Abstract
In an inducible oncogenesis model, the miR-200 family is inhibited during CSC formation but not transformation, and inhibition of miR-200b increases CSC formation. Interestingly, miR-200b directly targets Suz12, a subunit of a polycomb repressor complex (PRC2). Loss of miR-200 during CSC formation increases Suz12 expression, Suz12 binding, H3-K27 tri-methylation, and Polycomb-mediated repression of the E-cadherin gene. MiR-200b expression or Suz12 depletion blocks the formation and maintenance of mammospheres, and in combination with chemotherapy suppresses tumor growth and prolongs remission in mouse xenografts. Conversely, ectopic expression of Suz12 in transformed cells is sufficient to generate CSCs. The miR-200b-Suz12-cadherin pathway is important for CSC growth and invasive ability in genetically distinct breast cancer cells, and its transcriptional signature is observed in metastatic breast tumors. The interaction between miR-200 and Suz12 is highly conserved, suggesting that it represents an ancient regulatory mechanism to control the growth and function of stem cells.
Keywords: microRNAs, polycomb, cancer stem cells, chemotherapy
INTRODUCTION
The cancer stem cell hypothesis suggests that tumors consist of tumor-forming, self-renewing, cancer stem cells (CSCs) within a large population of non-tumor-forming cancer cells (Ailles and Weissman, 2007; Grimshaw et al., 2008; Polyak and Weinberg, 2009). CSCs resist standard chemotherapy that reduces tumor mass by killing non-stem cells. During remission, CSCs can regenerate all the cell types in the tumor through their stem cell-like behavior, resulting in relapse of the disease. In accord with this hypothesis, the combination of chemotherapy and metformin, an anti-diabetic drug that selectively inhibits CSCs, blocks tumor growth and prolongs remission in mouse xenografts (Hirsch et al., 2009).
As implied by their name, CSCs resemble embryonic stem cells (ESCs) in several respects. CSCs and ESCs have ability to self-renew, differentiate, and cause tumors when injected in nude mice. Inflammatory mediators such as STAT3 are up-regulated in CSCs and ESCs (Bao et al., 2009; Iliopoulos et al., 2009), and let-7 microRNA is down-regulated in CSCs (Yu et al., 2007; Iliopoulos et al., 2009) and fetal stem cells (Nishino et al., 2008). Lin28, a repressor of let-7 microRNA, is activated in CSCs (Iliopoulos et al., 2009), and it is important for the induction of pluripotent stem cells (Hanna et al., 2009). More generally, an ESC gene expression signature including Oct4, Sox2, Klf4, and Nanog is observed in highly aggressive human tumors (Ben-Porath et al., 2008). Lastly, as discussed below, miR-200 microRNA and polycomb complexes that mediate transcriptional repression play important roles in both CSCs and ESCs. These observations suggest that factors contributing to stem cell renewal during normal development may play similar roles in CSCs and contribute to subsequent metastasis (Ben-Porath et al., 2008; Gotoh, 2009; Shimono et al., 2009).
Polycomb complexes directly regulate key developmental factors that maintain ESC self-renewal and pluripotency (Boyer et al., 2006; Lee et al., 2006), and they are commonly up-regulated in cancer types in a manner associated with the aggressiveness of the tumor (Squazzo et al., 2006; Xhong et al., 2007; Bracken and Helin, 2009; Hoenerhoff et al., 2009). Two distinct polycomb complexes, PRC1 and PRC2, are critical to maintain a repressed gene state that is stable throughout cell generations (Schwartz and Pirrotta, 2007; Margueron et al., 2009; Simon and Kingston, 2009). PRC2, which includes Suz12 and the catalytic subunit Ezh2, is responsible for di- and tri-methylation of lysine 27 on histone H3 (Cao et al., 2002; Koyanagi et al., 2005; Boyer et al., 2006) and initiating gene repression. PRC2 is important in the establishment and maintenance of aberrant silencing of tumor suppressor genes during cellular transformation (Villa et al., 2007; Herranz et al., 2008), and upregulation of Suz12 coincides with transformation and cancer (Herranz et al., 2008; Hussain et al., 2009; Pizzatti et al., 2009). PRC1 recognizes chromatin marked with methylated H3-K27, and it is believed to be the primary PcG complex that mediates transcriptional repression. Overexpression of the Bmi1 subunit of PRC1 is often observed in cancer tissues.
During metastasis, migrating breast CSCs undergo a loss of polarity leading to an epithelial-mesenchymal transition (EMT) analogous to that occurring in normal development. The miR-200 microRNA family regulates the EMT and cancer cell invasion and migration (Bracken et al., 2008; Burk et al., 2008; Gregory et al., 2008; Korpal et al., 2008), and it is also suppresses expression of stem cell factors in ESCs (Lin et al., 2009; Wellner et al., 2009). MiR-200 family members are down-regulated in different types of cancer (Adam et al., 2009; Olson et al., 2009; Bendoraite et al., 2010), and they are specifically down-regulated in breast CSCs in comparison to non-tumorigenic cancer cells (Shimono et al., 2009). Lastly, miR-200c strongly inhibits both the ability of normal mammary stem cells to form mammary ducts and the ability of breast CSCs to form tumors in vivo, indicating that down-regulation of miR-200c is a molecular link between CSCs and normal stem cells of the same developmental lineage (Shimono et al., 2009).
miR-200 RNAs regulate the EMT by directly inhibiting expression of Zeb1 and Zeb2, which are DNA-binding transcriptional repressors of the E-cadherin (Cdh1) gene (Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008). Loss of miR-200 results in increased Zeb1 and Zeb2 levels, leading to repression of Cdh1 and the EMT. siRNA-mediated inhibition of CDH1 expression is sufficient to induce the EMT (Onder et al., 2008). Interestingly, Zeb1 binding to a conserved pair of Zeb-type E-box elements upstream of the promoter inhibits the expression of miR-200 family RNAs, thereby generating a negative feedback loop that controls Cdh1 expression (Bracken et al., 2008; Burk et al., 2008). In cancer cells, Cdh1 repression also depends on the PRC2 complex, which is recruited to the Cdh1 promoter via the DNA-binding transcriptional repressor Snail (Herranz et al., 2008); it is unknown if this occurs in CSCs. In breast CSCs, miR-200c modestly reduces expression of the Bmi1, a subunit of PRC1, through direct targeting of its 3′UTR (Shimono et al., 2009; Wellner et al., 2009). However, this interaction is not well conserved throughout evolution, and its significance for Cdh1 expression and the EMT is unclear.
Recently, we described an inducible model of cellular transformation in which a transient inflammatory signal in a non-transformed breast epithelial cell line causes an epigenetic switch to the transformed state including the formation of a sub-population of CSCs (Iliopoulos et al., 2009; Hirsch et al., 2010). Using this experimental model, we identify the miR-200 microRNA family as a critical regulator for CSC growth and function. We show that miR-200b strongly inhibits expression of the Suz12 subunit of PRC2 through a direct and evolutionarily conserved interaction with the 3′UTR. Loss of miR-200 during CSC formation results in increased Suz12 binding and H3-K27 tri-methylation at the CDH1 promoter and repression of E-cadherin. This pathway is important for tumor formation and the response to cancer treatment in mouse xenografts, and its transcriptional signature is observed in metastatic human breast tumors. Thus, miR-200 acts as a tumor suppressor that blocks CSC formation by inhibiting the PRC2 polycomb complex, and hence preventing the repression of E-cadherin and other critical target genes.
