Abstract
Chemical cross-linking of reactive groups in native proteins and protein complexes in combination with the identification of cross-linked sites by mass spectrometry has been in use for more than a decade. Recent advances in instrumentation, cross-linking protocols, and analysis software have led to a renewed interest in this technique, which promises to provide important information about native protein structure and the topology of protein complexes. In this article, we discuss the critical steps of chemical cross-linking and its implications for (structural) biology: reagent design and cross-linking protocols, separation and mass spectrometric analysis of cross-linked samples, dedicated software for data analysis, and the use of cross-linking data for computational modeling. Finally, the impact of protein cross-linking on various biological disciplines is highlighted.
The concept of protein cross-linking as a (bio)chemical tool to infer structural information about protein conformations and protein-protein interactions in combination with mass spectrometry was introduced at the end of the 1990s (1). In a seminal paper, Young et al. (1) used chemical cross-linking of lysine residues in bovine basic fibroblast growth factor FGF-2 (heparin-binding growth factor 2) to provide distance constraints for the computational derivation of the fold of this small (17-kDa) protein. FGF-2 was cross-linked with bis(sulfosuccinimidyl) suberate, purified by size exclusion chromatography, and digested with trypsin. Cross-linked peptides were separated by HPLC and analyzed on line by ESI-TOF and off line by MALDI-TOF mass spectrometry. Putative cross-links were then assigned based on their precursor masses, and some of them were verified by MALDI postsource decay. The authors could identify 15 cross-links that did not bridge directly adjacent lysines and therefore provided information on the three-dimensional structure of the protein. These data were used to assign FGF-2 to the β-trefoil family by excluding calculated models that did not fit the distance constraints.
In the last decade, the application of protein cross-linking has expanded, first and foremost driven by developments in mass spectrometry as the method of choice for the high throughput identification of proteins and their modifications. Reviews by Back et al. (2), Sinz (3), and most recently Lee (4) give an overview on the evolution of the field. However, despite the progress that has undoubtedly been made, cross-linking is still considered a “niche” technique that has not (yet) lived up to its promises. High throughput generation of data supporting protein fold prediction and the determination of protein-protein interactions have not been realized routinely. There may be several reasons for that such as the necessity of access to high end mass spectrometers, the requirement of specialized reagents, and the need for tailored software. However, recent years have seen an increased interest in this technique, which is reflected in the literature and by the emergence of new reagents and software tools.
Here, we present an overview of recent developments in methodology, instrumentation, and bioinformatics related to chemical cross-linking of proteins and the analysis of cross-linked peptides by mass spectrometry. Other cross-linking areas such as protein-DNA cross-linking, photoinduced cross-linking, or the characterization of disulfide bonds will not be covered in detail in this paper. We critically discuss advantages and limitations of different concepts and look beyond the immediate outcome of cross-linking experiments (putative interactions and/or distance constraints) and examine the potential role of chemical cross-linking in the analysis of protein interaction networks and, more generally, for structural and systems biology.
CROSS-LINKING CHEMISTRIES AND LINKER DESIGN
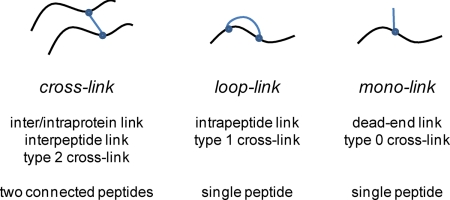
The major aim of the cross-linking reaction is the formation of a covalent bond between two spatially proximate residues within a single or between two polypeptide chains. Unfortunately, this is not the only possible reaction product; it is also possible and, depending on the sample, even more likely that only one end of the bifunctional cross-linker will react with the protein because the other end does not come into contact with another cross-linkable residue, or the second reactive group is deactivated, e.g. by hydrolysis, before forming a cross-link. Therefore, different products of the cross-linking reaction, which are summarized in Fig. 1, may be observed. The nomenclature of these species varies between authors, and in this article, we will classify them either as monolinks, loop-links, and cross-links. A more detailed discussion about nomenclature of cross-linking products can be found in a paper by Schilling et al. (5).
Fig. 1.
Nomenclature of common products of chemical cross-linking reactions. The terminology, cross-link, loop-link, and monolink, used in this article is shown.
Over the years, a large number of chemical cross-linking reagents have been developed. Broadly, they may be classified in several categories according to their reactivity (e.g. amine- or thiol-reactive and homo- and heterobifunctional) or the incorporation of additional functional groups (e.g. cleavable sites and affinity tags). In the following, we will discuss both conventional and functionalized reagents.
Conventional Cross-linking Reagents
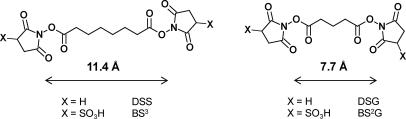
This group of chemical cross-linking reagents consists of two reactive sites connected through a spacer or linker region, typically an alkyl chain. Most commonly, the reactive groups of cross-linkers target the primary amino group of lysine (and the protein N termini). For this purpose, N-hydroxysuccinimidyl or sulfosuccinimidyl esters are almost exclusively used. These “active esters” have high reaction rates but are at the same time also susceptible to rapid hydrolysis in aqueous solutions with half-lives at a time scale of tens of minutes under typical reaction conditions (pH > 7, 25–37 °C). The competing hydrolysis reaction limits the possible reaction time and makes it difficult to obtain good cross-linking yields for low protein concentrations. Common succinimide-type linkers are disuccinimidyl suberate (DSS;1 six-carbon linker) and disuccinimidyl glutarate (DSG; three-carbon linker) as well as their sulfo analogs bis(sulfosuccinimidyl) suberate (BS3) and bis(sulfosuccinimidyl) glutarate, which are more soluble in purely aqueous solutions. DSS and DSG, in contrast, require prior dissolution in small volumes of polar organic solvents such as N,N-dimethylformamide or DMSO before addition to the sample. Structures are shown in Fig. 2.
Fig. 2.
Structures of most commonly used amine-reactive cross-linking reagents: DSS, BS3, DSG, and bis(sulfosuccinimidyl) glutarate (BS2G).
Lysine cross-linking has several advantages, including the high prevalence of Lys residues (about 6%) and relatively high reaction specificity. Side reactions of N-hydroxysuccinimide esters with other amino acids usually do not occur at relevant levels under carefully controlled reaction conditions (pH, reaction times, and reagent excess), although they have been reported in the literature (6, 7). Similar specific cross-linking reactions can be carried out when targeting cysteine residues, e.g. by maleimides, but the low abundance of Cys (<2%) makes this less attractive. Other cross-linking chemistries are not frequently used either because the reactions cannot be performed under appropriate (“native”) conditions or because reaction products are instable or inhomogeneous. Examples include arginine-specific cross-linking or acidic cross-linking (8, 9).
In addition to homobifunctional cross-linkers, several heterobifunctional linkers have been described. These may incorporate two different reactive groups, e.g. Lys- and Cys-reactive, or may combine different cross-linking concepts, e.g. chemical and photoinduced cross-linking. However, these approaches pose additional difficulties to data analysis.
A notable exception to the general linker design is formaldehyde, which only contains a single aldehyde group but is able to connect two amino acid side chains via a two-step reaction. Formaldehyde is a less specific reagent, although lysine and tryptophan residues are primarily targeted (10, 11). Coupling reagents, for example carbodiimides such as ethyl diisopropyl carbodiimide, are only involved in an intermediate reaction step but do not introduce additional atoms into the molecule. The result is a so-called “zero-length” cross-link in the form of an amide bond between Lys and Asp/Glu residues that, however, requires very close spatial proximity. Furthermore, it poses additional difficulties in that cross-links between two sites that are near each other in the primary sequence may be difficult to discriminate from missed cleavages during the course of mass spectrometric analysis.
Functionalized Cross-linking Reagents
To facilitate the analysis of the products of cross-linking reactions by mass spectrometry, different types of functionalized cross-linking reagents have been proposed. These include linkers carrying stable isotope labels, affinity tags, or moieties that give characteristic fragmentation patterns in tandem mass spectrometry experiments. The use of stable isotope-labeled cross-linkers was first reported by Müller et al. (12) in 2001. By using a mixture of a cross-linker containing only natural (“light”) isotopes and a “heavy” (usually deuterated) form of the reagent, reaction products carry a unique isotopic signature. This feature is used for detecting peptides carrying mono- or cross-links among a large excess of unmodified peptides in enzymatic digests of complex samples but also facilitates the interpretation of MS/MS spectra of cross-linked peptides provided that the heavy and light form of the cross-linked peptides are sequenced. This is possible because only fragment ions containing the cross-link site are shifted in mass between light and heavy forms. Different stable isotope-labeled cross-linking reagents such as DSS or BS3 are now commercially available from suppliers such as Creative Molecules and the Pierce division of Thermo Scientific, and more complex reagents have also been prepared in labeled form. Alternatively, an isotopic signature may also be introduced into cross-linked peptides by digestion in H218O (13, 14), which allows the differentiation of cross-linked peptides from all other classes of peptides (including monolinks) because 18O is incorporated into both tryptic C termini, ideally resulting in a mass shift of 8 Da for these peptides. However, achieving quantitative 18O labeling is difficult even for linear peptides.
