Abstract
Many fundamental processes in the cell are performed by complex macromolecular assemblies that comprise a large number of proteins. Numerous macromolecular assemblies are structurally rather fragile and may suffer during purification, resulting in the partial dissociation of the complexes. These limitations can be overcome by chemical fixation of the assemblies, and recently introduced protocols such as gradient fixation during ultracentrifugation (GraFix) offer advantages for the analysis of fragile macromolecular assemblies. The irreversible fixation, however, is thought to render macromolecular samples useless for studying their protein composition. We therefore developed a novel approach that possesses the advantages of fixation for structure determination by single particle electron microscopy while still allowing a correlative compositional analysis by mass spectrometry. In this method, which we call “electron microscopy carbon film-assisted digestion”, macromolecular assemblies are chemically fixed and then adsorbed onto electron microscopical carbon films. Parallel, identically prepared specimens are then subjected to structural investigation by electron microscopy and proteomics analysis by mass spectrometry of the digested sample. As identical sample preparation protocols are used for electron microscopy and mass spectrometry, the results of both methods can directly be correlated. In addition, we demonstrate improved sensitivity and reproducibility of electron microscopy carbon film-assisted digestion as compared with standard protocols. We show that sample amounts of as low as 50 fmol are sufficient to obtain a comprehensive protein composition of two model complexes. We suggest our approach to be an optimization technique for the compositional analysis of macromolecules by mass spectrometry in general.
An increasing number of proteins are nowadays recognized to fulfill their cellular tasks as part of macromolecular machines (1). Recent advances in the isolation procedures allow purification of many of these assemblies for further biochemical and structural analysis (2–4). Thereby, MS of peptides derived by digestion of the proteins of the assemblies (5, 6) or MS of intact assemblies (7, 8) is used to determine the protein composition, whereas electron microscopy (EM)1 can be used to provide information about the structure of the assembly using computational averaging and reconstruction techniques (9). Available protocols to combine information derived by MS and EM therefore either analyze the endoproteolytic digestion products (10–13) or the intact assembly (14). However, the sample preparation protocols typically differ between EM and MS. Although for MS the sample is either directly digested in solution (6), subsequent to gel electrophoretic separation of the sample (15–17) or used in an intact form (7, 8), EM typically requires the adsorption of the sample to carbon film (13, 18–20). In the case of sample heterogeneity, the utilization of these different sources of material (i.e. in-solution versus carbon-adsorbed particles) may be associated with the analysis of different particle populations in MS and EM due to for example the individual kinetics of adsorption, thus making a direct correlation of the EM and MS data difficult.
To combine the information obtained by both methods, we thus set out to correlate MS and EM. To do this would require identical material and conditions for the EM and MS analyses to ensure that identical particle populations are measured by both techniques. To meet this need, we developed an approach for digestion of the assemblies directly on EM carbon film followed by an analysis of the generated peptides by MS. Our method differs from established protocols for sample preparation in that identical conditions and input material are used for the analysis of the composition by MS and of the structure by EM.
The key to our approach is that disintegration of fragile assemblies can often be reduced by stabilization of the interacting components to reduce sample heterogeneity caused by the sample preparation to a minimum. Chemical fixation with e.g. formaldehyde or glutaraldehyde is known to cross-link fragile and transient assemblies (21) and thus greatly improves the homogeneity of carbon-adsorbed macromolecular samples in EM (3). Optimized sample integrity and a minimum of background (the latter including contaminants and degraded or aggregated components) can be attained by the use of suitable fixation protocols (3). Mild fixation, for example, is achieved by exposure to low concentrations of glutaraldehyde (≤0.1%), which cross-links the accessible primary amines of lysines and protein N termini (21). The improved sample quality seen in the EM led us to hypothesize that the mild fixation used might leave enough protein regions unmodified for them to be cleaved into peptides for sequencing by MS and identification by standard database searches. In that way, the advantage of fixation (improved structural integrity) would be retained, whereas the potential disadvantage of too strong fixation (excessive structural cohesion) would be avoided.
Here we describe this new technique, which we call “electron microscopy carbon film-assisted digestion” (ECAD). It involves mild fixation of the macromolecular assemblies to obtain improved structural integrity of fragile macromolecular assemblies and to allow EM- and MS-based characterization of identical, parallel samples. This adds to the repertoire of approaches for interactome analysis of macromolecular complexes and is especially useful for composition studies of fragile assemblies. ECAD allows a correlation of composition and structure of a macromolecular assembly as discrepancies that may be imposed by differences in the preparation of the respective samples for EM and MS are avoided. We further suggest that the improved integrity of a macromolecule upon ECAD of chemically stabilized macromolecular assemblies can also be an advantage for MS analysis in general independent of the EM analysis.
EXPERIMENTAL PROCEDURES
Supplies
All chemicals and reagents including the lyophilized mixture of GroEL-GroES and Glu-fibrinogen were purchased from Sigma-Aldrich unless otherwise specified. A 25% EM grade glutaraldehyde solution was obtained from Electron Microscopy Sciences (Hatfield, PA) and was used no more than 1 month after initial puncture of the seal to ensure sufficient activity of the cross-linker. Uranyl formate was obtained from Polysciences Inc. (Warrington, PA). RNases A and T1 were purchased from Ambion (Austin, TX). Trypsin (sequencing grade trypsin, modified) was from Promega (Madison, WI).
Biochemical Methods
GroEL-GroES Complexes
A lyophilized mixture of GroEL-GroES was dissolved in buffer G (50 mm HEPES, pH 8.0, 50 mm KCl, 50 mm MgCl2) supplemented with 2.5 mm ATP to a concentration of 1 mg/ml. GroEL-GroES (125 μg) was loaded onto a 10–30% glycerol gradient (also containing a gradient from top to bottom of 2.5–0 mm ATP) in buffer G, and ultracentrifugation was performed in a Sorvall TH-660 rotor (Thermo Scientific, Waltham, MA) over 17 h at 30,000 rpm and 4 °C.
25 S U4/U6.U5 Tri-snRNP Particles
Native human 25 S tri-snRNPs were isolated from HeLa nuclear extract by H-20 immunoaffinity chromatography and subsequent glycerol gradient centrifugation as described previously (22). Ultracentrifugation was performed in a Sorvall TH-660 rotor over 17 h at 30,000 rpm and 4 °C. The protein and RNA composition of fractions containing U snRNPs were determined by electrophoresis on denaturing polyacrylamide gels with SDS (for proteins) or urea (for RNA). Proteins were stained with Coomassie Brilliant Blue, and RNA was stained with silver (23, 24).
Fractionation and Cross-linking with Glutaraldehyde
All particles were prepared by using ultracentrifugation in a glycerol gradient as the final purification step before sample preparation for EM and MS. The glycerol gradients were fractionated from the bottom by using a peristaltic pump connected to a fraction collector (3). A 25% glutaraldehyde solution was added to the peak fractions to a final concentration of 0.075% (v/v), and the peak fractions were incubated for 18 h at 4 °C before EM and MS analysis.
Electron Microscopy Methods
Preparation of Carbon Films for EM and ECAD
Thin (∼7–10-nm) carbon films were made in a carbon evaporator (E12E, Boc Edwards, Kirchheim, Germany) by evaporation of spectral carbon (Ringsdorff Werke GmbH, Bonn, Germany) onto freshly cleaved mica (Plano GmbH, Wetzlar, Germany) using a standard indirect coating setup. Before adsorption of particles, the coated mica was freshly cleaved into 2 × 4-mm pieces (except for the analysis of 3.5 fmol of GroEL-GroES where a single 1 × 2-mm piece was used).
