Abstract
Sequence polymorphisms in a 58kb interval on chromosome 9p21 confer a markedly increased risk for coronary artery disease (CAD), the leading cause of death worldwide 1,2. The variants have a substantial impact on the epidemiology of CAD and other life-threatening vascular conditions since nearly a quarter of Caucasians are homozygous for risk alleles. However, the risk interval is devoid of protein-coding genes and the mechanism linking the region to CAD risk has remained enigmatic. Here we show that deletion of the orthologous 70kb noncoding interval on mouse chromosome4 affects cardiac expression of neighboring genes, as well as proliferation properties of vascular cells. Chr4Δ70kb/Δ70kb mice are viable, but show increased mortality both during development and as adults. Cardiac expression of two genes near the noncoding interval, Cdkn2a and Cdkn2b, is severely reduced in chr4Δ70kb/Δ70kb mice, indicating that distant-acting gene regulatory functions are located in the noncoding CAD risk interval. Allele-specific expression of Cdkn2b transcripts in heterozygous mice revealed that the deletion affects expression through a cis-acting mechanism. Primary cultures of chr4Δ70kb/Δ70kb aortic smooth muscle cells exhibited excessive proliferation and diminished senescence, a cellular phenotype consistent with accelerated CAD pathogenesis. Taken together, our results provide direct evidence that the CAD risk interval plays a pivotal role in regulation of cardiac Cdkn2a/b expression and suggest that this region affects CAD progression by altering the dynamics of vascular cell proliferation.
Each day, cardiovascular disease causes 2,400 deaths in the United States alone, more than cancer, accidents and diabetes combined 3. The largest proportion of this mortality is due to coronary artery disease (CAD), which causes approximately 1 of every 5 deaths in the United States. CAD has a complex etiology and there is strong evidence that both environmental and genetic factors are major determinants of disease risk 3. However, identifying the genomic loci associated with increased CAD susceptibility has been a challenge, and most of the known risk loci explain only small proportions of CAD cases (e.g., ref. 4). Genome-wide association studies have recently identified common sequence variants on human chromosome 9p21 that confer an increased risk for CAD and myocardial infarction 1,2. These associations have been confirmed in multiple additional cohorts 5–9 and were extended to other severe arterial diseases 10. Even in homozygous individuals, the variants increase the relative risk for CAD only moderately by a factor of 1.3 to 2. However, since the risk alleles are very common, they contribute substantially to the epidemiology of CAD. Between 20% and 25% of Caucasians are homozygous for risk alleles, resulting in estimates of 10%to31% population attributable risk, depending on cohort and cases considered 1,2.
Despite compelling genetic evidence for association, the mechanism by which 9p21 sequence polymorphisms confer an increased CAD risk is unknown, preventing the development of pharmacological or behavioral intervention strategies. The variants are not associated with established CAD risk factors such as plasma lipoprotein levels, hypertension, or diabetes, suggesting that they influence CAD pathogenesis through a previously unappreciated pathway 1,2. The CAD-associated SNPs are located within a 58kb linkage disequilibrium block on chromosome 9p21.3 that does not contain any known protein-coding genes. Several expressed sequence tags of apparently noncoding transcripts, including a proposed long noncoding RNA, have been mapped to the risk interval, but their functional relevance remains elusive 11,12. Gene expression studies in human peripheral blood cells and in vitro reporter assays have provided support for the notion that gene regulatory elements might be located within the risk interval, but there is conflicting evidence whether the CAD risk variants are associated with increased 13 or decreased 14 regulatory activity. More importantly, it is unclear whether altered gene regulation would result in cellular or physiological phenotypes that are relevant to CAD pathogenesis, highlighting the need to study the function of this noncoding interval in a suitable in vivo system.
