Abstract
HK97 is a double-stranded DNA bacteriophage that undergoes dramatic conformational changes during viral capsid maturation and for which x-ray structures, at near atomic resolution, of multiple intermediate and mature capsid states are available. Both amide H/2H exchange and crystallographic comparisons between the pre-expanded Prohead II particles and the expanded Head II of bacteriophage HK97 revealed quaternary interactions that remain fixed throughout maturation and appear to maintain intercapsomer integrity at all quasi- and icosahedral 3-fold axes. These 3-fold staples are formed from Arg and Glu residues and a metal binding site. Mutations of either Arg-347 or Arg-194 or a double mutation of E344Q and E363A resulted in purification of the phage in capsomer form (hexamers and pentamers). Mutants that did assemble had both decreased thermal stability and decreased in vitro expansion rates. Amide H/2H exchange mass spectrometry showed that in the wild type capsid some subunits had a bent “spine” helix (highly exchanging), whereas others were straight (less exchanging). Similar analysis of the never assembled mutant capsomers showed uniform amide exchange in all of these that was higher than that of the straight spine helices (characterized in more mature intermediates), suggesting that the spine helix is somewhat bent prior to capsid assembly. The result further supports a previously proposed mechanism for capsid expansion in which the delta domains of each subunit induce a high energy intermediate conformation, which now appears to include a bent helix during capsomer assembly.
The viral capsid, particularly in double-stranded DNA bacteriophage, requires a highly stable macromolecular structure capable of encapsulating genome at near liquid crystalline density. Viral capsids are composed of hundreds to thousands of individual subunits that efficiently assemble into a closed capsid form often of a highly symmetrized icosahedral geometry, avoiding kinetic traps that would result in increased off-pathway assemblies. Recent studies have proposed that capsid assembly is mediated by weak intersubunit interactions that nucleate larger assembly intermediates, resulting in a considerably more stable capsid form due to a favorable geometry with a more constrained network of interactions. Measurements in systems such as cowpea chlorotic mottle virus, hepatitis B virus, and the bacteriophages P22 and HK97 have estimated the association energy of the initial assembly interaction between two subunits at 2–5 kcal/mol, which is seemingly low for a robust assembly product (1–5). An entropically driven process based on burial of hydrophobic surfaces was considered the driving force for the initial weak interactions with subsequent nucleation and elongation reactions leading to assembly of the full capsid (2, 6). Most complex viruses undergo a staged assembly process involving conformational transitions that occur after the initial assembly of a procapsid (7). The process is known as virion maturation. The interplay between interactions necessary for the initial assembly of capsomers into the procapsid and those that facilitate capsid maturation have been poorly understood, but recent crystal structures of procapsid and mature capsid states of HK97 allowed us to evaluate the structural properties that may facilitate maturation.
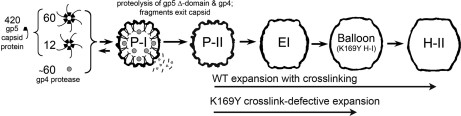
HK97 is an amenable system for the study of capsid assembly and maturation. Symmetric procapsid particles can be assembled in Escherichia coli with the expression of just two gene products, gp4 (protease) and gp5 (capsid subunit). Maturation can then be followed in vitro by lowering the pH or chemically perturbing the procapsids. Unlike bacteriophages such as P22 that assemble their capsids directly from individual monomeric subunits, HK97 subunits initially assemble into capsomers composed of six-subunit (hexamers) or five-subunit (pentamers) oligomers. Twelve pentamers and 60 hexamers then assemble to form an icosahedral capsid with a triangulation number of 7 laevo, although a portal complex substitutes one of the pentamers during in vivo assembly. Residues 2–103 at the N terminus of the subunit, referred to as the delta domain, is thought to serve the same role as the scaffolding proteins identified for other phage in the assembly process (8). Capsomers then assemble, packaging the protease (gp4), to form the initial procapsid, Prohead I (P-I).1 If the expression is done without the protease or with an inactive (by mutation) protease, this step is reversible (Fig. 1). The equilibrium of this assembly can be controlled in vitro with specific buffers and concentrations that favor either the capsomer or the capsid form (9). Expression with an active protease leads to proteolysis of the delta domains in the assembled P-I state followed by autodigestion of the protease and diffusion of the fragments from the particle. P-I then undergoes subtle structural adjustments, resulting in the Prohead II state composed entirely of the cleaved gp5* subunits (10, 11). At this stage of assembly in vivo, concatameric double-stranded DNA is packaged through a portal complex (composed of gp3 subunits) that fits into a single 5-fold vertex of the capsid. We used an HK97 construct that lacks gp3, so the purified Prohead II capsid is icosahedrally symmetric and cannot package DNA. Purified P-II can be matured in vitro using low pH and other chemical perturbation methods. During maturation, conformational changes in the subunits and their interactions result in large scale expansion and morphological changes in the capsid. The diameter of the capsid shell increases from 540 Å in P-II to 660 Å in Head II (H-II), the fully expanded particle form (12, 13). Intermediate particle forms can be trapped during the expansion and were previously characterized with a variety of biophysical techniques including cryo-EM microscopy (14, 15), x-ray crystallography (12, 13, 16), and small angle x-ray scattering (16–18). During the expansion process, self-catalyzed covalent cross-links are formed through isopeptide bond formation between Lys-169 and Asn-356 of different subunits situated on adjacent capsomers (19). The reaction is promoted by Glu-363, which is adjacent to the bonding residues and functions as a proton acceptor. Cross-linking during maturation was previously shown by differential scanning calorimetry (DSC) to greatly enhance the thermal stability of HK97 (5). In addition to covalent bonding, the H-II has significantly more buried surface area than P-II as seen in the highly intercalated intersubunit interactions depicted in the previous 3.44-Å structure of Head II (13, 20). A cross-link-defective mutant, K169Y, stills undergoes particle expansion, reaching the penultimate particle form, termed Head I (H-I), which has nearly identical conformations of hexamer capsomers but less extruded pentamers than H-II (16). H-I was used for all H/2H exchange studies instead of H-II because the cross-links in H-II dramatically affect the efficiency of proteolysis required for the mass spectrometry-based experiment (12, 20).
Fig. 1.
HK97 assembly and expansion pathway. The schematic diagram depicts the assembly and expansion of HK97 in an E. coli expression system lacking the portal protein and other machinery required for genome packaging. 42-kDa subunits assemble into hexamer and pentamer capsomers, which then assemble into an initial icosahedral procapsid shell, P-I. Proteolytic cleavage of the delta domain of each subunit results in the formation of the metastable intermediate form P-II, which is able to undergo in vitro maturation when perturbed by various chemical agents. WT expansion proceeds through EI, balloon, and ultimately H-II forms, an expansion process that involves covalent cross-linking. K169Y mutant P-II proceeds through EI to the H-I form without any cross-linking occurring. Other than the lack of cross-links, H-I is identical to balloon.