EM Sample Preparation by Negative Staining
EM grids for negative staining were prepared using the sandwich method (19) except for the freezing step for cryoimaging. The protein concentrations of the samples used for adsorption to the carbon films were in the range suitable for electron microscopy, i.e. 10–200 μg/ml. During preparation, the sample was kept at 4 °C by ice-cooling the sample preparation block. Briefly, the carbon film was floated on the surface of 25 μl of particle solution, allowing adsorption of particles to the carbon film, over a period of 1 min to several hours. Adsorption times were adjusted to yield a total adsorbed particle amount of 50 fmol on a total of up to 10 carbon films as judged by counting the particles on the carbon film in the electron microscope. For negative staining, the carbon film was incubated with an aqueous ∼2% uranyl formate solution for 2 min and subsequently attached to a 400 mesh EM copper grid on which a carbon film containing holes of ∼1-μm diameter had already been mounted. A second carbon film incubated with uranyl formate solution was then used to form a sandwich, enclosing the specimen in a layer of staining solution. The grids were stored under dry conditions at room temperature until image acquisition.
Unstained Cryo-EM Preparation by Vitrification
For cryo-EM in a stain-free buffer, the buffer of the glycerol gradient fractions was exchanged to a glycerol-free buffer using a spin column (Zeba, Pierce). During preparation, the sample was kept at 4 °C. Particles were adsorbed on a piece of carbon film for a defined period of time (1 min to several hours depending on the particle concentration). Subsequently, the carbon film was attached to an EM copper grid covered by a carbon film containing holes. The EM grid was carefully blotted and plunge-frozen in liquid ethane (25). Specimens were stored in liquid nitrogen until imaging in the EM.
Electron Microscopy
The specimens were imaged at room temperature (negative staining) or at cryogenic conditions (unstained cryo-EM) under low dose conditions on a 200-kV transmission electron microscope equipped with a field emission gun (CM200 FEG, Philips/FEI, Eindhoven, The Netherlands) operated at 160 kV. Images were taken on a 4096 × 4096 charge-coupled device camera (TemCam-F415, TVIPS, Gauting, Germany) at a magnification of 122,000× and 2× binning of the charge-coupled device pixels using an electron dose of ∼20 e−/Å2 (26).
Mass Spectrometry Methods
In-solution Digestion of Purified Particles
The protein concentration in the glycerol gradient fractions containing GroEL-GroES or tri-snRNP was determined using Bradford assays to c = 0.19 mg/ml and c = 0.2 mg/ml, respectively. The samples were diluted to the following absolute sample amount prior to in-solution digest: for GroEL-GroES, the sample amounts were 3.5 fmol (3.1 ng), 7 fmol (6.1 ng), and 50 fmol (43.7 ng) (each in a 30-μl volume); for tri-snRNP, we used sample amounts of 10, 20, 50 (85.ng), 100, 150, 250, and 500 fmol (each in a 30-μl volume). Dilution was performed by adding to each sample 7.2 mg of solid urea to obtain a final concentration of 4 m in a 30-μl volume and by adjusting the final volume with 50 mm Tris-HCl, pH 7.5. In the case of glutaraldehyde treatment (see below), samples were incubated with glycine (0.1 m final concentration) for 30 min at room temperature prior to the addition of urea to block residual activity of the cross-linker unless otherwise specified. Samples were incubated for 5 min at room temperature by vigorous shaking. The samples were subsequently diluted with 50 mm Tris-HCl, pH 7.5 to give a final urea concentration of 1 m. The volume of the denatured samples subjected to digestion was thus 120 μl. For tri-snRNP, U4/U6 and U5 small nuclear RNAs were digested for 2 h at 52 °C by adding 0.1 μg of RNase A and 0.16 μg of RNase T1. Samples were chilled briefly on ice, and proteins were digested with 1 μg of trypsin at 37 °C overnight. Digestion was stopped by adding TFA to a final concentration of 0.1% (v/v). Samples were stored at −20 °C until MS analysis. In-solution digests were performed in five technical replicates with the exception of 10 and 20 fmol of tri-snRNPs that were only analyzed twice.
Digestion of Macromolecular Samples Attached to Carbon Film (ECAD)
Fixed macromolecular samples (see above) were adsorbed to the carbon film exactly as performed for the EM analysis, i.e. by putting the mica with the carbon film on top of the particle solution so that the carbon film floats on the solution. After adsorption, the piece of mica is lifted using tweezers and extensively blotted with filter paper (Whatman) without destroying the carbon film. Optionally, the carbon film with absorbed particles can be washed with a washing buffer (e.g. the glycerol-free and glutaraldehyde-free sample buffer). Subsequently, the carbon film is transferred to a reaction tube (Eppendorf) containing 60 μl of a buffer comprising 4 m urea, 50 mm Tris-HCl, pH 7.5, 0.1 m glycine to neutralize the chemical activity of glutaraldehyde. Of note, experiments in which the mica was removed indicated that the MS results are similar irrespective of the presence or absence of the mica. Subsequent to 30-min incubation at room temperature for inactivation of the residual glutaraldehyde activity, samples were sonicated for 15 min at 4 °C to disrupt the carbon film, and the sample volume was adjusted to a concentration of 1 m urea by dilution with 50 mm Tris-HCl, pH 7.5. The volume of the denatured samples subjected to digestion was thus 240 μl. Of note, digestion was performed in the presence of disrupted (i.e. sonicated) carbon film. Hydrolyses of RNA and protein were performed as described above. The samples were centrifuged for 5 min at 13,000 rpm, and the supernatant was collected. Peptides were extracted from the carbon film pellet by adding 120 μl of 60% (v/v) acetonitrile in water containing 0.2% (v/v) formic acid and shaking in a thermomixer for 15 min at 37 °C. The supernatant was dried in a SpeedVac and pooled with the supernatant containing 1 m urea (see above). TFA was added to the pooled supernatants to give a final TFA concentration of 0.1% (v/v). Samples were stored at −20 °C until the MS measurement. Unfixed samples subjected to ECAD were treated likewise except that no glutaraldehyde and no glycine were added to the sample. Different sample amounts for ECAD (e.g. 3.5, 7, and 50 fmol of GroEL-GroES complex and 10–50 fmol of tri-snRNP) were prepared by using a smaller piece of carbon (down to 1 × 2 mm) or, more usually, a greater number of carbon films; in our experience, up to 7–10 carbon films can be processed in a single reaction tube containing 60 μl of the 4 m urea solution. All ECAD experiments using GroEL-GroES and tri-snRNP (50 fmol) were performed five times; the other samples were analyzed only once.
Off-line Nano-LC Analysis
Samples were injected into a nano-LC system (Dual Gradient, Dionex, Idstein, Germany) in six (in-solution) or 12 (ECAD) loading cycles using a 20-μl injection volume for each cycle. The system was equipped with precolumns working in back-flush mode (25 × 0.15 mm packed in house with C18, 5 μm, 300 Å; number 218TP5215, Vydac, Hesperia, CA). Each sample loading cycle was performed for 10 min with a flow rate of 5 μl/min in loading solvent A (3.5% (v/v) ACN containing 0.1% (v/v) TFA in water). After loading of the sample, the precolumn was flushed with loading solvent for 15 min at 5 μl/min between successive loading cycles. Peptides were eluted from the precolumn in back-flush mode and separated on an analytical column packed in house (200 × 0.075 mm, C18, 5 μm, 300 Å; number 218TP5215, Vydac) by a standard gradient from 10% solvent B to 60% solvent B over 3 h (solvent A, 0.1% (v/v) TFA in water; solvent B, 80% (v/v) ACN, 0.1% (v/v) TFA in water) at a flow rate of 300 nl/min. The eluted peptides were mixed in a T-piece (MicroTEE, 6 μl dead volume, Upchurch Scientific Inc., Oak Harbor, WA) with 10 mg/ml α-cyano-4-hydroxycinnamic acid matrix (Sigma-Aldrich) containing 10 fmol/μl Glu-fibrinogen as internal standard in 70% (v/v) ACN, 0.1% (v/v) TFA delivered at a flow rate of 0.9 μl/min. Every 15 s the eluate mixed with matrix was spotted onto a stainless steel MALDI target (Opti-TOFTM LC/MALDI insert, Applied Biosystems) by a Probot Spotter (Dionex). Per gradient, ∼600 peptide-containing fractions were collected.
