upper AAA-ATPase cavity and translocated to the CP where degradation occurs.
Fig. 5.
Schematic representation of protein degradation through the 26S proteasome. Ubiquitylated substrates first bind to the Ub receptors Rpn10 and Rpn13. As the substrates proceed, they get deubiquitylated by Rpn11, then unfolded in the upper ring of the AAA-ATPase hexamer, and translocated to the CP by the lower AAA-ATPase domains. The AAA-ATPase subunits Rpt2 and Rpt5 open the gate to the CP where substrates get cleaved by CP subunits β1, β2, and β5 located in the CP inner cavity (8).
The mathematical framework of “modeling by satisfaction of spatial restraints” allows a systematic integration of all available data to obtain, in principle, all subunit configuration(s) consistent with the input data (18). Specifically, the RP requires a platform that can represent subunits at different levels of detail, reflecting the varying structural characterization of subunits. The Integrative Modeling Platform (IMP) may provide the required functionality for the exciting endeavor of localizing the RP subunits (for a review, see Lasker et al. (113) in this issue; see also Refs. 18 and 85). However, we emphasize that the quality of any model is ultimately limited by the quality and amount of input data. To obtain a most accurate model, it will be important to remove erroneous protein-protein interaction data. For example, recent in vitro redesign of the CP-AAA-ATPase interface suggests that Rpt5 binds to the α3/α4 CP pocket (86), whereas the data underlying our CP-AAA-ATPase model suggest that Rpt2 binds to α4 (87, 88), and genetic data indicate interaction of Rpt2 with α3 (89). Thus, taking into account the data from Yu et al. (86), two conflicting models are possible where either Rpt2 or Rpt5 interact with the α3/α4 CP pocket. Again, the identification of cross-linked peptides is probably the most promising technology to identify only those protein-protein interactions that occur in the fully assembled complex and in turn filter out erroneous data (for a review, see Leitner et al. (112) in this issue). In addition, the CP-AAA-ATPase model can be validated using cryo-EM of the 26S proteasome with specifically labeled AAA-ATPase subunits.
Elucidation of proteasomal dynamics will undoubtedly be the next challenge after obtaining a draft of the static architecture of the 26S proteasome. In particular, cryo-EM may be uniquely suitable for unraveling the sequence of events during substrate degradation similar to the role of cryo-EM in elucidating ribosomal protein synthesis (90). More detailed insights into the temporal behavior of proteasomal degradation are then to be expected from systematically incorporating such temporal data into the molecular modeling approach (91).
Acknowledgments
We thank Eri Sakata for insightful discussions.
* This work was supported by a 3D Repertoire grant and a PROSPECTS grant within the Research Framework Programs 6 and 7 (FP6 and FP7) of the European Commission, respectively, and by the Deutsche Forschungsgemeinschaft Cluster of Excellence “Munich-Centre for Advanced Photonics.” This work was also supported, in part, by National Institutes of Health Grants R01 GM54762, U54 RR022220, PN2 EY016525, and R01 GM083960 (to A. S.).
1 The abbreviations used are:
- CP
- core particle
- RP
- regulatory particle
- EM
- electron microscopy
- Rpt
- regulatory particle triple A-ATPase
- Rpn
- regulatory particle non-ATPase
- UCH
- ubiquitin C-terminal hydrolase
- PAN
- proteasome-activating nucleotidase
- PC
- proteasome cyclosome
- NPC
- nuclear pore complex
- eIF
- eukaryotic initiation factor
- TPR
- tetratricopeptide repeat
- DUB
- deubiquitylating enzyme
- BRCC
- BRCA1/BRCA2-interacting complex
- UIM
- ubiquitin interaction motif
- Ub
- ubiquitin
- PRU
- pleckstrin-like receptor of ubiquitin.
