Abstract
The ribosomal stalk complex plays a crucial role in delivering translation factors to the catalytic site of the ribosome. It has a very similar architecture in all cells, although the protein components in bacteria are unrelated to those in archaea and eukaryotes. Here we used mass spectrometry to investigate ribosomal stalk complexes from bacteria, eukaryotes, and archaea in situ on the ribosome. Specifically we targeted ribosomes with different optimal growth temperatures. Our results showed that for the mesophilic bacterial ribosomes we investigated the stalk complexes are exclusively pentameric or entirely heptameric in the case of thermophilic bacteria, whereas we observed only pentameric stalk complexes in eukaryotic species. We also found the surprising result that for mesophilic archaea, Methanococcus vannielii, Methanococcus maripaludis, and Methanosarcina barkeri, both pentameric and heptameric stoichiometries are present simultaneously within a population of ribosomes. Moreover the ratio of pentameric to heptameric stalk complexes changed during the course of cell growth. We consider these differences in stoichiometry within ribosomal stalk complexes in the context of convergent evolution.
Ribosomes universally translate the genetic code into proteins. They consist of two asymmetric subunits between which mRNA is decoded and amino acids are added to a growing peptide chain. On the large subunit there is a noticeable protrusion, observable by electron microscopy, known as the stalk complex (also denoted as L8) (1). This complex is involved in the binding and orientation of translation factors and exists with variable composition throughout all three domains of life. In bacteria we and others have shown previously that it is composed of either two or three dimers of the protein L12 (termed L7 when N-acetylated) attached to a single copy of the scaffolding protein L10 (2, 3). These assemblies of stalk proteins, either L10(L7/L12)4 or L10(L7/L12)6, are referred to as pentameric or heptameric stalk complexes hereafter. In eukaryotes there is an identical arrangement for the stalk complex but of unrelated proteins with no sequence homology to L10/L12. In this case P0 is the L10 equivalent scaffolding protein, and two different but related proteins (P1 and P2) take the place of L12 (nomenclature according to Ref. 4). In plants P3 occurs in addition to P1 and P2 (5). The P-proteins are named after their propensity for phosphorylation when attached to the ribosome. In yeast, but not in higher eukaryotes, P1 and P2 have both evolved into two α and β proteins (6). In archaea the stalk complex constituents, although named L10 and L12, share sequence homology with the P-proteins (7). L12 and its P1/P2 counterparts are the only ribosomal proteins that have acidic pI values, do not interact directly with rRNA, and are present in multiple copies on the ribosome.
We have shown previously that by applying a combination of MS and tandem MS approaches to intact MDa particles such as ribosomes we can obtain precise information, especially regarding the overall stoichiometry and composition of different stalk complexes (2, 8–11). This is due to the fact that the stalk complex is readily observable in mass spectra of intact ribosomes (Fig. 1). This is in contrast to the situation in most crystallographic investigations where the dynamics and heterogeneity of the stalk prevent its high resolution structure determination. Because it dissociates readily in the mass spectrometer as an intact, oligomeric species we can exploit this property, and previously we have shown that for the mesophilic bacteria Bacillus subtilis and Escherichia coli the stalk complex is unequivocally a pentamer comprising L10 and four copies of L7/L12 (2). For ribosomes of the extreme thermophilic bacteria Thermus thermophilus and Thermatoga maritima we demonstrated that the stalk complex was in fact a heptamer, rather than the anticipated pentamer, comprising one L10 and six copies of L12 (2). This led us to speculate that heptameric stalk complexes were likely to be present on ribosomes from species growing at higher temperatures. Although classification of the species has changed since their discovery, the generally accepted consensus at present separates them into four distinct categories based on their optimal growth temperatures (OGTs)1: thermophiles (>55 °C), moderate thermophiles (>65 °C), extreme thermophiles (>75 °C), and hyperthermophiles (>85 °C) (12).
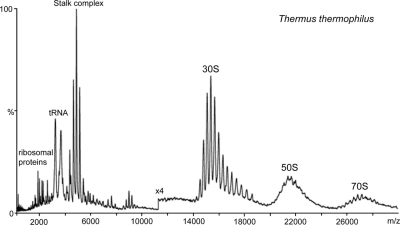
Fig. 1.
Mass spectrum of intact ribosomes from T. thermophilus. The mass spectrum of intact ribosomes from T. Thermophilus in 1 m ammonium acetate showing well resolved charge states for the 30 S subunit at 14,000–18,000 m/z and 70 S subunit at 25,000–28,000 m/z is shown. The 50 S subunit lacks charge state resolution probably because of the heterogeneity associated with the partial loss of the stalk complex, which appears as a series of well resolved charge states at 4000–6000 m/z. Individual ribosomal proteins and tRNA are observed at the lower m/z regions of the spectrum.
Here we report on investigations of ribosome stalk compositions from a range of species from all three domains of life, including examples with different OGTs. For bacteria we extended our earlier findings by including in our analysis the thermophile Bacillus stearothermophilus (OGT 55 °C) and compared it with the extreme thermophile Thermus aquaticus (OGT of 70–75 °C). For eukaryotes we compared ribosomes from three animals (brine shrimp, silkworm, and rabbit) and a thermophilic red alga, Galdieria sulphuraria, the eukaryote with the highest known OGT of 56 °C. Within the archaea we targeted the mesophilic methanogens Methanococcus vannielii, Methanococcus maripaludis, and Methanosarcina barkeri with OGTs of 35, 35–40, and 37 °C, respectively. Methanogens, capable of producing methane, are the most common and widely dispersed of the archaea. M. maripaludis is a model species among the methanogenic archaea. M. barkeri, unlike most methanogens, which only use carbon dioxide, is able to ferment a variety of carbon sources (13). Surprisingly stalk complexes of different stoichiometries were present simultaneously on ribosomes from these mesophilic archaea. Our MS study of ribosomes isolated from M. vannielii and M. barkeri harvested at different stages of growth showed that the ratio of pentameric versus heptameric stalks changes during cell growth with pentameric species being predominant at the early stages and during the lag phase and the proportion of heptameric complexes increasing toward the latter stages of cell growth. Overall therefore in this study we widened our previous MS investigations into ribosomal stalk complexes and targeted species with different OGTs within bacterial, eukaryotic, and archaeal domains.
