Abstract
Despite over a century of study of malaria parasites, parts of the Plasmodium falciparum life cycle remain virtually unknown. One of these is the early gametocyte stage, a round shaped cell morphologically similar to an asexual trophozoite in which major cellular transformations ensure subsequent development of the elongated gametocyte. We developed a protocol to obtain for the first time highly purified preparations of early gametocytes using a transgenic line expressing a green fluorescent protein from the onset of gametocytogenesis. We determined the cellular proteome (1427 proteins) of this parasite stage by high accuracy tandem mass spectrometry and newly determined the proteomes of asexual trophozoites and mature gametocytes, identifying altogether 1090 previously undetected parasite proteins. Quantitative label-free comparative proteomics analysis determined enriched protein clusters for the three parasite developmental stages. Gene set enrichment analysis on the 251 proteins enriched in the early gametocyte proteome revealed that proteins putatively exported and involved in erythrocyte remodeling are the most overrepresented protein set in these stages. One-tenth of the early gametocyte-enriched proteome is constituted of putatively exported proteins, here named PfGEXPs (P. falciparum gametocyte-exported proteins). N-terminal processing and N-acetylation at a conserved leucine residue within the Plasmodium export element pentamotif were detected by mass spectrometry for three such proteins in the early but not in the mature gametocyte sample, further supporting a specific role in protein export in early gametocytogenesis. Previous reports and results of our experiments confirm that the three proteins are indeed exported in the erythrocyte cytoplasm. This work indicates that protein export profoundly marks early sexual differentiation in P. falciparum, probably contributing to host cell remodeling in this phase of the life cycle, and that gametocyte-enriched molecules are recruited to modulate this process in gametocytogenesis.
The burden imposed by malaria protozoan parasites to human populations worldwide is due to the severity of the disease, particularly when caused by the species Plasmodium falciparum, and to the efficient transmission of the parasite between humans and the Anopheles mosquito vectors. Gametocytes, the precursors of male and female gametes, are the Plasmodium developmental stages critical to the transmission of the parasites from the human blood stream to the mosquito gut when the insect bites an infected individual. In the human host and in in vitro cultures, P. falciparum gametocytogenesis proceeds in about 10 days through the five classically described morphological stages I to V (1). The typical elongated form of gametocytes in this species appears around day 2 of maturation in the crescent-shaped stage II gametocyte, which progressively further elongates during maturation. In P. falciparum infections, only mature stage V gametocytes are detectable in the peripheral circulation, whereas earlier stages are sequestered in internal organs (2).
Although malaria parasites were discovered over a century ago, knowledge on parts of the Plasmodium life cycle is still very poor. One such stage is the early stage I gametocyte, which is morphologically very similar to a pigmented asexual trophozoite. The molecular characterization of this stage was for long restricted to the identification of two abundant gametocyte-specific proteins, Pfs16 (PFD0310w) and Pfg27 (PF13_0011), up-regulated, respectively, from 24 and 30 h postinvasion (3, 4). Recently, transcriptome analyses comparing isogenic parasites able and unable to produce gametocytes identified genes up-regulated at the onset of gametocytogenesis (5, 6) and characterized four gametocyte-specific proteins significantly up-regulated in the first 30–40 h of sexual differentiation, Pfmdv-1/peg3 (PFL0795c) (5, 7), Pfpeg4 (PF10_0164) (5), and Pfg14.744 and Pfg14.748 (PF14_0744 and PF14_0748) (6). Interestingly, all such proteins and Pfs16 are localized in the gametocyte parasitophorous vacuole (PV)1, and some also are localized in the gametocyte-infected erythrocyte cytoplasm (6, 7). This suggests that, despite the overall similarities with an asexual trophozoite, the early gametocyte utilizes sexual stage-specific molecules to modify the PV and the host cell during gametocytogenesis. Compared with early gametocytes, purified mid- (III-IV) and mature (V) gametocytes stages were more deeply characterized in microarray and proteomics analyses, which altogether indicated that progression of sexual differentiation is accompanied by the up-regulation of 200–300 gametocyte-specific transcripts (8) and proteins (9, 10). Studies on the rodent malaria parasite Plasmodium berghei also indicated that a high proportion of the gametocyte proteins is specific for the proteome of the female or of the male gametocytes (11).
The aim of this work was therefore to obtain a genome-wide molecular characterization for the early gametocyte stages of P. falciparum by a quantitative comparative proteomics analysis of trophozoites and early and mature gametocytes. We developed a purification procedure for the isolation of early gametocytes that achieved purity better than 95% and allowed us to determine its proteome by liquid chromatography tandem mass spectrometry. Functional differences between early and late gametocytes were revealed by a gene set enrichment analysis (12) on semiquantitative expression data computed by an established peptide counting method (exponentially modified protein abundance index (emPAI)) (13). This work not only provided the first characterization of such virtually unknown stages of the parasite life cycle but also identified key cellular processes and molecules enriched in this early phase of gametocytogenesis.
EXPERIMENTAL PROCEDURES
Parasites
P. falciparum clones 3D7 (14) and F12 (15) were cultured in human 0+ erythrocytes provided from Prof. G. Girelli, Università La Sapienza, Rome, Italy, as described in Olivieri et al. (16). To obtain transgenic parasites, d-sorbitol-synchronized ring stage parasites were transfected as described in Olivieri et al. (16).
Plasmids
pHHMC*/3R0.5 plasmid (17), kindly provided by Prof. A. Cowman, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia, was modified to insert a genomic fragment of the pfg27 promoter and the TAA-less pfg27 coding sequence PCR-amplified with primers n.1 and .2 in supplemental Table S5. The gfp coding sequence and the pfg27 genomic downstream region were cloned from plasmid p27GFP (16), yielding plasmid pPfg27:GFP (supplemental Fig. S1). The plasmid for expressing a GST-fused fragment of the PfGEXP10 protein in bacteria was obtained by cloning a 297-bp intronless genomic region of the pfgexp10 (PFA0670c) gene coding sequence in a BamHI/NotI-digested pGEX-6P-3 vector (Amersham Biosciences) with primers pfGEXP10 dir (GGATCCACTTCGGTATCTTATAGTGATGAAGG) and pfGEXP10 rev (GCGGCCGCTCTATATCTCTTTTCAGTGGATGCC relevant restriction sites are underlined in the primer sequences). Insertion in the final expression vector pGST-GEXP10 was controlled by sequencing. PCR amplification was carried out with Accuzyme DNA polymerase (Bioline) with conditions described in Olivieri et al. (16).
Purification of Parasite Stages
3D7/pPfg27:GFP Percoll-purified schizonts were used to start a culture in which gametocytogenesis was induced by a 5–8% increase in hematocrit. One day after stressed parasites appeared, a multilayer Percoll gradient (4) was used to eliminate schizonts and mature gametocytes. The resulting erythrocytes, ring stages, and early gametocytes were passed through a MACS (CS Miltenyi) column to retain the hemozoin-containing stage I-II gametocytes, which, eluted from the column, were fluorescence-activated cell sorter (FACS)-sorted to purify the fluorescent stage I-II gametocytes, detecting GFP emission by a 530-nm band pass filter, and gating on forward/side light scatter. Routinely 7 × 106–1 × 107 parasites were purified; centrifuged in PBS, 2% BSA at 2000 rpm for 20 min; and frozen at −80 °C. For the trophozoite sample, synchronous trophozoites of gametocyte-less clone F12 (15) from a high parasitemia culture were MACS-purified. 2.9 × 107 NF54 stage V gametocytes, cultured in a semiautomated culture system (18), were purified by Percoll gradient centrifugation (4).
Sample Preparation for LC-MS/MS Experiments
Cells were lysed by a freeze/thawing procedure as described in Lasonder et al. (9) and divided in a soluble, a wash, and a pellet fraction. Protein mixtures samples were solubilized in SDS-PAGE loading buffer and separated by SDS-PAGE on a 10% gel. Gels were stained with Coomassie G-250 (Bio-Rad) and cut into 10 slices per cellular fraction. Gel slices were treated with DTT and iodoacetamide and digested by trypsin as described in Lasonder et al. (9). Digested samples were acidified to a final concentration of 0.1% TFA and purified by stop and go extraction tips (19).