REFERENCES
- 1.Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 2.Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 3.Voges D., Zwickl P., Baumeister W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 [DOI] [PubMed] [Google Scholar]
- 4.Jentsch S., Haendler B. (2009) The Ubiquitin System in Health and Disease, Springer, Heidelberg, Germany: [PubMed] [Google Scholar]
- 5.Pühler G., Weinkauf S., Bachmann L., Müller S., Engel A., Hegerl R., Baumeister W. (1992) Subunit stoichiometry and three-dimensional arrangement in proteasomes from Thermoplasma acidophilum. EMBO J. 11, 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268, 533–539 [DOI] [PubMed] [Google Scholar]
- 7.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 8.Tanaka K. (2009) The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 12–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nickell S., Beck F., Scheres S. H., Korinek A., Förster F., Lasker K., Mihalache O., Sun N., Nagy I., Sali A., Plitzko J. M., Carazo J. M., Mann M., Baumeister W. (2009) Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 106, 11943–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davy A., Bello P., Thierry-Mieg N., Vaglio P., Hitti J., Doucette-Stamm L., Thierry-Mieg D., Reboul J., Boulton S., Walhout A. J., Coux O., Vidal M. (2001) A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2, 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrell K., Wilkinson C. R., Dubiel W., Gordon C. (2000) Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem. Sci. 25, 83–88 [DOI] [PubMed] [Google Scholar]
- 12.Sharon M., Taverner T., Ambroggio X. I., Deshaies R. J., Robinson C. V. (2006) Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 4, e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu H., Reis N., Lee Y., Glickman M. H., Vierstra R. D. (2001) Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20, 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y. (2009) Toward an atomic model of the 26S proteasome. Curr. Opin. Struct. Biol. 19, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson C. V., Sali A., Baumeister W. (2007) The molecular sociology of the cell. Nature 450, 973–982 [DOI] [PubMed] [Google Scholar]
- 16.Sali A., Glaeser R., Earnest T., Baumeister W. (2003) From words to literature in structural proteomics. Nature 422, 216–225 [DOI] [PubMed] [Google Scholar]
- 17.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Rout M. P., Sali A. (2007) Determining the architectures of macromolecular assemblies. Nature 450, 683–694 [DOI] [PubMed] [Google Scholar]
- 18.Alber F., Förster F., Korkin D., Topf M., Sali A. (2008) Integrating diverse data for structure determination of macromolecular assemblies. Annu. Rev. Biochem. 77, 443–477 [DOI] [PubMed] [Google Scholar]
- 19.Finley D., Tanaka K., Mann C., Feldmann H., Hochstrasser M., Vierstra R., Johnston S., Hampton R., Haber J., Mccusker J., Silver P., Frontali L., Thorsness P., Varshavsky A., Byers B., Madura K., Reed S. I., Wolf D., Jentsch S., Sommer T., Baumeister W., Goldberg A., Fried V., Rubin D. M., Toh-e A. (1998) Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem. Sci. 23, 244–245 [DOI] [PubMed] [Google Scholar]
- 20.Xie Y., Varshavsky A. (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. U.S.A. 98, 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 22.Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. (2009) Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 23.Stone M., Hartmann-Petersen R., Seeger M., Bech-Otschir D., Wallace M., Gordon C. (2004) Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J. Mol. Biol. 344, 697–706 [DOI] [PubMed] [Google Scholar]
- 24.Hölzl H., Kapelari B., Kellermann J., Seemüller E., Sümegi M., Udvardy A., Medalia O., Sperling J., Müller S. A., Engel A., Baumeister W. (2000) The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 150, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson P. I., Whiteheart S. W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 26.Frickey T., Lupas A. N. (2004) Phylogenetic analysis of AAA proteins. J. Struct. Biol. 146, 2–10 [DOI] [PubMed] [Google Scholar]
- 27.Djuranovic S., Hartmann M. D., Habeck M., Ursinus A., Zwickl P., Martin J., Lupas A. N., Zeth K. (2009) Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol. Cell 34, 580–590 [DOI] [PubMed] [Google Scholar]