EXPERIMENTAL PROCEDURES
Ribosome Preparation
Bacterial ribosomes were prepared using a protocol described previously (14) and adapted recently (15). The 50 S subunit was isolated from preparations of the 70 S by hydrophobic interaction chromatography from sucrose gradient ultracentrifugation as described previously (15). Artemia salina 60 S subunits were isolated and purified as described (16). Ribosomes from M. barkeri were prepared according to Ref. 17 with minor modifications. Briefly, cells were collected at A600 = 0.65 (lag phase), 1.07 (mid-log phase), and 1.4 (late log phase) and passed through the French press twice in buffer A (10 mm Tris, pH 7.5, 10 mm MgCl2, 200 mm NH4Cl, 6 mm β-mercaptoethanol). After eliminating cell debris by centrifugation, ribosomes were pelleted at 39,000 rpm in an SW 40 Ti rotor (Beckman Coulter) for 3 h at 4 °C. After resuspension ribosomes were additionally purified through a 30% sucrose cushion in buffer A by centrifugation at 39,000 rpm for 18 h at 4 °C. The ribosomal pellet was washed with buffer B (10 mm Tris, pH 7.5, 10 mm MgCl2, 50 mm NH4Cl, 6 mm β-mercaptoethanol). Ribosomes were dissolved in buffer B, aliquoted, and stored at −80 °C. M. maripaludis ribosomes were purified following the same procedure. Other ribosomal samples were kind gifts. Immediately prior to mass spectrometry aliquots of ribosomes were buffer-exchanged using Bio-Rad Bio-Spin columns into 200 mm–1 m ammonium acetate solution at pH 7.5 and stored on ice.
Mass Spectrometry
Aliquots (∼2 μl) of 1 m ammonium acetate-buffered solutions containing 1–10 μm bacterial 50 and 70 S or eukaryotic 60 and 80 S ribosomes were introduced via nanoflow capillaries into a Q-ToF 2 mass spectrometer (Micromass UK Ltd.) modified for high mass operation and equipped with a nanoflow Z-spray source (18). The following experimental parameters were used: capillary voltage, 1.7 kV; cone gas, 100 liters/h; sample cone, 90–100 V; extractor cone, 0–10 V; collision energy, up to 200 V; ion transfer stage pressure, 4.0 × 10−3 millibar; multichannel plate, 2.4 kV. In tandem MS the relevant m/z was selected in the quadrupole and subjected to acceleration, up to 200 V, in the collision cell.
Alternatively a high mass Q-TOF-type instrument adapted for a QSTAR XL platform (MDS Sciex) was used (19) for acquiring spectra of archaeal 70 S ribosomes in 200–250 mm ammonium acetate-buffered solutions. In this instrument a flow-restricting sleeve is installed in the front part of the first quadrupole guide to increase the pressure locally and allow collisional cooling of heavy ions. Additionally in both spectrometers the low frequency extended mass range of the second quadrupole permits the isolation of ions up to 35,000 m/z.
RESULTS
Bacterial Ribosomal Stalk Complexes Suggest Temperature-dependent Stoichiometry
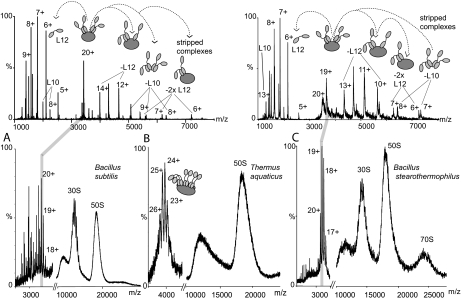
The mass spectrum recorded from an aqueous solution of 70 S ribosomes from B. subtilis is shown in Fig. 2A. Although not as well resolved as the spectrum for ribosomes from T. thermophilus (Fig. 1) in which the 70, 50, and 30 S ribosomes all showed some charge state resolution, the peaks at ∼18,000 m/z by analogy were assigned to the intact 50 S subunit and correspond to this 1.5-MDa particle. The peak for the 30 S subunit at ∼14,000 m/z showed some resolution, allowing tentative charge state assignments for this ∼800-kDa species. At about 4000 m/z we observed ions that are characteristic of the intact stalk complex. We optimized conditions to resolve this feature in the mass spectrum. Its molecular mass was determined from the observed charge states, and from the established masses of the component proteins we calculated the most likely overall stoichiometry. In this case the measured mass was consistent with one copy of L10 and four copies of L12 in a pentameric complex (see Table I).
Fig. 2.
Mass spectra of 70 S ribosomes from B. subtilis, T. aquaticus, and B. stearothermophilus ribosomes and tandem MS of two pentameric stalk complexes. Mass spectra and tandem MS spectra of 70 S ribosomes from B. subtilis (A), T. aquaticus (B), and B. stearothermophilus (C) ribosomes in 1 m ammonium acetate, pH 7.5 are shown. Conditions were optimized for observation of the stalk complex. Individual charge states of two stalk complexes from B. subtilis and B. stearothermophilus were then isolated in the quadrupole and accelerated in the collision cell to analyze their components (MS/MS or tandem MS) (upper panels). At m/z values lower than the isolated peak individual, highly charged proteins are observed that have been released from the complex. At m/z values higher than the isolated charge state peaks are assigned to complexes “stripped” of these released proteins. The schematics at the top of the figure illustrate this process: L10 is represented in dark gray, and L12 composed of two domains connected via a flexible linker is represented in light gray. MS/MS spectra above 3000 m/z have been magnified by a factor of 5 in both cases.
Table I. Measured and calculated masses for proteins from stalk complexes of various species together with stoichiometry defined by MS/MS.
Where masses of the component proteins were not known a priori, the molecular mass of the stalk complex was calculated from the measured masses of the constituent proteins.