Column Packing
Nanoanalytical columns were packed as follows. A 2-mm frit at one end of a fused silica capillary (360-μm outer diameter, 75-μm inner diameter; Polymicro Technologies, Phoenix, AZ) was generated by polymerizing Kvasil1TM (PQ Europe, Amersfoort, The Netherlands). Column material was packed into the capillaries at 50 bars by using a high pressure chamber (Brechbühler) working with helium. The analytical column was connected to the system by using a PEEK polymer tubing and stainless steel nuts and ferrules (PEEK, gray, × 0.015; 400-μm inner diameter; Upchurch Scientific Inc.). Precolumns manufactured in house were packed with fused silica capillaries (375-μm outer diameter, 150-μm inner diameter, 500–750 mm in length; Polymicro Technologies). Precolumns were cut into lengths of 25 mm, and both ends were sealed with fittings (Inline MicroFilters, Upchurch Scientific Inc.) by using MicroTight Sleeves (0.0155 × 0.025; Upchurch Scientific Inc.) and Inline MicroFilters (PEEK, 0.5 μm; Upchurch Scientific Inc.).
MALDI-MS and MSMS Analysis
MS analysis was performed on a MALDI-TOF/TOF 4800 analyzer (Applied Biosystems/Sciex MDS, Foster City, CA) equipped with a neodymium-doped yttrium aluminium garnet (Nd:YAG) laser (355-nm wavelength and 200-Hz repetition rate). For MS spectra in positive ion mode, a total of 800 shots were generated, and for MSMS a maximum of 2000 shots were accumulated for each precursor; dynamic stop criteria depending on the spectral quality were used. Job-wide interpretation of the MS data allowed the 15 highest intensity peptides of every spot to be sequenced in MSMS mode. Peak lists were created using “TS2Mascot open” or the “Peak to Mascot” tool of the 4000 Series Explorer software v3.5.3 (release date, February 2007). Collision energy in MSMS mode was set to 1 × 10−6 torr with the potential difference between the source II accelerator and the collision cell set at 1 kV.
Protein Identification
Proteins were identified by searching fragment spectra against the databases RefSeq (taxonomy, human; release date, February 29, 2008) with 70,679 sequences used for the actual database search or NCBInr (release dates, August 28, 2006 and October 8, 2007) with 128,611 and 194,779 sequences, respectively, used for the actual database search using Mascot (27) v2.2.06 as search engine with the following parameters: taxonomy, human; specificity of trypsin considered; two missed cleavages allowed; oxidation (Met) and carbamylation (Lys and N termini) as variable modifications; no fixed modification; MS mass tolerance set to 100 ppm; and MSMS mass tolerance set to 600 millimass units. For data evaluation with Mascot, only “bold red” peptides with a peptide score ≥20 were considered (except for the analysis of overall peptide hits and average peptide score shown in Fig. 9, A and B). Of note, this particular score for a single “red bold” peptide was chosen as cutoff because our experience with the 4800 MALDI-TOF/TOF instrument revealed that this particular score is still valid for protein identification under the condition that the corresponding product ion spectra (MSMS) are manually validated. Keratins were removed from the protein identification list. Ambiguities caused by the redundancy in the databases as well as by protein isoforms or protein families with shared sequences were corrected on the basis of visual inspection of the Mascot database result. Thereby, the protein entry with the higher Mascot score was kept in the list.
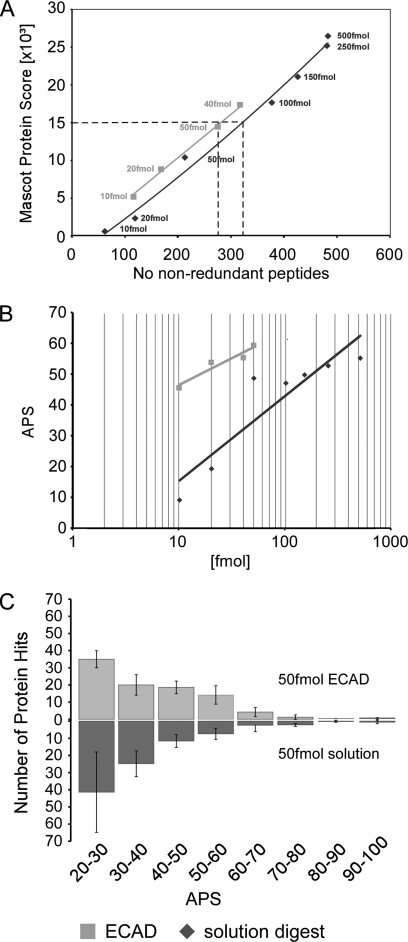
Fig. 9.
Peptide scores. A, plot of the Mascot protein score versus the number of non-redundant peptides for ECAD (light grey) and the in-solution digest (dark grey). Overall, ECAD achieves a higher Mascot score based on fewer non-redundant peptides, i.e. ECAD detects selected peptides with a better score. A Mascot protein score of 15,000 is obtained from ∼280 non-redundant peptides in ECAD, whereas ∼320 non-redundant peptides would be required to achieve this particular Mascot protein score in the in-solution digest (dashed lines). B, semilogarithmic plot of the APS for tri-snRNP proteins as a function of sample amount (light grey, ECAD; dark grey, in-solution digest). C, histogram of the number of database protein hits from digests according to APS determined from five repeated experiments using 50 fmol of sample for the in-solution digest (dark grey) and ECAD (light grey).
Statistics
Statistical Analysis
p values were computed by paired, two-sided Wilcoxon tests as implemented in the software package R (28).
Generation of Color-coded Three-dimensional Models
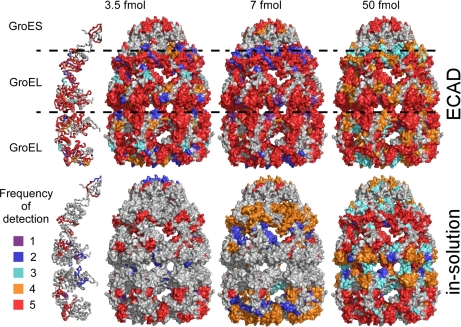
Peptide location maps in the protein three-dimensional model were generated by combining the data sets from the five independent MALDI-MS (MSMS) analyses. Peptides found in all five independent analyses are shown in red, peptides found four times in the five analyses are shown in orange, peptides found three times are shown in turquoise, peptides found twice are shown in dark blue, and peptides found once are shown in purple. Peptides with their corresponding color code are visualized in the three-dimensional structure of GroEL-GroES in complex with seven molecules of ADP (Protein Data Bank code 1svt (29)). For the color coding of the surface of the GroEL-GroES structure, we used scripting and visualization tools provided by the PyMOL software (DeLano Scientific LLC, Palo Alto, CA).
RESULTS
ECAD Approach
Our approach was designed to determine the protein composition of macromolecular complexes using the same sample preparation protocol for both EM and MS analysis (the work flow is summarized in Fig. 1). In particular for EM of low concentrated samples, the assemblies are required to be adsorbed onto carbon film to accumulate sufficient amounts for EM analysis (3, 13). As different assemblies (e.g. broken assemblies, aggregates, or contaminating complexes) may adsorb with individual kinetics, the particle population seen in the electron microscope and in MS can differ. The approach presented herein eliminates such possible differences because the same sample handling is used for EM and MS analyses. To test our approach, we used well characterized macromolecular assemblies with a level of fixation similar to that obtained by using a previously established protocol (3) so that separation of individual proteins by SDS-PAGE was prevented (Fig. 2).
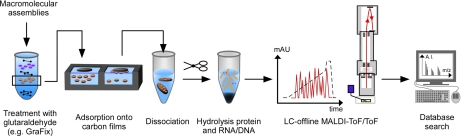
Fig. 1.
The work flow of ECAD. Purified macromolecular assemblies are adsorbed onto EM carbon film using the same sample preparation protocol as used for EM analysis to ensure identical particle populations during the analysis. Typically, the complexes are mildly chemically cross-linked using glutaraldehyde, but also untreated samples can be subjected to ECAD. After inactivation of residual cross-linker using an excess of glycine (in the case that fixed samples were used) and a denaturing step, the protein and, where applicable, RNA/DNA moieties are hydrolyzed. Generated peptides are subjected to MS-based sequencing using a MALDI or ESI instrument. Proteins are identified by database search of the non-glutaraldehyde-modified peptides. A.I., absolute intensity; mAU, milliabsorbance units.