Liquid Chromatography Tandem Mass Spectrometry
Peptide identification experiments were performed using a nano-HPLC Agilent 1100 flow system connected on line to a 7 Tesla linear ion trap (LTQ) ion cyclotron resonance FT mass spectrometer (Thermo Fisher, Bremen, Germany). Digested samples were measured up to three times to increase the number of low abundance protein identifications. Peptides were separated on 15 cm 100 μm-inner diameter PicoTip columns (New Objective, Woburn, MA) packed with 3 μm Reprosil C18 beads (Dr. Maisch GmbH, Ammerbuch, Germany) using a 90 min gradient from 12% buffer B to 40% buffer B (buffer B contained 80% acetonitrile in 0.5% acetic acid) with a flow rate of 300 nl/min. Peptides eluting from the column tip were electrosprayed directly into the mass spectrometer with a spray voltage of 2.0–2.2 kV. Data acquisition with the LTQ-FT instrument was performed in a data-dependent mode to automatically switch between MS and MS2. Full-scan MS spectra of intact peptides (m/z 350–1500) with an automated gain control accumulation target value of 1,000,0000 ions were acquired in the Fourier transform ion cyclotron resonance cell with a resolution of 50,000. The four most abundant ions were sequentially isolated and fragmented in the linear ion trap by applying collisionally induced dissociation using an accumulation target value of 10,000, a capillary temperature of 100 °C, and a normalized collision energy of 27%. A dynamic exclusion of ions previously sequenced within 180 s was applied. All unassigned charge states and singly charged ions were excluded from sequencing. A minimum of 500 counts was required for MS2 selection.
Peptide Identification by Mascot Database Searches
Raw spectrum files were converted into Mascot generic peak lists by MaxQuant (20) version 1.0.12.22. Proteins were identified by searching peak lists containing fragmentation spectra with Mascot version 2.2 (Matrix Science) against a the P. falciparum database (version 5.5, 5635 sequences) supplemented with the human International Protein Index database (version 3.52, 73,928 sequences) and frequently observed contaminants and concatenated with reversed copies of all entries. Mascot search parameters for protein identification specified an initial mass tolerance of 30 ppm for the parental peptide and 0.5 Da for fragmentation spectra and a trypsin specificity allowing up to three miscleaved sites. Carbamidomethylation of cysteines was specified as a fixed modification, and oxidation of methionine and deamidation of glutamine and asparagine were set as variable modifications. The required minimal peptide length was set at 6 amino acids. Internal mass calibration of measured ions and peptide validation by establishing false discovery rates (FDRs) was performed by MaxQuant as described in Cox and Mann (20). A final absolute recalibrated mass tolerance for the parental peptide was determined at 10 ppm. We accepted proteins with an FDR better than 1% and multiply charged peptides with an FDR better than 1% and not more than three variable modifications based on the number of accepted identifications of the reversed protein sequences. For proteins identified by one peptide, we applied a more stringent threshold and accepted unmodified peptides with a Mascot score threshold of 40 that were unambiguously assigned to one protein by a Mascot peptide delta score larger than 10. Assigned fragmentation spectra of single peptide hits were extracted using the MSQuant software package and are provided in supplemental Fig. S4. Assembling of peptide sequences on protein groups was performed by maximum parsimony where proteins were considered identified by detection of at least one unique peptide. In only a few cases (15 of 2400), P. falciparum peptide sets were assigned to protein groups with more than one protein. Validated P. falciparum peptides were mapped to protein sequences of PlasmoDB version 5.5 using the software package Protein Coverage Summarizer to retrieve positional information of the peptides.
Identified Peptide Count Analysis to Determine Protein Abundance Index Values
To determine protein abundance values in our samples, validated mass spectrometric data were quantified by a peptide count analysis that computed emPAI values for all proteins (13). emPAI values were calculated according to the following formula.
The number of “observable” peptides per protein was calculated from the output of the program Protein Digestion Simulator, which computes peptide masses and hydrophobicities of simulated digests of protein databases. Protein emPAI abundance values were calculated from merged peptide lists for three samples (trophozoites, stage I-II gametocytes, and stage V gametocytes) and were normalized between different life cycle stages by the median emPAI abundance value as reported in Lasonder et al. (21).
Self-organizing Tree Algorithm (SOTA) Clustering Analysis
Relative emPAI expression profiles of proteins detected by at least two peptides in one of the life cycle stages (2211 of 2400 identified proteins) were clustered using The Institute of Genomic Research MultiExperiment Viewer (MeV4.2) software package (22). A SOTA by Euclidean distance was used with the following settings: for growth termination criteria: maximum cycles, 10; maximum epochs/cycle, 1000; maximum cell diversity, 0.01; minimum epoch error improvement, 0.0001; for centroid migration and neighborhood parameters: winning cell migration weight, 0.01; parent cell migration weight, 0.005; sister cell migration weight, 0.001, neighborhood level, 5; and for cell division criteria: cell variability, p = 0.05.
Gene Set Enrichment Analysis (GSEA)
GSEA statistically tests whether expression of a predefined group of genes (gene sets) correlates between cellular states and does not define subsets based on expression levels prior to the statistical test (12); therefore, it is more sensitive than gene ontology (GO) enrichment tests. In total, 433 gene sets were collected from different sources with a minimal number of 15 genes per set. Gene sets were constructed from P. falciparum GO annotations. Gene ontology v1.2.obo and gene_association.GeneDB_Pfalciparum_20080928 were downloaded from the GO consortium web site for mapping genes to GO terms and their parental GO terms using Ontologizer. GO terms based on ontology-based pattern identification clusters obtained from Plasmodium expression data and literature mining (23) were included in the gene sets as well. In addition, manually curated Plasmodium pathways were retrieved from PlasmoDB and added to the gene sets. Finally, the predicted conserved Plasmodium exportome from Sargeant et al. (24), the enriched gametocyte stage I-II proteins, and the enriched gametocyte stage V proteins identified in the above SOTA analysis were added. Expression data were log2-transformed to obtain a normal distribution required for the Pearson metric ranking method built into the GSEA v2.0 package.
The goal of the GSEA method is to determine whether members of a predefined gene set are randomly distributed around the ranked expression list or whether they are largely located around the top or bottom of that list. The enrichment score (ES) of a gene set is calculated by increasing a running sum statistic when one of the gene set members is found on the ranked list and by decreasing the running sum statistic when it is not encountered. We used a Pearson metric for ranking genes combined with a weighted method in the running sum statistics. For each gene set, the statistical significance (nominal p value) of the ES is estimated by a gene set-based permutation test procedure repeated 1000 times to generate a null distribution for the ES. The nominal p value is then calculated relative to this null distribution. To account for multiple testing of 433 gene sets, the significance level is adjusted by calculation of an FDR or a family-wise error rate (FWER) of normalized enrichment scores. We applied the most conservative multiple testing correction by a significance threshold or FWER < 0.05 because of the low number of expression data sets: one data set of gametocyte stage I-II and one data set of gametocyte stage V. For the gene sets that passed the significance threshold, leading edge subsets, which define members participating particularly in biological processes, were calculated in the GSEA software package by the integrated leading edge analysis tool. Leading edge subsets are defined as those genes in the tested gene set that appear at the top or bottom in the ranked list before the point where the running sum reaches its absolute maximum. The leading edge subset is seen as the core of a gene set that accounts for the enrichment signal in GSEA and is used to assess overlap between gene sets in the enrichment analysis.
Determination of N-terminal Acetylated Plasmodium Export Element (PEXEL) Motif-containing Peptides
Putative exported proteins that contain PEXEL (RX(L/I)X(D/E/Q)) motifs (25) were downloaded from PlasmoDB (1444 proteins). A refined protein set of putatively exported PEXEL motif-containing proteins based on Plasmodium species conservation (475 proteins) was obtained from Sargeant et al. (24), and a second refined data set based on proteins with a predicted PEXEL motif plus a preceding hydrophobic region (411 proteins) was taken from Hiss et al. (26).
Mascot search parameters for acetylated peptide identification specified an initial mass tolerance of 30 ppm for the parental peptide and 0.5 Da for fragmentation spectra and a semitrypsin specificity allowing up to two miscleaved sites. Carbamidomethylation of cysteines was specified as a fixed modification, and acetylation of lysine and the peptide N terminus, oxidation of methionine, and deamidation of glutamine and asparagine were set as variable modifications. Only peptides that contain a C-terminal tryptic site were considered for subsequent verification analysis by the MaxQuant software package. The assignment in MS2 spectra of individual modifications sites was scored automatically using the posttranslational modification algorithm (27) also implemented in MaxQuant and finally verified by manual inspection of MS2 spectra (spectra are provided in supplemental Fig. S5). N-terminally acetylated peptide sequences were mapped on protein sequences that contain PEXEL motifs, and the set of N-terminal acetylated peptides within the PEXEL domains represent PEXEL proteins that show an N-acetyltransferase substrate sequence as reported for PfEMP2 (28).