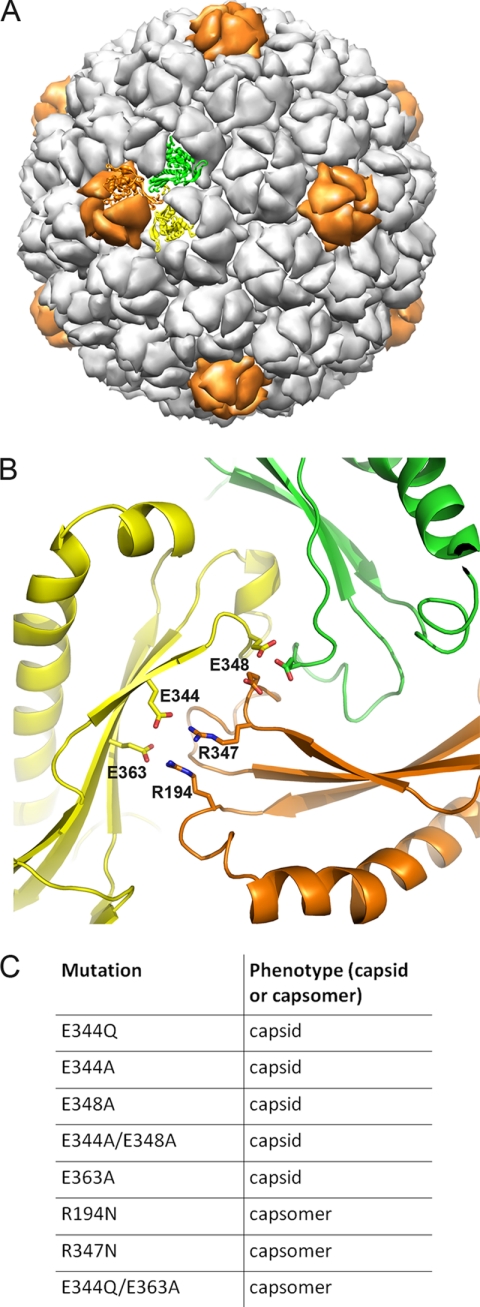
It was hypothesized that for highly intercalated mature capsid forms such as that seen in bacteriophage HK97 early procapsid intermediates are necessary for initial positioning of subunits before conformational changes can facilitate a protein architecture with increased stability. We recently showed with amide H/2H exchange and crystallographic comparisons between the pre-expanded P-II particles and the mature H-II that maturation is probably guided by tertiary structure twisting and secondary structure changes around a fixed set of intercapsomer interactions that surround all quasi- and icosahedral 3-fold axes in the capsid shell (12). The major interactions that appear to facilitate these “3-fold staples” include two sets of salt bridges and a putative metal binding site (Fig. 2). The salt bridge interactions are between residues Glu-344 and Arg-194 and between residues Glu-363 and Arg-347. Glutamate 363 serves dual roles as it is involved in both a salt bridge with Arg-347 and serves as a proton acceptor that catalyzes the isopeptide bond formation (21). The metal binding site is formed by 3-fold related glutamates at position 348 interacting with a sphere of electron density at high σ level in the P-II crystal structure (12). Although comparable density for metal ions is not present at the equivalent position in crystal structures of the late intermediates, the positions of the glutamates are nearly identical, indicating a stable interaction with some mechanism for neutralizing the negative charge repulsion. In contrast to the near identical conformations of the residues at the 3-fold interface, the rest of the subunit was shown to undergo a large scale twisting motion, causing hinging in all three P-domain β-strands (see Fig. 8A for domain nomenclature). These data imply that interactions at the 3-fold interface may be crucial in assembling the capsid from individual capsomers as well as providing a fixed point from which subunits bend while maintaining intercapsomer contacts.
Fig. 2.
Importance of 3-fold intercapsomer contacts. A, P-II capsid from previously solved 3.65-Å crystal structure rendered in low resolution in chimera. Two hexon subunits (subunits a and f, yellow and green, respectively) and one penton subunit (orange) that form a quasi-3-fold interaction are shown as ribbons. B, zoomed in view of quasi-3-fold interaction between the two hexamer subunits and one pentamer subunit as highlighted in A. The view is from inside the capsid, 180° rotated from the view shown in A. Residues involved in salt bridges as well as a putative metal binding site (Glu-348) are labeled accordingly. C, table identifying various mutations made to perturb 3-fold contacts. The phenotypes following protein expression are identified. Mutants are distinguished as to whether they were purified as capsids or capsomers (hexamers and pentamers) following protein expression. Data for the Glu-363 mutants are from Dierkes et al. (21).
Fig. 8.
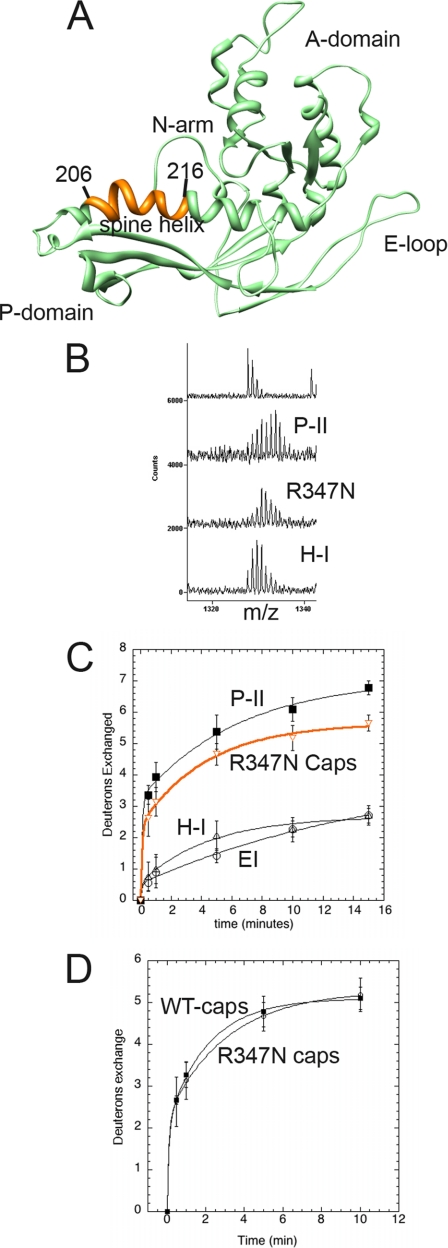
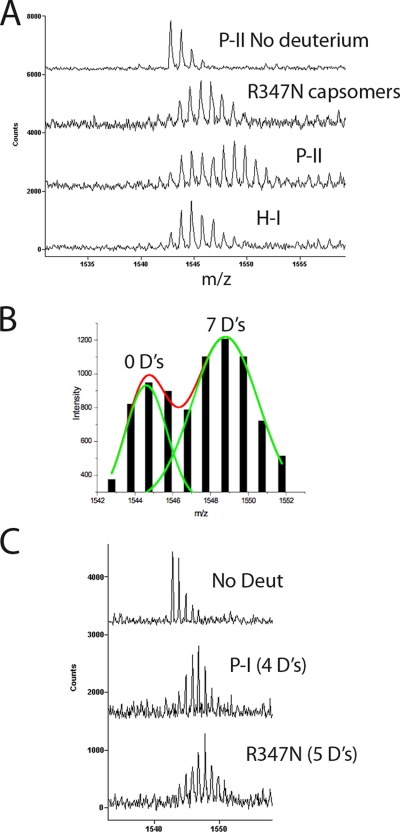
Solvent accessibility of R347N capsomer spine helix. A, subunit C of Prohead II is shown with the major domains labeled. Residues 206–216 of the spine helix are colored orange. B, mass envelopes for P-II and H-I particle forms as well as the R347N capsomers following 5 min of exchange. The top spectrum is non-deuterated P-II. C, H/2H exchange results of the residues colored orange are plotted for the R347N capsomers (orange curve) and compared with the solvent accessibility curves for the same fragment in the P-II capsid state, EI, and the nearly mature H-I capsid form. D, the solvent accessibility of the same spine helix fragment is shown for both the R347N capsomers and WT capsomers that were disassembled from the P-I state.