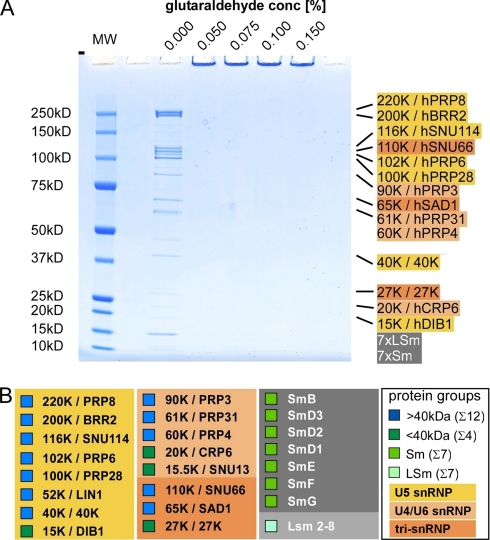
Fig. 2.
Gel electrophoresis of non-cross-linked and cross-linked macromolecular assemblies. A, SDS-PAGE of 10 μg of tri-snRNP loaded without (0.0%) glutaraldehyde and after incubation with 0.05, 0.075, 0.1, and 0.15% (v/v) glutaraldehyde for 18 h at 4 °C as indicated above each lane. Treatment of the macromolecular assemblies with these low concentrations of glutaraldehyde prevents separation of tri-snRNP proteins by denaturing SDS-PAGE. The regular position of the tri-snRNP-specific proteins (30), in the case of no fixation, is indicated on the right. The seven Sm proteins and seven LSm proteins are listed as Sm and LSm, respectively. The color coding is as follows: yellow, U5 snRNP-specific proteins; light orange, U4/U6 snRNP-specific proteins; orange, tri-snRNP-specific proteins; gray, Sm and LSm proteins. MW, molecular weight marker. Under these conditions, a monodisperse population of intact, individual tri-snRNP particles can be visualized by single particle EM. B, overview of all tri-snRNP proteins with classification into proteins of at least 40 kDa (blue), smaller than 40 kDa (dark green), Sm proteins (green), and LSm proteins (light green) as indicated in small squares in front of the protein name. In addition, the proteins are assigned to the subcomplexes U5 snRNP (yellow), U4/U6 snRNPs (light orange), tri-snRNP-specific proteins (orange), Sm proteins (dark gray), and LSm proteins (light gray).
In our procedure (Fig. 1), we adsorbed the particles onto carbon films of 7–10-nm thickness produced by evaporating carbon onto a freshly cleaved piece of mica. To do this, the films were floated on the particle solution for a defined period (1 min to several hours depending on sample concentration). One of the films was mounted on a standard EM grid and subjected to EM analysis. By single particle EM using negatively stained or vitrified (cryo-EM) specimens (19, 25), the particles on the complete carbon film were counted by extrapolation from several representative areas of the specimen (see Fig. 3A for a negatively stained specimen and Fig. 3B for a native cryospecimen in vitrified buffer). Single particle image processing was performed to determine the structure of the particle (Fig. 3, C and D). The other carbon films were then transferred to a reaction tube containing 60 μl of 4 m urea and an excess of glycine to block residual activity of glutaraldehyde. After denaturation, digestion of the samples with endoproteinases and ribonucleases was performed directly on the EM carbon film (see Fig. 1). Of note, an excess of enzymes was used (that was above the recommended enzyme-to-substrate ratio) to ensure that putative adsorption of the enzymes onto the carbon film did not result in too low amounts of enzymes for the digestion of the sample. Digestion products were separated and analyzed by nano-LC off-line MALDI-MSMS or nano-LC ESI-MSMS, and proteins were then identified by searching fragment spectra of unfixed, i.e. non-glutaraldehyde-modified, peptides against a database.
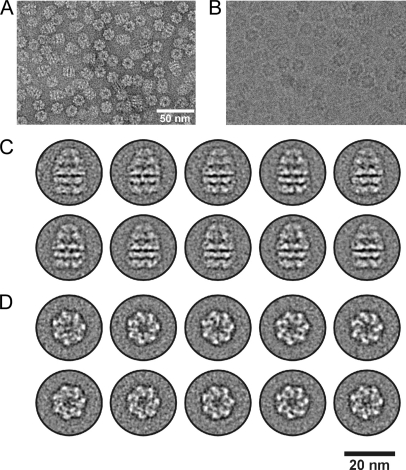
Fig. 3.
Electron microscopic analysis of GroEL-GroES. A, overview of negatively stained GroEL-GroES particles. B, unstained cryospecimen of GroEL-GroES in vitrified ice. The scale in B is the same as that in A. C, representative class averages comprising ∼20 particles per average based on a data set of ∼7000 particle images. The averages show side views of the particles with the GroES subunit at the top. D, same as in C except that selected class averages of top views are depicted.
Required Amount for ECAD
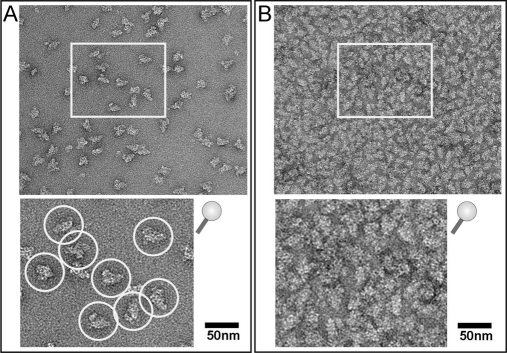
In general, we noted that an amount of ∼2–25 fmol of various macromolecular assemblies on a piece of EM carbon film (2 × 4 mm) showed a particle density suitable for structure determination by single particle EM analysis (Figs. 3 and 4A; Fig. 4B shows an overloaded carbon film that is not suitable for image processing). A correlation of structure and composition using identically prepared samples for both techniques therefore requires that the MS approach also works with low sample quantities, and we show here that our method is indeed applicable for quantities as low as 50 fmol. To establish and validate our approach, we first used a symmetric complex of well known composition, the bacterial GroEL-GroES (containing 14 copies of GroEL and seven copies of GroES (29)). We then examined a more complex assembly, the human U4/U6.U5 tri-snRNP (30) (comprising 29 core proteins of which most occur singly and a few occur as double copies).
Fig. 4.
Electron microscopic analysis of human U4/U6.U5 tri-snRNP complexes. A, negatively stained EM image of ∼5 fmol of tri-snRNP per specimen. Individual particles showing the typical triangular shape of tri-snRNP can be distinguished (upper panel, overview image; bottom panel, subwindow at a higher magnification as indicated in the upper panel). Such a particle density is suitable for single particle image processing. Carbon films with adsorbed particles of this density were subjected to ECAD analysis. B, negatively stained EM image of tri-snRNP showing a particle density too high for image processing; exact counting of the particles is not possible at this high density (upper panel, overview image; bottom panel, subwindow at a higher magnification as indicated in the upper panel). Such a particle density is unsuitable for single particle image processing. These overloaded carbon films can in principle be analyzed by ECAD to increase the sample amount subjected to digestion; however, such overloading is not suitable for a correlation of the MS and EM analyses.