Production of GST Fusion Proteins and Mouse Immunization
Expression of GST from plasmid pGEX-6P-3 and of the GST-GEXP10 fusion protein from pGST-GEXP10 was induced in Escherichia coli BL21 by 1 mm isopropyl β-d-galactopyranoside for 2h at 37 °C. BugBuster Protein Extraction Reagent (Novagen) was used for protein extraction. Purification of the GST fusion protein from soluble fraction was achieved using glutathione-Sepharose 4 Fast Flow resin (Amersham Biosciences) according to manufacturer's instructions. SDS-PAGE analysis showed that the GST fusion protein could be eluted as a highly purified protein. Immunizations were carried out with BALB/c mice under the guidelines of Istituto Superiore di Sanità, conforming to European Commission directive 86/609 regulating ethical issues on laboratory animal treatment. Two mice were subsequently immunized intraperitoneally with 40, 20, 20, and 10 μg of purified fusion protein in a total volume of 300 μl formulated in Freund's adjuvant on days 0, 28, 42, and 56, respectively. On day 0, prebleed samples were taken as preimmune sera. The final bleed was 7 days after the last immunization.
Western Blot Analysis
Protein extracts from Percoll-purified gametocytes (107/lane) were obtained by lysis in 10 mm Tris-Cl, pH7.5, 50 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.05% SDS, 0.1 mm PMSF supplemented with Complete protease inhibitor mixture (Roche Applied Science). Soluble and insoluble fractions were separated by gradient gel electrophoresis on Bis-Tris 4–12% gels (Novex). Transfer of separated proteins onto a nitrocellulose filter (Protran, 0.2 μm) was performed by electroblotting for 1 h at 350 mA. Membranes were incubated for 1 h with the anti-PfGEXP10 antibody at a 1:500 dilution. Horseradish peroxidase-conjugated secondary antibody against mouse IgGs was diluted 1:30,000, and the chemiluminescent reaction was revealed with the Super Signal West Substrate kit (Pierce).
Immunofluorescence Analysis
Parasites were fixed in suspension with 4% paraformaldehyde, 0.075% glutaraldehyde and permeabilized with 0.1% Triton X-100 as described in Tonkin et al. (29). After a 30-min preincubation in 1% BSA, parasites were incubated with the anti-PfGEXP10 or the anti-GST mouse antibody (1:100) and the anti-Pfmdv-1/peg3 rabbit antiserum (7). After incubation and washes in 1× PBS, parasites were incubated with an affinity-purified, rhodamine-conjugated secondary antibody against mouse or rabbit IgGs diluted 1:200 and Hoechst at 1 μg/ml for nuclear staining. After the final washes, samples were observed at 100× magnification in a Leitz DMR fluorescence microscope equipped with the following filters: BP 340–380 (Hoechst), BP 515–560 (rhodamine), and BP 470–490 (fluorescein). Images were collected with a Leica cooled charge-coupled device camera and deconvolved using the AutoDeblur software (AutoQuant Imaging, Inc.).
RESULTS
Preparation of Purified, Stage-specific P. falciparum Samples
As reliable blood stage proteome determination critically depends on sample purity, targeted strategies were separately applied to obtain highly pure synchronized parasites for the three blood stages. To obtain gametocyte-free trophozoites of P. falciparum, we used parasite clone F12, a 3D7A derivative unable to produce the earliest detectable gametocyte stages (15). Synchronous trophozoites from F12 high parasitemia cultures were MACS-purified, and 8 × 107 parasites were analyzed by mass spectrometry. To obtain mature stage V gametocytes, routine Percoll purification of mature gametocyte cultures previously treated with N-acetylglucosamine to clear residual asexual parasites (30) produced a sample of 2.9 × 107 parasites free of asexual stages.
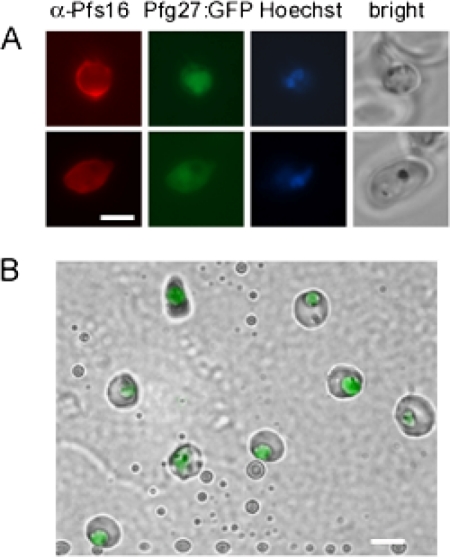
To obtain purified early gametocytes, a P. falciparum 3D7A line was obtained that expresses GFP specifically in gametocytes and from an early stage of sexual differentiation. The GFP coding sequence was fused with that of the early gametocyte protein Pfg27 (4) under the control of pfg27 genomic flanking sequences in plasmid pPfg27:GFP (supplemental Fig. S1). Parasite line 3D7/pPfg27:GFP was obtained after transfection and selection of WR99210-resistant parasites. The pfg27 flanking sequences in the plasmid produced, as expected (16), GFP fluorescence only in morphologically recognizable gametocytes and in round shaped stage I gametocytes from ∼40 h post invasion. Immunofluorescence analysis (IFA) confirmed the identity of the latter cells as all GFP fluorescent round shaped parasites were also positive for the early gametocyte marker Pfs16 (Fig. 1A). To achieve the purity required for proteome analysis, a three-step protocol was developed to purify fluorescent early gametocytes from the 3D7/pPfg27:GFP line. As diagrammatically shown in supplemental Fig. S2, a Percoll gradient was used to eliminate mid-/late stage gametocytes and schizonts. Passage of the remaining cells through a MACS column retained pigmented early gametocytes and small trophozoites from ring stages and uninfected red blood cells. Finally, FACS purified the GFP-positive young sexual stages from the non-fluorescent trophozoites. This protocol routinely yielded ≥95% pure preparations of stage I/early stage II gametocytes with less than 5% contamination of asexual parasites (ring or trophozoite stages) or uninfected red blood cells as determined by cytofluorimetric (supplemental Fig. S2) and microscopic (Fig. 1B) examination. Two samples of 7 × 106 and 1.8 × 107 cells were prepared with this protocol and subjected to mass spectrometry analysis.
Fig. 1.
Fluorescent P. falciparum early gametocytes for proteomics analysis. A, stage I (top) and II (bottom) gametocytes expressing the Pfg27-GFP fusion protein (green) stained with a monoclonal antibody specific for the early gametocyte protein Pfs16 (red fluorescence). Nuclei were stained with Hoechst. Bar, 5 μm. B, sample of FACS-sorted parasites showing highly purified stage I and II gametocytes. Bar, 10 μm.
Liquid Chromatography-Tandem Mass Spectrometry Determination of Three Parasite Blood Stage Proteomes
Protein samples from the above parasite preparations were analyzed by high mass accuracy nano-LC-MS/MS, previously applied to analysis of infected blood (9) and mosquito stages (21), with a more sensitive and accurate mass spectrometer. To accomplish in-depth proteomes, proteins were size-fractionated by one-dimensional SDS-PAGE into 10 gel slices per cellular fraction (either pellet or cytosolic), and tryptic digests of these slices were measured by LC-MS/MS in triplicate. We used an LTQ-FT ICR mass hybrid mass spectrometer to acquire parent ion masses with mass accuracies smaller than 30 ppm in the ICR cell and to acquire fragment ions within 400 ms in the ion trap. Iterative calibration algorithms were used by the MaxQuant software package to achieve an average absolute mass accuracy of 2.00 (s.d. 1.74) ppm for all identified peptide ions. The acquired MS/MS spectra were searched against a combined database of all possible predicted tryptic peptides derived from all P. falciparum and human proteins. Stringent validation criteria for protein identifications established a protein false discovery rate less than 1% by reverse database searches.
