Abstract
Cytotoxic T lymphocyte (CTL)-mediated death of virus-infected cells requires prior recognition of short viral peptide antigens that are presented by human leukocyte antigen (HLA) class I molecules on the surface of infected cells. The CTL response is critical for the clearance of human respiratory syncytial virus (HRSV) infection. Using mass spectrometry analysis of complex HLA-bound peptide pools isolated from large amounts of HRSV-infected cells, we identified nine naturally processed HLA-B27 ligands. The isolated peptides are derived from six internal, not envelope, proteins of the infective virus. The sequences of most of these ligands are not conserved between different HRSV strains, suggesting a mechanism to explain recurrent infection with virus of different HRSV antigenic subgroups. In addition, these nine ligands represent a significant fraction of the proteome of this virus, which is monitored by the same HLA class I allele. These data have implications for vaccine development as well as for analysis of the CTL response.
The recognition of short viral peptides associated with human histocompatibility complex (human leukocyte antigen (HLA)1) class I molecules on the cell surface allows cytotoxic T lymphocytes (CTLs) to recognize and kill virus-infected cells (1). These peptides are generated by proteolytic processing of newly synthesized viral proteins in the cytosol by the combined action of proteasomes, ERAAP (endoplasmic reticulum aminopeptidase associated with antigen processing), and in some cases other peptidases (2). This degradation of viral proteins generates peptides of 8–11 residues that are translocated to the endoplasmic reticulum lumen by transporters associated with antigen processing. These short peptides then assemble with the HLA class I heavy chain and β2-microglobulin. Usually, two major anchor residues in the antigenic peptide, at position 2 and the C terminus (3, 4), must be deeply accommodated into specific pockets of the antigen recognition site of the HLA class I molecule to stabilize the nascent complexes (5, 6) and allow for their subsequent transport to the cell membrane where they are exposed for CTL recognition (7).
Human respiratory syncytial virus (HRSV) (8), a member of the Paramyxoviridae family, is the single most important cause of bronchiolitis and pneumonia in infants and young children (9–11). Infections of this virus occur in people of all ages, but although usually mild infections are reported in healthy adults, HRSV poses a serious health risk in immunocompromised individuals (12, 13) and in the elderly (14, 15). The single-stranded, negative-sense RNA genome of this enveloped virus codes for 11 proteins.
Although the immune mechanism involved in HRSV disease and protection is not well understood, specific CD8+ T lymphocytes are required for the clearance of virus-infected cells (16). Previously, several HRSV epitopes restricted by different HLA class I molecules were identified using CTLs from seropositive individuals (17–21). However, these experiments were performed with synthetic peptides against individual proteins. In contrast, only one published study attempted to elucidate the nature and diversity of the possible array of HRSV ligands restricted by individual HLA molecules (22). In this study, virus-infected cells were cultured with stable, isotope-labeled amino acids, which were expected to act as anchor residues for the HLA allele of interest. The MHC molecules were then immunoprecipitated, and mass spectrometry analysis was performed. This study identified one HRSV ligand for each of the HLA-A2 and -B7 class I molecules (22). Therefore, is only one HRSV ligand restricted by a single HLA molecule exposed on the cell membrane surface as suggested by this study? Conversely, could a particular HLA molecule bind several ligands of this small virus simultaneously? To answer these questions, we compared HLA-B27 ligands isolated from large amounts of healthy or HRSV-infected cells without any methodological bias (selection of individual protein, use of HLA consensus scoring algorithms, etc.). This analysis demonstrated the existence of diverse, naturally processed HLA-B27 ligands from six different HRSV proteins in infected cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Antibodies (Abs)
B27-C1R is a transfectant (23) of the human lymphoid cell line HMy2.C1R (C1R) that expresses its endogenous HLA class I antigens at a low level (24, 25). RMA-S is a transporter associated to antigen processing (TAP)-deficient murine cell line that expresses the mouse H-2b haplotype (26). The RMA-S transfectant cells expressing HLA-B27 were described previously (27). All cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 5 μm β-mercaptoethanol. The Abs used in this study were the polyclonal FITC-anti-HRSV Ab (which recognizes HRSV F and G proteins) (Chemicon International, Single Oak Drive Temecula, CA), the monoclonal W6/32 Ab (which is specific for a monomorphic HLA-A, -B, -C determinant) (28), and the ME1 Ab (which is specific for HLA-B27, -B7, -Bw22) (29).
Synthetic Peptides
Peptides were synthesized in a peptide synthesizer (model 433A, Applied Biosystems, Foster City, CA) and purified by reverse-phase HPLC. The correct molecular mass of peptides was established with MALDI-TOF MS, and the correct composition of HRSV peptides was determined by quadrupole ion trap micro-HPLC.