Bacterial GroEL-GroES as Simple Model System
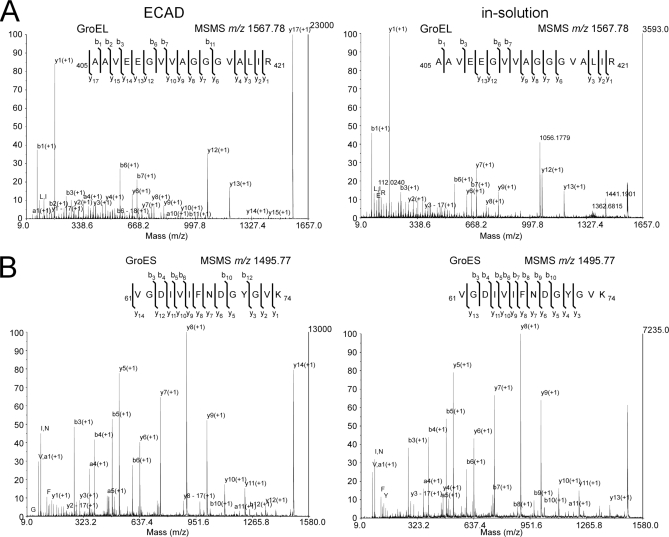
GroEL-GroES contains 14 copies of GroEL and seven copies of GroES (29) and forms two stacked homoheptameric GroEL subcomplexes that are covered by a homoheptameric GroES lid (Fig. 3). We compared the ECAD performance of fixed material with standard in-solution digest of unfixed material. Examples of MSMS spectra are given in Fig. 5. For the GroEL peptide AAVEEGVVAGGGVALIR (residues 405–421), the absolute intensity and thus signal-to-noise ratio was comparable or even higher using ECAD as compared with the in-solution digest (Fig. 5A). ECAD showed an almost complete coverage of the y-type ion series in MSMS. Likewise, the GroES peptide VGDIVIFNDGYGVK (residues 61–74) showed a comparable or even higher absolute ion intensity (Fig. 5B). Together these results indeed suggest an improved quality of the spectra using the ECAD approach.
Fig. 5.
MSMS spectra of GroEL-GroES using ECAD of fixed sample and in-solution digest of unfixed material. Shown is a collection of MSMS spectra that were derived from GroEL- (A) and GroES (B)-specific peptides upon fragmentation by CID. The spectra were selected from two comparable experiments using either 50 fmol of non-fixed material for in-solution digestion (right) or 50 fmol of glutaraldehyde-treated sample for ECAD (left). Both peptides displayed here were identified five times in five technical replicates. A, the MSMS-generated sequence information of the GroEL peptide AAVEEGVVAGGGVALIR (residues 405–421) is compared. The absolute intensity (and the signal-to-noise ratio) is higher by a factor of ∼6.5. Sequencing of the ECAD-generated peptide derives an almost complete coverage of the y-type ion series. B, similar results were observed for the GroES-specific peptide VGDIVIFNDGYGVK (residues 61–74); the absolute ion intensity was higher by a factor of 1.8.
For GroEL-GroES, we analyzed ∼3.5–7 fmol of particles to account for the multiple occurrence of the two constituent proteins (see also supplemental Table S1 for a comprehensive list of identified peptides in the respective analyses and supplemental Table S2 for a summary of the data). With such low amounts, the sensitivity and reproducibility of MS might become an issue. Therefore, we performed five independent ECAD experiments using adsorbed, glutaraldehyde-fixed GroEL-GroES, taking the frequency of identification of a given non-glutaraldehyde-modified peptide in these experiments as a measure of reproducibility (see also supplemental Table S1), whereas in a parallel set of experiments, we digested unfixed samples in solution following a standard protocol. In a range of 3.5–50 fmol of adsorbed, fixed GroEL-GroES complexes, the detected peptides still covered a large portion of the protein sequence (Fig. 6; see also supplemental Table S1), and the reproducibility (five times in five replicates indicated by red colored peptides in Fig. 6; see also supplemental Table S1) was surprisingly good even at 3.5 fmol. Overall, the sequence coverage of ECAD using 3.5 fmol of particles was comparable with that obtained from 50 fmol in in-solution digests (Fig. 6).
Fig. 6.
Performance of ECAD applied to GroEL-GroES. The number of times a given peptide was sequenced by MS and identified by database search in five independent digests is illustrated by coloring the respective peptide stretch of the crystallographic model (29) where red indicates best reproducibility (see key at the bottom). The sequence coverage of glutaraldehyde-treated samples processed according to the ECAD protocol is good even at very low sample amounts of 3.5 or 7 fmol, whereas the sequence coverage is poor for the standard in-solution digest of unfixed sample.
Sensitivity and Reproducibility Using Complex Macromolecular Assembly
We next examined a more complex macromolecular assembly, the human U4/U6.U5 tri-snRNP that comprises three small RNAs and 29 core proteins ranging in size from 8.5 to 274 kDa (30). Most of its proteins occur singly, and a few (the Sm proteins) occur as double copies (see also Fig. 2B). We therefore classified the proteins into a group of particle-specific proteins larger than 40 kDa, a group of particle-specific proteins smaller than 40 kDa, and into two groups of the single copy LSm and the double copy Sm proteins that both form a rigid seven-membered ring (31, 32). In the EM, human tri-snRNP adopted a typical triangular shape (Fig. 4) like that observed in our previous study (33).
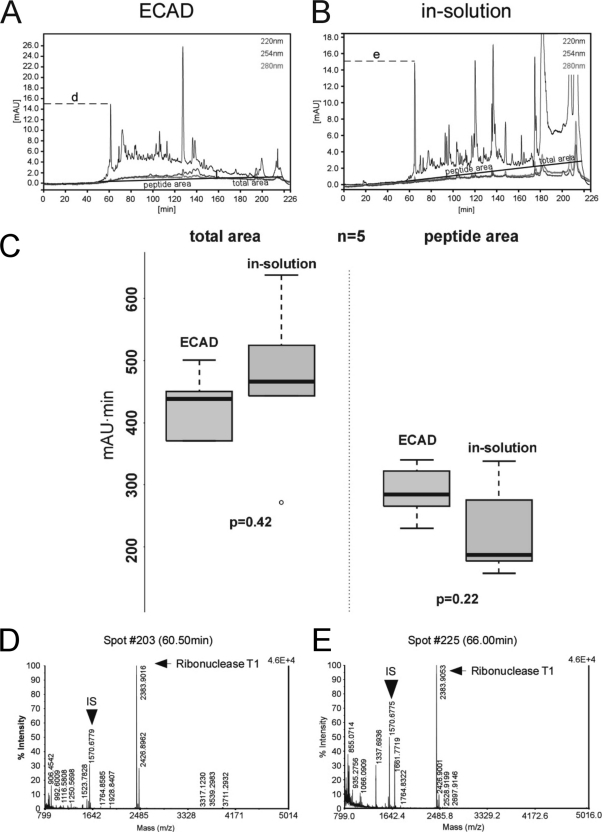
We first performed MS analyses of 10, 20, and 50 fmol of peptides derived from an in-solution digest of tri-snRNPs to confirm that instrument sensitivity of the nano-LC off-line MALDI-TOF/TOF system used is not a limiting factor for sequencing of 20 fmol of peptides (data not shown). Next, we compared the UV chromatograms and MS and MSMS spectra of peptides derived from the ECAD procedure with those derived from in-solution digested samples (50 fmol of tri-snRNPs each). We observed a similar and surprisingly strong UV absorbance for both ECAD and in-solution digestion procedures (Fig. 7, A and B). The UV chromatograms indicate that equal amounts of sample were separated by LC before spotting with MALDI matrix (Fig. 7C). The visible peaks originated from the sample itself as well as from partial digestion of the added RNases (during in-solution digestion) or complete digestion of these RNases (during ECAD), contaminating keratins (data not shown), and unspecific background contamination as revealed by “mock” experiments without protein sample (i.e. tri-snRNP). The latter clearly showed that both procedures gave rise to species that showed an absorption of UV light of up to 20 milliabsorbance units (data not shown). These experiments revealed that the added RNases were only (if at all) partially digested during in-solution digests and eluted from the LC in three strong peaks (at ∼120, ∼138, and ∼160 min; see also Fig. 7B). Of note, RNases are surprisingly stable against endoproteolytic digestion (34, 35). However, the RNases were not present in the elution profile of the ECAD samples (Fig. 7A and mock experiments (data not shown)), suggesting that the RNases were hydrolyzed by trypsin or were adsorbed by the carbon films. Of note, we always used a large excess of enzyme (higher than the usual enzyme-to-substrate ratio; see “Experimental Procedures”) to ensure that a sufficient amount of enzymes is present in the sample and not completely adsorbed by the carbon film.