RESULTS
MiR-200b is Selectively Down-regulated in CSCs, and its Inhibition Results in Enrichment of CSCs
Treatment of non-transformed breast epithelial cells (MCF-10A) carrying an inducible Src oncogene (ER-Src) with tamoxifen induces cellular transformation in 24–36 hours (Hirsch et al., 2009; Iliopoulos et al., 2009; Hirsch et al., 2010). A sub-population of these transformed cells are cancer stem cells (CSCs), as defined by expression of the CD44 marker, mammosphere formation, and the ability to cause tumors in nude mice. In a genetic screen to be described elsewhere, we identified miR-200 family members (miR-200b, miR-200a, miR-429, miR-200c) as being important for CSC growth.
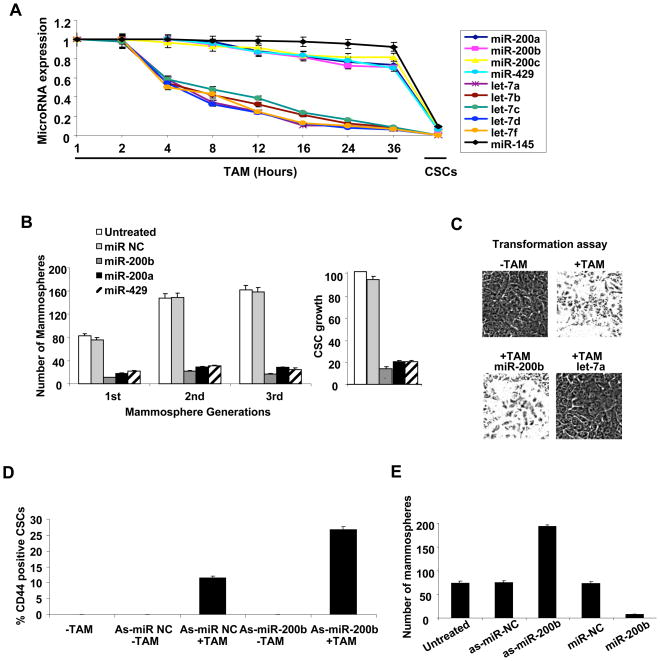
Interestingly, miR-200 family members are down-regulated in CSCs, but their expression levels are unchanged during the transition between non-transformed and transformed cells (Figure 1A). The specific down-regulation in CSCs contrasts with the behavior of let-7 family members, which are strongly down-regulated during the cellular transformation process and even further down-regulated in CSCs (Iliopoulos et al., 2009), suggesting a specific role of miR-200 in CSC function. In accord with this suggestion, individual expression of miR-200 family members blocks growth of CSCs and mammospheres (Figure 1B), but it does not affect the ability of non-transformed cells to become transformed upon Src induction (Figure 1C). Because of the redundancy between miR-200 family members and the slightly stronger effect of miR-200b on inhibiting CSC and mammosphere growth, subsequent experiments have been performed with miR-200b.
Figure 1. MiR-200b Regulates Cancer Stem Cell Growth.
(A) Expression (mean ± SD) of the indicated microRNAs at the indicated times after induction of transformation with tamoxifen (TAM) and in CSCs purified by flow cytometry.
(B) CSCs and 1st, 2nd, 3rd passage mammospheres were transfected with the indicated microRNAs and examined for the percentage of CSC growth (relative to untransfected CSCs) and number of mammospheres/1000 cells.
(C) Transformation of TAM-treated ER-Src cells transfected with miR-200b or let-7a microRNAs. Data are presented as mean ± SD.
(D) Flow cytometry analysis of CD44/CD24 profiles and percentage of CSCs (mean ± SD) in the population (mean ± SD) after transfection with the indicated anti-sense microRNAs.
(E) Number of mammospheres/1000 cells ((mean ± SD) after transfection of the indicated antisense microRNAs.
Inhibition of miR-200b expression (via antisense RNA) results in enrichment of the CSC population (Figure 1D. S1A) and increases the efficiency of ER-Src transformed cells to form mammospheres (Figure 1E). Thus, CSC and mammosphere growth is blocked by overexpression of miR-200b and improved by inhibiting miR-200b. Taken together with the specific down-regulation in CSCs in comparison to non-stem cancer cells (NSCCs) in the same population, these observations indicate that miR-200b is an important and specific regulator of CSC formation and growth.
MiR-200b Directly Regulates Suz12 Expression in CSCs
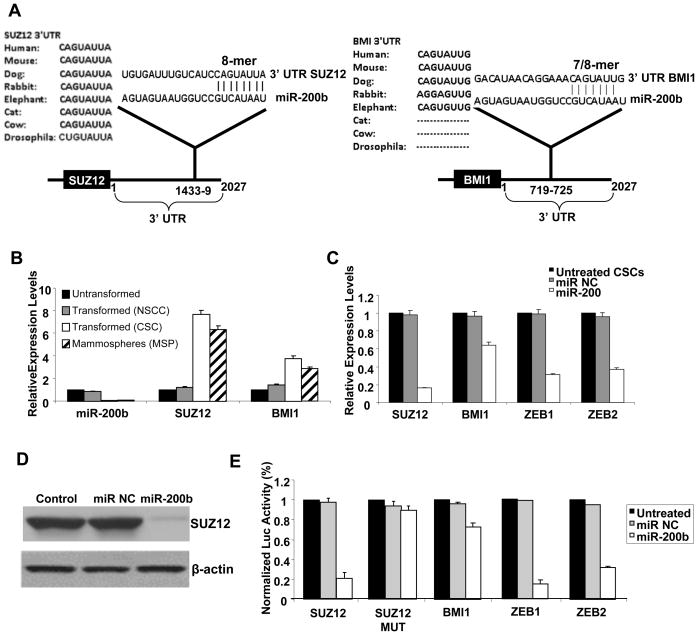
Mir-200 targets the Zeb1 and Zeb2 transcriptional repressors (Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008) and Bmi1 (Shimono et al., 2009; Wellner et al., 2009), a component of the PRC1 polycomb complex. MiR-200 also targets Fog2 (Hyun et al., 2009), a co-factor for GATA transcription factors essential for heart morphogenesis and coronary vasculature (Tevosian et al., 2000). Based on sequence complementarity and phylogenic conservation, we identified Suz12, a component of the PRC2 polycomb complex that methylates H3-K27, as a potential gene target of miR-200b (Figure 2A). Specifically, there is perfect complementarity between miR-200b seed sequence and Suz12 3′UTR sequence that is highly conserved among species including fruit flies. In contrast, the complementarity between the miR-200b seed sequence and Bmi1 3′UTR is imperfect (mismatch in first nucleotide), and this interaction is not well conserved (not found in rabbit, cat, and cow), suggesting that Suz12 is the more important target of miR-200b.
Figure 2. Direct Targeting of Suz12 by MiR-200b Through an Evolutionarily Conserved Interaction.
(A) Predicted miR-200b binding sites in the 3′UTRs of Suz12 and Bmi1 with sequence complementarity and phylogenic conservation of 8-nt seed sequence indicated. Zeb1 and Zeb2 show similar complementarity as Suz12 with the exception of Drosophila.