To create a mouse model for investigating the function of the human 58kb noncoding CAD risk interval, we sought to generate a severe (null) allele for this locus by its targeted removal from the mouse genome. Human-mouse orthology could be unambiguously established since 50% of the basepairs in the human region are alignable to mouse 15 and synteny with flanking genes is preserved (Fig. 1a–c). The mouse interval is 70kb in size and thus 20% longer than the human orthologous region, partially due to increased repetitive sequence content. Due to the large size of the interval of interest, its targeted deletion was accomplished through a sequential double-targeting strategy followed by Cre-mediated recombination (Fig 1d; Suppl. Figures 1–3). Chr4Δ70kb/Δ70kb mice are viable and fertile. While the majority of live-born homozygous animals survive to weaning and beyond without obvious morphological or behavioral phenotypes, we did observe reduced embryonic, post-natal, and adult survival due to the deletion (for details see Suppl. Material and Suppl. Fig. 4). Clinical analysis of adult animals at seven months of age showed no significant general aberrations in urine and blood chemistry markers, differential blood cell counts, or histopathological appearance of internal organs including heart, liver, lung, kidney, spleen, and gastrointestinal tract. However, in a larger cohort of chr4Δ70kb/Δ70kb mice on which gross necropsy was performed between 7 and 14 months of age (or at time of premature death), 9 of 20 (45%) animals were found to have internal neoplasms or tumors of various types (see Suppl. Material), compared to none in a cohort of age-matched wild-type controls (P=0.0012, FET). In addition to this increased tumor incidence, both male and female chr4Δ70kb/Δ70kb mice that were fed standard mouse chow ad libitum gained weight significantly faster than wild-type controls, resulting in a 17% increased body mass by 30 weeks of age(Suppl. Fig. 5).
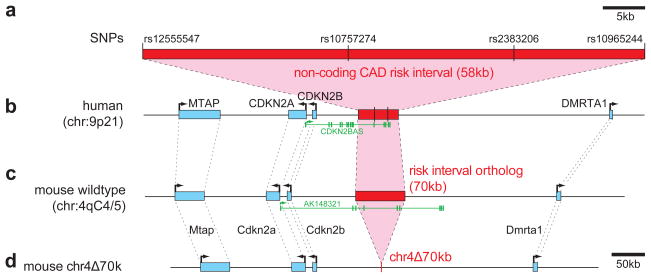
Figure 1. Deletion of the noncoding region orthologous to the 58kb CAD risk interval on human chromosome 9p21.
a) Noncoding CAD risk interval with SNPs found to be most significantly associated with CAD in genome-wide association studies 1,2 and SNPs defining the boundaries of the linkage disequilibrium block. b)Overview of the human locus including neighboring genes (blue, intron/exon structure not shown). A noncoding RNA of unknown function transcribed from this locus 12 is shown in green (CDKN2BAS, also known as ANRIL, GenBank NR_003529.3). c)Orthologous region on mouse chromosome 4. The exon structure of a noncoding transcript of unknown function, AK148321, is shown in green. d)Chr4Δ70kb after successful targeting and deletion of the 70kb CAD risk interval.
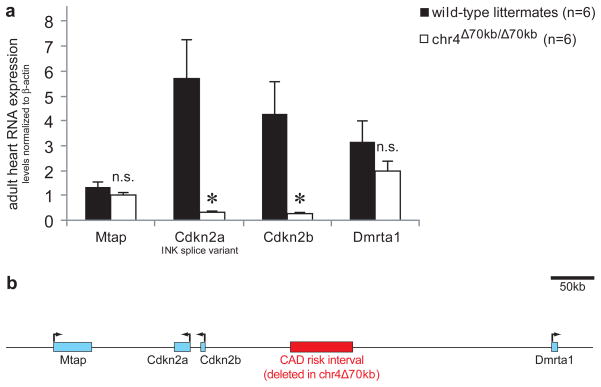
To investigate the effects of the deletion in more detail, we examined the possibility that the CAD risk interval is required for distant-acting regulation of gene expression. To compare mRNA expression levels of surrounding genes between wild-type and chr4Δ70kb/Δ70kb mice, we isolated mRNA from hearts and other adult mouse tissues and performed reverse transcription followed by quantitative PCR. In heart tissue, chr4Δ70kb/Δ70kb mice had substantially depressed expression levels of the neighboring Cdkn2a and Cdkn2b genes, but no significant alteration of expression levels of two other neighboring genes, Mtap and Dmrta1 (Fig. 2a). Cardiac expression of Cdkn2a and Cdkn2b was more than ten-fold decreased compared to wild-type controls. These results indicate that the CAD risk interval is required for appropriate expression of Cdkn2a and Cdkn2b in the heart. Due to the known roles of these genes in several disease-related pathways 16–19, these results support the possibility of a regulation-mediated mechanism by which the chr4Δ70kb deletion might impact on cellular, physiological and pathological processes.
Figure 2. Deletion of the CAD risk interval affects expression of neighboring genes Cdkn2a and Cdkn2b.
a) Expression levels of Cdkn2a (INK splice variant) and Cdkn2b are significantly reduced in heart tissue of chr4Δ70kb/Δ70kb mice. In contrast, expression of the neighboring genes Mtap and Dmrta1 is not significantly altered. b) Overview of locus with locations of genes. Error bars are standard error of the mean; asterisks indicate P ≤ 0.005 (t-test, one-tailed).