Fig. 7.
Assessment of digested sample amounts loaded onto LC system for ECAD and in-solution digests. A and B, UV chromatograms of the off-line LC analysis of 50 fmol of tri-snRNP particles treated by ECAD (A) and by in-solution digestion (B). To analyze the area below the curve at 220 nm, a peptide region (indicated as “peptide area”) is defined where mainly small digestion products suitable for MS analysis are expected. This transition occurs after an elution time of ∼160–180 min as judged visually and by inspection of the corresponding MS spectra. Furthermore, the total integral (indicated as “total area”) from 0 to 226 min is calculated for the five independent runs. For ECAD (A), a “hill” formed by many individual peaks can be seen in the peptide area, which indicates a broad separation of species and is usually favorable for the MS analysis; few undigested products appear at the washout of the LC run. For in-solution digests (B), however, more non-digested material is observed at the washout of the run. C, box-and-whisker plots of the areas under the five 220 nm curves as described above. There is no significant difference in the distribution of the total areas as judged by unpaired, two-sided Wilcoxon tests (left panel; p = 0.42). Also, there is no significant difference in the distribution of the peptide areas (right panel; p = 0.22), indicating that similar amounts of material were loaded for ECAD and for in-solution digests, supporting the conclusion of good comparability between these two experimental series. D and E, technical comparison of the ECAD and in-solution MS analysis based on representative MS spectra (for the corresponding peak in the LC analysis indicated by the letters “d” and “e,” compare A and B). IS denotes the Glu-fibrinogen peptide contained within the MALDI matrix at a concentration of 10 fmol/μl for internal standardization to ensure constant quality of the MS spectra. Furthermore, a representative ribonuclease T1 peak shows that the proteolytic digestion conditions and the detection sensitivity of equal amounts of enzyme added in the later step of the digestion protocol were similar during the ECAD and in-solution measurements. Thus, the UV chromatograms and MS and MSMS spectra do not argue for any significant difference between ECAD-treated and in-solution digested samples in terms of the input amount, so that the amounts that were initially used for both methods of hydrolysis are well comparable. mAU, milliabsorbance units.
We then evaluated the performance of ECAD in comparison with in-solution digests with respect to the minimum sample amount for the MS analysis required to obtain a comprehensive proteome (Fig. 8, A and B; compare also supplemental Table S3). Based on these results, we conclude that a sample amount of 50 fmol is sufficient for MS analysis using ECAD (Fig. 8A). This is also a reasonable sample amount that can be prepared on (usually 5–10) EM carbon films. As this advantage of ECAD was apparently not effected by accidentally applying incomparable amounts of the sample (Fig. 7), this finding demonstrates the sensitivity and reproducibility of ECAD.
Fig. 8.
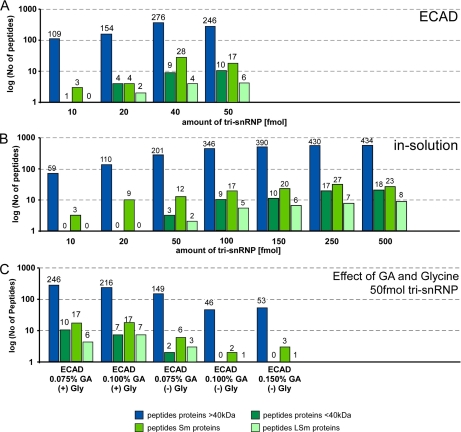
Sensitivity of ECAD versus in-solution digest. U4/U6.U5 tri-snRNP-specific proteins were classified in groups of proteins of at least 40 kDa, smaller than 40 kDa, Sm proteins (present in two copies), and LSm proteins (present in one copy per particle) (compare also Fig. 2B for a detailed list of tri-snRNP proteins). In each panel, the number of identified peptides classified into these four protein groups is shown in a logarithmic scale. A, sensitivity of ECAD using glutaraldehyde-treated tri-snRNP in terms of peptides identified at four sample amounts of tri-snRNP (10, 20, 40, and 50 fmol). Sample amounts of 100 fmol and above are technically not feasible with ECAD. A good performance of ECAD at low sample amounts of 40–50 fmol was observed. B, same as A but for standard in-solution digests of unfixed tri-snRNP (10, 20, 50, 100, 150, 250, and 500-fmol sample amounts; the amounts of 100–500 fmol in this series are technically not feasible with ECAD). Compared with ECAD of glutaraldehyde-treated sample, fewer peptides are detected upon in-solution digestion. C, the effect of the glutaraldehyde (GA) concentration and presence of glycine on the peptide yield using ECAD. Increasing concentrations of glutaraldehyde lead to detection of fewer peptides. Without the addition of glycine to glutaraldehyde-containing samples, a low number of peptides are generated, whereas upon addition of glycine, the number of detectable peptides was considerably higher.
Effect of Different Cross-linking Levels on MS Analysis
Subsequently, we were interested in the effect of different cross-linking levels on the performance of the approach and tested this by selecting different glutaraldehyde concentrations (Fig. 8C). Glutaraldehyde concentrations in the range of about 0.075–0.15% are typically used in the molecular EM field. At these concentrations, a separation of proteins by SDS-PAGE was not possible (compare Fig. 2A). A concentration of 0.075% (v/v) of fixative as, for example, used in a typical GraFix sample was compatible with the ECAD digest, whereas higher concentrations resulted in a decrease of the detection of peptides (compare +Gly conditions in Fig. 8C).
Effect of Glycine
The activity of glutaraldehyde can efficiently be inactivated by incubation with an excess of glycine. We thus analyzed the necessity to block the residual activity of glutaraldehyde by adding or omitting glycine (0.1 m glycine, pH 7.9) to the ECAD and in-solution samples. Accordingly, prior inactivation of the fixative by addition of glycine markedly improved the detection and sequencing of peptides in ECAD (compare +Gly and −Gly conditions in Fig. 8C) and in-solution digest (supplemental Fig. S1). However, the presence of glycine in the fixed in-solution digested sample was not sufficient to show an effect comparable with ECAD (supplemental Fig. S1).
Peptide Scores
The improved performance of ECAD is finally supported by the peptide scores of in-solution digests of unfixed specimens and ECAD digests of fixed material. For the U4/U6.U5 tri-snRNP, we observed that higher Mascot protein scores were generated from non-redundant peptides by ECAD as compared with the in-solution digests (Fig. 9A). For example, a Mascot protein score of 15 × 103 was achieved from ∼280 non-redundant peptides in ECAD, whereas ∼320 non-redundant peptides derived from in-solution digest would be required to obtain this particular Mascot protein score.
We therefore selected the average peptide score (36) (APS; defined as the ratio of the protein score to the number of non-redundant peptides) using Mascot as a measure for the confidence of identification as the peptide score indicates the spectrum quality. In the range of 10–50 fmol, the APS for the tri-snRNP proteins varied only marginally between 45.3 and 58.9 for ECAD, whereas it was substantially lower for the in-solution digest (Fig. 9B; see also supplemental Table S3). By interpolation of the values, it can be estimated that ∼400 fmol of sample would be required for the in-solution digest to achieve an average APS comparable with 50 fmol using ECAD. Additionally, the scattering of the average APS values was substantially higher for in-solution digests compared with ECAD, indicating a better reproducibility of ECAD. Moreover, when a list of proteins was generated by a Mascot search for proteins with an APS of ≥20, more proteins showed a high APS of 40–70 when ECAD was applied; in contrast to an exponential decrease in the APS histogram in the case of the in-solution digest, a distinct plateau ranging from an APS of 30 to 60 was seen for ECAD (Fig. 9C).
DISCUSSION
The objective of the work presented herein was to develop an approach that enables us to determine the protein composition of exactly the particle population imaged in the electron microscope and thus used for further structure determination. Such an approach allows correlation of both the results of MS and EM and thus overcomes uncertainties that arise from different specimens being analyzed by MS and EM. Our ECAD approach addresses this issue by using the same sample preparation protocol for both MS and EM and by digesting the proteins directly from the EM carbon film. We demonstrated that peptides can be generated by tryptic digestion of proteins in a quality and quantity sufficient to identify the protein components of macromolecular complexes. Furthermore, our approach also showed an improved sensitivity and reproducibility of detection of peptides as compared with standard in-solution digestion.