(B) MiR-200b, Suz12 and Bmi1 RNA levels (mean ± SD) in untransformed, transformed non-stem cancer cells (NSCCs), CSCs, and mammospheres (MSP).
(C) RNA levels (mean ± SD) of the indicated genes after transfection with miR-200b or control microRNAs in CSCs.
(D) Suz12 and β-actin protein levels after transfection with miR-200b or control microRNAs in CSCs.
(E) Relative luciferase activity (mean ± SD) mediated by reporter constructs harboring the 3′UTR of the indicated genes (and mutated version of Suz12) upon transfection with miR-200b or control microRNAs.
Several lines of evidence indicate that miR-200b targets directly Suz12 and Bmi1 mRNA through binding in their 3′UTRs with the effect on Suz12 being more robust. First, in comparison to non-transformed cells, NSCCs, CSCs and mammospheres derived from ER-Src transformed cells show lower levels of miR-200b and higher levels of Suz12 and Bmi1 mRNA expression levels (Figure 2B). Second, upon miR-200b overexpression in CSCs, Suz12 RNA levels are reduced 6-fold (Figure 2C) and Suz12 protein levels are correspondingly reduced (Figure 2D). Mir-200b also causes a 1.6-fold reduction in Bmi1 RNAs levels and a 3-fold reduction in Zeb1 and Zeb2 RNA levels in CSCs (Figure 2C). Third, miR-200b overexpression inhibits the activities of luciferase reporter constructs containing the Suz12, Bmi1, Zeb1, and Zeb2 3′UTRs, with the effect on Bmi1 being significantly weaker than on the other target genes (Figure 2E). Importantly, mutation of the putative target site in the Suz12 3′UTR abolishes repression by miR-200b (Figure 2E), thereby validating the functional significance of the miR-200-Suz12 interaction.
Suz12 is Important for the Function of Cancer Stem Cells
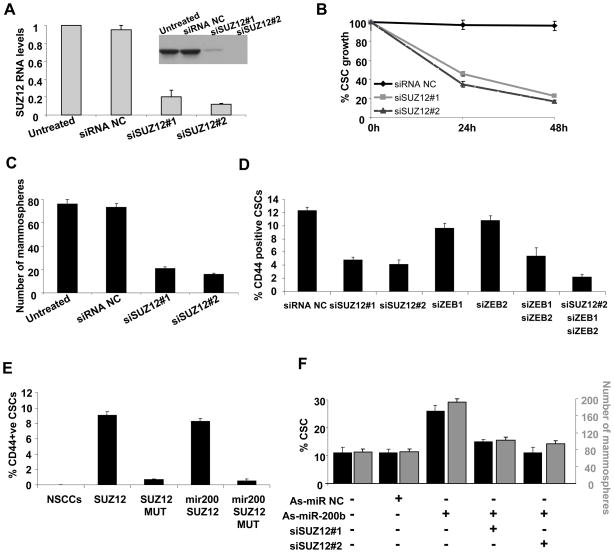
MiR-200 targets Suz12 and is important for CSC growth, suggesting the importance of Suz12 in the function of CSCs. In accord with this suggestion, reduction of Suz12 expression by two different siRNAs (5–10-fold; Figure 3A) inhibits CSC growth (Figure 3B), mammosphere formation (Figure 3C), and the proportion of CSCs in the population of transformed ER-Src cells (Figures 3D, S1B). The effect of Suz12 depletion on the proportion of CSCs is slightly greater than achieved by simultaneous depletion of Zeb1 and Zeb2 (individual depletion of the Zeb proteins has only a modest effect). Simultaneous depletion of Suz12, Zeb1, and Zeb2 reduces the proportion of CSCs below the level of occurring upon Suz12 depletion alone. Thus, Suz12 and the combination of Zeb1 and Zeb2 make critical and independent contributions to CSC function.
Figure 3. Suz12 is Required for CSC and Mammosphere Growth Mediated by Loss of MiR-200.
(A) Suz12 RNA and protein (inset) levels (mean ± SD) in CSCs treated with 2 different siRNAs against Suz12 or control siRNA (NC).
(B) Percentage of CSCs obtained by sorting ER-Src cells treated with TAM for 36h were transfected with Suz12 or control siRNAs, and examined for the percentage of CSC growth (relative to untransfected cells) 24h and 48h afterwards.
(C) Number of mammospheres/1000 cells (mean ± SD) 48h after transfection with Suz12 or control siRNAs.
(D) Percentage CSCs (mean ± SD) of ER-Src transformed cells after transfection with siRNAs against the indicated genes.
(E) NSCCs obtained by sorting an ER-Src transformed cell population were transfected with Suz12 or control expression vectors in the presence or absence of co-transfected miR-200 were analyzed for the percentage of CSCs by flow cytometry (mean ± SD).
(F) Percentage of CSCs (black) and number of mammospheres (gray) after transfection with antisense-miR negative control (as-miR NC), antisense-miR-200b (as-miR-200b), or Suz12 siRNAs. Data are shows as mean ± SD.
Strikingly, ectopic expression of Suz12 in NSCCs results in CSC formation (Figures 3E, S1C). In accord with the absence of the 3′UTR in the Suz12 expression construct, expression of miR-200 does not reduce the level of CSC formation achieved by ectopic expression of Suz12. The level of ectopically expressed Suz12 RNA in NSCCs is 5-fold beyond that observed in the parental (non-transfected) cell line (Figure S1D), which is comparable to that in CSCs. Conversely, inhibition of Suz12 expression suppresses the effect of antisense-miR-200b in formation of the CSC population and mammospheres (Figure 3F). The level of Suz12 expression in Suz12-depleted CSCs is comparable to that observed in NSCCs in the same population, indicating that the phenotypes observed upon depletion are physiologically relevant. Taken together, these results strongly argue that regulation of Suz12 levels by miR-200 is critical for CSC function.
MiR-200b Regulates E-cadherin Expression Through Direct Suz12 Binding and H3-K27 Tri-methylation
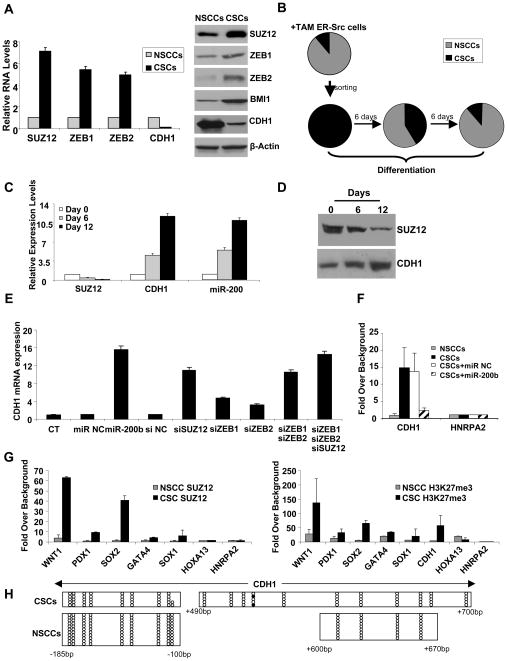
To address how Suz12 affects the process of CSC formation, we analyzed its role in regulation of the E-cadherin (Cdh1) gene, an important Suz12 target in ES cells (Herranz et al., 2008). In comparison to NSCCs, CSCs in the same population show strongly increased Suz12 levels and dramatically reduced Cdh1 levels (Figure 4A). Furthermore, differentiation of ER-Src CSCs into NSCCs (Figure 4B) results in inhibition of Suz12 and up-regulation of Cdh1 at both the mRNA (Figure 4C) and protein level (Figure 4D). In addition, miR-200 RNA levels are strongly induced upon differentiation (Figure 4C). Both miR-200 and Suz12 are important for CDH1 expression, because miR-200b overexpression or siRNA inhibition of Suz12 results in up-regulation of Cdh1 expression levels in CSCs (Figure 4E). The level of Cdh1 up-regulation upon inhibition of Suz12 is comparable to that observed for the combined inhibition of both Zeb1 and Zeb2, as expected from their role in CSC formation (Figure 3D).