Isolation of HLA-bound Peptides
HLA-bound peptides were isolated from 4 × 1010 healthy or HRSV-infected B27-C1R transfectant cells. Cells were lysed in 1% Igepal CA-630 (Sigma), 20 mm Tris/HCl buffer, 150 mm NaCl, pH 7.5 in the presence of a protease inhibitor mixture. HLA-peptide complexes were isolated by affinity chromatography of the soluble fraction with the W6/32 monoclonal Ab. HLA-bound peptides were eluted at room temperature with 0.1% aqueous TFA and concentrated with a Centricon 3 column (Amicon, Beverly, MA) as described previously (30, 31).
Electrospray Ion Trap Mass Spectrometry Analysis
Peptide mixtures recovered after the ultrafiltration step were concentrated with Micro-Tip reverse-phase columns (C18, 200 μl, Harvard Apparatus, Holliston, MA). Each C18 tip was equilibrated with 80% acetonitrile in 0.1% TFA, washed with 0.1% TFA, and then loaded with the peptide mixture. The tip was then washed with an additional volume of 0.1% TFA, and the peptides were eluted with 80% acetonitrile in 0.1% TFA. Peptide samples were then concentrated to about 18 μl using vacuum centrifugation.
Recovered HLA class I peptides were analyzed in three different HPLC runs by μLC-MS/MS using an Orbitrap XL mass spectrometer (Thermo Electron, San Jose, CA) fitted with a capillary HPLC column (Eksigent, Dublin, CA). The peptides were resolved on homemade Reprosil C18 capillary columns (75-μm inner diameter) (32) with a 7–40% acetonitrile gradient for 2 h in the presence of 0.1% formic acid. The seven most intense masses that exhibited single, double, and triple charge states were selected for fragmentation from each full mass spectrum by CID.
Database Searches
Pep-Miner (33) was used for peak list generation of the μLC-MS/MS data. The peaks were identified using multiple search engines: Pep-Miner, Proteome Discoverer 1.0 SP1 (Thermo) combining the results of Sequest 3.31 and Bioworks Browser 3.3.1 SP1 (ThermoFisher) (34), and Mascot (server 2.2, Matrix Science) (35) using the human and the virus parts of the NCBI database (June 2008) including 574,003 proteins. The search was not limited by enzymatic specificity, the peptide tolerance was set to 0.01 Da, and the fragment ion tolerance was set to 0.5 Da. This search was not limited by any restriction bias (selection of individual protein, use of HLA consensus scoring algorithms, etc.). Identified peptides were selected if the following criteria exist: Pep-Miner score above 75, Mascot score above 20, Sequest Xcorr above 2, P(pep) less than 1 × 10−4 with Bioworks Browser, Proteome Discoverer score higher than 20, and mass accuracy of 0.005 Da (see Table I). When the MS/MS spectra fitted more than one peptide, only the highest scoring peptide was analyzed.
Table I. Summary of HRSV ligands detected by MS/MS analysis.
| Nominal massa | Experimental massa | ΔMassb | m/z | Sequencec | Protein | Position |
|---|---|---|---|---|---|---|
| 614.299 | 614.297 | 0.002 | 2+ | HRQDINGKEM | Nucleoprotein | 100–109 |
| 409.868 | 409.868 | 0.000 | 3+ | HRQDINGKEM | Nucleoprotein | 100–109 |
| 672.862 | 672.858 | 0.004 | 2+ | RRANNVLKNEM | Nucleoprotein | 184–194 |
| 444.278 | 444.277 | 0.001 | 3+ | KRYKGLLPKDI | Nucleoprotein | 195–205 |
| 488.766 | 488.764 | 0.002 | 2+ | SRSALLAQM | Matrix | 76–84 |
| 536.809 | 536.807 | 0.002 | 2+ | VRNKDLNTL | Matrix | 169–177 |
| 525.770 | 525.769 | 0.001 | 2+ | GRNEVFSNK | Polymerase | 2089–2097 |
| 389.924 | 389.922 | 0.002 | 3+ | KRLPADVLKK | Matrix 2-22k | 150–159 |
| 445.563 | 445.562 | 0.001 | 3+ | LRNEESEKMAK | Phosphoprotein | 198–208 |
| 404.893 | 404.891 | 0.002 | 3+ | HRFIYLINH | Non-structural protein 2 | 37–45 |
a Mass of monoisotopic ion in amu.
b Difference between nominal and experimentally detected monoisotopic ions.
c The HLA-B27 anchor motifs are underlined.
Electrospray Ion Trap Mass Spectrometry Analysis of Synthetic peptides
In addition, the corresponding synthetic peptide was made, and its manually identified MS/MS spectrum was used to confirm the assigned sequence of the HRSV ligand. Sequencing of the synthetic peptides was carried out by quadrupole ion trap electrospray MS/MS in a Deca XP LCQ instrument (Thermo Electron) coupled to μLC (Thermo Electron) with a 7–40% acetonitrile gradient for 24 min in the presence of 0.5% acetic acid. An MS/MS mode was used that focused on each hypothetical parental peptide with an isolation width (m/z) of 1.5 Da (36). The charge and mass of the ionic species were determined by high resolution sampling of the mass/charge rank. Collision energy and ion precursor resolution were improved to optimize the fragmentation spectrum.