We developed ECAD for the compositional analysis of mildly chemically fixed samples as they are obtained e.g. by using the recently introduced GraFix protocol (3). GraFix combines mild chemical fixation of macromolecular assemblies with high pressure conditions that occur during gradient ultracentrifugation and results in improvements of the structural integrity among other favorable effects. GraFix has been used to improve the sample quality for single particle EM of a growing number of different assemblies including small nuclear ribonucleoproteins (18, 37) and spliceosomes (3, 38), a box C/D small ribonucleoprotein (39), RNA editing complexes (13), survival of motor neuron complexes (40), the pre-mRNA 3′ processing complex (41), the RNA-induced silencing complex (RISC)-loading complex (42), the RNA polymerase (10), the DNA gyrase (10), the DNA topoisomerase (10), a multifunctional replication protein (43), basal transcription factors (44), and p53 (45).
So far, an analysis of the composition of these samples could only be performed indirectly either on aliquots of the eluate prior to fixation (i.e. an aliquot of the eluate was used for a compositional analysis by MS, whereas another aliquot was loaded on the GraFix gradient for EM) or by running a parallel gradient without the addition of glutaraldehyde and subjecting these to MS analysis and/or SDS-PAGE/Western blotting. Such approaches, however, do not ensure that identical particle populations are analyzed in the GraFix-treated and the untreated sample. Moreover, some of these complexes such as the RNA editing machinery (13) isolated from trypanosomes suffer from ultracentrifugation in non-cross-linking gradients. Thus, insufficient amounts of particles are obtained for a compositional analysis of the unfixed material that excludes the application of standard protocols for analysis of their composition. Likewise, we showed here that a gel electrophoretic analysis of the fixed assemblies is not possible as it does not result in a separation of the cross-linked proteins. Finally, also the eluate can represent a mixture of different assembly stages, thus limiting the value of a compositional analysis of the eluate. Although the protein composition of intact macromolecular assemblies can also be determined (46–48), such an approach has not been applied to carbon-adsorbed macromolecular assemblies and might also impose significant challenges in the assignment of peaks to particle populations due to the complex nature of the chemical reaction profile of glutaraldehyde in aqueous solution (21).
In contrast, the protocol presented herein offers a unique possibility to determine the protein composition of fixed macromolecular assemblies. We observed an increase in sensitivity and reproducibility by using the ECAD approach as compared with standard processing of fixed (and also unfixed; see below) material. Thus, our approach overcompensates for a possible decrease in the sensitivity that might be caused by the cross-linking of lysine residues and the resulting inhibition of the enzymatic cleavage by the lysine- and arginine-specific endoproteinase trypsin. In this respect, it should be noted that we searched and detected non-glutaraldehyde-modified peptides, indicating that despite cross-linking a sufficient quality and quantities of non-cross-linked peptides are still generated by endoproteinase digestion.
Experiments using unfixed material for ECAD showed likewise a reproducible detection of peptides spread over the entire sequence including peptides that were not detected in the MS analysis of fixed sample,2 indicating that ECAD is also applicable for unfixed macromolecular samples. However, an EM analysis and correlation of MS and EM might be challenging in the case that the sample integrity suffers from the adsorption to carbon film without prior stabilization of the complexes by mild chemical cross-linking (3). In these cases, already the identification of intact particles versus disintegration products and thus counting of particles on the carbon film using EM images might become an issue.
Together, our results suggest that the MS analysis of macromolecular complexes benefits from the usage of carbon film. Possible explanations for the improvement of the MS analysis include an increased activity of the endoprotease in the presence of carbon as reported previously for other enzymes (49) and/or the prevention of aggregation of adsorbed, partly digested proteins in which the more hydrophobic protein core has been exposed upon endoproteolytic digestion. In a solution, such partly digested particles may be prone to form aggregates due to the (partial) uncovering of hydrophobic residues. This may in turn interfere with the further digestion of the sample. In contrast, the presence of glycine in the fixed in-solution digested sample is not sufficient to show an effect comparable with ECAD.
In conclusion, hydrolysis of macromolecular complexes on EM carbon films and the subsequent analysis of the proteins by MS confer several advantages upon structure-related proteomics studies. The ECAD approach closes a gap between current state-of-the-art single particle EM and MS analysis. The MS analysis of macromolecular assemblies benefits from mild chemical fixation that is in turn required for high quality imaging of fragile assemblies by single particle EM. Fixation of assemblies with formaldehyde, which can be reversed by heating before MS, has previously been used to trap loosely associated proteins in complexes (50). Also, the proteome of formalin-fixed paraffin-embedded samples has been investigated (51). However, in these studies, the modification is reversed prior to proteomics analysis, and thus, none of these studies were able to address the question of how to process samples in a concentration and amount suitable for a correlative MS/EM approach. With the ECAD approach, we have a tool at hand to study the protein composition of fixed and also of unfixed macromolecular samples with improved sensitivity in general.
Supplementary Material
Acknowledgments
We thank M. Raabe and U. Plessmann for expert technical assistance in MS; T. Conrad, P. Kempkes, and H. Kohansal for expert technical help with cell cultures; and N. Fischer for help with mock experiments.
* This work was supported in part by a Young Investigator Program grant of the European Alternative Splicing Network of Excellence (EURASNET) of the 6th European Union framework (to H. U.) and the Bundesministerium für Bildung und Forschung and the 6th European Union framework program “3D Repertoire” (to H. S.).
 This article contains supplemental Fig. S1 and Tables S1–S3.
This article contains supplemental Fig. S1 and Tables S1–S3.
2 M. M. Golas, F. M. Richter, B. Sander, H. Urlaub, and H. Stark, submitted manuscript.
1 The abbreviations used are:
- EM
- electron microscopy
- APS
- average peptide score
- ECAD
- electron microscopy carbon film-assisted digestion
- snRNP
- small nuclear ribonucleoprotein
- PEEK
- polyetheretherketone.