Figure 4. MiR-200b Functions through Suz12 and H3K27 Tri-methylation in CSCs.
(A) Suz12 and E-cadherin (Cdh1) mRNA (left) and protein (right) levels (mean ± SD) in NSCCs and CSCs.
(B) Percentage (mean ± SD) of CSCs (black) and NSCCs (gray) at the indicated times after plating CSCs (obtained from sorting ER-Src transformed cells) under differentiation conditions.
(C) Suz12, Cdh1, and miR-200 RNA levels (mean ± SD) during differentiation of CSCs.
(D) Suz12 and Cdh1 protein levels during differentiation of CSCs.
(E) Cdh1 RNA levels (mean ± SD) in CSCs after transfection of miR-200b or siRNAs against the indicated genes.
(F) Chromatin immunoprecipitation showing association of Suz12 at CDH1 and control HNRPA2 gene in NSCCs and CSCs expressing miR-200 or a control microRNA. Data are presented as mean ± SD.
(G) Suz12 association and H3-K27 trimethylation (mean ± SD) at the indicated loci in NSCCs and CSCs.
(H) DNA methylation analysis of regions within the CDH1 promoter in CSCs and NSCCs. For each horizontal line, open circles indicate non-methylated residues and black circles indicated methylated residues in a given clone that was sequenced after bisulfite conversion.
Chromatin immunoprecipitation analysis reveals that Suz12 binds strongly (10–14 fold enrichment) to the CpG island near the CDH1 mRNA initiation site in ER-Src derived CSCs, but not in NSCCs in the same population (Figure 4F). Importantly, overexpression of miR-200b strongly reduces Suz12 binding to the Cdh1 CpG island (Figure 4F). As expected from this regulated Suz12 binding, H3-K27 trimethylation at the Cdh1 region is strongly enhanced in the CSC population, but not in NSCCs (Figure 4G). This Polycomb-mediated repression appears to occur in the absence of DNA methylation, because the CDH1 target site in unmethylated in CSCs (Figures 4H, S2).
Taken together, these observations suggest that direct binding of Suz12 and H3-K27 tri-methylation represses Cdh1 expression, which is important for the maintenance of the CSC phenotype. Interestingly, increased Suz12 binding and H3-K27 methylation is not restricted to the CDH1 locus, but rather is observed at all Suz12 targets tested (Figure 4G). This suggests that the increased Suz12 association with the CDH1 locus is due to increased Suz12 levels and not to alterations in the protein(s) that recruit Suz12 to this locus.
MiR-200b–Suz12 Interaction is Important for the Growth and Invasive Ability of Cancer Stem Cells Derived from Different Cancer Cell lines
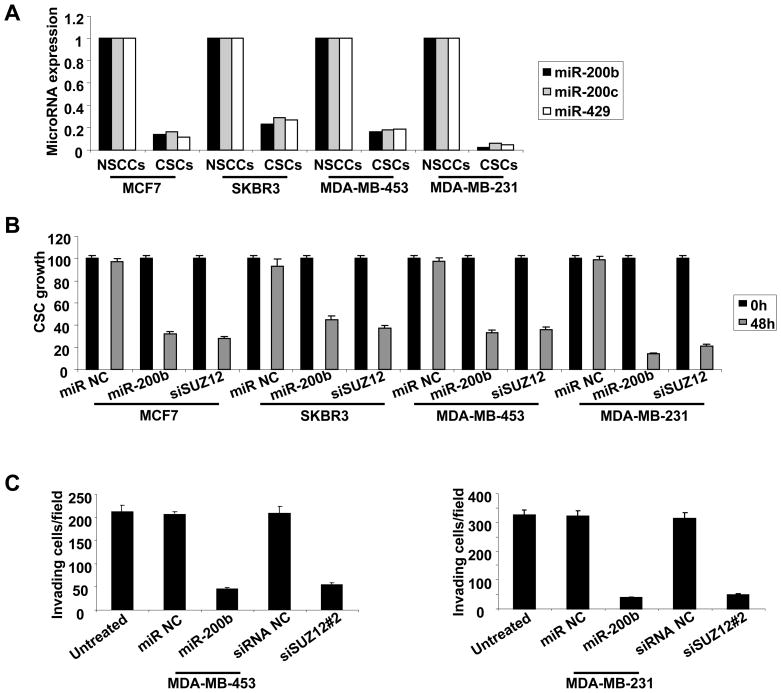
To address whether the miR-200b-Suz12 interaction is important for CSCs in other cancer cell lines, we examined CD44high/CD24low CSC populations derived from MCF7, SKBR3, MDA-MB-453 and MDA-MB-231 cancer cells that were isolated from genetically different types of breast tumors. In all cases, the miR-200 family members (miR-200b, miR-200c, miR-429) were highly down-regulated in CSCs (Figure 5A), and miR-200b overexpression or Suz12 inhibition significantly affect CSC growth (Figure 5B). We also analyzed CSCs derived from MDA-MB-231 and MDA-MB-453 cells for their ability to infiltrate through Matrigel in a modified Boyden chamber assay using 5% horse serum as a chemoattractant. Under these conditions, miR-200b overexpression or Suz12 inhibition significantly reduces the invasive ability of both MDA-MB-453 and MDA-MB-231 cells (Figure 5C). Thus, the miR-200b-Suz12 interaction is important for growth and invasive ability of CSCs derived from genetically distinct breast cancer cell lines.
Figure 5. MiR-200b-Suz12 Pathway Affects Growth and Invasive Ability of CSCs Derived from Genetically Distinct Breast Cancer Cell Lines.
(A) MiR-200b, miR-200a and miR-429 expression levels in NSCCs and CSCs cells derived from the indicated breast cancer cell lines.
(B) Growth of CSCs derived from the indicated cell lines 48h post transfection with miR-200b or siRNA against Suz12. Data are presented as mean ± SD.
(C) Invasion assays in CSCs derived from MDA-MB-435 and MDA-MB-231 cells 12h post transfection with miR-200b or siRNA against Suz12. Data are presented as mean ± SD.