The introduction of affinity tags into protein chemistry reagents has found widespread use for quantitative proteomics as well as for the characterization of post-translational modifications (15, 16). Along this line, affinity-tagged cross-linking reagents have also been introduced by several research groups. Most frequently, biotin is used as the affinity group, allowing the isolation of modified peptides by avidin affinity chromatography (17, 18). A different approach introduced by de Koster and co-workers (19) uses an azide-containing cross-linking reagent to capture cross-links on a cyclooctyne resin involving a “click chemistry”-type reaction. Yan et al. (20) reported a lysine-reactive linker with a protected thiol group that is used for enrichment after performing the cross-linking step. The SH group then reacts with beads that carry an iodoacetyl group for capture and a photocleavage site for elution.
Affinity cross-linkers, however, need to be custom synthesized and are typically considerably more bulky than conventional reagents. This may affect their reactivity because of steric hindrance, and the accuracy of spatial constraints is reduced. So far, the application of this group of cross-linkers has been largely restricted to model studies.
Another variety of functionalized reagents uses linkers with specially designed fragmentation properties. Most frequently, these linkers contain labile bonds that are easily cleaved during collision-induced dissociation as first proposed by Bruce and co-workers (21). Their concept, termed protein interaction reporter (PIR), also involves the generation of a diagnostic ion from the cross-linker upon fragmentation. Therefore, the primary fragmentation products of this type of cross-linker are the two connected peptides with part of the linker attached. Sequence information for the individual peptides may be obtained from subsequent MS3 experiments. Again, a major limitation is the design of gas-phase cleavable reagents without making them excessively large or poorly soluble. Also, MS3 experiments require more analysis time, and sensitivity is compromised. A different strategy has recently been proposed by Adkins and co-workers (22). In their approach, a diagnostic neutral loss of NO2 is produced from a cross-linking reagent containing a nitro group. From the presently available data, however, it is difficult to estimate whether this diagnostic pattern is generally applicable for a large number of cross-linked peptides. In contrast, Bruce and co-workers (23) have described different applications for PIR cross-linkers, including in vivo cross-linking in Shewanella oneidensis. In this case, they reported more than 20 cross-links mainly involving membrane proteins.
In summary, the “ideal” cross-linking reagent is stable, reactive, and sufficiently soluble under the relevant biological conditions that favor protein (complex) stability; does not fragment under conditions that induce peptide bond cleavage to allow identification; and does not exceed a certain cross-linking distance to generate meaningful spatial information. The major challenge has not been the actual design or chemical synthesis of novel cross-linking reagents that exhibit the majority of these properties. Rather, it has been the difficulty to find conditions that combine appropriate reaction conditions for cross-linking of native proteins and the physicochemical properties of complex multifunctional cross-linkers that have limited progress in the field.
Cross-linking Protocols
Regardless of the particular reagent used, an essential step is the refinement and optimization of the cross-linking protocol. A prerequisite to a successful experiment is that the reaction proceeds under conditions that preserve the native state of the protein or protein complex. Therefore, the pH of the buffer should ideally be in the range of 6.5–8.5. Obviously, the buffer used must not contain any functional groups that interfere with the reaction such as amines in the case of succinimide cross-linking. The use of high protein concentrations (in the mg ml−1 range) is highly desirable although not always achievable for relevant targets. Reactions may be carried out at different temperatures ranging from 4 to 37 °C. The reaction time depends on the reagent and ranges from minutes to 1 h or more. When reactive compounds are still present after that period of time, they are usually quenched before the sample is processed further, e.g. by a pH shift or the addition of scavenging reagents.
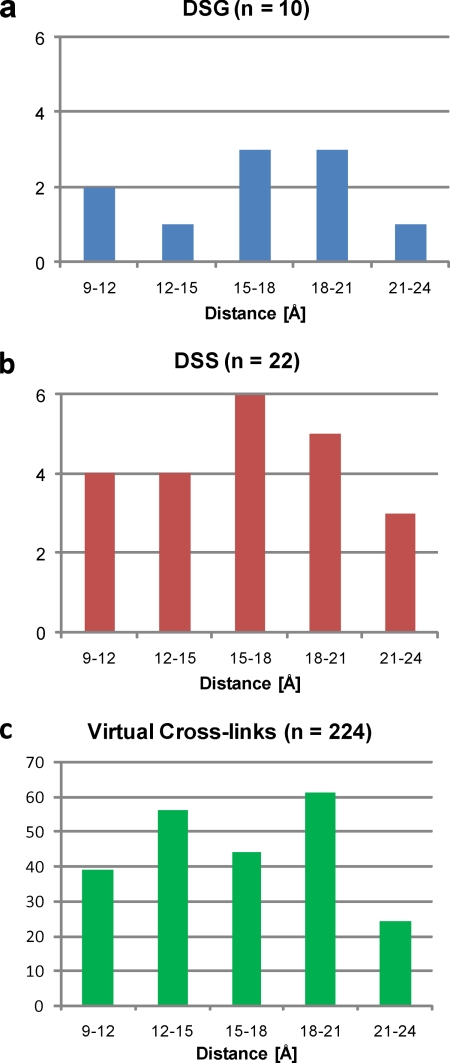
The chain length of the spacer determines many essential properties of the reagent, including its hydrophobicity (and therefore solubility) and the maximum distance between cross-linked residues. The longer the spacer, the more likely it is that two cross-linkable reactive groups are within the distance range of the reagent. However, this comes at the price of a reduced accuracy in determining the spatial distance between the cross-linked residues. Even with relatively small linkers spanning about 10 Å, residues in a distance of up to 25 Å (measured from the backbone α-carbons) may be linked because of the length and the flexibility of the amino acid side chains that are involved. With more complex cross-linker designs, this value may become much larger, eventually making such reagents useless for obtaining even low resolution structural information, particularly for small proteins. In contrast, the use of very short linkers (such as formaldehyde) or zero-length cross-linkers such as ethyl diisopropyl carbodiimide requires almost direct contact of the cross-linkable sites. In the literature, it is frequently proposed that using cross-linking reagents that differ only in their spacer length (e.g. DSS and DSG) may be used to refine the distance constraints. In practice, the difference of three -CH2- groups between DSS and DSG contributes less than 3 Å to the total span of the cross-link, so the main effect is that fewer cross-links are observed with DSG. For example, in a mixture of seven proteins with known three-dimensional structure, the absolute number of non-redundant cross-links we observed decreased from 22 using DSS to 10 using DSG, whereas, interestingly, the average distances as determined from the Protein Data Bank data hardly differed, averaging around 17 and 16 Å, respectively (Fig. 3, a and b).2 It is also evident that the number of experimentally observed cross-links is significantly smaller than the theoretical cross-links shown in Fig. 3c, although the distribution of distance constraints for DSS follows a similar distribution. This suggests that only a fraction of the theoretically possible cross-links are confidently identified. Existing cross-links may not be observed for several reasons, including their low abundance; unfavorable chromatographic, ionization, and fragmentation properties; and unsuitable peptide length (see also “Analysis of Cross-linked Samples by Mass Spectrometry”).
Fig. 3.
Comparison of cross-linking using DSS and DSG. Shown are the distances (Nε-Nε) between two cross-linked lysine residues in different model proteins determined from published three-dimensional structures for DSG (a) and DSS (b) and the theoretically possible (“virtual”) (c) cross-links. Chemical cross-linking was performed as described previously (26); details about the calculations can be found in the supplemental method details for simulations.
One issue that is sometimes raised in connection with cross-linking experiments is the generation of artifacts and ensuing false positive identifications. Although stringent criteria for data analysis are essential to avoid random assignments in MS data sets (discussed below), the chances of generating cross-links from randomly “interacting” partners are actually very low. One has to consider that for a cross-link to be formed reactive sites must be in close proximity for a sufficient period of time for a substantial fraction of the population of interacting protein molecules. Random contacts, by definition, do not fulfill this requirement and result, if at all, only in very small amounts of a particular link that will likely be below the limit of detection.
ANALYSIS OF CROSS-LINKED SAMPLES BY MASS SPECTROMETRY
Separation and Enrichment of Cross-linked Peptides
Based on the reaction principles of chemical cross-linking, reaction products are present in low abundance compared with unreacted protein. With few exceptions (24), cross-linked protein samples are subjected to proteolytic digestion prior to mass spectrometric analysis (“bottom-up” approach). In such samples, cross-linked peptides are again rare and of low abundance compared with unmodified peptides. Typically, therefore, the complexity of the resulting sample requires the use of chromatographic or electrophoretic separation steps to reduce the interference of unmodified proteins or peptides and/or to enrich for the targeted cross-linked peptides. Most frequently, one-dimensional SDS-PAGE or reversed-phase HPLC is used for this purpose.
In the gel-based strategy, bands corresponding to cross-linked protein (as determined by the mass shift) are cut from the gel, digested in gel, and analyzed either directly by MALDI-MS or subjected to LC-MS/MS analysis. Clearly, this approach is limited to the analysis of cross-linked products of individual proteins or very simple mixtures in which the cross-linked products are observed as discrete bands in the gel. Efficient recovery of cross-linked peptides from the gel may be impaired because of their size. On the other hand, the gel-based workflow removes the large excess of unreacted protein or protein carrying only monolinks. In general, SDS-PAGE is an attractive method to optimize cross-linking protocols even for complex samples. In this case, an increased extent of cross-linking results in the appearance of diffuse bands on the gel caused by the formation of a large number of heterogeneous monolinked intermediates and cross-link products. With increasing degree of cross-linking, high mass aggregates become visible on the top of the gel. The formation of such heavily cross-linked species should be avoided as the products may be difficult to digest or poorly soluble.