REFERENCES
- 1.Alberts B. (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 [DOI] [PubMed] [Google Scholar]
- 2.Forler D., Köcher T., Rode M., Gentzel M., Izaurralde E., Wilm M. (2003) An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 21, 89–92 [DOI] [PubMed] [Google Scholar]
- 3.Kastner B., Fischer N., Golas M. M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., Uchtenhagen H., Urlaub H., Herzog F., Peters J. M., Poerschke D., Lührmann R., Stark H. (2008) GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 [DOI] [PubMed] [Google Scholar]
- 4.Oeffinger M., Wei K. E., Rogers R., DeGrasse J. A., Chait B. T., Aitchison J. D., Rout M. P. (2007) Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods 4, 951–956 [DOI] [PubMed] [Google Scholar]
- 5.Yang W., Steen H., Freeman M. R. (2008) Proteomic approaches to the analysis of multiprotein signaling complexes. Proteomics 8, 832–851 [DOI] [PubMed] [Google Scholar]
- 6.Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 7.Heck A. J. (2008) Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods 5, 927–933 [DOI] [PubMed] [Google Scholar]
- 8.Sobott F., McCammon M. G., Hernández H., Robinson C. V. (2005) The flight of macromolecular complexes in a mass spectrometer. Philos. Transact. A Math. Phys. Eng. Sci. 363, 379–391 [DOI] [PubMed] [Google Scholar]
- 9.van Heel M., Gowen B., Matadeen R., Orlova E. V., Finn R., Pape T., Cohen D., Stark H., Schmidt R., Schatz M., Patwardhan A. (2000) Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 [DOI] [PubMed] [Google Scholar]
- 10.Kühner S., van Noort V., Betts M. J., Leo-Macias A., Batisse C., Rode M., Yamada T., Maier T., Bader S., Beltran-Alvarez P., Castaño-Diez D., Chen W. H., Devos D., Güell M., Norambuena T., Racke I., Rybin V., Schmidt A., Yus E., Aebersold R., Herrmann R., Böttcher B., Frangakis A. S., Russell R. B., Serrano L., Bork P., Gavin A. C. (2009) Proteome organization in a genome-reduced bacterium. Science 326, 1235–1240 [DOI] [PubMed] [Google Scholar]
- 11.Nickell S., Beck F., Scheres S. H., Korinek A., Förster F., Lasker K., Mihalache O., Sun N., Nagy I., Sali A., Plitzko J. M., Carazo J. M., Mann M., Baumeister W. (2009) Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 106, 11943–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golas M. M., Böhm C., Sander B., Effenberger K., Brecht M., Stark H., Göringer H. U. (2009) Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 28, 766–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poliakov A., van Duijn E., Lander G., Fu C. Y., Johnson J. E., Prevelige P. E., Jr., Heck A. J. (2007) Macromolecular mass spectrometry and electron microscopy as complementary tools for investigation of the heterogeneity of bacteriophage portal assemblies. J. Struct. Biol. 157, 371–383 [DOI] [PubMed] [Google Scholar]
- 15.Lei Z., Elmer A. M., Watson B. S., Dixon R. A., Mendes P. J., Sumner L. W. (2005) A two-dimensional electrophoresis proteomic reference map and systematic identification of 1367 proteins from a cell suspension culture of the model legume Medicago truncatula. Mol. Cell. Proteomics 4, 1812–1825 [DOI] [PubMed] [Google Scholar]
- 16.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 17.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 18.Häcker I., Sander B., Golas M. M., Wolf E., Karagöz E., Kastner B., Stark H., Fabrizio P., Lührmann R. (2008) Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat. Struct. Mol. Biol. 15, 1206–1212 [DOI] [PubMed] [Google Scholar]
- 19.Golas M. M., Sander B., Will C. L., Lührmann R., Stark H. (2003) Molecular architecture of the multiprotein splicing factor SF3b. Science 300, 980–984 [DOI] [PubMed] [Google Scholar]
- 20.Gabashvili I. S., Agrawal R. K., Spahn C. M., Grassucci R. A., Svergun D. I., Frank J., Penczek P. (2000) Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell 100, 537–549 [DOI] [PubMed] [Google Scholar]
- 21.Migneault I., Dartiguenave C., Bertrand M. J., Waldron K. C. (2004) Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques 37, 790–796, 798–802 [DOI] [PubMed] [Google Scholar]
- 22.Behrens S. E., Lührmann R. (1991) Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes Dev. 5, 1439–1452 [DOI] [PubMed] [Google Scholar]
- 23.Fazekas de St Groth S., Webster R. G., Datyner A. (1963) Two new staining procedures for quantitative estimation of proteins on electrophoretic strips. Biochim. Biophys. Acta 71, 377–391 [DOI] [PubMed] [Google Scholar]
- 24.Whitton J. L., Hundley F., O'Donnell B., Desselberger U. (1983) Silver staining of nucleic acids. Applications in virus research and in diagnostic virology. J. Virol. Methods 7, 185–198 [DOI] [PubMed] [Google Scholar]
- 25.Adrian M., Dubochet J., Lepault J., McDowall A. W. (1984) Cryo-electron microscopy of viruses. Nature 308, 32–36 [DOI] [PubMed] [Google Scholar]
- 26.Sander B., Golas M. M., Stark H. (2005) Advantages of CCD detectors for de novo three-dimensional structure determination in single-particle electron microscopy. J. Struct. Biol. 151, 92–105 [DOI] [PubMed] [Google Scholar]
- 27.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 28.R Development Core Team (2008) R, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 29.Chaudhry C., Horwich A. L., Brunger A. T., Adams P. D. (2004) Exploring the structural dynamics of the E. coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states. J. Mol. Biol. 342, 229–245 [DOI] [PubMed] [Google Scholar]
- 30.Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298, 2205–2208 [DOI] [PubMed] [Google Scholar]
- 31.Achsel T., Brahms H., Kastner B., Bachi A., Wilm M., Lührmann R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18, 5789–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomeranz Krummel D. A., Oubridge C., Leung A. K., Li J., Nagai K. (2009) Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458, 475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sander B., Golas M. M., Makarov E. M., Brahms H., Kastner B., Lührmann R., Stark H. (2006) Organization of core spliceosomal components U5 snRNA loop I and U4/U6 di-snRNP within U4/U6.U5 tri-snRNP as revealed by electron cryomicroscopy. Mol. Cell 24, 267–278 [DOI] [PubMed] [Google Scholar]
- 34.Urlaub H., Kruft V., Bischof O., Müller E. C., Wittmann-Liebold B. (1995) Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 14, 4578–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urlaub H., Thiede B., Müller E. C., Brimacombe R., Wittmann-Liebold B. (1997) Identification and sequence analysis of contact sites between ribosomal proteins and rRNA in Escherichia coli 30 S subunits by a new approach using matrix-assisted laser desorption/ionization-mass spectrometry combined with N-terminal microsequencing. J. Biol. Chem. 272, 14547–14555 [DOI] [PubMed] [Google Scholar]
- 36.Chepanoske C. L., Richardson B. E., von Rechenberg M., Peltier J. M. (2005) Average peptide score: a useful parameter for identification of proteins derived from database searches of liquid chromatography/tandem mass spectrometry data. Rapid Commun. Mass Spectrom. 19, 9–14 [DOI] [PubMed] [Google Scholar]
- 37.Karaduman R., Dube P., Stark H., Fabrizio P., Kastner B., Lührmann R. (2008) Structure of yeast U6 snRNPs: arrangement of Prp24p and the LSm complex as revealed by electron microscopy. RNA 14, 2528–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herold N., Will C. L., Wolf E., Kastner B., Urlaub H., Lührmann R. (2009) Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 29, 281–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bleichert F., Gagnon K. T., Brown B. A., 2nd, Maxwell E. S., Leschziner A. E., Unger V. M., Baserga S. J. (2009) A dimeric structure for archaeal box C/D small ribonucleoproteins. Science 325, 1384–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chari A., Golas M. M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. (2008) An assembly chaperone collaborates with the SMN complex to generate spliceosomal snRNPs. Cell 135, 497–509 [DOI] [PubMed] [Google Scholar]
- 41.Shi Y., Di Giammartino D. C., Taylor D., Sarkeshik A., Rice W. J., Yates J. R., 3rd, Frank J., Manley J. L. (2009) Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H. W., Noland C., Siridechadilok B., Taylor D. W., Ma E., Felderer K., Doudna J. A., Nogales E. (2009) Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 16, 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez M., Drechsler M., Stark H., Lipps G. (2009) DNA translocation activity of the multifunctional replication protein ORF904 from the archaeal plasmid pRN1. Nucleic Acids Res. 37, 6831–6848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papai G., Tripathi M. K., Ruhlmann C., Werten S., Crucifix C., Weil P. A., Schultz P. (2009) Mapping the initiator binding Taf2 subunit in the structure of hydrated yeast TFIID. Structure 17, 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres S. H., Melero R., Valle M., Carazo J. M. (2009) Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure 17, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernández H., Dziembowski A., Taverner T., Séraphin B., Robinson C. V. (2006) Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 7, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharon M., Taverner T., Ambroggio X. I., Deshaies R. J., Robinson C. V. (2006) Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 4, e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou M., Sandercock A. M., Fraser C. S., Ridlova G., Stephens E., Schenauer M. R., Yokoi-Fong T., Barsky D., Leary J. A., Hershey J. W., Doudna J. A., Robinson C. V. (2008) Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. U.S.A. 105, 18139–18144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovalenko G. A., Perminova L. V. (2008) Immobilization of glucoamylase by adsorption on carbon supports and its application for heterogeneous hydrolysis of dextrin. Carbohydr. Res. 343, 1202–1211 [DOI] [PubMed] [Google Scholar]
- 50.Vasilescu J., Guo X., Kast J. (2004) Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 4, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 51.Hood B. L., Conrads T. P., Veenstra T. D. (2006) Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics 6, 4106–4114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.