MiR-200b and Suz12 Regulate Tumor Growth and Remission In Vivo
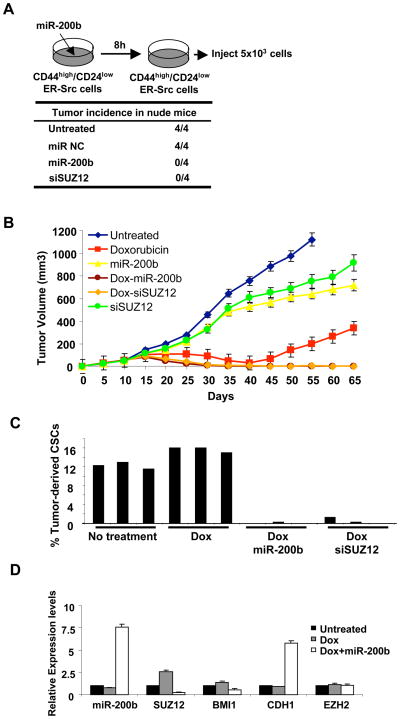
We examined the role of the miR-200b-Suz12 pathway for CSC function in vivo by performing xenograft experiments in which CSCs from ER-Src transformed cells were injected subcutaneously in nude (nu/nu) mice. As expected from the results obtained in cell lines, pre-treatment of CSCs with miR-200b or siRNA against Suz12 (but not a control siRNA) blocked tumor formation in nude mice (Figure 6A). More importantly, we examined mice with tumors (70 mm3) that arose 10 days after injection and were treated intraperitoneally with four cycles (days 10, 15, 20, 25) of doxorubicin, miR-200b, siRNA against Suz12, or combinations of doxorubicin with miR-200b or siRNA against Suz12 (Figure 6B). As expected, doxorubicin treatment caused significant regression of the tumor, but relapse of the disease occurred after 40 days. Treatment of either miR-200 or siRNA against Suz12 has only a very slight effect on tumor growth, presumably because these treatments did not affect NSCCs. Strikingly, the combinations of doxorubicin with either miR-200b or Suz12 depletion caused even stronger regression of tumor growth, and relapse was prevented. Thus, by analogy with metformin treatment (Hirsch et al., 2009) and in accord with the cancer stem cell hypothesis, these observations indicate that miR-200b and Suz12 are important for CSC function in vivo.
Figure 6. MiR-200b Expression or Suz12 Inhibition in Combination with Chemotherapy Prevents Tumor Relapse.
(A) Tumor incidence in nude mice injected with CSCs from ER-Src transformed cells that were pre-treated with miR-200b or siRNA against Suz12 for 8h.
(B) Tumor volume (mm3) in xenografts treated with doxorubicin, miR-200b, siSuz12#2 and combinations at days 10, 15, 20, and 25.
(C) Percentage of CSCs derived from ER-Src tumors treated with doxorubicin combined with miR-200b or siSuz12.
(D) Expression of the indicated RNAs (mean ± SD) in CD44high/CD24low cells taken from tumors at day 25 treated as indicated.
To determine the basis for why combinatorial therapy of doxorubicin with miR-200b or siSuz12 were more effective than doxorubicin alone, we examined the populations of cells recovered from tumors. After three cycles of treatment (day 25), the CSC population was nearly absent from mice subjected to combinatorial therapy, while they were easily observed in tumors from mice treated with doxorubicin alone (Figure 6C). Furthermore, after two cycles of treatment, CSCs derived from tumors treated with doxorubicin and miR-200b showed reduced levels of Suz12 and (to a lesser extent) Bmi1 mRNAs and strong up-regulation of Cdh1 mRNA as compared to CSCs from untreated tumors. Interestingly, CSCs derived from doxorubicin-treated tumors had somewhat reduced levels of miR-200b and Cdh1 and somewhat increased levels of Suz12 and (to a lesser extent) Bmi1 (Figure 6D). This latter observation is likely explained by the selective killing of NSCCs. Taken together, these results suggest that the miR-200b-Suz12-Cdh1 pathway is essential to block the formation and function of CSCs. Loss of this pathway results in CSC formation and tumor growth, whereas restoration of this pathway to CSCs blocks tumor growth and prevents relapse in combination with standard chemotherapy.
MiR-200b–Suz12 Interaction Contributes to the Metastatic Phenotype of Human Mammary Adenocarcinomas
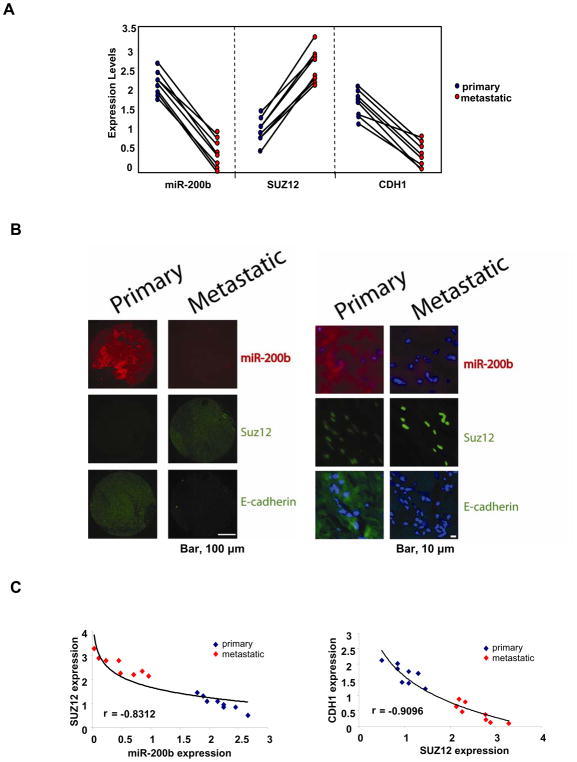
To address the relevance of the miR-200b-Suz12-Cdh1 pathway to human cancer, we measured the expression levels of miR-200b, Suz12 and Cdh1 in primary and metastatic tumor tissues from eight patients with breast cancer. For each patient, miR-200b and Cdh1 expression levels are reduced and Suz12 levels are increased in metastatic tumors relative to the primary tumors (Figure 7A). Furthermore, in-situ hybridization for miR-200b and immunofluorescence for Suz12 and Cdh1 identified the same relationships (Figure 7B). Lastly, there is a striking inverse relationship between Suz12 and miR-200b (r=−0.8312) or Cdh1 (r=−0.9096) in primary and metastatic breast tumors (Figure 7C), strongly arguing for a mechanistic relationship between miR-200b, Suz12 and Cdh1 in metastasis.
Figure 7. MiR-200b-Suz12-Cdh1 pathway in primary and metastatic human tumors.
(A) MiR-200b, Suz12 and Cdh1 expression levels (mean ± SD) in primary and metastatic breast tumor pairs.
(B) Microscopic images showing miR-200b RNA (in situ hybridization) as well as Suz12 and Cdh1 protein levels (immunohistochemistry) in primary and metastatic breast tumor pairs. Higher magnification shows the miR-200b cytoplsmic localization (red), Suz12 nuclear localization (green) and Cdh1 cytoplasmic localization (green); DAPI (blue) stained the nuclei.
(C) Correlation between miR-200b and Suz12 expression and Cdh1 and Suz12 expression in primary and metastatic breast tumors.