Here we confirmed this role for the 3-fold interactions by mutagenesis of relevant residues and characterized the resulting assembly products, thermal stabilities, and maturation kinetics. Some of the mutants did not assemble into particles following the formation of capsomers (e.g. R347N). Capsomers were then purified, and the amide exchange of the spine helices was analyzed with H/2H exchange coupled to mass spectrometry (22–24). Previous data illustrated a direct correlation between increased H/2H exchange and an increased bend in the helix conformation (12, 20). Amide exchange of the spine helix in the mutant capsomers was compared with previously characterized particle forms as well as P-I and WT capsomers disassembled from P-I.
EXPERIMENTAL PROCEDURES
Mutagenesis, Protein Expression, and Analysis
All 3-fold point mutations were introduced using site-directed mutagenesis. Protein expression and purification of all of the mutants were performed as described previously (7, 8) Purification of the R194N, R347N, and E344Q/E363A mutants was modified as no particles were seen after expression and lysis of the BL21(DE3)pLysS cells. Following lysis and subsequent removal of membrane and other cellular components by centrifugation, viral capsids are normally precipitated and pelleted after exposure to 0.5 m NaCl and 6% polyethylene glycol (7). The unassembled mutants were in the capsomer form after cell lysis and therefore did not precipitate during this step. The supernatant from the precipitation step was further purified by filtration in a Centricon (30-kDa molecular mass cutoff) filter followed by size exclusion chromatography using a Sephacryl S-300 column. A 20 mm Tris-buffered solution, pH 7.5, with 40 mm NaCl was used for all filtration and size exclusion chromatography steps. 4–12% SDS gels and Western blots using HK97 capsid subunit antibodies (raised in rabbit against either gp5* subunits or isolated delta domain protein) were used for verification of protein following expression. 1% agarose gels were used both for identification of particle forms and for time-dependent expansion assays. A 7.5% native acrylamide gel was used for distinguishing the presence of hexons and pentons in the R194N sample.
Hexon and Penton Interconversion
To produce pure hexons, protein concentrations between 1 and 3 mg/ml were treated with a final concentration of 20 mm Tris, pH 7.5, 0.5 m NaSCN (9). Samples were incubated at room temperature overnight. To produce pentons, the same protein concentration was incubated in 100 mm sodium citrate, pH 5.6, 200 mm guanidine HCl, 1% dimethylformamide overnight at room temperature. For comparison studies, wild type PH-I was disassociated with the hexon buffer above to initially disassemble the particles into capsomers.
Differential Scanning Calorimetry
P-II particles were diluted to 0.5 mg/ml with degassed 100 mm potassium phosphate, pH 7.5, 20 mm KCl. Protein samples were heated in the sample cell reservoir at a rate of 1.5 °C/min from 10 to 110 °C in a Microcal differential scanning calorimeter (VP-DSC microcalorimeter) The base line was subtracted from the respective buffer endotherm using Origin software version 5.0. The melting temperatures (Tm) were measured as the temperature associated to the peak of the major thermal event in each endotherm analyzed by Origin.
Electron Microscopy
The proteins at 0.02 mg/ml were absorbed on glow-discharged copper grids coated with carbon film (Electron Microscopy Sciences, catalog number CF300-Cu) for 2 min. The grids were subsequently washed twice with filtered distilled water, negatively stained with 2% uranyl acetate for 45 s, and air-dried completely. Specimens were imaged at a magnification of 100,000× at 2-μm defocus using an FEI Tecnai F20 Twin transmission electron microscope at an accelerating voltage of 120 kV. Images were recorded with a Gatan UltraScan 4k × 4k charge-coupled device camera.
Analytical Ultracentrifugation
Sedimentation velocity experiments on mutant capsid protein were performed at 20 °C with a protein concentration of 1 mg/ml in 20 mm Tris buffer, pH 7.5, containing 40 mm NaCl. The sedimentation at 40,000 rpm with an An-60 Ti rotor was followed by measuring the absorbance at 280 nm over time with a Beckman Optima XL-A analytical ultracentrifuge. The partial specific volume was calculated by amino acid sequence, and the solvent density was calculated according to the solvent composition using the SEDNTERP program. The sedimentation coefficient distributions were calculated with a continuous c(s) distribution model of Lamm equation solution implemented in the SEDFIT program. The diffusion coefficients were estimated by a weight average frictional ratio, and these two factors were related to the molecular weight using the Svedberg equation. The molecular weight distributions were derived with a continuous c(M) distribution model in the SEDFIT program.
Fluorescence Kinetics
A sample (3 μl of a 25 mg/ml solution) of particles was diluted with 50 mm sodium acetate, pH 4.0, 200 mm potassium chloride (expansion buffer) (7). The mixture was quickly transferred to a quartz cuvette followed by measurement using a Varian Eclipse fluorescence spectrometer. An excitation at 295 nm was followed by an emission scan ranging from 310 to 400 nm. The temperature in the cuvette was kept at 20 °C during the entire experiment. There was ∼30 s of dead time between mixing and the start of the emission scan. The data were written out in ASCII format and processed in KaleidaGraph (Synergy Software). The spectral center of mass was calculated by the following equation as described previously: Spectral center of mass = Σ(Fiλi)/ΣFi. The spectral center of mass is blue-shifted during expansion from P-II to the expansion intermediate (EI) capsid form as shown in the graph in Fig. 5. The shift is due to the burial of tryptophan residues and occurs at a rate similar to that of the overall morphological change in particle size as determined previously (17). The spectral center of mass was plotted versus time for each mutant and fit to an exponential decay curve using KaleidaGraph.
Fig. 5.
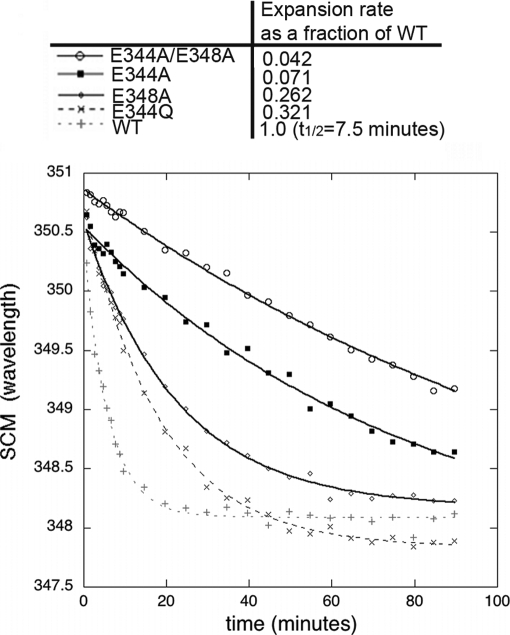
Expansion kinetics of 3-fold mutants monitored by fluorescence. P-II particles of the various mutants and WT were monitored for their change in spectral center of mass (SCM) (resulting from capsid expansion) following incubation with acidic buffer (pH 4.0). The time for half of the WT particles to expand (t½) is shown in the table as are the rates for mutant expansion as a fraction of the WT expansion rate.