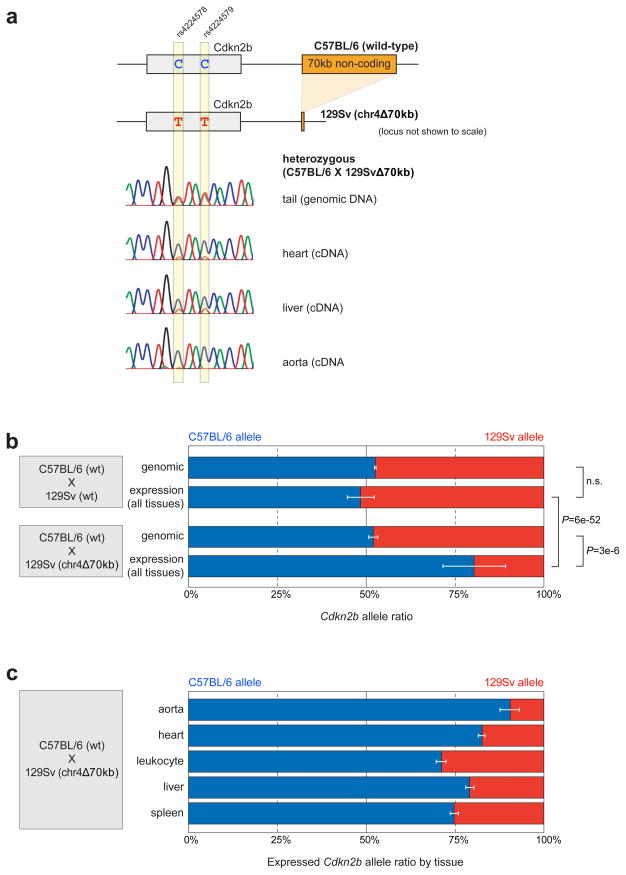
To test whether the observed regulatory effect on gene expression occurs through a cis- or trans-acting mechanism, we performed allele-specific expression analysis. We used for this purpose crosses of mice with the deletion linked to the Cdkn2b allele of the 129Sv strain (in which the deletion was originally created) and wild-type C57BL/6 strain mice. Strains C57BL/6 and 129Sv are distinguished by several transcribed SNPs in the Cdkn2b gene that can reveal quantitative differences in expression from the two alleles. Direct sequencing of PCR product from tail genomic DNA confirmed that the mice were heterozygous for the expected SNPs and the two alleles were detected at the expected 1:1 ratio (Fig. 3a,b). As a control, in cDNA derived from tissues of C57BL/6 (wt) × 129Sv (wt), the two alleles were also expressed at indistinguishable levels, confirming the absence of general strain-specific differences in Cdkn2b expression levels (Fig. 3b). In contrast, direct sequencing of RT-PCR product derived from RNA isolated from chr4+(C57BL/6)/Δ70kb(129Sv) hearts and other tissues revealed that Cdkn2b was predominantly expressed from the wild-type C57BL/6 allele and expression from the chr4Δ70kb 129Sv allele was strongly diminished (Fig. 3b). Among five organs and cell types examined, the most severe allele-specific down-regulation was observed for the heart and aorta (Fig. 3c). These results support that the CAD risk interval controls gene expression in cardiac and other tissues through a distant-acting cis-regulatory mechanism.
Figure 3. Deletion of the CAD risk interval affects gene expression through a cis-regulatory mechanism.
a) Two of seven transcribed single nucleotide polymorphisms (SNPs) that were used to distinguish expression of the C57BL/6 and 129Sv strain alleles of the Cdkn2b gene. Representative electropherograms from direct Sanger sequencing of PCR product are shown. b) No differences between alleles are observed in genomic DNA or tissue-derived cDNA in chr4+(C57BL/6)/+(129Sv) mice or in genomic DNA of chr4+(C57BL/6)/Δ70kb(129Sv) mice. In contrast, in tissues of chr4+(C57BL/6)/Δ70kb(129Sv) mice, the C57BL/6 allele is expressed four-fold higher than the 129Sv allele (all tissues combined). c) Allelic expression differences shown by individual tissues. Error bars indicate standard deviation (b) and standard error of means (c). P-values: t-test, two-tailed.