DISCUSSION
miR-200 Inhibition of Suz12, a PRC2 Subunit, Regulates the Formation and Growth of CSCs
MiR-200 targets the Zeb1 and Zeb2 transcriptional repressors (Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008) as well as the Bmi1 subunit of the PRC1 complex (Shimono et al., 2009; Wellner et al., 2009). Here, we show that miR-200 also targets the Suz12 subunit PRC2 complex through a direct interaction with a perfectly homologous and highly conserved region of the Suz12 3′UTR. In contrast, the interaction of miR-200 with the Bmi1 3′UTR has a mismatch at a key position in the seed sequence, and this region is not well conserved throughout evolution. In accord with this observation, miR-200 targets Suz12 more effectively than Bmi1, both in reporter constructs and in the natural genes.
Five lines of evidence indicate that PRC2 plays a critical role in the growth and function of CSCs. First, siRNA-inhibition experiments indicate that Suz12, and hence PRC2, is critical for CSC growth, invasion activity, and mammosphere formation in a variety of breast-derived cancer cell lines. Second, in accord with the importance of CSCs in tumor formation, PRC2 is critical for tumor growth in mouse xenografts. Third, Suz12 is up-regulated in CSCs via its interaction with miR-200, and it is required for increased CSC formation that occurs upon inhibition of miR-200. Fourth, PRC2 association and H3-K27 methylation at the Cdh1 promoter is strongly increased in CSCs, and this is responsible for repression of Cdh1, which is required for CSC function. Fifth, during differentiation of CSCs back to NSCCs, Suz12 levels drop and Cdh1 levels increase. These direct PRC2-mediated effects on Cdh1 expression are important, because Cdh1 is down-regulated during epithelial-mesenchymal transition (EMT) generating cells with properties of stem cells (Mani et al., 2008; Polyak and Weinberg, 2009).
Most importantly, regulation of Suz12 expression, and hence PRC2 function, by miR-200 is a critical step in the generation of CSCs, and this regulatory step is relevant for human cancer. As mentioned above, loss of miR-200 is important for CSC formation and growth, and this miR-200 function depends on Suz12. Furthermore, CSCs depleted for Suz12 lose all CSC functions (growth, mammosphere formation, tumor formation, Cdh1 repression), even though the level of Suz12 in such cells is comparable to that in NSCCs. Thus, Suz12 up-regulation in CSCs is critical for all CSC functions assayed, and this includes increased Suz12 occupancy at the Cdh1 promoter, which is essential for PRC2-based repression. Conversely, in NSCCs, ectopic expression of Suz12 at near-physiological RNA levels from a construct lacking the 3′UTR is sufficient to induce CSC formation in a manner that is not inhibited by miR-200. Lastly, the relevance of the miR-200-Suz12-Cdh1 pathway to human cancer and metastasis is demonstrated by the more pronounced CSC signature (low miR-200b, high Suz12, low Cdh1) in metastatic vs. primary breast tumors, and by the striking inverse relationship between Suz12 and either miR-200b or Cdh1 in both primary and metastatic breast tumors. This suggests that the miR-200b-PRC2 pathway is a strong causal factor in metastasis and a driving force in maintaining the CSC population.
The strong inhibitory effect mediated by the evolutionarily conserved interaction between miR-200 and Suz12 argues that miR-200 acts in CSCs to block PRC2 function. As PRC2-mediated methylation of H3-K27 is important for repression by PRC1, the interaction between miR-200 and Suz12 is effectively blocking the function of both polycomb complexes. Nevertheless, miR-200 also functions through the Bmi1 subunit of PRC1, indicating that tight regulation polycomb-based repression is critical for controlling the growth of CSCs. As the miR-200 interaction with Bmi1 is less conserved and less inhibitory, we suggest that mir-200 regulation of PRC1 expression serves an auxiliary role that may be dispensable in some organisms. In addition to its effects on polycomb-based repression, miR-200 also inhibits the Zeb1 and Zeb2 repressors that function independently of PRC2 at Cdh1 (Herranz et al., 2008), and our results indicate that PRC2- and Zeb-mediated repression are independently and comparably critical for CSC growth.
Regulation of miR-200 inhibition of Suz12 might be a conserved and specific function of stem cells
The induction of Suz12 in CSCs derived from NSCCs provides further evidence that CSCs are analogous to ESCs, in which PRC2 directly represses genes encoding developmental regulators. Conversely, differentiation of CSCs into NSCCs is associated with a dramatic reduction in Suz12 levels and PRC2-mediated repression of Cdh1, which is analogous to loss of polycomb-based repression of developmental regulators in differentiating ESCs. Taken together, these observations suggest the possibility that miR-200 inhibition of Suz12 might be an important regulatory step in ESCs, and that up-regulation of miR-200 during early development might be important to alleviate PRC2 repression and permit differentiation.
The interaction between miR-200 and Suz12 is conserved in Drosophila (the homologous microRNA is termed miR-8); 7 out of 8 nucleotides in the microRNA seed are complementary to the Suz12 3′UTR, and the single mismatch is at a position that is typically of modest functional importance. Interestingly, Drosophila miR-8 is expressed at much lower levels in early embryonic development (up to 8h post-fertilization) than at later times (Ruby et al., 2007). Furthermore, Suz12 expression is expressed in a reciprocal fashion, with much higher levels during the first 8h than at later times (Mod-ENCODE database). This decrease in Suz12 expression after the early stage of embryogenesis is consistent with genetic analysis showing that the maternal function of Suz12 is more important than the zygotic function (Birve et al., 2001). Lastly modulation of Suz12 levels in Drosophila suggests that Suz12 levels are limiting for H3-K27 methylation and polycomb-mediated repression (Chen et al., 2008). These observations prompt the speculation that the highly conserved interaction between miR-200 and Suz12 might represent an ancient regulatory mechanism to control the growth and function of stem cells.
Further Evidence Supporting the Cancer Stem Cell Hypothesis
The cancer stem cell hypothesis for the progression of human disease was originally based on the differential tumor-forming properties and chemotherapeutic responses of cancer stem cells and non-stem cancer cells. This hypothesis has been controversial, especially in the absence of experiments testing its validity. A prediction of this model is that agents that selectively inhibit cancer stem cells should function synergistically with chemotherapeutic drugs to delay relapse. In accord with this hypothesis, metformin selectively kills cancer stem cells, and tumor-bearing mice treated with the combination of metformin and doxorubicin remain in remission for extended times (Hirsch et al., 2009). However, as metformin also blocks the transition between non-transformed and transformed cells (Hirsch et al., 2009), similar experiments involving different types of agents that selectively kill CSCs would be useful.
Here, we show that miR-200 behaves similarly to metformin in that it selectively kills CSCs and acts in combination with doxorubicin to block tumor growth and prolong remission. Unlike metformin, which can block cellular transformation, miR-200 function is likely to be specific for CSCs, because its expression is unaffected during the cellular transformation process. Furthermore, down-regulation of Suz12, a critical target of miR-200 has the same effect on remission in combination with chemotherapy. Thus, for at least certain types of cancers, these observations provide very strong experimental validation of the cancer stem cell hypothesis.