H/2H Exchange Coupled to MALDI Mass Spectrometry
Concentrated HK97 capsomers (∼10 mg/ml) were diluted 7-fold in an 85% final D2O concentration buffered with 20 mm Tris, pH 7.5, containing either 200 mm sodium chloride or 200 mm potassium chloride. Samples were incubated in D2O at pH 7.5 for various time periods followed by addition of pH 2.5 non-deuterated quench solution (final D2O concentration of 9.0%). Following the quench, all samples were handled on ice at 4 °C. Protein was digested with 50 μl of pepsin-coated beads for 5 min. The beads were removed by centrifugation, and the supernatant was flash frozen in liquid N2. Samples were thawed individually, mixed 1:1 with α-cyano-4-hydroxycinnamic acid matrix solution, and vacuum-crystallized on a prechilled MALDI target followed by analysis on a MALDI-TOF mass spectrometer according to previously published protocols (25). All pepsin digest products were identified by sequencing using MS/MS using a Q-STAR-XL mass spectrometer or a MALDI TOF-TOF (ABI 4800) mass spectrometer. Back-exchange was calculated as 42% using the N-terminal peptide (residues 117–126), which exchanged amide protons for deuterium completely within 20 s. The sodium adduct of this peptide was present in much greater abundance in the spectra. All future H/2H measurements of this fragment were therefore performed on the adducted species. The total number of deuterons exchanged was calculated by subtracting the centroid of the mass envelope from the non-deuterated control from the centroid of each deuterated mass envelope. The error (standard deviation) was estimated from the average of three independent experiments with two to three measurements recorded for each experiment (a total of six to nine measurements for each time point). Data analysis and kinetic plots were obtained as described previously (24).
H/2H exchange reactions were performed for up to 15 min, enabling measurement of amide protons exchanging at both a fast (>1 min−1) and an intermediate rate (0.01–1 min−1) (26). Longer incubations measuring slow exchange rates (<0.01 min−1) were not performed in this study. Amide protons exchanging at intermediate to slow rates are generally a result of solvent protection due to either secondary structure or protein-protein interactions. The measured data for all fragments was best fit to either a single or two-exponential model accounting for deuterons exchanging at only a fast rate or both a fast and intermediate rate, respectively. The following equation represents the two-exponential fit,
where D is the total number of deuterons exchanged at time t; Nfast is the number of deuterons exchanging at a fast rate, kfast; and Ninter is the number of deuterons exchanging at an intermediate rate, kinter. The fast exchanging amide protons had nearly all exchanged by the first time point, so kfast was estimated as described previously (24). The mass envelope of the helix fragment spanning residues 204–216 could be resolved as a combination of two Gaussian distributions at early time points. The fitting to multiple Gaussian distributions as seen in Fig. 9 was performed in the software package Origin.
Fig. 9.
Bimodal versus unimodal exchange in spine helix. A, the mass envelopes of the spine helix fragment spanning residues 204–216 are shown for various HK97 intermediate forms following 1 min of solvent exchange with deuterium. The top trace represents the non-deuterated control with a mass distribution based on natural isotopic abundance. B, the mass envelope of P-II at 1 min of exchange showed a bimodal distribution that was fit to two Gaussian distributions with the program Origin. Following back-exchange calculation, the first envelope corresponded to an exchange of zero deuterons (D's), whereas the large envelope corresponded to seven deuterons. C, comparison of mass envelopes following 10 min of exchange in the R347N capsomers and the P-I particle form. Although the capsomers show a slightly greater exchange, neither mass envelope shows a bimodal distribution as in P-II. Deut, deuterons.
RESULTS
3-Fold Mutants That Still Form Particles
All of the residues at the “3-fold staple” were mutated: E348A, E344Q, E344A, R194N, R347N, and double mutants E348A/E344A and E344Q/E363A. Expression of E348A, E344A, E344Q, and E344A/E348A in E. coli resulted in the formation of normal icosahedral particles, whereas mutants R194N, R347N, and E344Q/E363A (Q/A) mutants were purified as free capsomers with no detectable capsids present. The mutants that formed particles, the so-called “capsid mutants,” showed no significant population of capsomers in preparations. The capsid mutants produced predominately P-II particles similarly to WT except for E344A, which had a significant fraction (∼65%) of expanded particles following the final high speed ultracentrifugation step (35 krpm for 2 h) used for pelleting the particles. It was determined that successive high speed pelleting steps were responsible for inducing the spontaneous maturation beyond P-II and the expansion of E344A (supplemental Fig. 1). E344Q also showed a very slight inclination to prematurely expand due to pelleting (<5%) but in very low amounts compared with E344A. The Glu-344 mutants therefore show an increased sensitivity to mechanical perturbation that leads to particle expansion. All mutant particles were isolated by anion exchange chromatography, and only a homogenous fraction of P-II particles was used for both DSC experiments and expansion assays as described below.
Differential Scanning Calorimetry
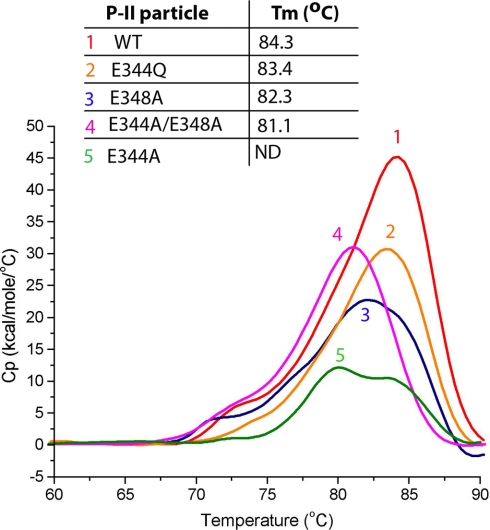
Previous calorimetry experiments compared the thermal stabilities of various expansion states of HK97 (5, 27). The studies showed that H-II has a significantly higher melting temperature than that of both P-II and P-I particles. The studies also revealed that the cross-links in WT H-II greatly stabilize the particle with a significantly higher denaturation temperature compared with that of the mature form of a cross-link-defective K169Y mutant (H-I). The P-II structure strongly suggests that the 3-fold interactions are critical to particle stability and that perturbation to these interactions would result in weaker intercapsomer association, resulting in a less stable particle. We performed differential calorimetry scans on all of the 3-fold mutations (Fig. 3). The results show that all of the mutants have endotherms with lower temperatures of denaturation. The endotherms of the mutants all have a shape similar to that of WT P-II except for the E344A mutant. The endotherm of E344A corresponds to P-II melting but appears to be a combination of two highly overlapped peaks with the center of the first measured at 80°. Although this measurement appears to indicate a lower thermal stability for E344A as compared with the other constructs, a strict comparison cannot be made because of the differences in the shape of the endotherm, which may indicate a modified mechanism of thermal denaturation. The other three mutant endotherms as well as WT P-II have a small shoulder in the beginning of the endotherm indicative of a conformational change or other event preceding the major melting and unfolding transitions. Previous DSC studies of WT P-II showed no such shoulder but did show a very broad peak (5). The faster heating rate of the previous study (1.98 °C/min) as compared with the current study (1.5 °C/min) may have blurred the shoulder that we can now resolve. As shown in supplemental Fig. 2, the shoulder of the major endotherm corresponds to an irreversible event that proceeds the melting of the P-II particles. Stopping the reaction at the temperature corresponding to the shoulder followed by cooling and then another DSC scan resulted in a single endotherm with the same melting temperature (Tm) as that determined in initial scans. All of the mutants tested had melting temperatures of ∼1–3 °C lower than that of WT P-II, indicating lower thermal stabilities. Under the peak of each endotherm, multiple irreversible transitions occurred, mainly the disassembly of the virus particles and the denaturation of both capsomers and individual subunits. These transitions occurred at very similar temperatures and could not be resolved.