| Species, particle studied, optimal growth temperature | Mass of the stalk complex (Da) | Masses of protein released in MS/MS (Da) | Predicted mass (Da) | Identity | Stoichiometry |
|---|---|---|---|---|---|
| Bacteria | |||||
| B. subtilis, 70 S, 38.8 °C | 68,501.3 ± 18.0 | 68,374 | Pentamer | 4:1 | |
| 12,623.1 ± 0.15 | 12,619 | L12-Met | |||
| 17,952.79 ± 0.4 | 18,029 | L10-Met | |||
| 55,836.7 ± 7.2 | 55,755 | Pentamer-L12 | |||
| 43,204.0 ± 3.4 | 43,136 | Pentamer-2 L12 | |||
| T. aquaticus, 50 S, 70–75 °C | 97,658.4 ± 24.2 | 95,883 | Heptamer | 6:1 | |
| 12,937.1 ± 0.3 | 12,908a | L12 | |||
| 18,435 ± 0.5 | 18,509a | L10 | |||
| B. stearothermophilus, 70 S/50 S, 55 °C | 69,481.25 ± 26.4 | 4:1 | |||
| 12,835.60 ± 1 | 12,780 | L12 | |||
| 17,863.79 ± 0.2 | L10 | ||||
| 56,441.96 ± 1.4 | Pentamer-L12 | ||||
| 51,511.66 ± 2 | Pentamer-L10 | ||||
| Eukaryotes | |||||
| A. salina brine shrimp, 60 S, 28 °C | 80,479.25 ± 9.3 | Pentamer | 4:1 | ||
| 34,309.48 ± 7.9 | P0 | ||||
| 11,440.38 ± 1.39 | 11,407 | P1-Met | |||
| 11,522.16 ± 0.7 | 11,503 | P2 | |||
| 68,737.2 ± 13.3 | Stalk-P1/P2 | ||||
| 57,390.5 ± 22.9 | Stalk-2(P1/P2) | ||||
| 46,073.1 ± 16.3 | Stalk-P0 | ||||
| O. cuniculus rabbit, 60 S, 39 °C | 81,115.89 ± 48.3 | Pentamer | 4:1 | ||
| 34,298.25 ± 1.95 | 34,216b | P0 | |||
| 11,810.15 ± 7.98 | 11,651b | P1 | |||
| 11,433.35 ± 0.48 | 11,475b | P2 | |||
| 11,510.63 ± 3.72 | 11,525b | P2c | |||
| 69,135 ± 60.0 | Pentamer-P1 | ||||
| 69,604.3 ± 32.7 | Pentamer-P2 | ||||
| Galdieria sulphuraria red alga, 60 S, 56 °C | 86,244 ± 32.1 | 4:1 | |||
| 82,880 ± 14.43 | 81,675 | Pentamer | |||
| 34,312.9 ± 12.7 | 34,763c | P0 | |||
| 11,778.4 ± 5.8 | 11,615c | P1 | |||
| 11,902.6 ± 1.7 | 11,835c | P2 | |||
| 70,283.7 ± 6.7 | Pentamer-P1/P2 | ||||
| 58,0431.5 ± 61.0 | Pentamer-2 P | ||||
| Archaea | |||||
| M. vannielii, 50 S, 35 °C | 75,342.7 ± 20.7 | 75,223 | Pentamer | 4:1 | |
| 9,819.71 ± 2.4 | 9,818 | L12 | |||
| 65,588.71 ± 31 | 65,405 | Pentamer-L12 | |||
| 55,729.02 ± 9.9 | 55,587 | Pentamer-2 L12 | |||
| 95,405.27 ± 23.6 | 94,859 | Heptamer | 6:1 | ||
| 9,818.93 ± 0.04 | 9,818 | L12 | |||
| 85,359.78 ± 25.53 | 85,041 | Heptamer-L12 | |||
| 75,445.09 ± 38.5 | 75,223 | Heptamer-2 L12 | |||
| M. barkeri, 70 S, 37 °C | 80,124.39 ± 18.0 | 80,040 | Pentamer | 4:1 | |
| 10,677.21 ± 0.41 | 10,677 | L12 | |||
| 69,455.71 ± 2.63 | 69,363 | Pentamer-L12 | |||
| 101,650.02 ± 30.7 | 101,394 | Heptamer | 6:1 | ||
| 90,762.69 ± 18.55 | 90,717 | Heptamer-L12 | |||
| M. maripaludis, 70 S, 35–40 °C | 74,814.80 ± 4.99 | 74,785 | Pentamer | 4:1 | |
| 9,745.39 ± 1.5 | 9,745 | L12 | |||
| 65,090.93 ± 6.77 | 65,040 | Pentamer-L12 | |||
| 55,333.38 ± 4.04 | 55,295 | Pentamer-2 L12 | |||
| 95,159.52 ± 15.63 | 94,275 | Heptamer | 6:1 | ||
| 84,603.44 ± 9.93 | 84,530 | Heptamer-L12 |
a From Thermus thermophilus, a related species.
b From Mus musculus (mouse), a related species.
c From Cyanidioschyzon merolae, a related species.
The stoichiometry of the stalk complex was then confirmed by isolating in the quadrupole defined m/z values that encompass a single charge state of the stalk complex and applying collision energy to induce their dissociation. This process yielded two series of products: the individual proteins L12 and L10 at low m/z values and the stalk complex stripped of highly charged proteins at higher m/z values (Fig. 2A, upper panel). Interestingly three stripped complexes were observed in this case, two with successive losses of L12 and one in which L10 was lost. Although successive loss of L12 proteins is anticipated under these conditions (20), loss of L10 was a surprising observation. This implies that four copies of L12 are sufficiently stable to maintain interactions in the gas phase in the absence of the scaffolding protein L10. Although we think that this complex has no significance in the function or assembly of the ribosome, it may indicate electrostatic interactions between dimers that are enhanced in the gas phase. The molecular masses of the proteins that were released indicate that the entire L10 and L12 populations lacked the N-terminal methionine. No other posttranslational modifications were apparent.
Overall from the mass spectrum of B. subtilis and the tandem mass spectrum recorded for the stalk complex the salient features were the peaks for the 30 and 50 S subunits of the ribosome together with the charge states for the intact pentameric stalk complex. The important points to note are that although some heterogeneity often arises in the stalk complex as a result of incomplete posttranslational modifications of the component proteins (2) this was not the case here. Moreover only one stoichiometry of the stalk complex was observed for ribosomes from this species corresponding to (L12)4:L10.
To investigate a possible relationship between the stoichiometry of the stalk complex and the OGT of the species suggested previously (2) we investigated ribosomes from the extreme thermophile T. aquaticus. Comparing the mass spectrum of T. aquaticus with that of B. subtilis (Fig. 2, A and B) we note that the stalk complex has higher charge state m/z values for the thermophilic species. The molecular mass of this species together with tandem MS experiments (not shown) define the overall stoichiometry as (L12)6:L10. For T. aquaticus neither L12 nor L10 sequences have been reported in databases so consequently we assigned these proteins based on overall DNA sequence homology to that from T. Thermophilus (21) (see Table I). No heterogeneity was apparent due to partial posttranslational modifications of either stalk protein. The finding that ribosomes from T. aquaticus have a heptameric stalk complex is in line with our previous findings for the closely related extreme thermophiles T. thermophilus and T. maritima (2).
Because all three of the extreme thermophiles that we investigated have heptameric stalk complexes we reasoned that elevated growth temperature may be associated with the existence of stalk complexes with this stoichiometry. We therefore investigated B. stearothermophilus, which grows at elevated temperatures, albeit less extreme than those of the extreme thermophiles investigated above. The mass spectrum recorded for 70 S ribosomes from this species showed partially resolved charge states for the 30 S particle, broad peaks for the 50 S particle, and in this case broad peaks also for the intact 2.5-MDa 70 S particle at 26,000 m/z (Fig. 2C). Isolation and activation in the collision cell of the stalk complex yielded both L12 and L10. The mass of L12 was consistent with the presence of N-terminal methionine in contrast to the situation in B. subtilis. Surprisingly the stoichiometry of the stalk complex observed for thermophilic B. stearothermophilus ribosomes corresponds to (L12)4:L10 rather than the heptameric stoichiometry observed for the extreme thermophilic species.