MATERIALS AND METHODS
Cell Culture and Separation of CSCs from Non-stem Cancer Cells (NSCCs)
MCF-10A cells (Soule et al., 1990) containing the ER-Src fusion gene (Aziz et al., 1999) were grown in DMEM/F12 medium supplemented with 5% donor horse serum (HS), 20 ng/ml epidermal growth factor (EGF), 10 μg/ml insulin, 100 μg/ml hydrocortisone, 1 ng/ml cholera toxin, 50 units/ml pen/step, with the addition of puromycin; Src induction and cellular transformation was achieved by treatment of 1 μM 4-OH tamoxifen (TAM), typically for 36h (Iliopoulos et al., 2009). Other breast cancer cell lines (MCF7, SKBR3, MDA-MB-231, MDA-MB-435) were grown in DMEM, 10% FBS and pen/step. To separate CSCs from NSCCs, flow cytometric cell sorting was performed on single cell suspensions that were stained with CD44 antibody (FITC-conjugated) (555478, BD Biosciences) and with CD24 antibody (PE-conjugated) (555428, BD Biosciences). As used throughout this work, CSCs are defined by the minority CD44high/CD24low population, whereas NSCCs are defined by the majority CD44low/CD24high. For differentiation experiments, CSCs sorted from ER-Src transformed cells (TAM-treated for 36h) were plated at 105 cells/ml on 6-well plates pre-coated with collagen IV (BD BioSciences) in DMEM/F12 supplemented with 5% serum without growth factors and passaged when they reached >95% confluence. CSC differentiation was monitored every 6 days by flow cytometric analysis.
Mammospheres were generated by placing transformed cell lines in suspension (1000 cells/ml) in serum-free DMEM/F12 media, supplemented with B27 (1:50, Invitrogen), 0.4% BSA, 20 ng/ml EGF and 4 μg/ml insulin (Dontu et al., 2003). After 6 days of incubation, mammospheres were typically >75 mM in size with ~97% being CD44high/CD24low. For serial passaging, 6-day old mammospheres were harvested using a 70 μm cell strainer, whereupon they were dissociated to single cells with trypsin (Dontu et al., 2003), and then regrown in suspension for 6 days.
Genetic Analyses
For siRNA experiments, CSCs or 6-day old mammospheres were transfected with 100 nM of siRNAs from Ambion Inc. against Suz12 (s23967 and s23969), Zeb1 (s229972), Zeb2 (s19033) and negative control (AM4611), 100 nM microRNAs and controls, or 100 nM antisense RNAs against miR-200 or control using siPORT NeoFX transfection agent. The resulting cells were assayed 24h or 48h post-transfection for CSC growth (CCK colormetric assay), mammospheres/1000 cells, tumor formation in mice, or RNA levels. For the experiment involving ectopic expression of Suz12, NSCCs were transfected using lipofectamine 2000 with a CMV-Suz12-ECFP construct and a mutated derivative lacking the CMV promoter and the Suz12 start site (Furano et al., 2006). Importantly, both Suz12 constructs lack the 3′UTR. Transfected cells were assayed 24h later for the presence of CSCs (flow cytometry) as well as Suz12 and Ezh2 RNA levels.
RNA Analysis
MicroRNA expression levels were tested using the mirVana qRT–PCR miRNA Detection Kit and qRT–PCR Primer Sets, according to the manufacturer’s instructions (Ambion Inc, TX, USA). RNU48 expression was used as an internal control. For analyzing mRNAs, total RNA was reverse-transcribed to form cDNA, and the resulting material analyzed by quantitative PCR in real-time using β-actin levels as a normalization control. For analyzing patient samples, RNAs from eight primary tumors and their corresponding metastatic tumors were purchased by Biochain Inc.
Western Blot Analysis
Nitrocellulose membranes containing electrophetically separated proteins from NSCCs and CSCs were probed with rabbit antibodies against Suz12 (ab12073, Abcam Inc.), Zeb1 (sc-20572, Santa Cruz Biotechnology Inc.), Zeb2 (sc130436, Santa Cruz Biotechnologhy Inc.), Bmi1 (ab-14389, Abcam Inc.), Cdh1 (3195, Cell Signaling Technology Inc.), and β-actin (4967, Cell Signaling Technology Inc.), treatmeed with peroxidase-conjugated goat anti-rabbit Ig secondary antibody (Oncogene Research Product), and then visualized by chemiluminescence (Amersham, Inc.).
MicroRNA Target Prediction and Conservation
The miRNA database TargetScan version 5.1 (http://www.targetscan.org/index.html) was used to identify potential miRNA targets for miR-200 and to compare the miR-200 seed sequence with the 3′UTRs of Suz12 and Bmi1 from different species.
Luciferase assays to Validate microRNA Interactions
Using Lipofectamine 2000 (Invitrogen), ER-Src cells were transfected with firefly luciferase reporter constructs containing the 3′UTR of Suz12, Bmi1, Zeb1, and Zeb2 together with 100nM microRNA negative control or miR-200b. In addition, we analyzed a derivative of the Suz12 construct containing two substitutions (CAGTATTA to CACTACTA) in the seed sequence in the Suz12 3′UTR that was generated by Quick-Changell site directed mutagenesis (Stratagene, Inc). Cell extracts were prepared 24h after transfection, and the luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, WI, USA).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was carried out as described previously (Yang et al., 2006). Chromatin fragments from NSCCs and CSCs obtained by sorting ER-Src cells treated with TAM for 36hr were immunoprecipitated with 10 μg of antibody against SuZ12 (ab12073, Abcam, Inc.) and 15 μg of anti-Histone H3 trimethyl lysine 27 (#07-449 Millipore, MA, USA). DNA extraction was performed using Qiagen Purification Kit (Qiagen, MD, USA). The samples were analyzed by quantitative PCR in real time with primer pairs listed in Table S1.
DNA Methylation Assay
Genomic DNA (10 μg) from NSCCs and CSCs as treated with sodium bisulfite for 5h at 55oC in the dark, purified, and then desulfonated as described previously (Lindahl-Allen and Antoniou, 2007). PCR amplified regions of the bisulfite-converted DNA spanning CDH1 (see Supplementary Table ST1) were then sequenced.
Invasion Assays
CSCs derived from MDA-MB-231 and MDA-MB-453 cells were transfected with siRNAs against Suz12 or miR-200 along with appropriate controls. Invasion assays were performed 12h post-transfection using BDBioCoat growth factor reduced MATRIGEL invasion chambers (PharMingen) with 5% horse serum (GIBCO) and 20 ng/ml EGF as chemoattractants.
Xenograft experiments
Nude mice experiments were performed in accordance with Institutional Animal Care and Use Committee procedures and guidelines of Tufts University. In initial experiments 5×103 CSCs derived from ER-Src transformed cells were pre-treated with microRNA negative control (100 nM), miR-200b or siSuz12 for 8h and then injected subcutaneously in the right flank of athymic nude mice (Charles River Laboratories). Tumor growth was monitored daily until the detection of palpable tumors. To analyze the effect of Suz12 and miR-200 in combinatorial therapy, 5×106 ER-Src untreated and TAM-treated (36h) cells were injected subcutaneously into athymic nude mice, and tumor volume was monitored every five days as calculated by the equation V(mm3)=axb2/2, where a is the largest diameter and b is the perpendicular diameter. When the tumors reached a size of ~70mm3, mice were randomly distributed in 6 groups (5 mice/group) and treated intraperitoneally with doxorubicin (4mg), miR-200b, siSuz12, and combinations of doxorubicin and miR-200b or siSuz12. There were four cycles of treatment every 5 days (days 10, 15, 20, 25), and tumor volume was monitored at various times up to 65 days. In addition, cells obtained from the treated tumors were analyzed for the percentage of CSCs by flow cytometry, and for Suz12, Bmi1, Cdh1, and Ezh2 RNA levels using primers previously described (Metsuyanim et al., 2008).