More complex samples are typically digested in solution prior to reversed-phase LC separation and MS/MS analysis of the resulting peptide mixture either in on-line or off-line mode. Additional separation steps such as isoelectric focusing may be used. These separation methods reduce the complexity of peptide samples and thus facilitate the detection of cross-linked peptides and their selection for sequencing in data-dependent MS/MS; however, they do not specifically enrich for cross-linked species. Such specific enrichment may be achieved by affinity chromatography when affinity-tagged cross-linking reagents are used (see above), although this approach cannot discriminate between tags present on mono- and cross-link products. In contrast, two other chromatographic techniques are able to at least partly discriminate between these two types of peptides. Strong cation exchange (SCX) chromatography takes advantage of the difference in positively charged groups in non-cross-linked and cross-linked peptides: although “normal” tryptic peptides typically carry two positive charges, one at the N terminus and one at the side chain of the C-terminal amino acid, cross-linked peptides have twice the number of protonated sites. Thus, the latter are eluted from SCX material only at higher salt concentrations. In practice, the efficiency of this method is limited by missed cleavages in unmodified peptides, leading to higher charge states as well as poor chromatographic efficiency: peptides usually elute in groups according to their charge state in solution (2+, 3+, 4+, etc.) (25). Nevertheless, our group has successfully used SCX fractionation in the analysis of cross-linked peptides obtained from whole cell lysates (26). Alternatively, we are currently evaluating the use of size exclusion chromatography (SEC) for the enrichment of cross-linked peptides. Peptide-level SEC takes advantage of the higher molar mass of cross-linked peptides. In addition, these peptides are more bulky than their linear counterparts, resulting in a further shift to lower retention volumes in SEC.
Analysis by Mass Spectrometry
Various instrument types have been used for MS analysis of cross-linked peptides, but generally the availability of high mass accuracy detection at the MS1 level is essential. This is due to the necessity to constrain the enormous search space being generated by the combination of two peptides in a cross-link. For larger databases, these numbers easily reach millions or even billions. Issues connected to data analysis in protein cross-linking experiments are discussed further under “Analysis of Mass Spectrometry Data from Cross-linked Samples.”
Up to now, most of the work has been carried out on MALDI-TOF or -TOF/TOF and ESI-Q-TOF instruments, particularly for less complex samples, and more recently ESI-LIT-FTICR (26–28) and -Orbitrap (22, 29, 30) hybrid instruments. Especially when searching larger databases, mass accuracies <10 ppm are essential, even more so when only MS1-level information is used for data analysis. As a result of the presence of two peptide chains in the molecule, the fragmentation behavior of cross-linked peptides is much more complex than for linear peptides. Even if the cross-linker itself does not contribute any additional fragment ions, MS/MS spectra contain simple backbone ions from one of the two peptide chains as well as fragment ions containing the cross-linker. The fragment ion spectra of cross-linked peptides are further complicated by the presence of fragment ions of different charge states when ESI is used as the ionization technique as precursor ions of cross-linked peptides typically carry three or more positive charges. However, this property can be utilized during the selection of precursor ions for data-dependent MS/MS by excluding precursors of charge states 1+ and 2+.
Because of the lack of appropriate software to automatically interpret fragment ion spectra of cross-linked peptides, in most studies until recently, the data have been analyzed manually. In addition to the generic limitations of manual evaluation such as limited throughput, lack of reproducibility, and lack of objective scoring criteria, it is particularly problematic in the case of cross-linked peptides because their spectra are highly complex and often of low signal intensity. Over the last few years, innovative approaches toward the automated interpretation of MS/MS spectra of cross-linked peptides have been described in the literature (reviewed by Lee (4) and discussed in more detail below). For example, xQuest, a search engine developed in our laboratory, takes advantage of isotope-coded cross-linkers (described above) to aid in the interpretation of tandem mass spectrometry data (26). By comparing MS/MS spectra of cross-links containing the light and heavy cross-linkers, respectively, xQuest is able to create subspectra of “backbone” ions (corresponding to fragments from a single peptide chain only) that are identical in the two spectra and “cross-link” ions (fragment ions, including the cross-link site) that exhibit a mass shift according to the isotope label. This deconvolution step considerably reduces the complexity of MS/MS spectra, and consequently, the significance of the assignment of fragment ions to peaks is increased. Other programs, for example, treat a cross-link as a variable mass modification of one linked peptide on the other peptide to facilitate searches (see “Analysis of Mass Spectrometry Data from Cross-linked Samples”).
But even with the availability of advanced bioinformatics tools, spectral quality is essential for the successful identification of cross-links. The presence of a sufficient number of fragment ions from both connected peptides is a critical prerequisite that is not always fulfilled. In particular, the combination of two peptides of very different lengths in a cross-link rarely yields reliable identifications because of the absence of bond cleavages in the shorter chain. Because large data sets of cross-linked peptides are scarce, there is very little information available on the general fragmentation behavior of cross-linked peptides. CID has been predominantly used as the fragmentation technique in cross-linking studies but is known to provide limited sequence information in a number of cases (31). For example, dominant cleavages on the N-terminal side of proline are induced, resulting in reduced abundance of fragment ions representing cleavages at other sites. Electron-based fragmentation techniques, i.e. electron capture dissociation (32) and electron transfer dissociation (ETD) (33), promise increased sequence coverage because of their non-ergodic nature and are particularly suitable for more highly charged precursors. Until recently, the use of these techniques in protein cross-linking studies has been very limited due to the restriction of electron capture dissociation to the expensive FTICR platform and of ETD to low resolution ion traps. The commercial availability of ETD on LIT-Orbitrap and Q-TOF systems should make this fragmentation technique more attractive, and the first data on ETD of cross-linked peptides have recently appeared in the literature (22).
New generations of MS/MS instruments now generate high mass accuracy MS/MS spectra without compromising sequencing speed (34, 35). This is expected to further improve the interpretation of cross-link data by providing unambiguous charge state information for both precursor and fragment ions and increased mass accuracy.
ANALYSIS OF MASS SPECTROMETRY DATA FROM CROSS-LINKED SAMPLES
The analysis of MS data from cross-linking experiments is a challenging undertaking mainly because of the overwhelming numbers of possible combinations that have to be considered by the search engine. As listed in Table I, several different approaches have been developed in the last decade to solve this task and to assist in the automated identification of cross-linked peptides from MS data. Most of these algorithms are designed for specific cross-linking chemistries (conventional, amine-reactive cross-linkers, cleavable cross-linkers, etc.) and the MS workflow that is used for the analysis.
Table I. Listing and main features of software tools for analysis of MS and MS/MS data from cross-linking experiments.
“MS1” indicates assignment of cross-links based on MS1-level information only; “MS/MS” indicates functionality to assign peaks of MS/MS spectra to theoretical spectra of candidate cross-links. “Batch search” indicates whether consecutive searching of multiple MS/MS spectra is possible. “Database size” indicates whether the search space is restricted to a “small” number of sequences (<100) or whether it has been demonstrated that large sequence databases may be searched (“large”). In the “Cross-linker” column, “#” indicates that the generic mass shift of the cross-linker may be defined by the user, “PIR” indicates that it was designed for protein interaction reporter cross-linking reagents, and “iso” indicates that software may be used with isotopically labeled cross-linkers. GPMAW, General Protein/Mass Analysis for Windows; ASAP, Automated Spectrum Assignment Program; CLPM, Cross-Linked Peptide Mapping Algorithm for Mass Spectrometric Analysis. ✓, available/possible; —, not available/not possible.
| Algorithm | MS data |
Database size |
Cross-linker | Scoring | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| MS1 | MS/MS | Batch search | Small | Large | ||||
| ProteinXXX (GPMAW) | ✓ | ✓ | — | ✓ | — | # | — | 37 |
| X-links | — | — | ✓ | ✓ | ✓ | PIR | — | 78 |
| Links and MS2Links (formerly ASAP and MS2assign) | ✓a | ✓b | — | ✓ | — | # | — | 1, 5 |
| CLPM | ✓ | — | — | ✓ | — | # | — | 39 |
| Pro-CrossLink (for 16O/18O-labeled samples) | ✓ | ✓ | — | ✓ | — | # | — | 79 |
| xQuest, xBobcat | ✓ | ✓ | ✓ | ✓ | ✓ | #/iso | ✓ | 26 |
| XLink (for 16O/18O-labeled samples, LC-MALDI) | — | ✓ | ✓ | ✓ | — | # | — | 80 |
| Protein Prospector (open mass modification search) | ✓ | ✓ | ✓ | ✓ | — | # | ✓ | 41 |
| Popitam (CXMS) (open mass modification search) | — | ✓ | ✓ | ✓ | — | # | ✓ | 29 |
| X!Links | — | ✓ | ✓ | ✓ | — | # | ✓ | 28, 81 |
| XDB approach (+search engine), Phenyx | — | ✓ | ✓ | ✓ | — | # | ✓ | 27 |
a Links.
b MS2Links.