Fig. 3.
Differential scanning calorimetry of 3-fold mutants. DSC scans and corresponding endotherms of WT P-II (1), E344Q P-II (2), E348A P-II (3), E344A/E348A (double mutant) (4), and E344A (5). The scans are overlaid with corresponding melting temperatures shown above (Tm). The Tm for E344A is shown as not determined (ND) because of its unusual endotherm.
3-Fold Mutant Expansion
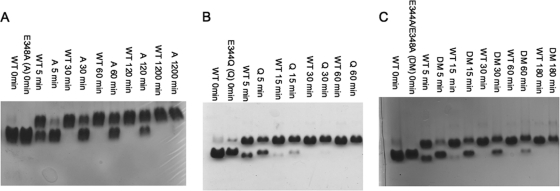
The rates of expansion of the P-II mutant particles and of WT P-II for comparison were measured by fluorescence spectroscopy as well as native agarose gel electrophoresis. Low pH has been the most common treatment used to induce in vitro particle expansion in previous kinetic and structural studies and was the only perturbation method used in this study (15, 19, 29). Gel analysis of low pH-treated mutant and WT P-II particles showed major differences in their expansion rates (Fig. 4). All particle forms were treated with sodium acetate, pH 4.0, for various amounts of time and then neutralized to pH ∼8.0 with Tris buffer. Upon reaching the EI state, HK97 particles can no longer revert to the P-II state. The assay therefore measures the fraction of particles converted to at least the EI form, which matures further to H-II upon neutralization (28, 29). Gel analysis showed that within 10 min nearly all of the WT P-II has expanded, whereas all the mutants require between 30 min and several hours for full conversion under acidic conditions. To more accurately quantify the expansion rate, fluorescence spectroscopy was used to monitor expansion under acidic conditions (Fig. 5). Previous studies have been able to quantify population kinetics of WT expansion by measuring the emission spectral center of mass, which is blue-shifted for EI as compared with P-II particles, a result of intrinsic tryptophan burial during the event. The emission maximum shifts from 351 nm in P-II to 348 nm in EI. The results show that the E348A, E344Q, E344A, and E344A/E348A mutants all mature more slowly than WT. E344Q is the next fastest followed by E348A, then E344A, and finally the slowest, E344A/E348A. The comparison of expansion rates shows that the more perturbing the mutation is to the 3-fold interactions, the slower the particle expands. For example, E344Q expands slower than WT but considerably faster than E344A. The Glu to Gln mutation likely allows a hydrogen bond to form with Arg-194 instead of the normal salt bridge, whereas the mutation to the alanine likely completely disrupts this interaction. The data also show that cumulative mutations at the 3-fold staple further slow the expansion process. E344A/E348A is slower than both the single mutations E344A and E348A.
Fig. 4.
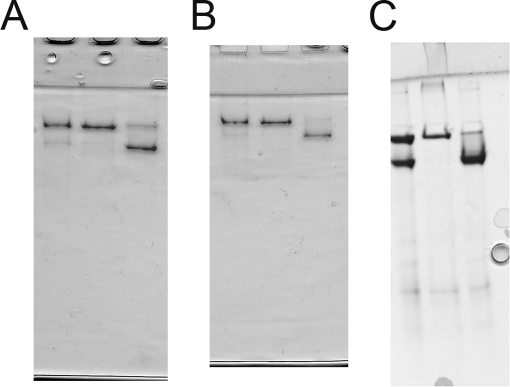
Low pH-induced expansion assays. Shown are agarose gels (1%) comparing change in particle forms as a function of time following incubation of samples in pH 4.0 buffer. The upper bands represent expanded particles, and the lower bands represents P-II. A, the gel compares WT P-II and E348A P-II (A) expansion. B, gel of WT P-II and E344Q (Q) expansion. C, gel of WT P-II and E344A/E348A (DM) expansion.
3-Fold Capsomer Mutants
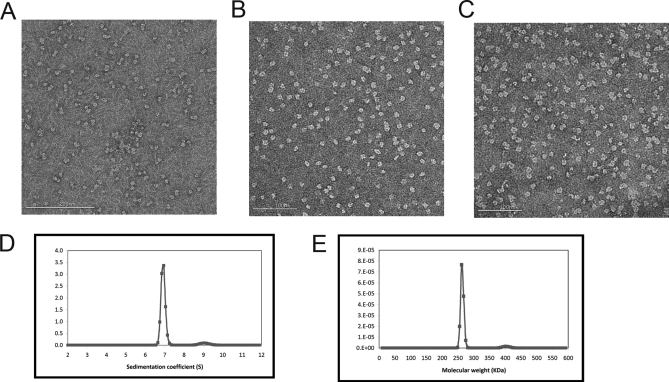
The mutants R194N, R347N, and Q/A were purified in pure capsomer form with no noticeable fraction of particles. These “capsomer mutants” did not pellet during the salt precipitation and subsequent centrifugation step that normally causes viral capsid precipitation. Analysis of the supernatant by SDS gel electrophoresis showed the presence of a ∼42-kDa band, indicative of the uncleaved gp5 subunit. All P-II preparations of the other mutants as well as WT resulted in a nearly pure population of gp5* (31 kDa), the product of the proteolytic cleavage of the delta domain by gp4 inside the assembled P-I particle form. The oligomeric state of the arginine and Q/A mutants was further evaluated by native acrylamide gel electrophoresis as done in previous studies (9) and additionally by analytical centrifugation and electron microscopy. Fig. 6 shows electron micrographs of WT capsomers and of the R347N and E344Q/E363A mutants. Analytical ultracentrifugation of the mutants also identified the oligomeric state as capsomers as seen in Fig. 6, D and E, which shows the Q/A mutant as having a sedimentation coefficient corresponding to a molecular mass of 262 kDa within 3% error of the true molecular mass of hexamers (253 kDa). Native acrylamide gel analysis resolved both hexamers and pentamers present in the capsomer mutants (Fig. 7). As with capsomers derived from disassembly of P-I, treatment of mutant capsomers with a Tris buffer, pH 7.5, containing 0.5 m NaSCN shifted the equilibrium to hexamers, whereas treatment with a citrate-buffered solution, pH 5.6, containing 0.2 m guanidine HCl and 1% dimethylformamide produced predominantly pentamers (9).
Fig. 6.
Mutant capsomers. Negative stained EM images of wild type protein capsomers (A), mutant Q/A (B), and mutant R347N (C). D, sedimentation velocity analysis of capsid protein mutant Q/A. The predominant species had a sedimentation coefficient of 6.9 S and a calculated molecular mass of 262 kDa (E). This value is within 3% error of the theoretical molecular mass of the hexameric capsomer (253 kDa) calculated from the sequence.