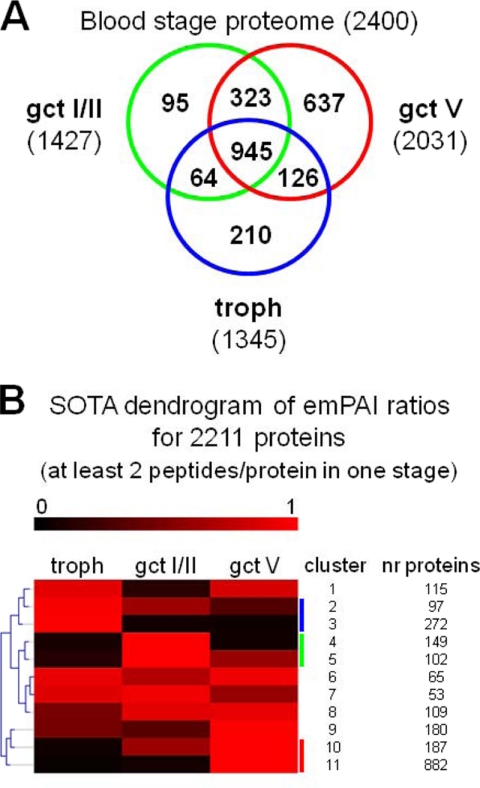
The proteomics analysis of P. falciparum trophozoites, early stage I-II gametocytes, and mature stage V gametocytes resulted in a total of 26,351 unique peptides mapping to 2400 non-redundant P. falciparum proteins across the three life cycle stages: 1345 in trophozoites, 1427 in early gametocytes, and 2031 in mature gametocytes (Fig. 2A and supplemental Table S1). The early stage gametocyte proteome was determined from two batches of FACS-purified parasites of 7 × 106 and 1.8 × 107 cells. These batches identified 653 and 1418 proteins, respectively; 644 proteins were shared. Only nine proteins (1.4% of the total) of the gametocyte batch of 7 × 106 cells were not found in the larger batch of 1.8 × 107cells, demonstrating a low degree of random sampling variation between samples. Such a random variation may thus account for at least some of the 95 proteins detected only in early gametocytes (Fig. 2A) that therefore may not be confidently considered as specific for such a stage solely on the basis of the proteomics analysis. Finally, this analysis identified a total of 1090 proteins not detected in previously published determinations of the proteomes of trophozoites and gametocytes restricted to the identification of fully tryptic peptides (9, 10).
Fig. 2.
Proteomes of early and mature gametocytes and of asexual trophozoites. A, Venn diagram of the proteins identified in the proteomes of the three parasite stages. B, dendrogram from the SOTA clustering analysis on the relative emPAI profiles of 2211 proteins distributed across the three developmental stages. Vertical colored bars mark clusters of the proteins enriched in the three parasite stages (blue, trophozoites (troph); green, early gametocytes (gct I/II); red, mature gametocytes (gct V)). nr, number.
Cluster Analysis of Protein Abundance Profiles
The majority of proteins (1459 of 2400) were detected in more than one life cycle stage (Fig. 2A). To reveal quantitative differences of protein levels between stages, we computed protein abundance values using the emPAI peptide counting method (13), previously successfully applied to analyze P. falciparum mosquito stage proteome data (21). To identify groups of proteins with similar relative expression profiles, we computed emPAI values for proteins detected by at least two peptides in at least one of the three parasite stages (2211 of 2400). Profiles of emPAI relative abundances for such proteins were analyzed by SOTA clustering (22), which identified 11 clusters. These were manually assembled in four categories containing proteins enriched in trophozoites (369 proteins from SOTA clusters 2 and 3), enriched in early gametocytes (251 proteins in clusters 4 and 5), enriched in mature gametocytes (1069 proteins in clusters 10 and 11), and shared between the three stages (522 proteins in clusters 1 and 6–9) (Fig. 2B and supplemental Table S2). In conclusion, this quantitative comparative analysis identified constitutive and enriched or possibly specific proteins produced in asexual trophozoites, in mature gametocytes, and, for the first time, in early gametocytes.
Validation of Young and Mature Gametocyte-enriched Proteomes
The proteomes of the early and the mature gametocytes were inspected for the presence of previously characterized gametocyte proteins. Proteins Pfg377 (PFL2405c), PfsMR5 (PfB0400w), and Pfs25 (PF10_0303), respectively produced from stage III gametocytes, stage V gametocytes, and gametes (31–33) were not detected in the early gametocyte proteome but only in that of mature stages. Conversely, proteins Pfg27 (4), Pfg14.744, Pfg14.748 (6), and Pfmdv-1/peg3 (5, 7), abundantly produced in stage I-II gametocytes, were predominantly represented in the early gametocyte proteome. Besides validating the stage specificity of the gametocyte samples, this analysis also provided information on the expression of other reference proteins. Protein Pfs16, expressed from early gametocytogenesis (3), the sexual stage-specific tubulin α-II (PFD1050w), and the fertilization proteins Pfs230 (PFB0405w) and Pfs45/48 (PF13_0247), reported to appear from stage II to stage III gametocytes (34, 35), were all overrepresented in the proteome of mature gametocytes, indicating that their early appearance in sexual differentiation is followed by a significant accumulation in later stages of gametocyte maturation. Analysis of eight gene products encoded by a subtelomeric region of chromosome 9 reported to be involved in cytoadherence, gametocytogenesis (36), and host cell remodeling (37) revealed that five, PFI1715w, PfRex-3 (PFI1755c), PfRex-4 (PFI1760w), PFI1770w, and PFI1800w, were enriched in early gametocytes, whereas PfRex-1 (PFI1735c), PfCLAG9 (PFI1730w), and PFI1780w were instead overrepresented in asexual trophozoites, possibly suggesting that such proteins play specific roles in such processes in the two developmental stages. Finally, of 106 gene products up-regulated in the transcriptome of late gametocytes annotated with GO function “Plasmodium sexual development” (8), 88.6% were overrepresented in (and 64% were specific for) mature gametocytes, confirming that functions of these genes are likely to be relevant for the late phases of gametocyte maturation.
Functional Characterization of Early Gametocyte Proteome by Gene Set Enrichment Analysis
To reveal pathways or processes associated specifically to the early phase of gametocytogenesis, a functional annotation of the 251 proteins overrepresented in this proteome was performed by GSEA, which is widely used for the analysis of microarray data. We applied this method as it is very sensitive to detect subtle differences between cellular states that may escape detection by other gene ontology enrichment analyses (12). The GSEA analysis identified six overrepresented gene sets in early gametocytes (Table I and supplemental Table S4) using a conservative significance threshold of FWER < 0.05. The most significantly represented set, besides the expected stage I-II gametocyte gene set control, was that of genes encoding predicted Plasmodium exported proteins (24), the second was that of chaperone proteins, and the third was that of gene products putatively involved in host cell remodeling (25). Therefore, the three most enriched gene sets were related to protein export, host cell remodeling, and chaperonins. A role for chaperonins in such processes was recently reported in Botha et al. (38). To independently confirm this finding, the presence of proteins containing the PEXEL domain or host targeting signal (24) was statistically analyzed in the early and mature gametocyte proteomes. A Fisher exact test showed a 2.2-fold enrichment of exported proteins in the enriched/specific proteome of stage I-II gametocytes compared with the total proteome of such stages (19 PEXEL-containing proteins in the 251 proteins of the early gametocyte-enriched/specific proteome versus a total of 50 PEXEL-containing proteins in the 1432 proteins detected in these stages; p = 0.009). In contrast, the same analysis showed a 3.2-fold depletion of such proteins in the stage V gametocyte-enriched/specific proteome (7 of 1069 in the enriched/specific proteome versus 42 of 2033 in the total proteome; p = 0.002). Thus, the above analyses altogether demonstrated that proteins predicted to be exported in the erythrocyte cytoplasm and putatively involved in host cell remodeling are predominantly represented in early sexual stages, providing evidence that this process marks the early phase of gametocytogenesis in P. falciparum.
Table I. GSEA of early gametocyte-enriched proteome.
NES, normalized enrichment score; LES, leading edge subset; gct I/II, gametocyte I/II.
| Name and description of gene seta | Sizeb | NES | p value | FDR q value | FWER p value | LES |
|---|---|---|---|---|---|---|
| % | ||||||
| gct I-II-enriched | 251 | −15.4829 | <0.001 | <0.001 | <0.001 | 100 |
| Exportome Plasmodium-conserved | 65 | −5.17703 | <0.001 | <0.001 | <0.001 | 75 |
| Genes coding for chaperones and their regulators | 74 | −3.24795 | <0.001 | <0.001 | <0.001 | 62 |
| OPI_GO:PM15591202_Trp: targeting malaria virulence and remodeling proteins to the host erythrocyte | 147 | −3.10504 | <0.001 | <0.001 | <0.001 | 65 |
| Genes coding for components involved in ribosome assembly | 33 | −2.75754 | <0.001 | 3.6 × 10−4 | 0.004 | 79 |
| OPI_GO:0004386: helicase activity | 90 | −2.56926 | <0.001 | 0.001 | 0.025 | 61 |
a See “Experimental Procedures” for gene set construction.
b Number of genes.