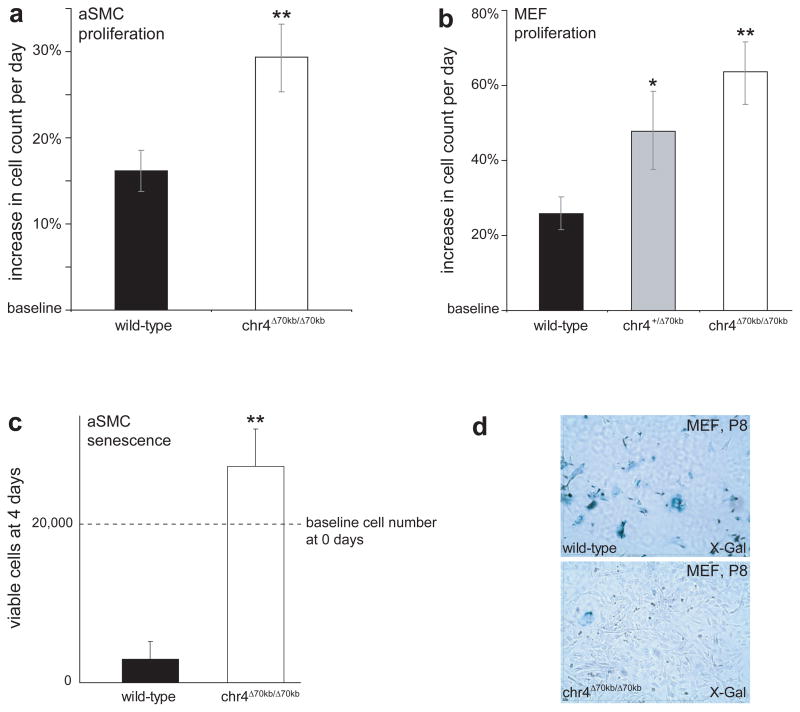
The proteins encoded by Cdkn2a, Cdkn2b and other cyclin-dependent kinase inhibitor genes have been implicated in cellular phenotypes including regulation of proliferation and cellular senescence 16,18,19. Given the severe impact of the chr4Δ70kb deletion on cardiac expression of Cdkn2a and Cdkn2b, we tested if cell proliferation and senescence are affected in chr4Δ70kb/Δ70kb mice. To test for an effect on cell proliferation, we measured the proliferation rates of primary cultures of aortic smooth muscle cells (aSMCs) and mouse embryonic fibroblasts (MEFs) during early passages. In both cell types, cells derived from chr4Δ70kb/Δ70kb mice proliferated excessively compared to wild-type controls, with daily proliferation rates nearly 2-fold increased in aSMCs and nearly 3-fold increased in MEFs (Fig. 4a,b). During later passages of these primary cultures, wild-type aSMCs and MEFs became senescent, whereas chr4Δ70kb/Δ70kb-derived cells that had been isolated and cultured under identical conditions continued to proliferate and did not show signs of senescence (Fig. 4c,d). These cellular phenotypes are consistent with known and proposed functions of Cdkn2a, Cdkn2b, and other cyclin-dependent kinase inhibitors 16,18–20.
Figure 4. Deletion of the CAD risk interval disrupts normal dynamics of cellular proliferation and senescence.
a) Increased proliferation of primary aSMC cultures. b) Increased proliferation of primary MEF cultures. c) Failure of normal cellular senescence of primary aSMC cultures in late passages. Cells derived from wild-type and chr4Δ70kb/Δ70kb mice were grown to senescence under identical conditions, seeded at equal densities and cell counts were determined after 4 days. d) Chr4Δ70kb/Δ70kb-derived MEFs fail to enter cellular senescence, as evident from absence of senescence staining by X-Gal in comparison to wild-type MEFs 30. Mean daily proliferation rates are shown in a) and b), viable cell count at day 4 is shown in c). Error bars indicate SEM. *, P<0.05; **, P<0.01; t-test.