MS1-based Algorithms
Algorithms that rely on MS1 information (e.g. from MALDI-TOF data) to identify cross-linked peptides use differential comparison of cross-linked and control samples (1, 36–39). Furthermore, labeling strategies such as tryptic digestion in 18O-labeled water (13) or mixing of unlabeled (14N) and 15N-labeled interacting proteins (40) have been used to identify cross-linked peptides at the MS1 level by introducing characteristic isotopic signatures. Candidate peptide combinations are assigned by peptide mass matching, similar to the peptide mass fingerprinting approach used in classic protein identification. However, MS1-based methods are very limited with regard to sample complexity, and cross-linked peptides cannot be identified unambiguously solely by MS1 information when searching larger databases even when high mass accuracy data are available. In theory, sub-ppm mass accuracy could limit the number of possible peptide cross-links even in large databases to a few candidates or even a single cross-link, especially if a PIR-type cross-linker would be used (23). In reality, however, even a low complexity protein sample gives rise to an enormous number of ions that cannot be assigned to any fully tryptic peptide. Missed and miscleavages and post-translational and artifactual modifications contribute to these “untypical” species, which are particularly relevant for cross-linked samples. Therefore, candidate cross-links identified by accurate mass data alone must be further confirmed at the MS/MS level using programs such as MS2Assign (5).
Identification of Cross-linked Peptides from LC-MS/MS Data
The development of instruments that are suitable for high throughput LC-MS/MS analysis (e.g. FTICR and Orbitrap hybrid instruments) and provide high mass accuracy now allows the analysis of more complex samples. These technological improvements raised the demand for novel data analysis software to deal with advanced cross-linking workflows (Table I).
The algorithms that were developed for LC-MS/MS data use precursor mass and fragment ion mass information to identify cross-linked peptides and follow a strategy similar to commonly used search engines for peptide identification. The assignments are based on (a) selection of candidate cross-links from the sequence database, (b) matching of theoretical MS/MS spectra against acquired MS/MS spectra, and (c) scoring of possible candidate/spectrum matches to separate true from false positive identifications. In addition to monolinks and loop-links, which may be considered variable modifications to a single peptide, all possible peptide pair combinations (cross-link candidates) have to be considered by the search engine. The number of possible peptide pairs is equal to the binomial coefficient kn + k − 1 where n is the number of peptides and k = 2 (binary combination). As a consequence, the search space grows exponentially with increasing numbers of peptides. A good estimate for the number of combinations is n2/2.
To illustrate this dramatic expansion of the search space with increasing sample complexity, we calculated the search space for proteome-level amine-reactive cross-linking using data taken from the UniProt/Swiss-Prot database (version 15.10). We selected only fully tryptic peptides, considering a maximum of two missed cleavages and a length of 5–45 amino acids for each peptide but no variable modifications such as phosphorylation or oxidation. Possible combinations amount to 7.3 × 109 for Escherichia coli, 8.2 × 1010 for Saccharomyces cerevisiae, and 7.5 × 1011 for Homo sapiens. The challenge that is posed by this combinatorial explosion becomes clear when the search space of a cross-linking experiment is compared with a conventional peptide/spectrum match search. Even for small proteomes such as E. coli (4367 proteins), the search space is roughly 60,000 times larger, whereas for the human proteome (20,333 proteins), the increase in complexity is ∼600,000-fold. This “explosion” of the search space is the reason why the identification of cross-linked peptides from large sequence databases is such a challenging task. Algorithms need to deal with the overwhelming numbers of possible candidates and the accompanying difficulty to separate true positives from false (random) matches.
Several strategies have been developed recently that use restricted databases to reduce the search space. So far, most cross-linking studies focus on samples of limited complexity such as purified proteins or small purified protein complexes. The composition of these samples is either known in advance or may be determined by common proteomics search strategies. In this case, the investigator is not restricted to a particular type of cross-linker and can choose from a wide variety of reagents.
The approach described by Maiolica et al. (27) is based on the generation of a database containing all possible linearized peptide pair permutations (XDB) where a single residue is considered a monolink site modified with the cross-linker mass. The rationale is that the two peptides present in a cross-link cover the entire set of possible single bond fragments of the cross-link. The advantage of this method is that the MS/MS data may be searched using one of the common search algorithms used in standard proteomics workflows, and this approach was recently introduced into the commercial search engine Phenyx. The method, although elegant, does not solve the search space problem, and manual data validation is still required. Furthermore, the scoring scheme of the search engine is not directly applicable to cross-links. For example, not all possible ion types for each peptide are considered.
Another strategy that, up to now, has only been used with restricted databases is implemented in the tools Batch-Tag, MS-Bridge, and MS-Product that are part of the Protein Prospector package (41) and Popitam (CXMS pipeline) (29). Both are based on the rationale that cross-linked peptides may be considered as a single linear tryptic peptide with a large variable mass modification that corresponds to the second peptide. These approaches identify one part of the cross-link at a time; therefore, the increase in search space is linear and should also allow the use of larger databases, although this has not yet been shown.
Batch-Tag performs the open mass modification search on a restricted subset of proteins that may be identified by using a conventional search engine strategy (42). Subsequently, this set of proteins/peptides is searched allowing a variable mass modification of up to 4000 Da. MS-Bridge then retrieves all candidate cross-links based on the precursor mass, whereas MS-Product matches the complete set of fragment ions of the candidates against the MS/MS spectrum. The recently reported open modification CXMS pipeline (29) makes use of high resolution MS/MS spectra acquired by an LTQ Orbitrap hybrid instrument and the open mass modification search engine Popitam (43), a search engine that is designed to identify peptides with variable modifications. The number of MS/MS spectra considered for the search is greatly reduced by considering only spectra of quadruply and higher charged precursors and the elimination of all spectra that are identified as linear peptides by a common search strategy. The remaining spectra are then searched by Popitam using a database containing the proteins of interest. Popitam uses sequence tags to identify peptides with variable modifications of up to 3 kDa. In a final step, the second peptide is retrieved from the sequence database corresponding to the mass of the modification, and search results have to be validated manually.
The recently developed tools to identify cross-linked peptides using restricted databases are very useful if single proteins or small protein complexes are studied. Nevertheless, these approaches cannot be used if the composition of the sample is largely unknown or the sample is more complex, e.g. in the case of whole proteomes or subcellular fractions.
Identification of Cross-links from Large Sequence Databases
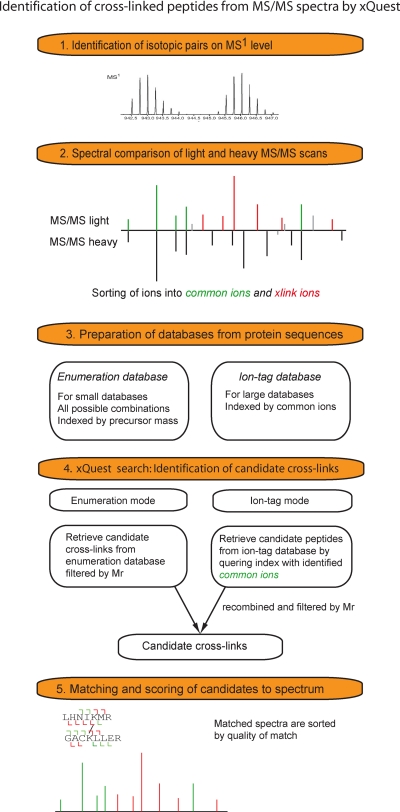
The recently developed, freely accessible software xQuest (26) makes use of isotopically coded cross-linkers to reduce the search space and to identify cross-linked peptides from large sequence databases (see Fig. 4). The software supports searches in two modes, the enumeration mode and the ion-tag mode. The enumeration mode (for databases of up to 100 proteins) considers all possible candidate cross-links, whereas the ion-tag mode may be used to search even larger sequence databases. The ion-tag mode uses a two-step approach: in the first pass possible candidate peptides are identified, and only in the second pass, these candidates are combined in a combinatorial way. This approach requires the use of isotopically coded cross-linkers and the generation of a fragment ion spectrum of both isotopic forms (see above). The presence of isotopically shifted feature pairs on the MS1 level allows the identification of modified peptides. Furthermore, by comparing the corresponding MS/MS spectra of light and heavy precursors, the fragment ions can be separated into sets of common ions (not shifted) and cross-link ions (isotopic shift). Using this information, the search space can be reduced to an extent that cross-links can even be identified from a complex mixture such as an E. coli total cell lysate.
Fig. 4.
Identification of cross-linked peptides from MS/MS spectra using information of isotopically coded cross-linkers and xQuest (26). 1, identification of isotopically shifted MS1 feature pairs. 2, comparison of light and heavy MS/MS spectra of isotope pairs and subsequent sorting of fragment ions into common ions (identical; indicated in green) and cross-link (x-link) ions (shifted; red). 3, preparation of either an “enumeration” mode database for small sequence databases of up to 100 proteins or an “ion-tag” mode database for large sequence databases. 4, xQuest search: retrieving all candidate cross-links filtered by mass if an enumeration mode database is used or retrieving all candidate peptides from the ion index utilizing information of common ions. All possible cross-link candidates are recombined and filtered by mass in a second step. 5, xQuest scoring: the software assigns scores to all candidate cross-links.
The use of xQuest with large sequence databases is based on the assumption that both peptides may be identified from the resulting common fragment ions. Therefore, it is designed for isotopically coded cross-linkers; this allows the identification of common ions but also restricts the choice of the cross-linker. In this mode, only pairs of light and heavy MS/MS spectra can be analyzed. The yield of such paired MS/MS spectra can be increased by directed sequencing of precursor ion pairs that show the expected isotopic shift using inclusion lists (44). This approach requires repeat injections after an initial LC-MS/MS run where isotopic pairs are detected on the MS1 level.