- 28.Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartmann-Petersen R., Tanaka K., Hendil K. B. (2001) Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 386, 89–94 [DOI] [PubMed] [Google Scholar]
- 30.Förster F., Lasker K., Beck F., Nickell S., Sali A., Baumeister W. (2009) An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem. Biophys. Res. Commun. 388, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang F., Wu Z., Zhang P., Tian G., Finley D., Shi Y. (2009) Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) Differential roles of the COOH termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26S proteasome. J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Förster A., Masters E. I., Whitby F. G., Robinson H., Hill C. P. (2005) The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 18, 589–599 [DOI] [PubMed] [Google Scholar]
- 35.Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupas A., Baumeister W., Hofmann K. (1997) A repetitive sequence in subunits of the 26S proteasome and 20S cyclosome (anaphase-promoting complex). Trends Biochem. Sci. 22, 195–196 [DOI] [PubMed] [Google Scholar]
- 37.Kajava A. V. (2002) What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26S proteasome. J. Biol. Chem. 277, 49791–49798 [DOI] [PubMed] [Google Scholar]
- 38.Effantin G., Rosenzweig R., Glickman M. H., Steven A. C. (2009) Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J. Mol. Biol. 386, 1204–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brohawn S. G., Partridge J. R., Whittle J. R., Schwartz T. U. (2009) The nuclear pore complex has entered the atomic age. Structure 17, 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobe B., Gleichmann T., Horne J., Jennings I. G., Scotney P. D., Teh T. (1999) Turn up the HEAT. Structure 7, R91–R97 [DOI] [PubMed] [Google Scholar]
- 41.Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Multiple Assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheel H., Hofmann K. (2005) Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics 6, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Z., Xue Y., Xu H., Gong W. (2006) Crystal structure of the C-terminal domain of S. cerevisiae eIF5. J. Mol. Biol. 359, 1–9 [DOI] [PubMed] [Google Scholar]
- 45.Dessau M., Halimi Y., Erez T., Chomsky-Hecht O., Chamovitz D. A., Hirsch J. A. (2008) The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell 20, 2815–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciccarelli F. D., Izaurralde E., Bork P. (2003) The PAM domain, a multi-protein complex-associated module with an all-alpha-helix fold. BMC Bioinformatics 4, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brennan R. G. (1993) The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74, 773–776 [DOI] [PubMed] [Google Scholar]
- 48.Krogan N. J., Lam M. H., Fillingham J., Keogh M. C., Gebbia M., Li J., Datta N., Cagney G., Buratowski S., Emili A., Greenblatt J. F. (2004) Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell 16, 1027–1034 [DOI] [PubMed] [Google Scholar]
- 49.Pick E., Hofmann K., Glickman M. H. (2009) PCI complexes: Beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell 35, 260–264 [DOI] [PubMed] [Google Scholar]
- 50.Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 51.Yao T., Cohen R. E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 52.Dong Y., Hakimi M. A., Chen X., Kumaraswamy E., Cooch N. S., Godwin A. K., Shiekhattar R. (2003) Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 12, 1087–1099 [DOI] [PubMed] [Google Scholar]
- 53.Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., Cohen R. E. (2009) K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanches M., Alves B. S., Zanchin N. I., Guimarães B. G. (2007) The crystal structure of the human Mov34 MPN domain reveals a metal-free dimer. J. Mol. Biol. 370, 846–855 [DOI] [PubMed] [Google Scholar]
- 55.Deveraux Q., van Nocker S., Mahaffey D., Vierstra R., Rechsteiner M. (1995) Inhibition of ubiquitin-mediated proteolysis by the Arabidopsis 26S protease subunit S5a. J. Biol. Chem. 270, 29660–29663 [DOI] [PubMed] [Google Scholar]
- 56.Mayor T., Graumann J., Bryan J., MacCoss M. J., Deshaies R. J. (2007) Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol. Cell. Proteomics 6, 1885–1895 [DOI] [PubMed] [Google Scholar]
- 57.Wang Q., Young P., Walters K. J. (2005) Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J. Mol. Biol. 348, 727–739 [DOI] [PubMed] [Google Scholar]