Proteins Putatively Involved in Host Cell Remodeling in Early Gametocyte Maturation
The above analysis altogether identified 63 putatively exported proteins in the full early gametocyte proteome (Table II). Of these, 37 were either shared with at least another stage or detected by one peptide only. SOTA analysis (described above) on the normalized emPAI values across the three stages indicated that 26 of these proteins were instead overrepresented or detected only in the early gametocyte proteome. This result suggested that these 26 proteins represent a subset of putatively exported protein candidates that play specific roles in host cell remodeling in early gametocytes, and they were therefore named P. falciparum gametocyte exported proteins (PfGEXPs). Importantly, these include proteins previously demonstrated to be exported in early gametocytes, Pfg14.744 and Pfg14.748 (6), or in early asexual parasites, PfRex-3 (37). Comparative analysis provided further clues on differences in protein export between asexual and sexual stages. As described for early gametocytes, also the proteome of asexual trophozoites, well known to actively remodel their host cell, contains 54 putatively exported proteins enriched or specific for such stages (in a total of 75 detected in this proteome), suggesting that protein export in asexual and early sexual development may present significant functional differences. In this respect, we noticed that five of the eight proteins required for the processing or the export of the polymorphic parasite adhesin PfEMP1 or the formation of adhesion-related knob structures in asexual parasites (39) were enriched in the trophozoite proteome with only MAL7P1.172 (PfGEXP11) overrepresented in the early gametocyte proteome. This suggests that such processes are predominantly related to asexual development despite previous work describing an asexual-like mechanism of cytoadherence and the presence of knobs in stage I gametocytes (40).
Table II. Putatively exported proteins in early gametocyte proteome.
| PlasmoDB ID | Exportome annotation | PEXEL start residue | PEXEL end residue | Motif (according to Ref. 24) | Stage enrichment | PfGEXP name (or previous annotation) |
|---|---|---|---|---|---|---|
| PFL2550w | dnaj_III | 35 | 59 | RILSD | Early gametocytes | PfGEXP01 |
| PF11_0037 | PHISTba | 65 | 89 | RNLYE | Early gametocytes | PfGEXP02 |
| PF14_0275 | Unique | 52 | 76 | RLLCT | Early gametocytes | PfGEXP03 |
| MAL13P1.475 | PHISTb | 81 | 105 | RSLLG | Early gametocytes | PfGEXP04 |
| PFI1770w | PHISTb | 73 | 97 | RKLSE | Early gametocytes | PfGEXP05 |
| PFA0675w | dnaj_III | 213 | 217 | RILYS | Early gametocytes | PfGEXP06 |
| MAL13P1.61 | hyp8 | 36 | 60 | RSLAE | Early gametocytes | PfGEXP07 |
| MAL7P1.178 | ab_hydb | Early gametocytes | PfGEXP08 | |||
| MAL8P1.2 | PHISTb | Early gametocytes | PfGEXP09 | |||
| PFA0670c | hyp8 | 42 | 66 | RWLSE | Early gametocytes | PfGEXP10 |
| MAL7P1.172 | PHISTc | 79 | 103 | RNLGE | Early gametocytes | PfGEXP11 |
| PF11_0503 | PHISTc | 35 | 59 | RTLAS | Early gametocytes | PfGEXP12 |
| MAL8P1.205 | 58 | 82 | RKLSE | Early gametocytes | PfGEXP13 | |
| PF11_0038 | Unique | 81 | 105 | RLLCE | Early gametocytes | PfGEXP14 |
| PF10_0310 | hyp2 | Early gametocytes | PfGEXP15 | |||
| PF14_0075 | Unique | 55 | 79 | RKLYQ | Early gametocytes | PfGEXP16 |
| PF14_0752 | PHISTa | 55 | 79 | RNLTE | Early gametocytes | PfGEXP17 |
| PFD0115c | Unique | 49 | 73 | RILVE | Early gametocytes | PfGEXP18 |
| PFA0550w | Unique | 106 | 130 | KRICK | Early gametocytes | PfGEXP19 |
| PFB0900c | PHISTc | 74 | 78 | Early gametocytes | PfGEXP20 | |
| PFC0080c | hyp1 | 131 | 135 | RLLAQ | Early gametocytes | PfGEXP21 |
| PFI1715w | Unique | 86 | 90 | RNLAE | Early gametocytes | PfGEXP22 |
| PF14_0744 | Unique | 53 | 57 | RSLSE | Early gametocytes | Pfg14-744 |
| PF14_0748 | PHISTa | 116 | 120 | RNLTE | Early gametocytes | Pfg14-748 |
| PFI1755c | Unique | 36 | 60 | RQLSE | Early gametocytes | Pfrex-3 |
| PFI1760w | Unique | 35 | 59 | RNLSE | Early gametocytes | Pfrex-4 |
| MAL7P1.171 | emp3 | 77 | 101 | RSLTE | Trophozoites | |
| MAL8P1.4 | PHISTc | 64 | 88 | RILYE | Trophozoites | |
| PF07_0087 | Unique | 51 | 75 | RKLLS | Trophozoites | |
| PF08_0137 | PHISTc | Trophozoites | ||||
| PF10_0023 | hyp16 | 77 | 101 | RFLSE | Trophozoites | |
| PF10_0025 | hyp16 | 47 | 71 | RLLSE | Trophozoites | |
| PF10_0159 | gbp130 | 74 | 98 | RILAE | Trophozoites | |
| PF11_0052 | Unique | 48 | 72 | RNITE | Shared | |
| PF11_0099 | Unique | 46 | 70 | RQLAK | Shared | |
| PF11_0508 | Unique | 76 | 100 | RVLYD | Trophozoites | |
| PF11_0509 | PHISTb_dnaj | 75 | 99 | RNLYC | Trophozoites | |
| PF13_0076 | Unique | 81 | 105 | RSLSE | Trophozoites | |
| PF13_0218 | Mature gametocytes | |||||
| PF13_0275 | Unique | 76 | 100 | RILTQ | Trophozoites | |
| PF13_0317 | Mature gametocytes | |||||
| PF14_0553 | Unique | 73 | 97 | RVLLE | Mature gametocytes | |
| PFA0110w | PHISTb_dnaj | 273 | 277 | Trophozoites | ||
| PFA0210c | Unique | 60 | 64 | Trophozoites | ||
| PFA0225w | Unique | 55 | 79 | RSILQ | Mature gametocytes | |
| PFA0300c | Unique | 119 | 123 | RLLGS | Shared | |
| PFB0085c | PHISTb_dnaj | 42 | 66 | RQLSE | Trophozoites | |
| PFB0090c | dnaj_I | 44 | 48 | Trophozoites | ||
| PFB0106c | Unique | 92 | 116 | RSLAE | Trophozoites | |
| PFB0915w | Unique | 88 | 92 | Trophozoites | ||
| PFB0920w | PHISTb_dnaj | 122 | 126 | RSLCE | Not determinedb | |
| PFC0085c | Unique | 76 | 100 | KSLAE | Trophozoites | |
| PFD0095c | PHISTb | 41 | 65 | RKLYH | Trophozoites | |
| PFD1035w | unique | 39 | 63 | KNLKS | Shared | |
| PFD1170c | PHISTb | 76 | 100 | RILSE | Trophozoites | |
| PFE0050w | Unique | 79 | 103 | RVLAE | Trophozoites | |
| PFE0055c | dnaj_I | 50 | 74 | RSLAE | Trophozoites | |
| PFE0060w | unique | 33 | 57 | RTLAD | Trophozoites | |
| PFE1170w | dnaj_III | Shared | ||||
| PFI1780w | PHISTc | 47 | 71 | KSLAE | Trophozoites | |
| PFL0040c | FIKK K | KCLSE | Not determined | |||
| PFL0600w | Unique | 26 | 50 | RILKE | Shared | |
| PFL2565w | PHISTa | 36 | 60 | RNLAQ | Not determined |
a PHIST; Plasmodium helical interspersed subtelomeric family.
b Not determined, these proteins showed less than two peptides in each of the stages and were not included in the emPAI-based comparative analysis.
Finally, the presence of the molecular machinery recently proposed to process (41, 42) and translocate (43) PEXEL-containing proteins in asexual parasites was also investigated. This revealed that protease Plasmepsin V (PF13_0133) was present in all stages and overrepresented in trophozoites and that four of five components of the putative translocon were present indeed in our proteome data: PTEX150 (PF14_0344) was enriched in the early gametocyte sample, HSP101 (PF11_0175) and EXP2 (PF14_0678) were enriched in the trophozoite sample, whereas TRX2 (MAL13P1.225) was shared across all stages (supplemental Table S2).