In the enumeration mode, xQuest is a generic tool suitable for most cross-linkers without the need for isotope coding. In addition, it offers an MS1 level-only search tool called xBobcat that assigns possible cross-links based on accurate precursor ion mass alone.
Future Perspectives for Cross-link Search Engines
In light of the increasing availability of computational power, it may soon be feasible to enumerate all possible combinations also from large sequence databases and to search them with one of the now available search tools. Even if this goal should be reached, unconstrained matching in such a huge search space will lead to many false positives, especially if low resolution MS/MS spectra are probed against all possible peptide combinations. This effect is comparable to the well known difficulty of searching data from conventional LC-MS experiments allowing a large number of variable modifications.
Currently, the biggest unresolved issue in computational approaches to cross-linking is the verification and validation of the results that the different algorithms provide without relying on manual validation. Several approaches have been developed to assess the quality of the match of a candidate cross-link spectrum to the theoretical spectrum. So far, cross-correlation scores, match ratio scores, and probabilistic (E-value-based) scores have been reported to separate true positive from false positive identifications (see Table I). In this respect, it also has to be considered that the likelihood of false positive identifications does not increase in a linear fashion with the database size but rather quadratically like the number of combinations itself.
A major source of false positive identifications arises from cases in which one peptide is identified correctly and the second peptide is an incorrect assignment. The proportion of such cases increases if one peptide is very short and fits the precursor mass that is required. In these cases, a candidate cross-link can receive a high score if one peptide alone generates the majority of detected fragment ions. Therefore, assignments that contain a very short peptide segment should be scrutinized very carefully because a random match of a single or even several theoretical fragment ions to spectral noise is easily possible. In addition, several other properties may lead to false positive identifications for short peptides. Typically, few ions that confirm the sequence identity are detectable, particularly in the case of ion trap analyzers that have a lower m/z limit for fragment ions. Therefore, b2, y1, and y2 ions are frequently lost. Furthermore, if the C-terminal amino acids of both peptides are identical (Arg/Arg or Lys/Lys), which is the case for ∼50% of tryptic cross-links, the bn − 1 ions are also identical when either one of the C-terminal amino acids is cleaved off. Therefore, these ions do not contribute sequence-specific information for the shorter peptide (see the example in the supplemental Figure S1 and Tables S1–S3). Finally, for larger databases, small peptides are not proteotypic and cannot be assigned to proteins unambiguously. For these reasons, we recommend using a minimum peptide length of five residues for either cross-linked peptide.
The reevaluation of published cross-linking data is typically very difficult. Frequently, the programs used for the analysis are no longer available or have never been released to the public. Also, experimental sections often lack details about the databases used and validation criteria. Although this is a general problem of the proteomics community, the enormous diversity of workflows in the cross-linking field makes it particularly problematic. In view of the issues raised above, more detailed descriptions of the data analysis procedure, standardized reporting formats similar to the minimum information about a proteomics experiment (45) concept, and public release of the relevant raw data would be highly beneficial. The recent emergence of more publically available algorithms is already a promising sign in that direction.
USE OF CROSS-LINKING DATA FOR MOLECULAR MODELING APPROACHES
Molecular Modeling of Proteins and Protein Complexes
The covalent cross-linking of protein complexes is a convenient technique to discover binding partners of proteins, determine the site of protein interactions, and construct protein-protein interaction networks. However, cross-linking data contain additional information. In particular, if the two amino acids that have been connected by the cross-linker are identified, this can provide information about the spatial distance between the amino acids on the surface of folded proteins. Such spatial distance indications are not highly accurate as they represent only a maximum distance given by the length of the cross-linker and are also influenced by conformational flexibility, but nevertheless they can be used as distance constraints for molecular modeling of protein folds and complex topologies, i.e. the arrangement of all complex constituents in space.
Common molecular modeling strategies usually consist of two steps. The first involves the creation of thousands or even millions of different protein or complex conformations where each protein or complex represents a sample point in the conformational space of the molecular system. In the second step, the energy of each conformation is evaluated using a scoring function that should ideally result in high rankings for conformations close to the native state and low rankings for conformations that are far off the native state. Both steps currently pose significant problems. The large number of degrees of freedom of a polypeptide chain and of protein-protein interactions, respectively, makes the search for the global energy minimum in the conformational space difficult. On the other hand, most currently applied scoring functions lack accuracy and are often unable to distinguish correct from incorrect conformations. The first problem might eventually be solved with increasing computing power. The second problem, however, is more difficult to address as it is based on the inability of current modeling software to treat all kinetic and thermodynamic aspects of binding events such as assessing the effects of water and pH on molecular interactions and incorporating conformational flexibility into calculations (46).
To overcome these challenges, experimental data can be exploited to guide molecular modeling approaches or to provide additional constraints discriminating true from false structures. Distance constraints can focus the conformational space toward the native protein or complex structure that in turn reduces the importance of having an accurate scoring function. Short distance constraints of around 6 Å can be obtained from the location of disulfide bonds and nuclear magnetic resonance spectroscopy experiments (47), whereas longer constraints can be inferred from covalently cross-linking segments of proteins or protein-protein complexes. But how useful are distance constraints for molecular modeling? Havel et al. (48) addressed this question and have put forward the following three rules. 1) Many imprecise distance constraints (e.g. cutoff distances or residue-residue contact information) are to be preferred over few constraints with precise distance information. 2) Distance constraints from residues that are widely separated in the primary sequence are preferred over sequentially nearby residues. 3) Distance constraints should involve as many different residues as possible.
Cohen and Sternberg (49) extended the above rules with the following three observations made on the myoglobin fold. 4) Distance information should be accurate to within a few Å. 5) A modeling software that is known to produce native-like conformations is to be preferred. 6) Dissimilarity between native and non-native structural models should be as large as possible.
For the particular case of predicting the structure of proteins, Havel et al. (48) stated that the number of distance constraints should be in the range of the number of residues in the protein structure. In fact, to determine the spatial coordinates of a protein structure to residue resolution, one would need 3n distance constraints where n is the number of residues in the protein (1). For determining the overall fold of a protein, the number of constraints was estimated to be in the range of n/10 (1, 2), which is likely the maximum yield of cross-links for a single protein structure. Hence, the determination of the protein structure from a cross-linking experiment alone seems to be out of reach but not the determination of its fold. An overview of studies on fold recognition using cross-linking and mass spectrometry can be found in the reviews by Sinz (3) and Lee (4).
In the case of guiding the discovery of protein complex topologies with distance constraints, it could be shown that sometimes as little as three constraints suffice for the deduction of native-like conformations of a protein complex (50). Compared with the folding of a protein, the degrees of freedom of a protein complex are reduced to six translational and three rotational degrees of freedom if the change in the backbone conformation upon complex formation can be neglected. Under such circumstances, up-to-date rigid body docking software is able to predict the correct topology with high probability (51, 52). Nevertheless, as we will show below, distance constraints from intermolecular cross-linking further improve the prediction and also help to validate the topology prediction. Using distance constraints from nuclear magnetic resonance experiments, Tang and Clore (47) could show that the prediction of the topology of two bacterial phosphotransferase complexes to 2-Å root mean square distance (r.m.s.d.) is possible with only three intermolecular cross-link constraints. Similarly, Balasu et al. (53) have put forward a topology model for the mitogen-activated protein kinase pathway enzyme ERK2 and its regulator PTP-SL. Also in this case, three constraints were sufficient to propose the model of the complex. Schulz et al. (54) managed to predict a model of an annexin complex with only four constraints, whereas Chu et al. (36) used the distance constraints from nine cross-links for a model of the signal recognition particle bound to its receptor. For further studies, see Refs. 55–57. A recent review of protein-protein docking methodologies is provided by Ritchie (50).
Quality and Quantity of Virtual Cross-links in Protein-Protein Complexes
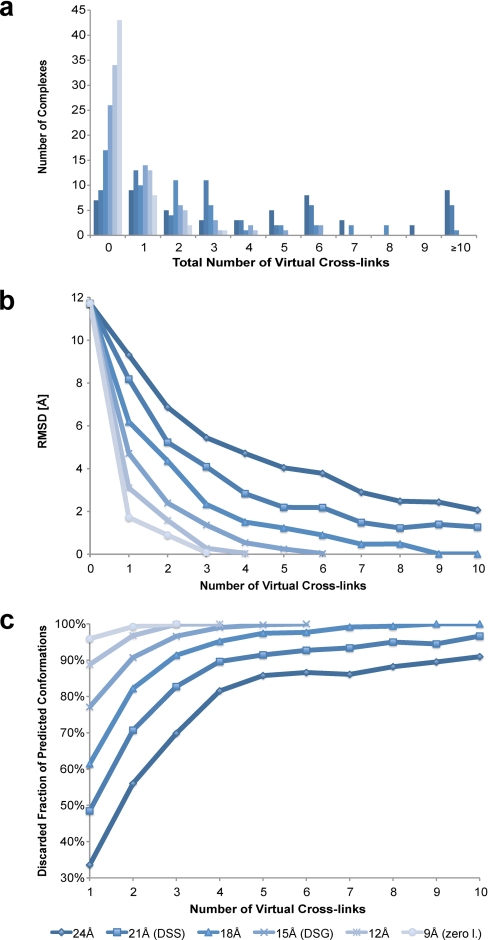
The applicability of cross-links has been addressed in the past as described above. However, all conclusions drawn from the cited studies originate mainly from fold prediction experiments or from experiments on single protein molecules or complexes using modeling software that can now be regarded as outdated. We have performed a theoretical analysis on the applicability of cross-linking-derived distance constraints on the 54 crystal structures of the first Protein-Protein Docking Benchmark set (58). These analyses address several important questions for cross-linking studies. 1) How many cross-links can be observed in native protein complexes? 2) Are distance constraints from cross-linking experiments useful for filtering out false positive predictions of protein complexes? 3) Which length should the ideal intermolecular cross-linker have? 4) How many cross-links are needed for a reasonable prediction of the complex topology?