- 58.Babbitt S. E., Kiss A., Deffenbaugh A. E., Chang Y. H., Bailly E., Erdjument-Bromage H., Tempst P., Buranda T., Sklar L. A., Baumler J., Gogol E., Skowyra D. (2005) ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell 121, 553–565 [DOI] [PubMed] [Google Scholar]
- 59.Kiss P., Szabó A., Hunyadi-Gulyás E., Medzihradszky K. F., Lipinszki Z., Pál M., Udvardy A. (2005) Zn2+-induced reversible dissociation of subunit Rpn10/p54 of the Drosophila 26S proteasome. Biochem. J. 391, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., Groll M. (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., Finley D. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 64.Lam Y. A., Lawson T. G., Velayutham M., Zweier J. L., Pickart C. M. (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416, 763–767 [DOI] [PubMed] [Google Scholar]
- 65.Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 66.Peth A., Besche H. C., Goldberg A. L. (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiu X. B., Ouyang S. Y., Li C. J., Miao S., Wang L., Goldberg A. L. (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao T., Song L., Jin J., Cai Y., Takahashi H., Swanson S. K., Washburn M. P., Florens L., Conaway R. C., Cohen R. E., Conaway J. W. (2008) Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell 31, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zediak V. P., Berger S. L. (2008) Hit and run: transient deubiquitylase activity in a chromatin-remodeling complex. Mol. Cell 31, 773–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 [DOI] [PubMed] [Google Scholar]
- 71.Besche H. C., Peth A., Goldberg A. L. (2009) Getting to first base in proteasome assembly. Cell 138, 25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walz J., Erdmann A., Kania M., Typke D., Koster A. J., Baumeister W. (1998) 26S proteasome structure revealed by three-dimensional electron microscopy. J. Struct. Biol. 121, 19–29 [DOI] [PubMed] [Google Scholar]
- 73.da Fonseca P. C., Morris E. P. (2008) Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 283, 23305–23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aloy P., Russell R. B. (2002) Potential artefacts in protein-interaction networks. FEBS Lett. 530, 253–254 [DOI] [PubMed] [Google Scholar]
- 75.Fields S. (2005) High-throughput two-hybrid analysis. The promise and the peril. FEBS J. 272, 5391–5399 [DOI] [PubMed] [Google Scholar]
- 76.Richmond C., Gorbea C., Rechsteiner M. (1997) Specific interactions between ATPase subunits of the 26S protease. J. Biol. Chem. 272, 13403–13411 [DOI] [PubMed] [Google Scholar]
- 77.Gorbea C., Taillandier D., Rechsteiner M. (2000) Mapping subunit contacts in the regulatory complex of the 26S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem. 275, 875–882 [DOI] [PubMed] [Google Scholar]
- 78.Sharon M., Robinson C. V. (2007) The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 76, 167–193 [DOI] [PubMed] [Google Scholar]
- 79.Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karpenahalli M. R., Lupas A. N., Söding J. (2007) TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson D., Hakala K., DeMartino G. N. (2009) Subcomplexes of PA700, the 19S regulator of the 26S proteasome, reveal relative roles of AAA subunits in 26S proteasome assembly, activation, and ATPase activity. J. Biol. Chem. 284, 24891–24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nickell S., Beck F., Korinek A., Mihalache O., Baumeister W., Plitzko J. M. (2007) Automated cryoelectron microscopy of “single particles” applied to the 26S proteasome. FEBS Lett. 581, 2751–2756 [DOI] [PubMed] [Google Scholar]
- 83.Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Striebel F., Kress W., Weber-Ban E. (2009) Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 19, 209–217 [DOI] [PubMed] [Google Scholar]
- 85.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 86.Yu Y., Smith D. M., Kim H. M., Rodriguez V., Goldberg A. L., Cheng Y. (2010) Interactions of PAN′s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 29, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Z., Torii N., Furusaka A., Malayaman N., Hu Z., Liang T. J. (2000) Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275, 15157–15165 [DOI] [PubMed] [Google Scholar]
- 88.Chen C., Huang C., Chen S., Liang J., Lin W., Ke G., Zhang H., Wang B., Huang J., Han Z., Ma L., Huo K., Yang X., Yang P., He F., Tao T. (2008) Subunit-subunit interactions in the human 26S proteasome. Proteomics 8, 508–520 [DOI] [PubMed] [Google Scholar]