Eukaryotic Ribosomal Stalk Complexes Exist with Only One Stoichiometry
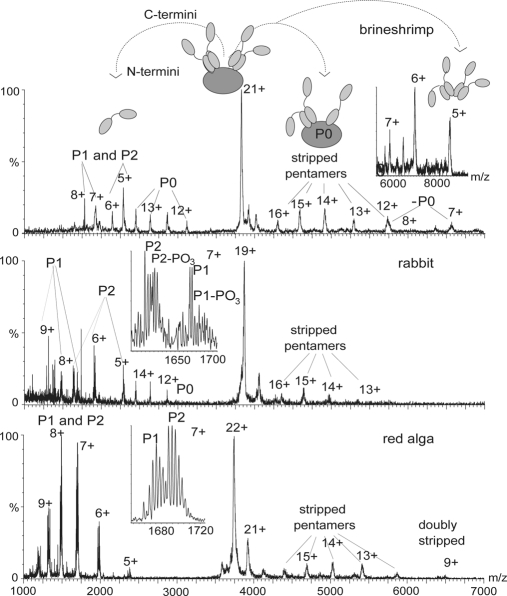
To represent a diverse range of eukaryotes we targeted ribosomes from the insect silkworm (Bombyx mori), the crustacean brine shrimp (A. salina), the mammal rabbit (Oryctolagus cuniculus), and the red alga G. sulphuraria, a plant. We obtained mass spectra and tandem mass spectra of each of the large (60 S) subunits and optimized conditions (22) for the observation of the various stalk complexes (Fig. 3). For the ribosomal stalk complex from brine shrimp (A. salina) the molecular masses of P1 and P2 proteins are established, and both proteins were reported to be partially phosphorylated (23). We assigned charge state series to P1 without the N-terminal methionine and P2 with the N-terminal methionine in place. The 34-kDa protein was assigned to P0 because it is associated with P1 and P2 although its sequence has not been reported. Our data showed no evidence of either protein being phosphorylated. There is, however, clear evidence for charge state series in the MS/MS spectra corresponding to the pentamer stripped of either P1 or P2 as well as loss of P0, confirming the overall pentameric stoichiometry.
Fig. 3.
Tandem mass spectra of eukaryotic ribosomes. Tandem mass spectra of ions of the stalk complexes isolated in the quadrupole from 60 S subunits of eukaryotic ribosomes in 1 m ammonium acetate, pH 7.5 are shown. The stripped complexes at m/z higher than the isolated charge state show pentamers, in all cases, from which one protein has dissociated (stripped pentamers). This reveals their universal pentameric nature. At lower m/z values individual, highly charged proteins released from the complex are observed. Insets show the 7+ charge states of P1 and P2 with magnesium adducts and, in the case of ribosomes from rabbit, phosphorylation of the P1 and P2 proteins.
For the rabbit P-proteins, only the 40 N-terminal amino acids of P0 have been reported in databases. This sequence is identical to that of its relatives the mouse Mus musculus and rat Rattus norvegicus, enabling us to assign P0. P1 and P2 were also assigned based on the masses of their rat and mouse counterparts, and phosphorylated forms of P1 and P2 were identified based on mass differences (Fig. 3, inset, middle panel). P1 proteins dissociate with higher charge states than P2, although they are very similar in mass and have close pI values, 4.28 and 4.42, respectively (based on mouse counterparts). This difference in charge states might be due to a higher degree of unfolding of P1 proteins, necessary for their dissociation from the stalk complex. The stripped complexes confirmed the overall stoichiometry of the rabbit stalk complex as pentameric.
For the silkworm B. mori more than one entry was found for P0 and P1, and this ambiguity means that it is not possible to assign precisely the posttranslational modifications on the basis of intact masses we observed. However, the overall mass of the stalk complex is only consistent with one possible stoichiometry, that of P0:(P1/P2)4 (data not shown). Similarly from mass spectra of the 60 S ribosomal subunit from G. sulphuraria we observed a complex of mass 82,145 Da (Fig. 3), and tandem MS of this complex gave rise to P1 and P2 peaks and at very low intensity to a third series assigned to P0. As no amino acid sequences of G. sulphuraria ribosomal proteins are known we based our assignment on the molecular masses of proteins from Cyanidioschyzon merolae, a related red alga for which the genome has been sequenced. The overall stoichiometry was again unambiguously pentameric.
Archaeal Stalk Complexes of Different Stoichiometries Co-exist on Ribosomes
Turning our attention to archaea we investigated ribosomes from three mesophilic archaeal species, M. vannielii, M. maripaludis, and M. barkeri. The 50 S subunit of M. vannielii ribosome was remarkably well resolved (cf. 50 S charge states in Fig. 4 with those in Fig. 2). Interestingly on close inspection of the region of 3000–5000 m/z, where typically we observed the stalk complex in mass spectra of 50 S particles, we found two series of overlapping charge states (Fig. 4, lower panel). Analysis of this region revealed two distinct complexes of L12:L10 with stoichiometries of 4:1 and 6:1. This surprising result was confirmed by tandem MS. Individual charge states from the two series of peaks were isolated independently, revealing losses of L12 and formation of stripped complexes containing three or five copies of L12 attached to L10, respectively (Fig. 4, upper and middle panels).
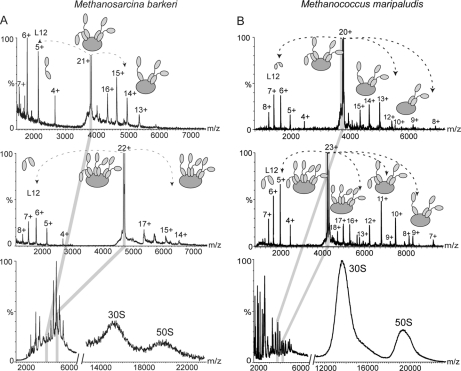
Fig. 4.