To address the first question, we have generated virtual, i.e. theoretically possible, cross-links of various lengths for the 54 protein complexes in the benchmark data set (see the supplemental method details for simulations). In general, one can observe that the number of intermolecular cross-links increases with the length of the cross-linker (Fig. 5a). A cross-linker with a maximum span of 24 Å (Nε-Nε distance) such as practically observed with DSS or DSG (compare Fig. 3) is able to produce more than 10 potential distance constraints, whereas a hypothetical 9-Å cross-linker is able to covalently link up to three intermolecular lysine pairs. However, as we have shown above, it is very difficult to practically achieve short distance constraints because the number of experimentally observed cross-links is usually much smaller than the number of theoretically possible cross-links.
Fig. 5.
Influence of distance constraints from Lys-Lys interprotein cross-links in benchmark protein complex data set. Approximate theoretical constraints for DSS (21 Å), DSG (15 Å), and zero-length cross-linking (9 Å) are highlighted in the legend. a, histogram of theoretically possible interprotein cross-links for various distance constraints. b, average r.m.s.d. between native and docked conformations for different numbers of virtual cross-links as constraints. c, fraction of 10,000 calculated structures that can be excluded dependent on the number and maximum length of distance constraints.
Nevertheless, shorter cross-links hold more structural information. Fig. 5b shows the difference between all 54 native complexes and their docked conformations in terms of r.m.s.d. values that were calculated on the Cα atoms of the smaller docking partner while keeping the larger binding partner fixed. The higher the number of cross-links observed for a protein complex, the less relevant the length of the cross-linker becomes. However, in particular, complexes with one or two cross-links of ≥21 Å often produce less accurate predictions with r.m.s.d. values above 9 or 5 Å, respectively.
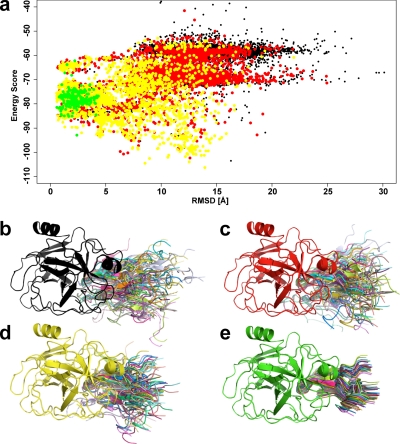
A similar tendency can be observed with the number of false positive structure predictions that can be filtered out from the pool of 10,000 calculated complex conformations (Fig. 5c). A larger number of cross-links or shorter distance constraints result in more non-native conformations being discarded. Three or four constraints of ≤18 Å are already sufficient to reduce the number of plausible topologies for a protein-protein complex to as few as 900 and 500 candidates, respectively. In the case of the bovine trypsin-inhibitor complex (Protein Data Bank code 1TAB), it was possible to discard all except 406 decoy complexes using three 21-Å distance constraints between Lys222E-Lys16I, Lys224E-Lys16I, and Lys60E-Lys31I where E stands for enzyme and I stands for inhibitor (Fig. 6). The inhibitor complex represents a special case in the benchmark data set for which pure computational calculations are unable to predict a native-like conformation on the basis of the calculated energy scores. However, once experimental data in the form of cross-link distance constraints are incorporated and the remaining predicted complex topologies are clustered, it becomes evident that the largest cluster contains almost entirely near-native conformations (Fig. 6a). Thus, it can be concluded that the incorporation of cross-link data into computational topology prediction of protein complexes significantly increases the likelihood of predicting native-like complex structures.
Fig. 6.
Refinement of protein-protein docking using distance constraints obtained from cross-linking data. a, conformational space of 10,000 calculated conformations for the bovine trypsin-inhibitor complex. Black, all conformations without use of distance constraints from cross-links; red and yellow, conformations remaining after consideration of one or two distance constraints from cross-links, respectively; green, largest cluster of conformations after consideration of three constraints and clustering. b–e, schematics of trypsin-inhibitor complex structures showing a superposition of 30 systematically sampled conformations from the respective groups in a. All three-dimensional structures were generated using PyMOL.
Our analysis of the large benchmark data set validated the conclusions previously made by Cohen and Sternberg (49) and Havel et al. (48). We could show that even for less accurate distance constraints three or four cross-links already give adequate information using appropriate modeling software. However, for the present data set, seven of the 54 protein complexes do not have any intermolecular lysine residues that can be cross-linked. Therefore, the development of complementary cross-linking chemistries is highly valuable.
It is evident that the combination of chemical cross-linking with computational methods is a powerful example for the benefits of combining experimental and computational approaches. The two methods complement each other. Chemical cross-links give confident distance constraints that are too few, however, to infer the topology of protein complexes by themselves. Computational docking can reveal atomic details on protein-protein complexes, but determining the correct (lowest energy) conformation is a major problem. The combination of both techniques provides results that are superior to those obtained by a single method. Further improvements could be achieved by including non-physicochemical data in the calculations. Currently, Rosetta's scoring function incorporates various types of physicochemical interactions, such as coulombic charge-charge interactions, hydrogen bonding, van der Waals interactions, desolvation energies, etc. (59, 60). In particular, information on the evolutionary conservation of surface residues could complement the current scoring function as residues that are part of the interface of a protein-protein complex are typically highly conserved (61).
GAINING BIOLOGICAL INSIGHTS FROM CROSS-LINKING EXPERIMENTS
Over the last decades, various cross-linking strategies have been used to preserve labile protein-protein interactions and to eventually identify the binding partners in macromolecular assemblies. Cross-linkers reactive against primary amines are added in the course of protein purification to physically and stably connect the proteins in a complex. The composition of the cross-linked complex is analyzed by gel-based assays, allowing the identification of its subunits (62, 63). To monitor transient enzyme-substrate or protein-ligand interactions in vitro and in vivo, a photocross-linking approach has been developed (64). The site-specific incorporation of a photoactivatable amino acid into recognition motifs and docking sites facilitates trapping of substrate and ligand proteins upon UV irradiation (65, 66). The biochemical isolation of photocross-linked proteins and their identification by mass spectrometry and other analytical techniques are clearly challenged by the fact that products of photocross-linking reactions are substoichiometric. Nevertheless, this method promises to offer a non-invasive way to study macromolecular interactions in a native environment (67). Other conceptually related approaches in the field of “bioorthogonal chemistry” (reviewed recently by Sletten and Bertozzi (68)) may well find applications in connection with cross-linking in the future.
Besides the investigation of protein-protein contacts, a method known as chromatin immunoprecipitation has shown to be invaluable for detailed analyses of protein-DNA interactions under native conditions (69). Cross-linking of intact cells (predominantly with formaldehyde) stabilizes protein-DNA complexes prior to selective immunoprecipitation, which has been used among others to map histone modifications or the in vivo position of transcription factors along target genes (70–72).
Advancing the analysis of protein-protein cross-links from merely detecting the interacting proteins to identifying the two physically connected amino acids in a polypeptide sequence by mass spectrometry allows for the first time the acquisition of low resolution structural information. The given length of the cross-linking reagent defines a measure for the maximum distance between the two linked residues. Up to now, cross-linking in conjunction with mass spectrometry has been preferentially applied to highly purified protein complexes and rarely to more complex samples (for reviews, see Refs. 3 and 4). The identification of a certain number of these distance constraints per protein complex will substantially support its analysis by classic structural methods. Two studies by Rappsilber and co-workers (27, 73) highlight the merit of cross-linking and mass spectrometry for the structural analysis of multisubunit protein complexes. Maiolica et al. (27) demonstrated that distance constraints derived from 25 non-redundant cross-links on a tetrameric Ndc80 complex provide data on the organization of the tetramerization domain and on the register of heterodimeric coiled coil stretches. These structural parameters guided engineering of optimized protein constructs that eventually yielded diffracting crystals of this four-protein complex (73). Apart from aiding protein crystallization, mass spectrometric detection of cross-links has been applied to protein complexes that are not amenable to crystallography. Recently, Rappsilber and co-workers (30) clarified the docking of initiation factor TFIIF subunits to the x-ray structure of the 12-subunit core of RNA polymerase II complex. Although the number and quality of cross-links identified on recombinantly expressed and highly purified protein complexes increase, the ultimate goal will be the acquisition of distance constraints from natively isolated protein assemblies on a routine basis.
Although the identification of cross-linked peptides by mass spectrometry imposes a considerable difficulty, the challenge to further develop this technology has been accepted by several research groups primarily because it promises to address pressing questions in cell, systems, and structural biology. The fitting of x-ray structures into cryoelectron microscopy (EM) maps of moderate resolution as well as the docking of protein interfaces tends to result in multiple spatial solutions that are not necessarily straightforward to discriminate (74). To this end, cross-linking in combination with mass spectrometry can provide spatial constraints to complement fitting and modeling algorithms as implemented e.g. in the Modeler software package (75). Thereby, the resolution gap between x-ray crystallography and single particle cryo-EM or EM tomography is bridged or at least reduced. In the case of large multicomponent assemblies not amenable to crystallization or even biochemical isolation, e.g. nuclear pores, centrosomes, or kinetochores, structural information of atomic detail is usually obtained of subcomplexes with a limited number of components. To create the big picture, namely atomic maps of entire assemblies, such “smaller pieces of the puzzle” need to be arranged in space. To this end, the identification of protein interfaces is critical and can be achieved by cross-linking MS workflows.