In situ microRNA hybridization
Sections of FFPE breast tumors were deparaffinized in xylene, 2×40 min on a 50 rpm shaker, followed by 5 min each in serial dilution of ethanol (100%, 100%, 75%, 50% and 25%) and followed by 2 changes of water. Slides were then submerged for 5 min in 0.2 N HCl, washed with DEPC-PBS, digested with proteinase K (40 μg/ml) for 60 min at 25°C, rinsed in 0.2% glycine/DEPC-PBS, 3XDEPC-PBS and postfixed with 4% formaldehyde in PBS for 10 min. Slides were then rinsed twice with DEPC-PBS, treated with acetylation buffer (300 μl acetic anhydride, 670 μl triethanolamine, 250 μl of 12 N HCl per 48 ml ddH2O) and then rinsed 4 times in DEPC-PBS followed by 2 rinses in 5xSSC. Slides were pre-hybridized at 49°C for 2 hrs in hybridization buffer (50% formamide, 5xSSC, 0.1% Tween-20, adjusted to pH 6.0 with 9.2 mM citric acid, 50 μg/ml heparin, 500 μg/ml yeast tRNA) in a humidified chamber (50% formamide, 5xSSC). Following pre-hybridization, slides were hybridized overnight at 49°C in a humidified chamber, using 20 nM of Mircury LNA Detection probe for mmu-miR-200b 3′-end labelled with DIG (Exiqon) in pre-warmed hybridization buffer. Sections were rinsed twice in 5xSSC, followed by 3 washes of 20 min at 49°C in 50% formamide/2xSSC. Sections were then rinsed 5 times in PBS/0.1% Tween-20 (PBST), and blocked for 1 hr in blocking solution (2% sheep serum, 2 mg/ml BSA in PBST). Anti-DIG-AP Fab fragments antibody (11093274910, Roche) was applied on sections overnight at 4°C. Next, slides were washed 2 times, in PBST for 10 min each and washed 3 times for 10 min each in 0.1 M Tris-HCl pH 7.5/0.15 M NaCl, followed by equilibration with 1 M Tris pH 8.2 for 10 min and the Fast Red (Roche) solution (1 tablet per 2ml of 0.1 M Tris-HCl pH 8.2). Following incubation for 30 min in the dark, slides were washed 3 times in PBST for 10 min and coverslipped in Vectashield mounting medium with Dapi (Vector Labs). Images were obtained using a Nikon Eclipse 80i microscope and a Spot charge-coupled device camera (Diagnostic Instruments). All photographs were processed using identical settings for capturing and further processing.
Immunfluorescence
Sections of FFPE breast tumors were deparaffinized in xylene, 3×5 min, followed by 10 min each in serial dilution of ethanol (100%, 100%, 95% and 95%) and followed by 2 changes of water. Antigen unmasking was achieved by boiling the slides (95–99°C) for 10 min, in 10 mM sodium citrate buffer pH 6.0. Sections were then rinsed 3 times in ddH2O, 1 time in PBS, and blocked for 1 hr in blocking solution (5% goat serum, 300 μl Triton-X 100 in 100 ml PBS). E-cadherin or Suz12 antibodies were diluted (1:200) and applied on sections overnight at 4°C. Next, slides were washed 3 times, in PBS for 5 min each, blocked for 1 hr in blocking solution, and incubated with Cy2-conjugated anti-goat antibody diluted 1:500 for 1 hr at room temperature in the dark. Slides were washed 3 times, in PBS for 5 min each, and coverslipped in Vectashield mounting medium with Dapi. Images were obtained as described above.
Highlights.
Inhibition of miR-200 family is required for cancer stem cell (CSC) formation
Suz12, a direct target of miR-200b, represses Cdh1 and is required for CSC growth
Ectopic expression of Suz12 induces CSC proliferation
MiR-200b and Suz12 expression inversely correlated in patient tumors
Supplementary Material
Acknowledgments
We thank Ken Yamamoto for providing the Suz12 expression construct, Fabio Petrocca for helpful discussions on the cell sorting and mammosphere assays, and Gary Struhl and Welcome Bender for comments on Drosophila. This work was supported by postdoctoral fellowships from the Leukemia and Lymphoma Society (C.P.), American Cancer Society (H.A.H.) and research grants from the National Institutes of Health to P.N.T. (CA 57486) and K.S. (CA 107486).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–466. doi: 10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Aziz N, Cherwinski H, McMahon M. Complementation of defective colony-stimulating factor 1 receptor signaling and mitogenesis by Raf and v-Src. Mol Cell Biol. 1999;19:1101–1115. doi: 10.1128/mcb.19.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O’Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol. 2010;116:117–125. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Muller J. Su(z)12, a novel Drosophila polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128:3371–3379. doi: 10.1242/dev.128.17.3371. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer research. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chen S, Birve A, Rasmuson-Lestander A. In vivo analysis of Drosophila SU(Z) function. Mol Genet Genomics. 2008;279:159–170. doi: 10.1007/s00438-007-0304-3. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano K, Masatsugu T, Sonoda M, Sasazuki T, Yamamoto K. Association of polycomb group SUZ12 with WD-repeat protein MEP50 that binds to histone H2A selectively in vitro. Biochem Biophys Res Comm. 2006;345:1051–1058. doi: 10.1016/j.bbrc.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gotoh N. Control of stemness by fibroblast growth factor signaling in stem cells and cancer stem cells. Curr Stem Cell Res Ther. 2009;4:9–15. doi: 10.2174/157488809787169048. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Grimshaw MJ, Cooper L, Papazisis K, Coleman JA, Bohnenkamp HR, Chiapero-Stanke L, Taylor-Papadimitriou J, Burchell JM. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, Tsichlis PN, Liu XS, Struhl K. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells and acts together with chemotherapy to blocks tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenerhoff MJ, Chu I, Barkan D, Liu ZY, Datta S, Dimri GP, Green JE. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28:3022–3032. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, Caragacianu D, Schrump DS. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kB, lin 28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl-Allen M, Antoniou M. Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE) Epigenetics. 2007;2:227–236. doi: 10.4161/epi.2.4.5231. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsuyanim S, Pode-Shakked N, Schmidt-Ott KM, Keshet G, Rechavi G, Blumental D, Dekel B. Accumulation of malignant renal stem cells is associated with epigenetic changes in normal renal progenitor genes. Stem Cells. 2008;26:1808–1817. doi: 10.1634/stemcells.2007-0322. [DOI] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3653. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzatti L, Binato R, Cofre J, Gomes BE, Dobbin J, Haussmann ME, D’Azambuja D, Bouzas LF, Abdelhay E. SUZ12 is a candidate target of the non-canonical WNT pathway in the progression of chronic myeloid leukemia. Genes Chromosomes Cancer. 2009 doi: 10.1002/gcc.20722. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cells traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature Reviews. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortallized human breast epithelial cell line, MCF10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Xhong X, Li Y, Peng F, Huang B, Lin J, Zhang W, Zheng J, Jiang R, Song E, Ge J. Identification of tumorigenic retinal stem-like cells in human solid retinoblastomas. Int J Cancer. 2007;121:2125–2131. doi: 10.1002/ijc.22880. [DOI] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.