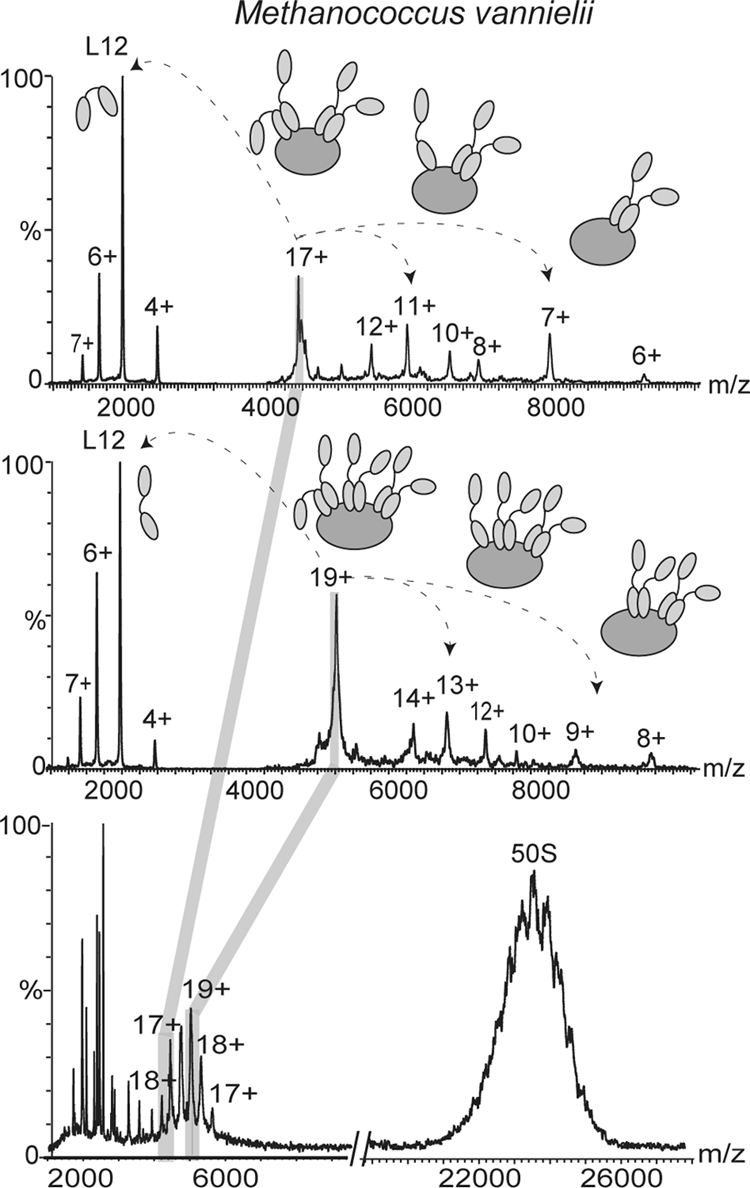
Mass spectra of M. vannielii. Mass spectra of the 50 S subunit of M. vannielii in 1 m ammonium acetate, pH 7.5 showing partial resolution of the charge states of the 50 S subunit are shown. Between m/z 4000 and 6000 (lower panel) overlapping series of charge states are observed that correspond to two different stoichiometries of the stalk complex. Tandem mass spectra of isolated charge states from the two series confirm the 6:1and 4:1 stoichiometry (middle and upper panels, respectively).
To investigate whether two stalk populations were present in other mesophilic archaeal species we also studied ribosomes from the extreme anaerobes M. maripaludis (OGT of 35–40 °C) and M. barkeri (OGT of 37 °C). The stalk complexes in both species were well resolved in the region of 3500–5500 m/z and could be assigned to two series of overlapping charge states (Fig. 5, A and B, respectively) corresponding in mass to both pentamers and heptamers. Isolation of the individual charge states from these series in a tandem MS experiment confirmed the presence of two populations corresponding to 4:1 and 6:1 L12:L10 (Fig. 5, A and B, upper and middle panels). Our investigations of the archaeal stalk complexes therefore showed that all three mesophilic methanogens studied here have both pentameric and heptameric stalk complexes simultaneously present within population of ribosomes.
Fig. 5.
Mass spectra of archaeal ribosomes. A, mass spectra of M. barkeri 70 S particle in 250 mm ammonium acetate, pH 7.5 showing overlapping series of charge states at 3500–5500 m/z corresponding to two different stoichiometries of the stalk complex. Tandem mass spectra of isolated charge states from the two series confirm the 6:1 and 4:1 stoichiometry (middle and upper panels, respectively). B, mass spectra of M. maripaludis 70 S particle in 250 mm ammonium acetate, pH 7.5 showing overlapping series of charge states at 3500–5000 m/z corresponding to two different stoichiometries of the stalk complex. Tandem mass spectra of isolated charge states from the two series confirm the 6:1 and 4:1 stoichiometry (middle and upper panels, respectively).
Stoichiometry of Stalk Complex Changes during Growth in Archaeal Mesophiles
We anticipated that the ratio of the two stalk populations in archaeal species could change as a function of different stages of cell growth, analogous to the acetylation of L12 in E. coli (11, 24). To investigate this possibility we purified ribosomes from both M. vannielii and M. barkeri at different stages of cell growth. Ribosomes purified from M. barkeri cells harvested during the lag phase revealed stalk complexes with predominantly 4:1 L12:L10 stoichiometry (Fig. 6, lower panel). During later stages of cell growth tandem MS spectra confirmed the presence of stripped complexes with a composition of 3:1 and 5:1 L12:L10 arising from the pentameric and heptameric stoichiometries, respectively (Fig. 5). A similar trend was observed for ribosomes from M. vannielii with an initial predominance of pentameric stalk complexes and an increase in the population of heptameric stalk complexes during cell growth (data not shown). The overall trend for both mesophilic archaeal species investigated here indicates that the proportion of heptameric stalk complexes versus pentameric stalk complexes increased during the progression of cell growth.
Fig. 6.
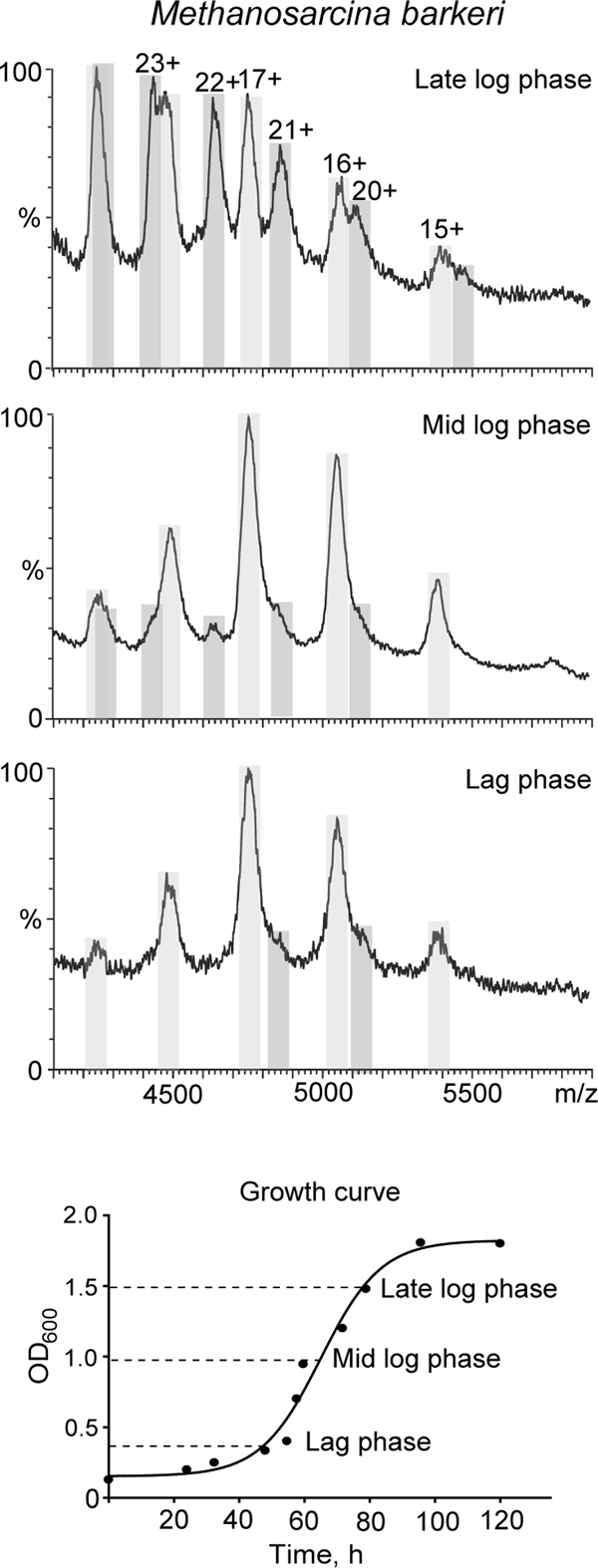
Mass spectra of archaeal stalk complexes during course of cell growth. Expansions of the region of mass spectra of M. barkeri ribosomal stalk complex at different stages of cell growth showing the change in the ratio of pentameric (light gray) versus heptameric (dark gray) species as a function of cell growth are shown. The lower panel shows the changes in optical density as a function of cell growth (min), and the times at which ribosomes were harvested are indicated. Prior to introducing them into the mass spectrometer ribosomes were exchanged into 200 mm ammonium acetate, pH 7.5.