Mass Spectrometric Identification of PEXEL Processing and N-Acetylated Termini of Endogenous Parasite Proteins
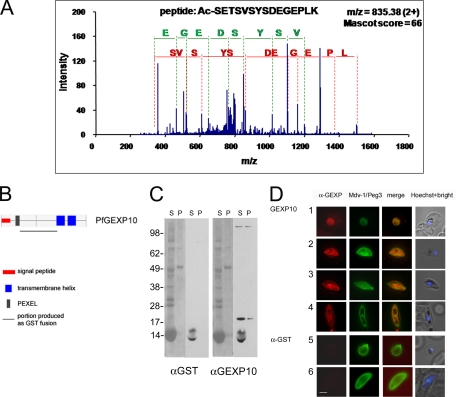
Most exported proteins in Plasmodium contain the conserved PEXEL motif (RXLX(E/Q/D)) that has been shown to contain a classical N-acetyltransferase substrate sequence by mass spectrometry (28, 44). To detect in our data set endogenous acetylated protein N termini cleaved at the PEXEL motif, the LC-MS/MS data were inspected using Mascot with semitrypsin enzyme specificity. Importantly, we re-searched a global data set that enabled finding a small fraction of all processed protein N termini. This analysis identified 14 proteins (Table III, supplemental Table S3, and supplemental Fig. S5) in which, in all cases, N-acetylation takes place at position 4 of the PEXEL motif, providing a mass spectrometry-based evidence of this modification in endogenous protein and confirming the identification of N-acetylated PEXEL peptides recently reported for proteins PFI1780w, PFE0050w, and PfEMP2 (PFE0040c) (28, 44). Interestingly, all modified proteins were detected in the early gametocyte and/or in the trophozoite proteomes, and none were detected in that of mature gametocytes. Three of the identified modified proteins were enriched in early gametocytes: Pf14.744, PfRex-3, and PfGEXP10, whose CID fragmentation spectrum is shown in Fig. 3A. As mentioned above, export has been reported for Pfg14.744 in gametocytes (6) and for PfRex-3 in ring stage parasites (37), and our detection of PEXEL cleavage and N-acetylation of these proteins provides a further insight into the molecular mechanism responsible for their trafficking in the host cytoplasm.
Table III. Detected N-acetylated protein termini within PEXEL motif.
troph, trophozoites; gct I-II, early stage I-II gametocytes.
| PlasmoDB ID | Annotation | Peptide mapping on PEXEL motifa | PEXEL position | Sample | Ref. |
|---|---|---|---|---|---|
| PF14_0744 | Pfg14.744 | RSLSElvlanypssnyafsnyasanysssnypsskm | 53–57 | gct I-II | |
| PFI1755c | PfRex-3 | RQLSEpvveeqdlkkt | 45–49 | gct I-II | |
| PFA0670c | PfGEXP10 | RWLSEtsvsysdegeplky | 51–55 | gct I-II | |
| PFI1780w | PHISTb domain protein | KSLAEaspeehnnlrs | 57–61 | troph and gct I-II | 31 |
| PFE0050w | One transmembrane PEXEL protein | RVLAEqedqyirn | 88–92 | troph and gct I-II | 31 |
| PF11_0503 | Hypothetical protein | RTLASamqhslrg | 45–49 | troph and gct I-II | |
| PF10_0023 | Protein with predicted PEXEL motif | RFLSEpllefdtvfdtflkn | 86–90 | troph and gct I-II | |
| PFE0060w | PIESP2 erythrocyte surface protein | RTLADfndmfanqkn | 42–46 | troph | |
| PFA0210c | Hypothetical protein, conserved | RILKEnkeesletaavnentkd | 60–64 | troph | |
| PF10_0023 | Protein with predicted PEXEL motif | RFLSEpllefdtvfdvfkd | 86–90 | troph | |
| RFLSEpllefdtvfdvfkdtflkn | |||||
| PF14_0013 | DnaJ protein, putative | RNLSEsqenhnssftrt | 83–87 | troph | |
| PFE0040c | PfEMP2 | RILSEteppmsleeimrt | 74–78 | troph | 32 |
| MAL7P1.174 | PHIST domain protein | RILSEveslqssnysevmkn | 51–55 | troph | |
| MAL8P1.206 | P. falciparum hypothetical protein, conserved | RLLTEimencslngfrl | 46–50 | troph |
a See “Experimental Procedures” for PEXEL prediction sources. In bold, PEXEL motif; underlined, identified N-acetylated peptide.
b PHIST; Plasmodium helical interspersed subtelomeric family.
Fig. 3.
PfGEXP10 processing and export in gametocytogenesis. A, mass spectrometry identification of the N-acetylated PfGEXP10 polypeptide after cleavage at its PEXEL motif. The y-ion fragment series are color-coded in green, and the b-ions are in red. B, primary structure of the PfGEXP10 protein according to PlasmoDB and Eukaryotic Linear Motif (ELM) database annotations. The PfGEXP10 portion produced as the GST fusion for generating specific antibodies is shown. C, Western blot analysis with anti-PfGEXP10 antibody on gametocyte extracts. Soluble (S) and insoluble (P) fractions from gametocyte extracts were electrophoresed, blotted, and detected with Ponceau S staining (left side of each panel). Blots incubated with the control anti-GST and the anti-PfGEXP10 antibodies are shown on the right side of each panel. D, IFA with anti-PfGEXP10 antibody on early and mid-stage gametocytes. Fixed, permeabilized 3D7 gametocytes were simultaneously incubated with the serum against the PfGEXP10 protein or a control anti-GST antiserum (red fluorescence) and a serum against the gametocyte PVM marker PfMdv-1/peg3 (green fluorescence). Nucleus-specific Hoechst staining is shown on the right. 1 and 5, stage I; 2, stage II; 3 and 6, stage III; 4, late stage IV. Bar, 5 μm.
A novel Parasite Protein Exported in Gametocyte Development
To investigate the possible export of the third gametocyte-enriched protein, PfGEXP10 (PFA0670c), shown above to be cleaved and N-acetylated at its PEXEL motif, we raised antibodies on a portion of the protein fused to GST (supplemental Fig. S3) and used them to characterize its production and localization in gametocyte development. The anti-PfgGEXP10 serum and a control serum against GST were used in Western blot and IFAs on 3D7 gametocytes. In Western blot, anti-GST antibody failed to detect any parasite specific band, whereas the anti-PfGEXP10 serum recognized a band at 17–20 kDa, predominantly in the parasite soluble fraction, at a molecular mass compatible with that of the protein processed at its PEXEL sequence (Fig. 3, B and C). The anti-PfGEXP10 antibody was also used in IFA on paraformaldehyde/glutaraldehyde-fixed, permeabilized gametocytes at different stages of maturation. To visualize the possible export of PfGEXP10, the gametocyte PV membrane (PVM) was simultaneously labeled with an antiserum specific for the gametocyte PVM protein Pfmdv-1/peg3 (5, 7). Unlike the anti-GST control antiserum, the anti-PfGEXP10 antiserum reacted with stage I and mid-stage gametocytes and clearly showed a punctate fluorescence pattern beyond the PV compartment (Fig. 3D). Specific staining in the erythrocyte cytoplasm was barely observable in stage I gametocytes, and it was far more evident in later maturation stages, indicating that the PfGEXP10 protein is indeed exported in the erythrocyte cytoplasm during gametocyte development.
DISCUSSION
This work provides the first comprehensive proteomics characterization of a largely unexplored part of the life cycle of P. falciparum, the early gametocyte stages at about 40–48 h of maturation after red blood cell invasion. This result could be achieved because we specifically developed a protocol able to obtain highly pure (>95%) preparative samples of stage I/early stage II gametocytes based on cell sorting of GFP-tagged parasites. Previous protocols obtained purification efficiencies of such trophozoite-like cells around 80% (8) by tight parasite synchronization, controlled overgrowth, and clearance of asexual parasites with N-acetylglucosamine, which were not necessary in the present protocol. In the past, genome-wide analyses of P. falciparum early gametocytes were achieved comparing gene expression in parallel cultures of parasite able and unable to enter gametocytogenesis (5, 6). Intrinsic biological variability and the unavoidable residual presence of unhealthy trophozoites were largely responsible for the poor overlaps between the gene lists obtained in those experiments. The possibility to obtain highly purified early gametocyte samples of P. falciparum and the present proteomics characterization now open the way to further cellular, molecular, and biochemical analyses of this part of the parasite life cycle.