The risk interval affects human CAD through a mechanism that appears to be independent of plasma lipid levels and other known risk factors 1,2. To study possible in vivo effects of the chr4Δ70kb allele on plasma lipids and early stages of atherogenesis, we placed 40 chr4Δ70kb/Δ70kb mice and 40 wild-type controls in an isogenic 129Sv background on a high-fat, high-cholesterol (“Western”) diet for 20 weeks 21. As expected, this diet caused substantial alterations in plasma lipid levels, however, no significant differences in this physiological response were observed between wild-type and chr4Δ70kb/Δ70kb mice (Suppl. Figure 6; see Suppl. Material for details). In addition, we did not observe significant differences in fatty lesion formation (Suppl. Figure 7). Nevertheless, the high-fat, high-cholesterol nutrition caused substantially increased mortality among chr4Δ70kb/Δ70kb mice compared to isogenic wild-type controls, indicating an overall increased susceptibility to detrimental effects of this noxious diet (Suppl. Fig. 8). Studies in complementary background strains and additional genetic manipulations of the lipid metabolism in chr4Δ70kb/Δ70kb mice maybe required to mimic the full course of human atherosclerosis 21–23. Such data could also help to distinguish if diet-induced increase in mortality is due to cardiovascular phenotypes other than the aortic fatty lesions examined here, or to extra-cardiovascular phenotypes that may be present in chr4Δ70kb/Δ70kb mice. Irrespective of the underlying etiology, these observations indicate that the CAD risk interval is not required for maintenance of normal plasma lipid levels in mice, consistent with the observation that variation in the human interval influences CAD risk independent of altered lipid levels.
The chromosome 9p21 common haplotype linked to CAD represents an important but particularly puzzling risk interval and the present study provides key insights regarding the in vivo function of this noncoding region. We have demonstrated that the precise orthologous mouse interval, despite its large distance from any protein-coding genes, is critically required for normal cardiac expression of two cell cycle inhibitor genes, Cdkn2a and Cdkn2b. These observations raise the question as to what type of underlying molecular mechanism mediates these regulatory effects. Subregions of the 58kb noncoding risk interval increase transcriptional activity in cell-based in vitro transfection assays 13, but the location and function of distinct small enhancer sequences with relevant in vivo activities remains to be established (see Supplementary Material). Alternatively, our results are also consistent with models in which the noncoding interval shields the Cdkn2a/b genes from the influence of very distal negative regulatory elements, either by spacing effects or due to presence of insulator elements. In contrast, any mechanism mediated by freely diffusible RNA molecules is not expected to result in the allele-selective regulation observed in chr4Δ70kb/Δ70kb mice (Fig. 3). Support for an RNA-mediated mechanism 11,12,24 is therefore restricted to scenarios in which the transcriptional activity itself affects local chromatin state or in which RNA molecules remain tethered to the chromosome from which they are transcribed. We have also shown that the aberrations of in vivo expression of Cdkn2a and Cdkn2b coincide with abnormal regulation of vascular cell proliferation and senescence. These phenotypes are reminiscent of mouse models in which the Cdkn2a/b genes themselves have been deleted. Specifically, primary cultures of Cdkn2a- or Cdkn2b-deficient fibroblasts 16,19 and Cdkn2a-deficient aSMCs 25 exhibit elevated proliferation rates. Hence, a parsimonious explanation for the cellular phenotypes observed in chr4Δ70kb/Δ70kb mice is that the noncoding CAD risk interval affects vascular cell proliferation and senescence by modulating the expression levels of Cdkn2a and Cdkn2b. Altered proliferation rates of vascular cells resulting from genetic manipulation of other cyclin-dependent kinase inhibitors have been closely linked to the dynamics of CAD pathogenesis 26 and Cdkn2a deficiency causes altered vascular injury responses in a mouse model of CAD 25. Moreover, sequence polymorphisms in the promoters of at least two other human cyclin-dependent kinase inhibitor genes have been implicated in increased cardiovascular disease risk 27,28. Thus, variation in distant-acting regulatory sequences required for cardiovascular expression of CDKN2A and CDKN2B provides a plausible mechanistic model for the increased CAD risk associated with the 9p21 region independently of lipid levels and other known risk factors.
Methods Summary
Targeted deletion of the 70kb noncoding intervalwas performed by two sequential targeting steps, followed by Cre-mediated recombination (Suppl. Figures 1–3). See Suppl. Table 1 for primer sequences used for vector construction and genotyping. For quantitative real-time reverse transcription PCR (RT-PCR), total RNA was extracted from wild-type and knockout mouse tissues using Trizol Reagent (Invitrogen) following the manufacturer’s instruction. Total RNA was treated with RNase-free DNase and first-strand cDNA was synthesized by standard methods. RT-PCR was performed by standard methods; primer sequences are provided in Suppl. Table 2. For allele-specific expression profiling, RNA extraction from tissues, DNase treatment and first-strand cDNA synthesis were done as described above. Genomic DNA was extracted from the tails of the same mice used for RNA extraction by standard methods. PCR products were generated using gene-specific primers containing M13 primer tags (M13 -40 forward or M13 -20 reverse) and Platinum Taq DNA Polymerase (Invitrogen). Primer sequences are provided in Suppl. Table 2. PCR products were gel purified using QiaGen MinElute Gel Extraction Kit (Qiagen) and sequenced using M13 primers. For proliferation and senescence assays, primary mouse embryonic fibroblasts (MEFs) were isolated from embryonic day 12.5 to 14.5-day-old chr4Δ70kb/Δ70kb, wild-type and heterozygous littermates. Each embryo was disaggregatedin 0.25% trypsin, and fragments were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum. Aortic smooth muscle cells were isolated from thoracic aorta of 4-week old chr4Δ70kb/Δ70kb mice and wild-type littermates as previously described 29. Cells were counted at each passage and re-cultured in constant concentrations (2×104/well). For senescence assays, cells were grown to senescence (passage 12), trypsinized and plated at 2×104/well on day 0. Cell counts were determined after 4 days. Senescence staining by X-Gal was done as previously described 30.