We also investigated whether different posttranslational modifications of L12 in archaeal mesophiles could result in preferential formation of pentameric or heptameric stalk complexes, similar to our experiments to probe the effect of acetylation (11). We isolated charge states from the pentameric and heptameric complexes by tandem MS; however, no differences were observed in dissociated products. In all ribosomal preparations no posttranslational modifications of L12 protein were found that might have contributed to L10 binding. We can conclude therefore that although there are three binding sites for L12 dimers present in L10, two stalk complexes with distinct stoichiometries accommodating two or three L12 dimers exist in the archaeal mesophiles studied here.
DISCUSSION
We have shown here that ribosomal stalk complexes from a diverse range of species, including thermophilic and mesophilic bacteria as well as eukaryotes and archaea, have a closely similar composition. Small differences in posttranslational modifications of stalk proteins were observed within each domain, particularly acetylation and phosphorylation, which we have shown previously to be important in E. coli (11), thermophilic bacteria (2), and eukaryotes (25). The overall stoichiometry of the stalk complexes, however, shows distinctive differences. For eukaryotic ribosomes we found exclusively pentameric stalk complexes despite our investigation of ribosomes from one of the most thermophilic eukaryotes. Those from extreme thermophilic bacterial ribosomes were exclusively heptameric, whereas those from mesophilic bacteria existed in pentameric form. Comparing the region of the spectrum assigned to the intact stalk complex for B. stearothermophilus and T. thermophilus we found that only one population of the stalk complex exists in both cases, whereas for archaeal mesophile M. vannielii there are clearly two (Fig. 7). Our surprising finding of the co-existence of two populations of ribosomes with different stalk complex stoichiometries for the mesophilic archaea M. vannielii prompted us to explore the composition of the stalk complex of two other archaeal mesophilic anaerobes, M. maripaludis and M. barkeri. As in the case of M. vannielii, we found that two populations of ribosomes with either two or three L12 dimers attached to L10 exist in the stalk complexes of these species. Moreover the ratio of pentameric and heptameric stalk complexes changed during different growth stages. At the initial stages of cell growth ribosomes with pentameric stalk complexes dominated, whereas the amount of heptameric stalk complexes attached to the ribosome increased with time.
Fig. 7.
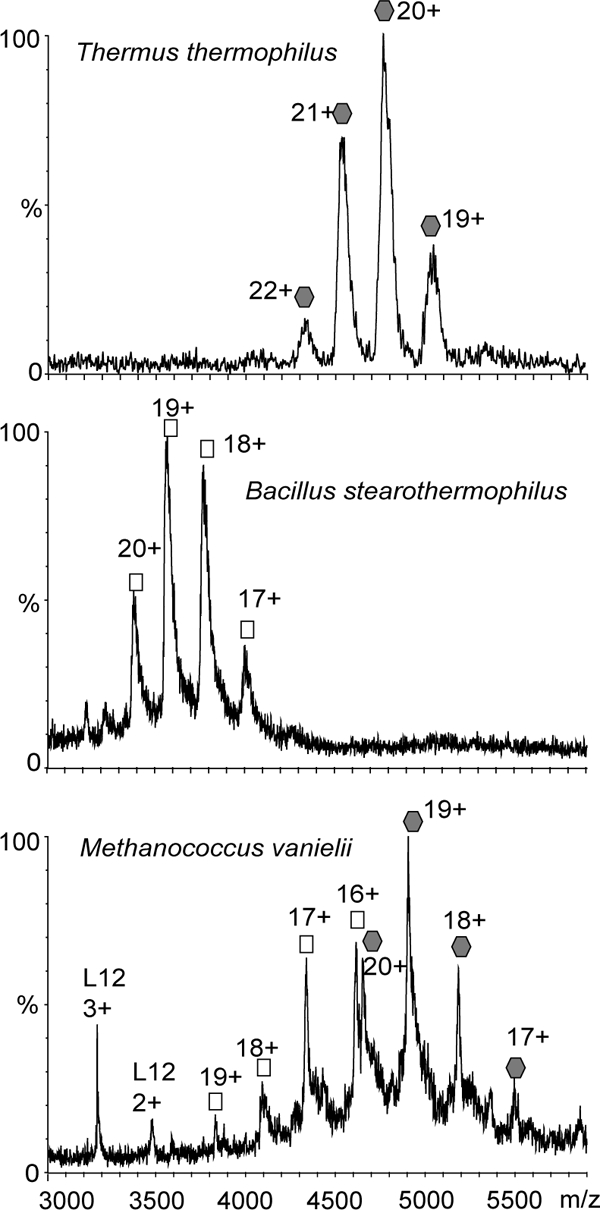
Mass spectra of ribosomal stalk complexes with either heptameric or pentameric stoichiometry and with both present simultaneously. M. vannielii is one of the species for which we observed both pentameric and heptameric ribosomal stalk complexes simultaneously (bottom panel). For other species strictly one stoichiometry was observed as illustrated by stalk complexes from B. stearothermophilus (pentamer; middle panel) and T. thermophilus (heptamer; top panel). All complexes were buffer-exchanged into 1 m ammonium acetate solution at pH 7.5 before introducing into the mass spectrometer.
We have previously established the stoichiometry of L12:L10 from bacterial ribosomes and P1/P2:P0 from Saccharomyces cerevisiae by application of electrospray MS (2 8). By alignment of bacterial L10 sequences we predicted that an eight-amino acid insertion at the C terminus of L10 is sufficient to accommodate binding of an extra L12 dimer (2). Alignment of the archaeal L10 protein sequences of M. vannielii, M. maripaludis, and M. barkeri with that of the archaeal hyperthermophile Pyrococcus horikoshii (26) (OGT of 98 °C) (supplemental Fig. 1) reported during our investigations (27) suggests that the stoichiometry of all stalk complexes would be 6:1. On the other hand the stoichiometry of the M. vannielii stalk complex estimated by scanning of SDS-PAGE gels stained with fluorescent dye is 4:1 (28). This contradiction may be explained by our finding that not all three binding sites are occupied, resulting in a dual stoichiometry of the stalk complexes.