- 89.Bajorek M., Glickman M. H. (2004) Keepers at the final gates: regulatory complexes and gating of the proteasome channel. Cell. Mol. Life Sci. 61, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kampmann M., Blobel G. (2009) Biochemistry. Nascent proteins caught in the act. Science 326, 1352–1353 [DOI] [PubMed] [Google Scholar]
- 91.Russel D., Lasker K., Phillips J., Schneidman-Duhovny D., Velázquez-Muriel J. A., Sali A. (2009) The structural dynamics of macromolecular processes. Curr. Opin. Cell Biol. 21, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takeuchi J., Tamura T. (2004) Recombinant ATPases of the yeast 26S proteasome activate protein degradation by the 20S proteasome. FEBS Lett. 565, 39–42 [DOI] [PubMed] [Google Scholar]
- 93.Wilkinson C. R., Wallace M., Seeger M., Dubiel W., Gordon C. (1997) Mts4, a non-ATPase subunit of the 26S protease in fission yeast is essential for mitosis and interacts directly with the ATPase subunit Mts2. J. Biol. Chem. 272, 25768–25777 [DOI] [PubMed] [Google Scholar]
- 94.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 95.Cagney G., Uetz P., Fields S. (2001) Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiol. Genomics 7, 27–34 [DOI] [PubMed] [Google Scholar]
- 96.Uetz P., Giot L., Cagney G., Mansfield T. A., Judson R. S., Knight J. R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Qureshi-Emili A., Li Y., Godwin B., Conover D., Kalbfleisch T., Vijayadamodar G., Yang M., Johnston M., Fields S., Rothberg J. M. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 97.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 99.Russell S. J., Sathyanarayana U. G., Johnston S. A. (1996) Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26S proteasome. J. Biol. Chem. 271, 32810–32817 [DOI] [PubMed] [Google Scholar]
- 100.Satoh K., Sasajima H., Nyoumura K. I., Yokosawa H., Sawada H. (2001) Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry 40, 314–319 [DOI] [PubMed] [Google Scholar]
- 101.Xie Y., Varshavsky A. (2000) Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 97, 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seeger M., Hartmann-Petersen R., Wilkinson C. R., Wallace M., Samejima I., Taylor M. S., Gordon C. (2003) Interaction of the anaphase-promoting complex/cyclosome and proteasome protein complexes with multiubiquitin chain-binding proteins. J. Biol. Chem. 278, 16791–1679612615927 [Google Scholar]
- 103.Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 104.Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., Cohen R. E. (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8, 994–1002 [DOI] [PubMed] [Google Scholar]
- 105.Tone Y., Tanahashi N., Tanaka K., Fujimuro M., Yokosawa H., Toh-e A. (2000) Nob1p, a new essential protein, associates with the 26S proteasome of growing Saccharomyces cerevisiae cells. Gene 243, 37–45 [DOI] [PubMed] [Google Scholar]
- 106.Isono E., Saeki Y., Yokosawa H., Toh-e A. (2004) Rpn7 Is required for the structural integrity of the 26S proteasome of Saccharomyces cerevisiae. J. Biol. Chem. 279, 27168–27176 [DOI] [PubMed] [Google Scholar]
- 107.Gandhi T. K., Zhong J., Mathivanan S., Karthick L., Chandrika K. N., Mohan S. S., Sharma S., Pinkert S., Nagaraju S., Periaswamy B., Mishra G., Nandakumar K., Shen B., Deshpande N., Nayak R., Sarker M., Boeke J. D., Parmigiani G., Schultz J., Bader J. S., Pandey A. (2006) Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat. Genet. 38, 285–293 [DOI] [PubMed] [Google Scholar]
- 108.Li T., Duan W., Yang H., Lee M. K., Bte Mustafa F., Lee B. H., Teo T. S. (2001) Identification of two proteins, S14 and UIP1, that interact with UCH37. FEBS Lett. 488, 201–205 [DOI] [PubMed] [Google Scholar]
- 109.Biegert A., Mayer C., Remmert M., Söding J., Lupas A. N. (2006) The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 34, W335–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 111.Lupas A., Van Dyke M., Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 112.Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lasker K., Phillips J. L., Russel D., Velázquez-Muriel J., Schneidman-Duhovny D., Tjioe E., Webb B., Schlessinger A., Sali A. (2010) Integrative Structure Modeling of Macromolecular Assemblies from Proteomics Data Mol. Cell. Proteomics 9, 1689–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]