Online Methods
Targeted Deletion of 70kb Noncoding Interval
Two targeting vectors were generated for the deletion. See Suppl. Table 1 for primer sequences used for vector construction and genotyping. One targeting vector, containing a homologous region at the 5′ end of the region of interest (proximal to Cdkn2a/b), was in the ploxPneoTK-2 vector. The second targeting vector, containing a homologous region at the 3′ end of the region (distal from Cdkn2a/b), was in a plox Phyg TK vector backbone. Both ploxPneoTK-2 and plox Phyg TK were generated in this laboratory.
Homologous arms were generated by PCR from W4/129S6 ES cell genomic DNA. The PCR product of the 5′ homologous arm (with Bam HI and EcoRI tags on primers) was cloned into ploxPneoTK-2 BamHI/EcoRI sites next to a loxP site, generating vector pCHD5′ neo for 5′ targeting. The PCR product of the 3′-homologous arm (with NotI and BglII tags on primers) was cloned into plox Phyg TK NotI/BamHI sites at the 3′ side of the PGK terminator of the PGK tk cassette, generating targeting vector pCHD3′ hyg for 3′ targeting.
Targeting vector pCHD5′ neo was electroporated into W4/129S6 ES cells (Taconic Farms). Neomycin-resistant clones were picked, screened by PCR and confirmed by Southern hybridization (Suppl. Fig. 1). Successfully 5′-targeted clones were pooled and electroporated with 3′ targeting vector pCHD3′ hyg. Hygromycin-resistant clones were picked, screened by PCR and confirmed by Southern hybridization (Suppl. Fig. 2).
Double-targeted clones (Hygr/Neor, note that the two loxP sites could be in cis or in trans) were pooled together, expanded and electroporated with about 20ug of Cre-recombinase-expressing plasmid pTURBO-Cre. When loxP sites were present in cis, the loxP bracketed sequence included: 1) the region chr4:89,054,800–89,126,878 (mm9) to be deleted, 2) the PGKhyg, PGKneo and HSV-tk cassettes, which can be deleted by Cre-recombinase-mediated loxP recombination (Suppl. Fig. 3). When loxP sites were present in trans, a translocation could be generated that resulted in one deleted and one duplicated allele of the region of interest. ES cells that underwent cis-recombination and deletion were identified by selecting for neomycin or hygromycin sensitivity and negative selection of HSV-tk for 1-(2-deoxy-2-fluoro-b-D-arabinofuransyl)-5-iodouracil (FIAU) resistance. Cells surviving this selection were screened by PCR for the predicted deletion using a primer outside the deleted region and T7 primer within the vector backbone left on the chromosome after deletion. The predicted deletion was further confirmed by PCR using primers outside the deleted region, negative PCR of Neo or Hyg primers, and by Southern blot analysis using a probe outside the deletion.
Assessment of Survival
To determine embryonic survival, embryos from timed pregnancies were dissected between E9.5 and E15.5. Embryos whose size and appearance was normal for the respective stage were considered as surviving and genotyped by PCR.
To determine the survival from birth to weaning, live-born pups resulting from wt × wt, chr4+/Δ70kb× chr4+/Δ70kb and chr4Δ70kb/Δ70kb× chr4Δ70kb/Δ70kb crosses were counted at day P0 or P1 and the same litters were counted again at weaning at day P20 or P21. Any dead or missing pups were considered as having died between birth and weaning. Pups from heterozygous crosses were counted regardless of their genotype since missing pups could generally not be recovered for genotyping.