It is interesting to speculate how this dual stoichiometry might arise. One possibility is that these two stalk complexes could be in equilibrium, the third binding site for an L12 dimer exchanging between a free and bound form. No such equilibrium, however, has been observed in the mass spectra of stalk complexes from ribosomes from other species. Moreover, it has been shown recently that the dissociation of L12 from ribosomes from T. maritima when “chased” by excess L12 is very slow, less than 10% per hour (3). Alternatively the copy number of L12 required for full heptameric stoichiometry may simply not be present in the archaeal species studied, leading to an insufficiency in the number of L12 proteins to satisfy the complete population of six copies of L12 on all copies of L10. This proposition is supported by our finding that the proportion of the stalk complex with three binding sites occupied which changed during cell growth, likely reflecting the changes in proportion of L10 and L12 expression levels. It is worth noting that in bacteria the genes of the stalk proteins L10 and L12 constitute their own operon, and L10 functions as a translational autoregulator of the operon binding to its own mRNA as part of the stalk complex (29). However, in archaea the genes encoding L10 and L12 proteins are in tandem as part of the L1 operon, and L10 does not have a regulatory function (30). This may explain insufficient L12 expression levels. The finding that B. stearothermophilus has an OGT of 55 °C and yet has a pentameric stalk complex indicates that the temperature at which it becomes imperative to have a higher stalk stoichiometry is greater than 55 °C. Evidence that growth temperatures below 55 °C do not require the heptameric stoichiometry was supplied by the most thermophilic eukaryote, the unicellular red microalga G. sulphuraria, which can represent up to 90% of the biomass in extreme habitats such as hot sulfur springs with temperatures of up to 56 °C. Although its thermophilicity may explain the quality of the MS data we obtained for this most thermophilic eukaryote, its pentameric stalk complex leads us to speculate that heptamer stoichiometry is unlikely to be found in eukaryotes.
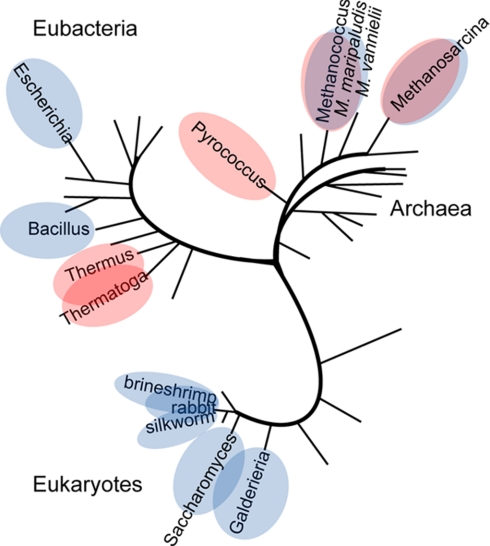
If we consider the evolutionary consequences of our findings, the fact that unrelated ribosomal stalk proteins in bacteria, archaea, and eukaryotes fulfill the same function in a very similar architecture of dimers bound to a scaffold protein (31) is a remarkable example of convergent evolution. The observation that both stoichiometries are seen in unrelated stalk complexes of bacteria and archaea as illustrated in the phylogenetic tree (Fig. 8) suggests that the stoichiometry of ribosomal stalk complexes is governed by adaptive selection rather than any accident of phylogenetic history. Adopted similar architecture of the stalk complex indicates that this must be a favored solution to providing the ribosome with a mechanism to increase the local concentration of elongation factors in the vicinity of the peptidyltransferase center. Therefore having six copies at very elevated temperatures must be thermodynamically advantageous for thermophiles compared with the favored four copies of the factor binding stalk proteins for moderate temperatures. The increase in protein dynamics at elevated temperatures might be compensated by the presence of the third pair of L12 dimers bringing translational factors into proximity of their binding sites on the ribosome. The evolutionary position of archaea descended directly from hyperthermophiles (32) may well be responsible for the dual stoichiometry of the stalk complex. It is tempting to speculate that at moderate temperatures it is not necessary to utilize the third pair of L12 proteins.
Fig. 8.
Schematic unrooted phylogenetic tree according to archaea model showing stoichiometry of ribosomal stalk complexes determined by MS. A (L12)6:L10 stoichiometry of the stalk complexes is indicated in red, (L12)4:L10 and (P1/P2)4:P0 stoichiometries are indicated in blue, and species in which both are found are shown with both blue and red.
More generally our results have shown that MS studies of large biological assemblies can give functional insights and determine the stoichiometry of subcomplexes in situ on large macromolecular assemblies (2). By careful analysis of mass spectra of ribosomes we were able to draw a wide variety of conclusions, ranging from the observation of posttranslational modifications to defining the overall stoichiometry of the stalk complex within different populations of ribosomes. It is doubtful that it would be possible to establish the coexistence of populations of ribosomes with different stoichiometries of the stalk complex by any other structural biology approach. Overall therefore we believe that this study enabled insight into the composition and associations of subunits within ribosomal stalk complexes from disparate biological sources with possible evolutionary implications.
Supplementary Material
Acknowledgments
We are very grateful to our many collaborators for the generous gifts of ribosome samples without which this study would not have been possible. In addition we acknowledge with thanks members of the Robinson group for useful comments throughout this research.
* This work was supported by the Biotechnology and Biological Sciences Research Council, the Royal Society, and European Commission 7th Framework Program Grant HEALTH-F4-2008-201648/PROSPECTS.
 This article contains supplemental Fig. 1.
This article contains supplemental Fig. 1.
1 The abbreviation used is:
- OGT
- optimal growth temperature.