Another field that will benefit from cross-linking is interaction proteomics (76, 77). Major challenges in the field are the discrimination of true interactors from false positive contaminants, distinguishing direct from indirect interactions, and stabilizing transient interactions for identification. Because spatial constraints such as maximum distances of amino acids derived from interprotein cross-links are complementary to interaction data, the combination of interaction proteomics with cross-linking/MS is rather obvious. The inclusion of probabilistic scoring models into current workflows is a very active field of research that promises to deal with the identification of false positives in large data sets. By computational modeling, spatial constraints can be translated into component positions of protein complexes (see the accompanying paper by Förster et al. (82)), and direct interactions are revealed. As discussed above, the success of such an approach depends on the number of spatial constraints identified per protein interface and the length of the cross-linker used. Contaminating proteins will not yield spatial constraints that connect to the bait and therefore are excluded from further analysis.
Each of the technical advances described above (the enrichment of cross-linked peptides, high accuracy mass spectrometry, and novel search algorithms) is incremental, but taken together, they comprise a considerable step forward. The analysis of isolated protein complexes has become feasible, and studying the interfaces of assemblies that contain a limited number of protein components is nowadays almost routine work. The analysis of complex mixtures, however, will be restricted to the identification of highly abundant cross-linked peptides from samples not more intricate than bacterial proteomes for the near future.
Supplementary Material
* This work was supported in part by funding from the European Union 7th Framework Program PROSPECTS (Proteomics Specification in Space and Time Grant HEALTH-F4-2008-201648) and from SystemsX.ch, the Swiss initiative for systems biology.
 This article contains supplemental Fig. S1, Tables S1–S3, and method details for simulations.
This article contains supplemental Fig. S1, Tables S1–S3, and method details for simulations.
2 A. Leitner, T. Walzthoeni, A. Kahraman, F. Herzog, O. Rinner, M. Beck, and R. Aebersold, unpublished data.
1 The abbreviations used are:
- DSS
- disuccinimidyl suberate
- BS3
- bis(sulfosuccinimidyl) suberate
- DSG
- disuccinimidyl glutarate
- EM
- electron microscopy
- ETD
- electron transfer dissociation
- LIT
- linear ion trap
- PIR
- protein interaction reporter
- r.m.s.d.
- root mean square distance
- SCX
- strong cation exchange
- SEC
- size exclusion chromatography
- XDB
- cross-link database
- CXMS
- chemical cross-linking coupled with mass spectrometry.
REFERENCES
- 1.Young M. M., Tang N., Hempel J. C., Oshiro C. M., Taylor E. W., Kuntz I. D., Gibson B. W., Dollinger G. (2000) High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 97, 5802–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back J. W., de Jong L., Muijsers A. O., de Koster C. G. (2003) Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 331, 303–313 [DOI] [PubMed] [Google Scholar]
- 3.Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 4.Lee Y. J. (2008) Mass spectrometric analysis of cross-linking sites for the structure of proteins and protein complexes. Mol. Biosyst. 4, 816–823 [DOI] [PubMed] [Google Scholar]
- 5.Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M. (2003) MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 [DOI] [PubMed] [Google Scholar]
- 6.Kalkhof S., Sinz A. (2008) Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal. Bioanal. Chem. 392, 305–312 [DOI] [PubMed] [Google Scholar]
- 7.Mädler S., Bich C., Touboul D., Zenobi R. (2009) Chemical cross-linking with NHS esters: a systematic study on amino acid reactivities. J. Mass Spectrom. 44, 694–706 [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q., Crosland E., Fabris D. (2008) Nested Arg-specific bifunctional crosslinkers for MS-based structural analysis of proteins and protein assemblies. Anal. Chim. Acta 627, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novak P., Kruppa G. H. (2008) Intra-molecular cross-linking of acidic residues for protein structure studies. Eur. J. Mass Spectrom. 14, 355–365 [DOI] [PubMed] [Google Scholar]
- 10.Sutherland B. W., Toews J., Kast J. (2008) Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J. Mass Spectrom. 43, 699–715 [DOI] [PubMed] [Google Scholar]
- 11.Toews J., Rogalski J. C., Clark T. J., Kast J. (2008) Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal. Chim. Acta 618, 168–183 [DOI] [PubMed] [Google Scholar]
- 12.Müller D. R., Schindler P., Towbin H., Wirth U., Voshol H., Hoving S., Steinmetz M. O. (2001) Isotope tagged cross linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal. Chem. 73, 1927–1934 [DOI] [PubMed] [Google Scholar]
- 13.Back J. W., Notenboom V., de Koning L. J., Muijsers A. O., Sixma T. K., de Koster C. G., de Jong L. Z. (2002) Identification of cross-linked peptides for protein interaction studies using mass spectrometry and O-18 labeling. Anal. Chem. 74, 4417–4422 [DOI] [PubMed] [Google Scholar]
- 14.Huang B. X., Kim H. Y. (2009) Probing Akt-inhibitor interaction by chemical cross-linking and mass spectrometry. J. Am. Soc. Mass Spectrom. 20, 1504–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitner A., Lindner W. (2006) Chemistry meets proteomics: the use of chemical tagging reactions for MS-based proteomics. Proteomics 6, 5418–5434 [DOI] [PubMed] [Google Scholar]
- 16.Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 17.Trester-Zedlitz M., Kamada K., Burley S. K., Fenyö D., Chait B. T., Muir T. W. (2003) A Modular Cross-Linking Approach for Exploring Protein Interactions. J. Am. Chem. Soc. 125, 2416–2425 [DOI] [PubMed] [Google Scholar]
- 18.Kang S., Mou L., Lanman J., Velu S., Brouillette W. J., Prevelige P. E., Jr (2009) Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun. Mass Spectrom. 23, 1719–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nessen M. A., Kramer G., Back J., Baskin J. M., Smeenk L. E., de Koning L. J., van Maarseveen J. H., de Jong L., Bertozzi C. R., Hiemstra H., de Koster C. G. (2009) Selective enrichment of azide-containing peptides from complex mixtures. J. Proteome Res. 8, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan F., Che F. Y., Rykunov D., Nieves E., Fiser A., Weiss L. M., Hogue Angeletti R. (2009) Nonprotein based enrichment method to analyze peptide cross-linking in protein complexes. Anal. Chem. 81, 7149–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X., Munske G. R., Siems W. F., Bruce J. E. (2005) Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77, 311–318 [DOI] [PubMed] [Google Scholar]
- 22.Chowdhury S. M., Du X., Toliæ N., Wu S., Moore R. J., Mayer M. U., Smith R. D., Adkins J. N. (2009) Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 81, 5524–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., Tang X., Munske G. R., Tolic N., Anderson G. A., Bruce J. E. (2009) Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol. Cell. Proteomics 8, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak P., Young M. M., Schoeniger J. S., Kruppa G. H. (2003) A top-down approach to protein structure studies using chemical cross-linking and Fourier transform mass spectrometry. Eur. J. Mass Spectrom. 9, 623–631 [DOI] [PubMed] [Google Scholar]
- 25.Dowell J. A., Frost D. C., Zhang J., Li L. (2008) Comparison of two-dimensional fractionation techniques for shotgun proteomics. Anal. Chem. 80, 6715–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maiolica A., Cittaro D., Borsotti D., Sennels L., Ciferri C., Tarricone C., Musacchio A., Rappsilber J. (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics 6, 2200–2211 [DOI] [PubMed] [Google Scholar]
- 28.Lee Y. J., Lackner L. L., Nunnari J. M., Phinney B. S. (2007) Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J. Proteome Res. 6, 3908–3917 [DOI] [PubMed] [Google Scholar]
- 29.Singh P., Shaffer S. A., Scherl A., Holman C., Pfuetzner R. A., Larson Freeman T. J., Miller S. I., Hernandez P., Appel R. D., Goodlett D. R. (2008) Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal. Chem. 80, 8799–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paizs B., Suhai S. (2005) Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 24, 508–548 [DOI] [PubMed] [Google Scholar]
- 32.Bakhtiar R., Guan Z. (2006) Electron capture dissociation mass spectrometry in characterization of peptides and proteins. Biotechnol. Lett. 28, 1047–1059 [DOI] [PubMed] [Google Scholar]