Fig. 7.
Conversion of mutant capsomers to hexamers or pentamers. The mutant, never assembled capsomers are shown by native acrylamide gels (7.5%) after conversion to either hexamer or pentamer forms. A, R347N capsomers. Lane 1 shows the capsomer sample after protein expression and purification, lane 2 shows capsomers after exposure to hexon buffer overnight, and lane 3 represents overnight treatment in penton buffer. Similar experiments were performed with E344Q/E363A capsomers (B) and R194N capsomers (C), both of which also never assembled into capsids.
H/2H Exchange of Mutant Capsomers
We examined the amide H/2H exchange of mutant capsomers to distinguish whether there were any major differences with WT capsomers and the previously characterized capsid particle forms. We particularly focused on the spine helix to determine whether the bent, highly exchanging form found in P-II was also present in the mutant capsomers. We analyzed two separate overlapping fragments of the helix; one spans residues 206–216 (Fig. 8), and the other spans residues 204–216 (Fig. 9). The fragment spanning residues 206–216 of the spine helix covers the N-terminal region, which is significantly bent for five of the seven subunits of the asymmetric unit in P-II (20) (Fig. 8). These data correspond to what is seen in the crystal structure of P-II where some of the spine helices are bent and others are straight. The presence of two different conformations of the spine helix represents a dramatic example of quasi-equivalence in the subunit population where the different subunits (A thru G) have distinguishable conformations within the asymmetric unit of the capsid despite the fact that they are identical gene products. Fig. 8B shows that the mass envelope is considerably wider for residues 206–216 in P-II as compared with both the mature particle form H-I and the capsomer mutant R347N. The envelope represents a combination of multiple exchange rates indicative of a certain percentage of subunits exchanging significantly less than others. The data acquired for the P-II, EI, and H-I particle forms were described previously (12, 20) but are shown along with the newly acquired capsomer data as a reference for the degree of amide exchange.
In Fig. 9A, we show a similar mass envelope for the fragment spanning residues 204–216 containing an extra two N-terminal residues than the other fragment. This fragment has an accentuated bimodality that could be more confidently deconvoluted than the smaller fragment. From the fitting of the bimodal Gaussian distribution (Fig. 9B), as published previously (20), it was calculated that the slower exchanging envelope corresponds to a net exchange of zero deuterons after 1 min of exchange, whereas the more pronounced envelope corresponds to a net exchange of seven deuterons after back-exchange correction as described under “Experimental Procedures.” Such a high level of amide exchange is not expected in canonical helices. Indeed, the amide exchange of residues 204–216 in later expansion intermediates such as H-I is much less, indicating that only straight helices are present in H-I.
Both the level of amide exchange and the bimodal exchange profile change drastically during initial expansion to EI and remain identical through to H-I when the helix undergoes a significant conformational change toward its straightened form with more extensive hydrogen bonding along its backbone. H-I, a nearly mature particle form, was shown by crystallography to have a straightened helix in comparison with P-II (16). When the amide exchange was measured as the centroid of each mass envelope and plotted against time, it appears that residues 206–216 from the R347N mutant capsomers exchange on average almost as many amides as in the P-II state (Fig. 8C) and are identical to WT disassembled capsomers (Fig. 8D). Interestingly, the R347N capsomers did not exhibit the bimodal behavior even though their helices were also significantly exchanging (Figs. 8B and 9A). The envelope of the R347N mutant appears to be nearly in the middle of the two Gaussian envelopes seen in P-II, accounting for their nearly similar exchange curves. P-I, the initial procapsid state formed by the assembly of capsomers, also showed a unimodal exchange profile (Fig. 9C). The level of amide exchange for the spine helix in P-I was slightly less than for free capsomers, but interestingly, a bimodal exchange was not seen for P-I unlike its succeeding intermediate, P-II. This is evidence that additional bending or distortion in some or all of the helices is occurring following the proteolytic cleavage event that results in the transition from P-I to P-II particle states.
DISCUSSION
Amide H/2H exchange and crystallographic comparisons between the pre-expanded and expanded states identified several major intercapsomer interactions that appeared to be crucial both for the assembly and the maturations properties of HK97. We mutated the residues in the critical 3-fold staple that persists throughout maturation to perturb the interactions to varying degrees. The study demonstrates that the salt bridges between Arg-194 and Glu-363 as well as Arg-347 and Glu-344 are critical to viral capsid assembly. Mutating either of the arginines to an asparagine prevented capsomers from assembling into capsids. Interestingly, mutating either of the glutamate residues individually did not prevent assembly, although mutating both did as seen in the E344Q/E363A mutant. This result is somewhat unexpected as breaking either residue of a respective salt bridge would be expected to be equivalently detrimental. An explanation may be that, because the two salt bridges are in close proximity to each other, mutating either glutamate may result in the free arginine repositioning to interact with the lone glutamate residue, thereby diminishing the cost of breaking the original salt bridge. When either of the arginines is mutated, the remaining arginine may not be in position to effectively interact with the second glutamate. The importance of having at least a single salt bridge, however, is clarified by the double glutamate mutant (E344Q/E363A), which was unable to assemble into a capsid. The double mutant E344A/E348A assembled normally as opposed to the E344Q/E363A mutant, which was isolated as free capsomers. These results suggest that Glu-348, which sits directly under the 3-fold axes (quasi and icosahedral) and was previously described to interact in a putative metal binding site with two other Glu-348 residues from neighboring capsomers, is not as crucial to capsid assembly as Glu-344 and Glu-363. The explanation may be that the salt bridges, not the metal binding site, are crucial for nucleating intercapsomer interactions or that the electron density for the metal binding site seen in the P-II crystal structure was an artifact of the crystallographic conditions and not physiologically relevant. Isothermal titration calorimetry experiments to probe binding of divalent cations may help answer the relevancy of the putative metal binding site. The results, more importantly, directly support previously proposed hypotheses that describe capsid assembly as facilitated by many weak interactions between individual components that gain strength in the overall contact energy through the oligomerization process that generates the capsid shell. Estimates of 2–5 kcal/mol association energy have been reported for intersubunit contacts during capsid assembly for several different viral systems; this is a range that is consistent with the range of free energy change associated with abrogating one to two salt bridge interactions, which in the case of HK97 tips the equilibrium of the assembly process to greatly favor the free capsomer form.
The 3-fold mutants that did assemble showed significant differences in both thermal stabilities and expansion kinetics in comparison with wild type P-II particles. The 1–3 °C decrease of melting temperature (Tm) indicates that the 3-fold salt bridges as well as the putative metal binding site are important to the stability of the capsids. The mechanism of capsid denaturation would likely involve a breakdown of quaternary interactions prior to complete subunit denaturation. In the case of HK97, the next highest order oligomerization state going from capsid or intermediate assembled particles is the capsomers. Weakening intercapsomer contacts would therefore lower the energy needed to disassemble a particle as seen in the differential scanning calorimetry data. Applying these 3-fold mutants to a complementation assay that would test for viable phage production following genome packaging would further address the relative importance of intercapsomer interactions to the ability of the capsid to withstand the high packaging forces concomitant with bacteriophage maturation (30).