For adult survival analysis on standard chow and on high-fat diet, animals that were found dead, met euthanasia criteria, or had an expected remaining survival time of less than 7 days (based on progression of pre-mortal symptoms in previously died animals, which included severe weight loss and general inactivity) when sacrificed for histological analysis were considered as died. Tick marks in survival plots indicate “censored” animals, which includes animals that were healthy when removed from the study for histological analysis. In order to exclude the possibility of bias in assessing euthanasia criteria, we also performed survival analysis for animals on the high-fat diet up to 120 days. All animals that died during this initial study phase were found dead and no animals were euthanized or removed for histological analysis prior to day 120. Consistent with the full study duration, a significantly increased mortality among chr4Δ70kb/Δ70kb animals on high-fat diet compared to wild-type controls was observed (P=0.006, Kaplan-Meier log-ranked survival test).
High-fat Diet, Plasma Lipid and Aortic Fatty Lesion Analysis
Deletion and wild-type mice at 6 weeks of age were fed with high-fat diet 21 containing about 15.8% fat, 1.25% cholesterol and 0.5% sodium cholate (Harlan, TD.88051). Mice were under the diet for 18–22 weeks. The control group was fed with chow diet containing about 6.5% fat (Labdiet, Formulab Diet 5008). For plasma lipid analysis, mice were fasted overnight (approximately 15–17 hours). Whole blood was collected into EDTA Capillary Blood Collection tube (Fisher) by tail bleeding. Blood cells were removed by centrifugation at 4 °C. Clear plasma was transferred to a new tube and frozen at −80 °C until analysis. Plasma lipids were measured by standard assays at the Cincinnati Mouse Metabolic Phenotyping Center. Aortic fatty lesion analysis was done as previously described 31. Briefly, mouse hearts were excised and the upper third, including the proximal aorta, was embedded in optimal cutting temperature (OCT) compound. Serial 10μm thick cryosections were cut in the region extending from the appearance to the disappearance of the aortic valves. Sections were mounted on Superfrost (VWR) slides, fixed in 10% neutral buffered formalin vapor and stained with Oil-Red O in PEG and Gill’s III hematoxylin. Lesion areas were determined using a calibrated eyepiece at 200 X magnification.
Real-time RT-PCR
Total RNA was extracted from wild-type and knockout mouse tissues using Trizol Reagent (Invitrogen) following the manufacturer’s instruction. Total RNA was treated with Promega RQ1 RNase-Free DNase. First-strand cDNA was synthesized using SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen). Real-time RT-PCR was performed using the Applied Biosystems SYBR Green PCR Master Mix and run on a 7500 Fast Real-Time PCR System (Applied Biosystems). Primer Sequences are provided in Suppl. Table 2.
Allele-specific Expression Profiling
RNA extraction from tissues, DNase treatment and first-strand cDNA synthesis were done as described above. Genomic DNA was extracted from the tails of the same mice used for RNA extraction. Tails were digested overnight at 50 °C in a buffer containing 1% SDS and 100–200 μg/ml Proteinase K. The lysate was heated at 95–100 °C for5 minutes, and diluted for PCR. PCR products were generated using gene-specific primers containing M13 primer tags (M13 -40 forward or M13 -20 reverse) and Platinum Taq DNA Polymerase (Invitrogen). Primer sequences are provided in Suppl. Table 2. PCR products were gel purified using QiaGen MinElute Gel Extraction Kit (Qiagen) and sequenced using M13 primers.
Histopathology, Blood and Urine Analysis
Complete clinical blood chemistry profiles, hematological analysis, urine analysis, general histopathology, and histopathological analysis of neoplasms and tumors were performed at Charles River Research Animal Diagnostic Services (Wilmington, MA).
Proliferation and Senescence Assays
Primary mouse embryonic fibroblasts (MEFs) were isolated from embryonic day 12.5 to 14.5-day-old chr4Δ70kb/Δ70kb, wild-type and heterozygous littermates. Each embryo was disaggregated in 0.25% trypsin, and fragments were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum. Aortic smooth muscle cells were isolated from thoracic aorta of 4-week old chr4Δ70kb/Δ70kb mice and wild-type littermates as previously described 29. Cell counts were determined at each passage using a hemocytometer and re-cultured in constant concentrations (2×104/well). Fig. 4a shows mean daily proliferation rates over seven early passages in aSMC cultures derived from three animals per genotype. Fig. 4b shows mean daily proliferation rates over four early passages of MEFs derived from four (heterozygous) to six (wild-type and homozygous) animals per genotype. For senescence assays, cells were grown to senescence, trypsinized and plated at 2×104/well on day 0. Fig. 4c shows senescence data from primary aSMC cultures in late passages (passage 12), derived from five animals per genotype. Cells were grown to senescence under identical conditions, seeded at equal densities and cell counts were determined after 4 days. Senescence staining by X-Gal in MEFs (Fig. 4d) was done as previously described 30 after 8 passages.