- 33.Mikesh L. M., Ueberheide B., Chi A., Coon J. J., Syka J. E., Shabanowitz J., Hunt D. F. (2006) The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta 1764, 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlister G. C., Phanstiel D., Wenger C. D., Lee M. V., Coon J. J. (2010) Analysis of tandem mass spectra by FTMS for improved large-scale proteomics with superior protein quantification. Anal. Chem. 82, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu F., Shan S. O., Moustakas D. T., Alber F., Egea P. F., Stroud R. M., Walter P., Burlingame A. L. (2004) Unraveling the interface of signal recognition particle and its receptor by using chemical cross-linking and tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 16454–16459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett K. L., Kussmann M., Björk P., Godzwon M., Mikkelsen M., Sørensen P., Roepstorff P. (2000) Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping—a novel approach to assess intermolecular protein contacts. Protein Sci. 9, 1503–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Koning L. J., Kasper P. T., Back J. W., Nessen M. A., Vanrobaeys F., Van Beeumen J., Gherardi E., de Koster C. G., de Jong L. (2006) Computer-assisted mass spectrometric analysis of naturally occurring and artificially introduced cross-links in proteins and protein complexes. FEBS J. 273, 281–291 [DOI] [PubMed] [Google Scholar]
- 39.Tang Y., Chen Y., Lichti C. F., Hall R. A., Raney K. D., Jennings S. F. (2005) CLPM: a cross-linked peptide mapping algorithm for mass spectrometric analysis. BMC Bioinformatics 6, Suppl. 2, S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taverner T., Hall N. E., O'Hair R. A., Simpson R. J. (2002) Characterization of an antagonist interleukin-6 dimer by stable isotope labeling, cross-linking, and mass spectrometry. J. Biol. Chem. 277, 46487–46492 [DOI] [PubMed] [Google Scholar]
- 41.Chu F., Baker P. R., Burlingame A. L., Chalkley R. J. (2010) Finding chimeras: a bioinformatic strategy for identification of cross-linked peptides. Mol. Cell. Proteomics 9, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., Burlingame A. L. (2008) In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics 7, 2386–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez P., Gras R., Frey J., Appel R. D. (2003) Popitam: towards new heuristic strategies to improve protein identification from tandem mass spectrometry data. Proteomics 3, 870–878 [DOI] [PubMed] [Google Scholar]
- 44.Schmidt A., Gehlenborg N., Bodenmiller B., Mueller L. N., Campbell D., Mueller M., Aebersold R., Domon B. (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol. Cell. Proteomics 7, 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor C. F., Paton N. W., Lilley K. S., Binz P. A., Julian R. K., Jr., Jones A. R., Zhu W., Apweiler R., Aebersold R., Deutsch E. W., Dunn M. J., Heck A. J., Leitner A., Macht M., Mann M., Martens L., Neubert T. A., Patterson S. D., Ping P., Seymour S. L., Souda P., Tsugita A., Vandekerckhove J., Vondriska T. M., Whitelegge J. P., Wilkins M. R., Xenarios I., Yates J. R., 3rd, Hermjakob H. (2007) The minimum information about a proteomics experiment (MIAPE). Nat. Biotechnol. 25, 887–893 [DOI] [PubMed] [Google Scholar]
- 46.Gilson M. K., Zhou H. X. (2007) Calculation of protein-ligand binding affinities. Annu. Rev. Biophys. Biomol. Struct. 36, 21–42 [DOI] [PubMed] [Google Scholar]
- 47.Tang C., Clore G. M. (2006) A simple and reliable approach to docking protein-protein complexes from very sparse NOE-derived intermolecular distance restraints. J. Biomol. NMR 36, 37–44 [DOI] [PubMed] [Google Scholar]
- 48.Havel T. F., Crippen G. M., Kuntz I. D. (1979) Effects of distance constraints on macromolecular conformation. 2. Simulation of experimental results and theoretical predictions. Biopolymers 18, 73–81 [Google Scholar]
- 49.Cohen F. E., Sternberg M. J. (1980) On the use of chemically derived distance constraints in the prediction of protein structure with myoglobin as an example. J. Mol. Biol. 137, 9–22 [DOI] [PubMed] [Google Scholar]
- 50.Ritchie D. W. (2008) Recent progress and future directions in protein-protein docking. Curr. Protein Pept. Sci. 9, 1–15 [DOI] [PubMed] [Google Scholar]
- 51.Janin J. (2005) Assessing predictions of protein-protein interaction: the CAPRI experiment. Protein Sci. 14, 278–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janin J., Wodak S. (2007) The third CAPRI assessment meeting Toronto, Canada, April 20–21, 2007. Structure 15, 755–759 [DOI] [PubMed] [Google Scholar]
- 53.Balasu M. C., Spiridon L. N., Miron S., Craescu C. T., Scheidig A. J., Petrescu A. J., Szedlacsek S. E. (2009) Interface analysis of the complex between ERK2 and PTP-SL. PLoS One 4, e5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulz D. M., Kalkhof S., Schmidt A., Ihling C., Stingl C., Mechtler K., Zschörnig O., Sinz A. (2007) Annexin A2/P11 interaction: new insights into annexin A2 tetramer structure by chemical crosslinking, high-resolution mass spectrometry, and computational modeling. Proteins 69, 254–269 [DOI] [PubMed] [Google Scholar]
- 55.D'Ambrosio C., Talamo F., Vitale R. M., Amodeo P., Tell G., Ferrara L., Scaloni A. (2003) Probing the dimeric structure of porcine aminoacylase 1 by mass spectrometric and modeling procedures. Biochemistry 42, 4430–4443 [DOI] [PubMed] [Google Scholar]
- 56.Kopp D. A., Berg E. A., Costello C. E., Lippard S. J. (2003) Structural features of covalently cross-linked hydroxylase and reductase proteins of soluble methane monooxygenase as revealed by mass spectrometric analysis. J. Biol. Chem. 278, 20939–20945 [DOI] [PubMed] [Google Scholar]
- 57.Lanman J., Lam T. T., Barnes S., Sakalian M., Emmett M. R., Marshall A. G., Prevelige P. E., Jr (2003) Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 325, 759–772 [DOI] [PubMed] [Google Scholar]
- 58.Chen R., Weng Z. (2002) Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins 47, 281–294 [DOI] [PubMed] [Google Scholar]
- 59.Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 60.Wang C., Schueler-Furman O., Baker D. (2005) Improved side-chain modeling for protein-protein docking. Protein Sci. 14, 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janin J., Bahadur R. P., Chakrabarti P. (2008) Probing the dimeric structure of porcine aminoacylase 1 by mass spectrometric and modeling procedures. Q. Rev. Biophys. 41, 133–18018812015 [Google Scholar]
- 62.Schraven B., Samstag Y., Altevogt P., Meuer S. C. (1990) Association of CD2 and CD45 on human T lymphocytes. Nature 345, 71–74 [DOI] [PubMed] [Google Scholar]
- 63.Mullins R. D., Pollard T. D. (1999) Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 9, 244–249 [DOI] [PubMed] [Google Scholar]
- 64.Brunner J. (1993) New photolabeling and cross-linking methods. Annu. Rev. Biochem. 62, 483–514 [DOI] [PubMed] [Google Scholar]
- 65.Hino N., Okazaki Y., Kobayashi T., Hayashi A., Sakamoto K., Yokoyama S. (2005) Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat. Methods 2, 201–206 [DOI] [PubMed] [Google Scholar]
- 66.Kraft C., Vodermaier H. C., Maurer-Stroh S., Eisenhaber F., Peters J. M. (2005) The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell 18, 543–553 [DOI] [PubMed] [Google Scholar]
- 67.Tanaka Y., Bond M. R., Kohler J. J. (2008) Photocrosslinkers illuminate interactions in living cells. Mol. Biosyst. 4, 473–480 [DOI] [PubMed] [Google Scholar]
- 68.Sletten E. M., Bertozzi C. R. (2009) Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. Engl. 48, 6974–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuo M. H., Allis C. D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein: DNA associations in chromatin environment. Methods 19, 425–433 [DOI] [PubMed] [Google Scholar]
- 70.Orlando V. (2000) Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25, 99–104 [DOI] [PubMed] [Google Scholar]
- 71.Bernstein B. E., Humphrey E. L., Liu C. L., Schreiber S. L. (2004) The use of chromatin immunoprecipitation assays in genome-wide analyses of histone modifications. Methods Enzymol. 376, 349–360 [DOI] [PubMed] [Google Scholar]
- 72.Park P. J. (2009) ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J. G., De Wulf P., Salek M., Rappsilber J., Moores C. A., Salmon E. D., Musacchio A. (2008) Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossmann M. G., Morais M. C., Leiman P. G., Zhang W. (2005) Combining x-ray crystallography and electron microscopy. Structure 13, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alber F., Förster F., Korkin D., Topf M., Sali A. (2008) Integrating diverse data for structure determination of macromolecular assemblies. Annu. Rev. Biochem. 77, 443–477 [DOI] [PubMed] [Google Scholar]
- 76.Köcher T., Superti-Furga G. (2007) Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 77.Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 78.Anderson G. A., Tolic N., Tang X., Zheng C., Bruce J. E. (2007) Informatics strategies for large-scale novel cross-linking analysis. J. Proteome Res. 6, 3412–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao Q., Xue S., Doneanu C. E., Shaffer S. A., Goodlett D. R., Nelson S. D. (2006) Pro-CrossLink. Software tool for protein cross-linking and mass spectrometry. Anal. Chem. 78, 2145–2149 [DOI] [PubMed] [Google Scholar]
- 80.Seebacher J., Mallick P., Zhang N., Eddes J. S., Aebersold R., Gelb M. H. (2006) Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5, 2270–2282 [DOI] [PubMed] [Google Scholar]
- 81.Lee Y. J. (2009) Probability-based shotgun cross-linking sites analysis. J. Am. Soc. Mass Spectrom. 20, 1896–1899 [DOI] [PubMed] [Google Scholar]
- 82.Förster F., Lasker K., Nickell S., Sali A., Baumeister W. (2010) Towards an integrated structural model of the 26 S proteasome. Mol. Cell. Proteomics 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.