In addition to lower thermal stabilities, the acid-induced in vitro expansion rates of the 3-fold mutants were significantly decreased in comparison with WT. Low pH has been the most commonly used method for expansion in previous HK97 studies. HK97 virions normally expand upon packaging of negatively charged DNA, which has been hypothesized to induce conformational changes when interacting with the negatively charged interior of the capsid (14). Increasing the proton ion concentration using acidic conditions may mimic this process, acting as a perturbant by either neutralizing a charged interaction of an acidic residue, protonating and charging a histidine, or perturbing an entire interface by changing the global electrostatics. The results indicated that increasingly perturbing the 3-fold interactions progressively slows capsid expansion. E344Q expands slower than WT but faster than E344A, whereas the double mutant E344A/E348A expands the slowest. Although we did not perform expansion assays on E363Q and E363A mutants, Dierkes et al. (21) previously showed by gel analysis that E363A P-II matures slower than WT during acid-induced expansion, whereas E363Q matures at nearly the same rate as WT. This result is similar to our Glu-344 mutations, which showed that the alanine mutation had a greater effect on slowing expansion than the glutamine mutation. There are several plausible explanations as to why these mutations affect expansion. The first is related to the cooperative mechanism of HK97 expansion in which the particle expands through a wavelike motion where triggering expansion in one region will translate through the capsid lattice (18). Because the 3-fold contacts appear to be crucial to stabilizing intercapsomer contacts, they may in fact be the key components in relaying motions from subunit to subunit between neighboring capsomers. Weakening the 3-fold contacts may therefore hinder the relaying of conformational changes through the lattice. Another possible explanation for the slow expansion rates of the mutants during acid incubation is that protonation of glutamates in the WT form is crucial for acid-induced expansion. In this case, a partial or momentary disruption or perturbation of a salt bridge for example would actually facilitate the subunit refolding and concomitant expansion. In addition, the 3-fold interactions that remain nearly identical between P-II and H-II would be more dynamic than originally hypothesized. An argument for this would be the behavior of the E344A mutant. During E344A P-II preparation, a significant portion of particles prematurely expanded during high speed pelleting, indicative of an increased sensitivity to mechanical perturbation. The fact that E344A mutant expands significantly more slowly than E344Q, however, would indicate that pH does not have a perturbing effect on the implicated glutamates because neither glutamine nor alanine is sensitive to protonation, yet the two mutants clearly have different expansion rates. Many solvent conditions have been previously shown to induce HK97 expansion, and although it is clear that the P-II state needs to be perturbed to undergo expansion, discovering the molecular triggers of expansion may be difficult in that they may vary from one perturbing solvent condition to another.
Some of the mutants fortuitously enabled the purification of previously never assembled capsomers, enabling us to investigate the protein conformational states by H/2H exchange coupled to mass spectrometry. The primary region of interest was the spine helix, which was implicated in previous studies to transition from a bent to a straight conformation upon maturation. Although capsomers disassembled from WT P-II showed high levels of amide exchange, similar to P-II, it was unclear whether the bent helix, which exchanges more readily, is a result of initial capsomer formation or the integration of capsomers into the full capsid form. The amide H/2H exchange data presented here for the mutants that remain in the capsomer form illustrate that, in fact, never before assembled capsomers have a conformation similar to that of WT disassembled capsomers in both the spine helix and other regions of the subunits.
The bimodal exchange behavior of the spine helix in P-II is caused by different subunits having different helix conformations, resulting in non-uniform exchange rates. Normally, such bimodal amide exchange distributions are attributed to a protein folding phenomenon where the folding of a portion of the protein is significantly slower than the intrinsic rate of chemical exchange for amide protons, also called exchange in the EX1 limit (31, 32). The bimodality we observed is clearly not the result of an EX1 mechanism of exchange but rather the result of quasi-equivalent subunits in the capsid assembly. Interestingly, neither the R347N capsomer mutant nor WT disassembled capsomers showed such bimodality. The unimodal mass envelope indicates that the helices in all of the subunits behave relatively similarly in the capsomer state. Although the average overall exchange of capsomers and P-II is similar, the mass envelopes are significantly different. The capsomer envelope appears close to the middle of the two Gaussian distributions in P-II, indicating that a portion of the helices becomes more well folded and another portion becomes less well folded.
H/2H exchange data of the helix in P-I (Fig. 9C) showed a slight increase in protection in the spine helix during assembly of capsomers into P-I. The increase is probably a result of intracapsomer changes and not a result of intercapsomer associations based on the previous crystal structure of P-II that indicated that all of the quaternary interactions of the spine helix occur between subunits of the same capsomer. This is likely the case in P-I also as low resolution data of P-I show the capsomer conformations to be much more similar to those of P-II than the mature particle forms (10, 11). In addition, cryo-EM experiments of P-I show that the delta domains from subunits of the same capsomer appear to interact with each other (and not with delta domains from other capsomers) directly underneath the capsomers (the internal region of the capsid).
Although the comparison of mass envelopes shows that the helix in the capsomers exchanges fewer amides than the helices of a few subunits in P-II, it still exchanges considerably more than the straight helix in the expanded particle forms EI and H-I. More importantly, the mass envelope was unimodal, indicating that all the spine helices in P-I are likely bent (Fig. 9C). These data further support the previously proposed hypothesis that the formation of capsomers, guided by quaternary contacts from the delta domains, is responsible for the bent subunit conformation. The bent conformation likely corresponds to a higher energy state of the HK97 subunit that may be facilitated by strong interactions between the delta domains. Proteolytic cleavage of the delta domains at the P-I to P-II transition most likely then relieves this strain, allowing the spine helices in most of the subunits to straighten into a lower energy state. This implies that the delta domains and perhaps other scaffolding domains or even scaffolding proteins in other viral systems are crucial not just for assembly but also might prime the energetic landscape of capsid maturation by trapping the procapsid in a higher energy state than what is present in the mature capsid form.
Double-stranded DNA phage such as P22 and λ as well as viruses such as herpes undergo significant conformational changes from procapsid to capsid during genome packaging and subsequent maturation. Some of these systems such as λ and T4 use accessory proteins, termed gpD and soc, respectively, that bind at the surface of the 3-fold vertices to stabilize the capsid (33, 34), whereas herpes uses the “triplex” protein at 3-fold interfaces (35) for a similar purpose. HK97 uses a combination of intercapsomer contacts in the form of salt bridges as documented in the current study as well as a unique form of self-catalyzed cross-links to enforce capsid stability. It therefore appears that strong interactions at 3-fold vertices are a common theme in many phage and viruses that may be crucial to both capsid assembly as shown for HK97 and maturation by providing a stable pivot point for conformational rearrangements.
Supplementary Material
Acknowledgments
We thank Joshua Price and Jeff Kelly for help with analytical ultracentrifugation. We thank Stephen Edgcomb and Jamie Williamson for help with DSC experiments. We thank Kelly Lee for assistance with in vitro capsid maturation protocols and fluorescence measurements. We also thank Lu Gan, Bob Duda, and Roger Hendrix for helpful discussions.
* This work was supported, in whole or in part, by National Institutes of Health Grant RO1 AI40101 and Training Grant GM08326.
 This article contains supplemental Figs. 1 and 2.
This article contains supplemental Figs. 1 and 2.
1 The abbreviations used are:
- P-I
- Prohead I
- DSC
- differential scanning calorimetry
- P-II
- Prohead II
- H-I
- Head I
- H-II
- Head II
- WT
- wild type
- Q/A
- E344Q/E363A
- EI
- expansion intermediate
- EM
- electron microscopy.