Supplementary Material
Acknowledgments
The authors thank Gary Owens for critical suggestions and discussion, Timothy Ley for providing vector pTURBO-Cre, Feng Chen and Zhong Wang for help with gene expression analysis and Dana Lee for help with plasma lipid analysis. L.A.P./E.M.R./J.C.C. were supported by the National Heart, Lung, and Blood Institute, and L.A.P. by the National Human Genome Research Institute. Research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California. Plasma lipid analysis at the University of Cincinnati Mouse Metabolic Phenotyping Center was supported by MMPC DK59630. All animal work was reviewed and approved by the LBNL Animal Welfare and Research Committee.
Footnotes
Author Contributions
A.V. and L.A.P. wrote the manuscript. All authors contributed to data collection and analysis and provided comments on the manuscript.
References (Main Text)
- 1.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316 (5830):1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 2.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316 (5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd-Jones D, et al. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119 (3):e21–181. doi: 10.1161/CIRCULATIONAHA.108.191261. [DOI] [PubMed] [Google Scholar]
- 4.Topol EJ, Smith J, Plow EF, Wang QK. Genetic susceptibility to myocardial infarction and coronary artery disease. Hum Mol Genet. 2006;15(Spec No 2):R117–123. doi: 10.1093/hmg/ddl183. [DOI] [PubMed] [Google Scholar]
- 5.Schunkert H, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117 (13):1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinohara K, et al. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008;53 (4):357–359. doi: 10.1007/s10038-008-0248-4. [DOI] [PubMed] [Google Scholar]
- 7.Lemmens R, et al. Variant on 9p21 strongly associates with coronary heart disease, but lacks association with common stroke. Eur J Hum Genet. 2009 doi: 10.1038/ejhg.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen GQ, et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008;53 (2):144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, et al. Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol. 2008;28 (11):2085–2089. doi: 10.1161/ATVBAHA.108.176065. [DOI] [PubMed] [Google Scholar]
- 10.Helgadottir A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40 (2):217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 11.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458 (7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasmant E, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67 (8):3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 13.Jarinova O, et al. Functional Analysis of the Chromosome 9p21.3 Coronary Artery Disease Risk Locus. Arterioscler Thromb Vasc Biol. 2009 doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4 (4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36 (Database issue):D773–779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serrano M, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85 (1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 17.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91 (5):649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 18.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23 (2):379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- 19.Latres E, et al. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19 (13):3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besson A, Assoian RK, Roberts JM. Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors? Nat Rev Cancer. 2004;4 (12):948–955. doi: 10.1038/nrc1501. [DOI] [PubMed] [Google Scholar]
- 21.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis. 1985;57 (1):65–73. doi: 10.1016/0021-9150(85)90138-8. [DOI] [PubMed] [Google Scholar]
- 22.Plump AS, et al. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71 (2):343–353. doi: 10.1016/0092-8674(92)90362-g. [DOI] [PubMed] [Google Scholar]
- 23.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258 (5081):468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 24.Broadbent HM, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17 (6):806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 25.Gizard F, et al. PPAR alpha inhibits vascular smooth muscle cell proliferation underlying intimal hyperplasia by inducing the tumor suppressor p16INK4a. J Clin Invest. 2005;115 (11):3228–3238. doi: 10.1172/JCI22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diez-Juan A, Andres V. The growth suppressor p27(Kip1) protects against diet-induced atherosclerosis. FASEB J. 2001;15 (11):1989–1995. doi: 10.1096/fj.01-0130com. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez P, et al. A single-nucleotide polymorphism in the human p27kip1 gene (-838C>A) affects basal promoter activity and the risk of myocardial infarction. BMC Biol. 2004;2:5. doi: 10.1186/1741-7007-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez I, et al. Role of the CDKN1A/p21, CDKN1C/p57, and CDKN2A/p16 genes in the risk of atherosclerosis and myocardial infarction. Cell Cycle. 2007;6 (5):620–625. doi: 10.4161/cc.6.5.3927. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida T, Kaestner KH, Owens GK. Conditional deletion of Kruppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ Res. 2008;102 (12):1548–1557. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92 (20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda Y, et al. Relationship between expression levels and atherogenesis in scavenger receptor class B, type I transgenics. J Biol Chem. 2000;275 (27):20368–20373. doi: 10.1074/jbc.M000730200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.