Comparative Proteome Analysis Provides Clues to Functional Processes in Early Gametocytogenesis
The improved performance of our mass spectrometry analysis with a severalfold increase in sensitivity, dynamic range, and sequencing speed compared with previous studies on sexual and asexual parasites (9, 10) allowed us to identify a total of 2400 non-redundant proteins in the three developmental stages, 1090 of which were undetected in previous parasite proteomics analyses, and to perform an in-depth quantitative comparative analysis of the proteome of the early gametocytes with those, newly determined here, of asexual trophozoites and mature gametocytes. Calculation of the relative protein abundance levels (emPAI values) for the proteins identified in the three stages and cluster analysis of the resulting profiles identified sets of proteins enriched in or possibly specific for the three developmental stages. The GSEA of the 251 gene products enriched in the early gametocyte proteome, against 400 gene sets from several general and Plasmodium-specific annotation sources, identified only five gene sets significantly overrepresented in this proteome. Remarkably, the three sets with the highest significance values were those of genes encoding (i) conserved Plasmodium exportome proteins (24), (ii) chaperonins and their regulators, and (iii) proteins involved in host cell remodeling (25), all referring to functionally linked processes of protein export and host cell remodeling. Indeed, more than 10% of the proteins enriched in the early gametocyte proteome are represented by the 26 putatively exported proteins here identified and named PfGEXPs. Published evidence and results obtained in this study indicate that three of them, Pfg14.744, PfRex-3, and PfGEXP10, are indeed cleaved at the conserved leucine residue within their PEXEL motif, N-terminally acetylated, and exported to the infected erythrocyte cytoplasm. Although further analysis is needed on other members of this gene group, these data strongly suggest that several such proteins are trafficked during gametocyte development.
Active and Specific Protein Export in Early Gametocytogenesis
This work reveals that the parasite protein export machinery is comparatively more active in the early phase of sexual differentiation than in mature gametocytes as PEXEL-containing proteins are overrepresented in the proteome enriched in early gametocytes and are, on the contrary, depleted in that of late gametocytes. Also, PEXEL processing and N-acetylation were detected only in the proteome data obtained from the early but not from the late gametocytes.
Our finding that protein export is highly active in early gametocytogenesis and recruits a significant set of gametocyte-enriched proteins opens questions on the physiological role of this process in sexual differentiation and on its differences with the analogous processes in asexual parasites required for parasite cytoadherence to the vascular endothelium and sequestration in various organs during infection (39). In this respect, two aspects of the gametocyte biology can be discussed where differences with asexual stage development are becoming increasingly evident: the role of the PV in gametocytogenesis and the sequestration properties of gametocytes. On the first issue, our results support at a genome-wide level previous hypotheses of major, stage-specific modifications of the parasite PV in early gametocytogenesis based on the observation that five of the six early gametocyte proteins identified prior to the present analysis, Pfs16 (3), Pfmdv-1/peg3 (5, 7), Pfpeg4 (5), Pfg14.744, and Pfg14.748 (6), are exported to or beyond the parasitophorous vacuole. It is conceivable that the proposed PV modifications are specific to P. falciparum where they may be necessary for the longevity of this compartment in the uniquely long gametocytogenesis of this parasite. In this respect, it is remarkable that of the 26 PfGEXP genes described here only four orthologs exist in the current gene annotation of rodent Plasmodium species (PB001016.01.0, PB000298.03.0, PB000716.00.0, and PB000835.03.0 in the case of P. berghei) in which gametocyte development is much faster, only slightly longer than the respective ∼24-h asexual cycles. On the second issue, the limited available studies suggest that sequestration of the P. falciparum developing gametocytes in internal organs, chiefly the bone marrow and the spleen, relies on adhesive properties different from those of asexual parasites (2). Although late trophozoites and schizonts adhere to various endothelial cell types through interactions of the knob-located PfEMP1 molecules with host ligands, such as ICAM-1 and CD36, the mid-stage gametocytes do not have knobs (45), bind poorly to such ligands, and generally show a much lower affinity to endothelial cells also if of bone marrow origin (46).2 Specific information on adhesive properties of early gametocytes is largely missing and partly controversial: one report showed that these stages show asexual-like adhesive properties and knobs (40), whereas the latter structures could not be detected in another study (45). In light of these observations, our data may suggest that the large use of gametocyte-enriched exported molecules may be at least in part functional in the yet undefined mechanisms of sexual-stage specific sequestration.
In conclusion, our work shows that at the onset of sexual differentiation the parasite specifically dedicates a relevant expression of its proteome to export proteins and to possibly modify the host erythrocyte with molecules enriched in this developmental stage. This observation and the genome-wide data produced here will be able to direct further work on these underexplored parasite stages and to unveil novel target mechanisms to interrupt gametocytogenesis in the frame of novel transmission-blocking strategies.
Supplementary Material
Acknowledgments
Prof. G. Girelli, University of Rome, and C. Currà and M. Ponzi, Istituto Superiore di Sanità, are acknowledged for the gifts of human red blood cells and for mouse immunization, respectively. Dr. P. Mortensen, Center for Experimental BioInformatics, University of Southern Denmark, is acknowledged for help generating MS/MS spectra of single peptide hits.
* This work was supported in part by European Union FP6 Network of Excellence Biology and Pathology of Malaria Parasite (BioMalPar) and by European Union FP7 Projects Signalling in Life Cycle Stages of Malaria Parasites (MALSIG) and European Virtual Institute for Malaria Research (EVIMalaR).
 This article contains supplemental Figs. S1–S5 and Tables S1–S5.
This article contains supplemental Figs. S1–S5 and Tables S1–S5.
2 F. Silvestrini, M. L. Tiburcio, L. Bertuccini, and P. Alano, unpublished results.
1 The abbreviations used are:
- PV
- parasitophorous vacuole
- MACS
- magnetic activated cell sorter
- emPAI
- exponentially modified protein abundance index
- GSEA
- gene set enrichment analysis
- PEXEL
- Plasmodium export element
- FACS
- fluorescence-activated cell sorter
- LTQ
- linear ion trap
- FDR
- false discovery rate
- SOTA
- self-organizing tree algorithm
- GO
- gene ontology
- ES
- enrichment score
- FWER
- family-wise error rate
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IFA
- immunofluorescence analysis
- PVM
- PV membrane
- BP
- band passfilter.