REFERENCES
- 1.Ceres P., Zlotnick A. (2002) Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry 41, 11525–11531 [DOI] [PubMed] [Google Scholar]
- 2.Katen S., Zlotnick A. (2009) The thermodynamics of virus capsid assembly. Methods Enzymol. 455, 395–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahadur R. P., Rodier F., Janin J. (2007) A dissection of the protein-protein interfaces in icosahedral virus capsids. J. Mol. Biol. 367, 574–590 [DOI] [PubMed] [Google Scholar]
- 4.Parent K. N., Suhanovsky M. M., Teschke C. M. (2007) Phage P22 procapsids equilibrate with free coat protein subunits. J. Mol. Biol. 365, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross P. D., Cheng N., Conway J. F., Firek B. A., Hendrix R. W., Duda R. L., Steven A. C. (2005) Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. EMBO J. 24, 1352–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zlotnick A. (1994) To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 241, 59–67 [DOI] [PubMed] [Google Scholar]
- 7.Duda R. L., Hempel J., Michel H., Shabanowitz J., Hunt D., Hendrix R. W. (1995) Structural transitions during bacteriophage HK97 head assembly. J. Mol. Biol. 247, 618–635 [DOI] [PubMed] [Google Scholar]
- 8.Duda R. L., Martincic K., Hendrix R. W. (1995) Genetic basis of bacteriophage HK97 prohead assembly. J. Mol. Biol. 247, 636–647 [DOI] [PubMed] [Google Scholar]
- 9.Xie Z., Hendrix R. W. (1995) Assembly in vitro of bacteriophage HK97 proheads. J. Mol. Biol. 253, 74–85 [DOI] [PubMed] [Google Scholar]
- 10.Conway J. F., Duda R. L., Cheng N., Hendrix R. W., Steven A. C. (1995) Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J. Mol. Biol. 253, 86–99 [DOI] [PubMed] [Google Scholar]
- 11.Conway J. F., Cheng N., Ross P. D., Hendrix R. W., Duda R. L., Steven A. C. (2007) A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J. Struct. Biol. 158, 224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gertsman I., Gan L., Guttman M., Lee K., Speir J. A., Duda R. L., Hendrix R. W., Komives E. A., Johnson J. E. (2009) An unexpected twist in viral capsid maturation. Nature 458, 646–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wikoff W. R., Liljas L., Duda R. L., Tsuruta H., Hendrix R. W., Johnson J. E. (2000) Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289, 2129–2133 [DOI] [PubMed] [Google Scholar]
- 14.Conway J. F., Wikoff W. R., Cheng N., Duda R. L., Hendrix R. W., Johnson J. E., Steven A. C. (2001) Virus maturation involving large subunit rotations and local refolding. Science 292, 744–748 [DOI] [PubMed] [Google Scholar]
- 15.Lata R., Conway J. F., Cheng N., Duda R. L., Hendrix R. W., Wikoff W. R., Johnson J. E., Tsuruta H., Steven A. C. (2000) Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell 100, 253–263 [DOI] [PubMed] [Google Scholar]
- 16.Gan L., Conway J. F., Firek B. A., Cheng N., Hendrix R. W., Steven A. C., Johnson J. E., Duda R. L. (2004) Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Mol. Cell 14, 559–569 [DOI] [PubMed] [Google Scholar]
- 17.Lee K. K., Gan L., Tsuruta H., Hendrix R. W., Duda R. L., Johnson J. E. (2004) Evidence that a local refolding event triggers maturation of HK97 bacteriophage capsid. J. Mol. Biol. 340, 419–433 [DOI] [PubMed] [Google Scholar]
- 18.Lee K. K., Tsuruta H., Hendrix R. W., Duda R. L., Johnson J. E. (2005) Cooperative reorganization of a 420 subunit virus capsid. J. Mol. Biol. 352, 723–735 [DOI] [PubMed] [Google Scholar]
- 19.Duda R. L. (1998) Protein chainmail: catenated protein in viral capsids. Cell 94, 55–60 [DOI] [PubMed] [Google Scholar]
- 20.Gertsman I., Komives E. A., Johnson J. E. (2010) HK97 maturation studied by crystallography and H/2H exchange reveals the structural basis for exothermic particle transitions. J. Mol. Biol. 397, 560–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dierkes L. E., Peebles C. L., Firek B. A., Hendrix R. W., Duda R. L. (2009) Mutational analysis of a conserved glutamic acid required for self-catalyzed cross-linking of bacteriophage HK97 capsids. J. Virol. 83, 2088–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Englander S. W., Sosnick T. R., Englander J. J., Mayne L. (1996) Mechanisms and uses of hydrogen exchange. Curr. Opin Struct. Biol. 6, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., Smith D. L. (1993) Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandell J. G., Baerga-Ortiz A., Akashi S., Takio K., Komives E. A. (2001) Solvent accessibility of the thrombin-thrombomodulin interface. J. Mol. Biol. 306, 575–589 [DOI] [PubMed] [Google Scholar]
- 25.Mandell J. G., Falick A. M., Komives E. A. (1998) Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Anal. Chem. 70, 3987–3995 [DOI] [PubMed] [Google Scholar]
- 26.Kang S., Prevelige P. E., Jr (2005) Domain study of bacteriophage p22 coat protein and characterization of the capsid lattice transformation by hydrogen/deuterium exchange. J. Mol. Biol. 347, 935–948 [DOI] [PubMed] [Google Scholar]
- 27.Ross P. D., Conway J. F., Cheng N., Dierkes L., Firek B. A., Hendrix R. W., Steven A. C., Duda R. L. (2006) A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J. Mol. Biol. 364, 512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K. K., Gan L., Tsuruta H., Moyer C., Conway J. F., Duda R. L., Hendrix R. W., Steven A. C., Johnson J. E. (2008) Virus capsid expansion driven by the capture of mobile surface loops. Structure 16, 1491–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan L., Speir J. A., Conway J. F., Lander G., Cheng N., Firek B. A., Hendrix R. W., Duda R. L., Liljas L., Johnson J. E. (2006) Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure 14, 1655–1665 [DOI] [PubMed] [Google Scholar]
- 30.Smith D. E., Tans S. J., Smith S. B., Grimes S., Anderson D. L., Bustamante C. (2001) The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 413, 748–752 [DOI] [PubMed] [Google Scholar]
- 31.Konermann L., Simmons D. A. (2003) Protein-folding kinetics and mechanisms studied by pulse-labeling and mass spectrometry. Mass Spectrom. Rev. 22, 1–26 [DOI] [PubMed] [Google Scholar]
- 32.Weis D. D., Wales T. E., Engen J. R., Hotchko M., Ten Eyck L. F. (2006) Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 17, 1498–1509 [DOI] [PubMed] [Google Scholar]
- 33.Lander G. C., Evilevitch A., Jeembaeva M., Potter C. S., Carragher B., Johnson J. E. (2008) Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 16, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fokine A., Chipman P. R., Leiman P. G., Mesyanzhinov V. V., Rao V. B., Rossmann M. G. (2004) Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 101, 6003–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newcomb W. W., Trus B. L., Booy F. P., Steven A. C., Wall J. S., Brown J. C. (1993) Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232, 499–511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.