REFERENCES
- 1.Frank J. (2001) Ribosomal dynamics explored by cryo-electron microscopy. Methods 25, 309–315 [DOI] [PubMed] [Google Scholar]
- 2.Ilag L. L., Videler H., McKay A. R., Sobott F., Fucini P., Nierhaus K. H., Robinson C. V. (2005) Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 8192–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaconu M., Kothe U., Schlünzen F., Fischer N., Harms J. M., Tonevitsky A. G., Stark H., Rodnina M. V., Wahl M. C. (2005) Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121, 991–1004 [DOI] [PubMed] [Google Scholar]
- 4.Wool I. G., Chan Y. L., Glück A., Suzuki K. (1991) The primary structure of rat ribosomal proteins P0, P1, and P2 and a proposal for a uniform nomenclature for mammalian and yeast ribosomal proteins. Biochimie 73, 861–870 [DOI] [PubMed] [Google Scholar]
- 5.Bailey-Serres J., Vangala S., Szick K., Lee C. H. (1997) Acidic phosphoprotein complex of the 60S ribosomal subunit of maize seedling roots. Components and changes in response to flooding. Plant Physiol. 114, 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalo P., Reboud J. P. (2003) The puzzling lateral flexible stalk of the ribosome. Biol. Cell 95, 179–193 [DOI] [PubMed] [Google Scholar]
- 7.Shimmin L. C., Ramirez C., Matheson A. T., Dennis P. P. (1989) Sequence alignment and evolutionary comparison of the L10 equivalent and L12 equivalent ribosomal proteins from archaebacteria, eubacteria, and eucaryotes. J. Mol. Evol. 29, 448–462 [DOI] [PubMed] [Google Scholar]
- 8.Hanson C. L., Videler H., Santos C., Ballesta J. P., Robinson C. V. (2004) Mass spectrometry of ribosomes from Saccharomyces cerevisiae: implications for assembly of the stalk complex. J. Biol. Chem. 279, 42750–42757 [DOI] [PubMed] [Google Scholar]
- 9.Videler H., Ilag L. L., McKay A. R., Hanson C. L., Robinson C. V. (2005) Mass spectrometry of intact ribosomes. FEBS Lett. 579, 943–947 [DOI] [PubMed] [Google Scholar]
- 10.Hanson C. L., Fucini P., Ilag L. L., Nierhaus K. H., Robinson C. V. (2003) Dissociation of intact Escherichia coli ribosomes in a mass spectrometer. Evidence for conformational change in a ribosome elongation factor G complex. J. Biol. Chem. 278, 1259–1267 [DOI] [PubMed] [Google Scholar]
- 11.Gordiyenko Y., Deroo S., Zhou M., Videler H., Robinson C. V. (2008) Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J. Mol. Biol. 380, 404–414 [DOI] [PubMed] [Google Scholar]
- 12.Heine M., Chandra S. B. C. (2009) The linkage between reverse gyrase and hyperthermophiles: a review of their invariable association. J. Microbiol. 47, 229–234 [DOI] [PubMed] [Google Scholar]
- 13.Maeder D. L., Anderson I., Brettin T. S., Bruce D. C., Gilna P., Han C. S., Lapidus A., Metcalf W. W., Saunders E., Tapia R., Sowers K. R. (2006) The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188, 7922–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spedding G. (1990) Ribosomes and Protein Synthesis: a Practical Approach (Spedding G. ed) pp. 1–27, Oxford University Press, Oxford [Google Scholar]
- 15.Clemons W. M., Jr., Brodersen D. E., McCutcheon J. P., May J. L., Carter A. P., Morgan-Warren R. J., Wimberly B. T., Ramakrishnan V. (2001) Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: purification, crystallization and structure. J. Mol. Biol. 310, 827–843 [DOI] [PubMed] [Google Scholar]
- 16.Uchiumi T., Wahba A. J., Traut R. R. (1987) Topography and stoichiometry of acidic proteins in large ribosomal subunits from Artemia salina as determined by crosslinking. Proc. Natl. Acad. Sci. U.S.A. 84, 5580–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christodoulou J., Larsson G., Fucini P., Connell S. R., Pertinhez T. A., Hanson C. L., Redfield C., Nierhaus K. H., Robinson C. V., Schleucher J., Dobson C. M. (2004) Heteronuclear NMR investigations of dynamic regions of intact E. coli ribosomes. Proc. Natl. Acad. Sci. U.S.A. 101, 10949–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobott F., Benesch J. L., Vierling E., Robinson C. V. (2002) Real-time monitoring of subunit exchange between small heat shock proteins using electrospray mass spectrometry. J. Biol. Chem. 277, 38921–38929 [DOI] [PubMed] [Google Scholar]
- 19.Chernushevich I. V., Thomson B. A. (2004) Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 76, 1754–1760 [DOI] [PubMed] [Google Scholar]
- 20.Benesch J. L., Robinson C. V. (2006) Mass spectrometry of macromolecular assemblies: preservation and dissociation. Curr. Opin. Struct. Biol. 16, 245–251 [DOI] [PubMed] [Google Scholar]
- 21.Williams R. A., Smith K. E., Welch S. G., Micallef J., Sharp R. J. (1995) DNA relatedness of Thermus strains, description of Thermus brockianus sp. nov., and proposal to reestablish Thermus thermophilus (Oshima and Imahori). Int. J. Syst. Bacteriol. 45, 495–499 [DOI] [PubMed] [Google Scholar]
- 22.Hernández H., Robinson C. V. (2007) Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2, 715–726 [DOI] [PubMed] [Google Scholar]
- 23.Amons R., Pluijms W., Möller W. (1979) The primary structure of ribosomal protein eL12/eL12-P from Artemia salina 80 S ribosomes. FEBS Lett. 104, 85–89 [DOI] [PubMed] [Google Scholar]
- 24.Deusser E., Wittmann H. G. (1972) Ribosomal proteins: variation of the protein composition in Escherichia coli ribosomes as function of growth rate. Nature 238, 269–270 [DOI] [PubMed] [Google Scholar]
- 25.Damoc E., Fraser C. S., Zhou M., Videler H., Mayeur G. L., Hershey J. W., Doudna J. A., Robinson C. V., Leary J. A. (2007) Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell. Proteomics 6, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 26.Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maki Y., Hashimoto T., Zhou M., Naganuma T., Ohta J., Nomura T., Robinson C. V., Uchiumi T. (2007) Three binding sites for stalk protein dimers are generally present in ribosomes from archaeal organism. J. Biol. Chem. 282, 32827–32833 [DOI] [PubMed] [Google Scholar]
- 28.Shcherbakov D., Dontsova M., Tribus M., Garber M., Piendl W. (2006) Stability of the ‘L12 stalk’ in ribosomes from mesophilic and (hyper)thermophilic Archaea and Bacteria. Nucleic Acids Res. 34, 5800–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnsen M., Christensen T., Dennis P. P., Fiil N. P. (1982) Autogenous control: ribosomal protein L10-L12 complex binds to the leader sequence of its mRNA. EMBO J. 1, 999–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer C., Köhrer C., Gröbner P., Piendl W. (1998) MvaL1 autoregulates the synthesis of the three ribosomal proteins encoded on the MvaL1 operon of the archaeon Methanococcus vannielii by inhibiting its own translation before or at the formation of the first peptide bond. Mol. Microbiol. 27, 455–468 [DOI] [PubMed] [Google Scholar]
- 31.Grela P., Bernadó P., Svergun D., Kwiatowski J., Abramczyk D., Grankowski N., Tchórzewski M. (2008) Structural relationships among the ribosomal stalk proteins from the three domains of life. J. Mol. Evol. 67, 154–167 [DOI] [PubMed] [Google Scholar]
- 32.Gribaldo S., Brochier-Armanet C. (2006) The origin and evolution of Archaea: a state of the art. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.