REFERENCES
- 1.Hawking F., Wilson M. E., Gammage K. (1971) Evidence for cyclic development and short-lived maturity in the gametocytes of Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. 65, 549–559 [DOI] [PubMed] [Google Scholar]
- 2.Alano P. (2007) Plasmodium falciparum gametocytes: still many secrets of a hidden life. Mol. Microbiol. 66, 291–302 [DOI] [PubMed] [Google Scholar]
- 3.Bruce M. C., Carter R. N., Nakamura K., Aikawa M., Carter R. (1994) Cellular location and temporal expression of the Plasmodium falciparum sexual stage antigen Pfs16. Mol. Biochem. Parasitol. 65, 11–22 [DOI] [PubMed] [Google Scholar]
- 4.Carter R., Graves P. M., Creasey A., Byrne K., Read D., Alano P., Fenton B. (1989) Plasmodium falciparum: an abundant stage-specific protein expressed during early gametocyte development. Exp. Parasitol. 69, 140–149 [DOI] [PubMed] [Google Scholar]
- 5.Silvestrini F., Bozdech Z., Lanfrancotti A., Di Giulio E., Bultrini E., Picci L., Derisi J. L., Pizzi E., Alano P. (2005) Genome-wide identification of genes upregulated at the onset of gametocytogenesis in Plasmodium falciparum. Mol Biochem. Parasitol. 143, 100–110 [DOI] [PubMed] [Google Scholar]
- 6.Eksi S., Haile Y., Furuya T., Ma L., Su X., Williamson K. C. (2005) Identification of a subtelomeric gene family expressed during the asexual-sexual stage transition in Plasmodium falciparum. Mol. Biochem. Parasitol. 143, 90–99 [DOI] [PubMed] [Google Scholar]
- 7.Furuya T., Mu J., Hayton K., Liu A., Duan J., Nkrumah L., Joy D. A., Fidock D. A., Fujioka H., Vaidya A. B., Wellems T. E., Su X. Z. (2005) Disruption of a Plasmodium falciparum gene linked to male sexual development causes early arrest in gametocytogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16813–16818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young J. A., Fivelman Q. L., Blair P. L., de la Vega P., Le Roch K. G., Zhou Y., Carucci D. J., Baker D. A., Winzeler E. A. (2005) The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol Biochem. Parasitol. 143, 67–79 [DOI] [PubMed] [Google Scholar]
- 9.Lasonder E., Ishihama Y., Andersen J. S., Vermunt A. M., Pain A., Sauerwein R. W., Eling W. M., Hall N., Waters A. P., Stunnenberg H. G., Mann M. (2002) Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature 419, 537–542 [DOI] [PubMed] [Google Scholar]
- 10.Florens L., Washburn M. P., Raine J. D., Anthony R. M., Grainger M., Haynes J. D., Moch J. K., Muster N., Sacci J. B., Tabb D. L., Witney A. A., Wolters D., Wu Y., Gardner M. J., Holder A. A., Sinden R. E., Yates J. R., Carucci D. J. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 [DOI] [PubMed] [Google Scholar]
- 11.Khan S. M., Franke-Fayard B., Mair G. R., Lasonder E., Janse C. J., Mann M., Waters A. P. (2005) Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687 [DOI] [PubMed] [Google Scholar]
- 12.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 14.Walliker D., Quakyi I. A., Wellems T. E., McCutchan T. F., Szarfman A., London W. T., Corcoran L. M., Burkot T. R., Carter R. (1987) Genetic analysis of the human malaria parasite Plasmodium falciparum. Science 236, 1661–1666 [DOI] [PubMed] [Google Scholar]
- 15.Alano P., Roca L., Smith D., Read D., Carter R., Day K. (1995) Plasmodium falciparum: parasites defective in early stages of gametocytogenesis. Exp. Parasitol. 81, 227–235 [DOI] [PubMed] [Google Scholar]
- 16.Olivieri A., Silvestrini F., Sanchez M., Alano P. (2008) A 140-bp AT-rich sequence mediates positive and negative transcriptional control of a Plasmodium falciparum developmentally regulated promoter. Int. J. Parasitol. 38, 299–312 [DOI] [PubMed] [Google Scholar]
- 17.O'Donnell R. A., Freitas-Junior L. H., Preiser P. R., Williamson D. H., Duraisingh M., McElwain T. F., Scherf A., Cowman A. F., Crabb B. S. (2002) A genetic screen for improved plasmid segregation reveals a role for Rep20 in the interaction of Plasmodium falciparum chromosomes. EMBO J. 21, 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ifediba T., Vanderberg J. P. (1981) Complete in vitro maturation of Plasmodium falciparum gametocytes. Nature 294, 364–366 [DOI] [PubMed] [Google Scholar]
- 19.Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips formatrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 20.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 21.Lasonder E., Janse C. J., van Gemert G. J., Mair G. R., Vermunt A. M., Douradinha B. G., van Noort V., Huynen M. A., Luty A. J., Kroeze H., Khan S. M., Sauerwein R. W., Waters A. P., Mann M., Stunnenberg H. G. (2008) Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog. 4, e1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y., Young J. A., Santrosyan A., Chen K., Yan S. F., Winzeler E. A. (2005) In silico gene function prediction using ontology-based pattern identification. Bioinformatics 21, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 24.Sargeant T. J., Marti M., Caler E., Carlton J. M., Simpson K., Speed T. P., Cowman A. F. (2006) Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marti M., Good R. T., Rug M., Knuepfer E., Cowman A. F. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 [DOI] [PubMed] [Google Scholar]
- 26.Hiss J. A., Przyborski J. M., Schwarte F., Lingelbach K., Schneider G. (2008) The Plasmodium export element revisited. PLoS One 3, e1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 28.Chang H. H., Falick A. M., Carlton P. M., Sedat J. W., DeRisi J. L., Marletta M. A. (2008) N-terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. (2004) Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 137, 13–21 [DOI] [PubMed] [Google Scholar]
- 30.Gupta S. K., Schulman S., Vanderberg J. P. (1985) Stage-dependent toxicity of N-acetyl-glucosamine to Plasmodium falciparum. J. Protozool. 32, 91–95 [DOI] [PubMed] [Google Scholar]
- 31.de Koning-Ward T. F., Olivieri A., Bertuccini L., Hood A., Silvestrini F., Charvalias K., Berzosa Díaz P., Camarda G., McElwain T. F., Papenfuss T., Healer J., Baldassarri L., Crabb B. S., Alano P., Ranford-Cartwright L. C. (2008) The role of osmiophilic bodies and Pfg377 expression in female gametocyte emergence and mosquito infectivity in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 67, 278–290 [DOI] [PubMed] [Google Scholar]
- 32.Eksi S., Williamson K. C. (2002) Male-specific expression of the paralog of malaria transmission-blocking target antigen Pfs230, PfB0400w. Mol. Biochem. Parasitol. 122, 127–130 [DOI] [PubMed] [Google Scholar]
- 33.Kaslow D. C., Quakyi I. A., Syin C., Raum M. G., Keister D. B., Coligan J. E., McCutchan T. F., Miller L. H. (1988) A vaccine candidate from the sexual stage of human malaria that contains EGF-like domains. Nature 333, 74–76 [DOI] [PubMed] [Google Scholar]
- 34.Rawlings D. J., Fujioka H., Fried M., Keister D. B., Aikawa M., Kaslow D. C. (1992) Alpha-tubulin II is a male-specific protein in Plasmodium falciparum. Mol. Biochem. Parasitol. 56, 239–250 [DOI] [PubMed] [Google Scholar]
- 35.Vermeulen A. N., van Deursen J., Brakenhoff R. H., Lensen T. H., Ponnudurai T., Meuwissen J. H. (1986) Characterization of Plasmodium falciparum sexual stage antigens and their biosynthesis in synchronised gametocyte cultures. Mol. Biochem. Parasitol. 20, 155–163 [DOI] [PubMed] [Google Scholar]
- 36.Dixon M. W., Thompson J., Gardiner D. L., Trenholme K. R. (2008) Sex in Plasmodium: a sign of commitment. Trends Parasitol. 24, 168–175 [DOI] [PubMed] [Google Scholar]
- 37.Spielmann T., Hawthorne P. L., Dixon M. W., Hannemann M., Klotz K., Kemp D. J., Klonis N., Tilley L., Trenholme K. R., Gardiner D. L. (2006) A cluster of ring stage-specific genes linked to a locus implicated in cytoadherence in Plasmodium falciparum codes for PEXEL-negative and PEXEL-positive proteins exported into the host cell. Mol. Biol. Cell 17, 3613–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Botha M., Pesce E. R., Blatch G. L. (2007) The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: regulating chaperone power in the parasite and the host. Int. J. Biochem. Cell Biol. 39, 1781–1803 [DOI] [PubMed] [Google Scholar]
- 39.Maier A. G., Rug M., O'Neill M. T., Brown M., Chakravorty S., Szestak T., Chesson J., Wu Y., Hughes K., Coppel R. L., Newbold C., Beeson J. G., Craig A., Crabb B. S., Cowman A. F. (2008) Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134, 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Day K. P., Hayward R. E., Smith D., Culvenor J. G. (1998) CD36-dependent adhesion and knob expression of the transmission stages of Plasmodium falciparum is stage specific. Mol. Biochem. Parasitol. 93, 167–177 [DOI] [PubMed] [Google Scholar]
- 41.Boddey J. A., Hodder A. N., Günther S., Gilson P. R., Patsiouras H., Kapp E. A., Pearce J. A., de Koning-Ward T. F., Simpson R. J., Crabb B. S., Cowman A. F. (2010) An aspartyl protease directs malaria effector proteins to the host cell. Nature 463, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russo I., Babbitt S., Muralidharan V., Butler T., Oksman A., Goldberg D. E. (2010) Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature 463, 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Koning-Ward T. F., Gilson P. R., Boddey J. A., Rug M., Smith B. J., Papenfuss A. T., Sanders P. R., Lundie R. J., Maier A. G., Cowman A. F., Crabb B. S. (2009) A newly discovered protein export machine in malaria parasites. Nature 459, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boddey J. A., Moritz R. L., Simpson R. J., Cowman A. F. (2009) Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic 10, 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinden R. E. (1982) Gametocytogenesis of Plasmodium falciparum in vitro: an electron microscopic study. Parasitology 84, 1–11 [DOI] [PubMed] [Google Scholar]
- 46.Rogers N. J., Hall B. S., Obiero J., Targett G. A., Sutherland C. J. (2000) A model for sequestration of the transmission stages of Plasmodium falciparum: adhesion of gametocyte-infected erythrocytes to human bone marrow cells. Infect. Immun. 68, 3455–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.