MHC-Peptide Complex Stability Assays
The following synthetic peptides were used as controls in complex stability assays: Flu NP (SRYWAIRTR, HLA-B27-restricted) (37) and C4CON (QYDDAVYLK, HLA-Cw4-restricted) (38). RMA-S B27 transfectants, a cell line deficient in TAP that expresses low amounts of MHC class I on the cell surface (27), were incubated at 26 °C for 16 h in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum. This allows the expression on the cellular membrane of empty MHC class I molecules (without antigenic peptide) that are stable only at 26 °C but not at 37 °C. Later, the cells were washed and incubated for 2 h at 26 °C with various concentrations of peptide in the same medium. Then, the MHC-specific peptides could bind and stabilize the empty MHC class I molecules. Later, the cells were transferred to 37 °C and collected for flow cytometry after 4 h. This allows the internalization of empty MHC class I molecules and thus can discriminate between bound and unbound peptides. MHC expression was measured using 100 μl of a hybridoma culture supernatant containing the ME1 (anti-HLA-B27) monoclonal Ab as described (39). Samples were acquired on a FACSCalibur flow cytometer (BD Biosciences) and analyzed using CellQuest Pro 2.0 software (BD Biosciences). Cells incubated without peptide had peak fluorescence intensities close to background staining with secondary Ab alone. The fluorescence index was calculated for each time point as the ratio of peak channel fluorescence of the sample to that of the control incubated without peptide. Binding of peptides was expressed as EC50, which is the molar concentration of the peptide at 50% of the maximum fluorescence obtained at a concentration range of 100–0.001 μm.
RESULTS AND DISCUSSION
Nine Viral HLA Ligands Were Differentially Detected in HRSV-infected Cells
B27-C1R cells were incubated with the Long strain of HRSV and assayed at different times for the presence of HRSV antigens by flow cytometry. The results indicate that the transfectant cell line incubated with the virus, but not the mock-infected control, expressed HRSV F and G proteins (110 ± 10 mean fluorescence intensity versus 4 ± 3 for the mock control cells). These cells continued to synthesize HRSV viral proteins and secrete infectious virus several months after infection (data not shown). Thus, a B27-C1R transfectant cell line persistently infected with HRSV was obtained in the same manner as previously reported for Epstein-Barr virus-transformed human B-cell lines (40).
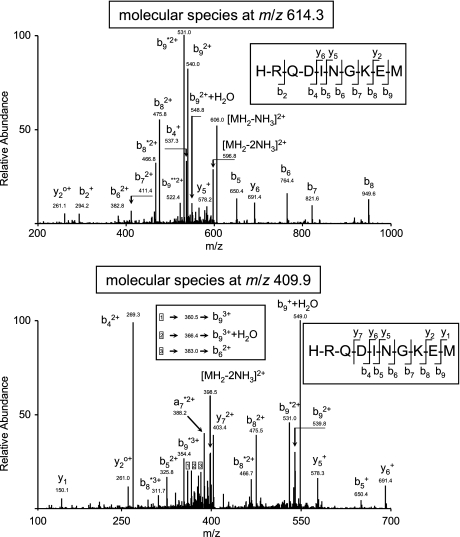
HLA-bound peptide pools were isolated from large amounts of either healthy or HRSV-infected cells. These peptide mixtures were subsequently separated by reverse-phase HPLC and analyzed by mass spectrometry. Using several software technologies (see “Experimental Procedures”), 209 fragmentation spectra were resolved as peptidic sequences of different human cellular proteins.2 Moreover, 10 fragmentation spectra present in the HRSV-infected HLA-bound peptidic pool, but absent in the control uninfected pool (data not shown), were resolved with high confidence parameters as peptides of HRSV viral proteins. Additionally, a human proteome database search failed to reveal the identity of these spectra as human protein fragments, confirming the viral origin of these peptides (data not shown). Two different ion peaks at m/z 409.9 and 614.3 were assigned to the same viral amino acid sequence. These ion peaks corresponded to double (Fig. 1, upper panel) and triple charge (Fig. 1, lower panel) states, respectively, of the peptide HRQDINGKEM, which spans residues 100–109 of the HRSV nucleoprotein (Table I). Virtually all significant fragments of both MS/MS spectra were assigned as daughter ions of the tentative peptidic sequence (Fig. 1). This theoretical assignment was confirmed by identity with the MS/MS spectrum of the corresponding synthetic peptide (supplemental Fig. 1). In addition, the eight other molecular ions were assigned as HLA-restricted viral ligands (Table I and supplemental Figs. 2–9), and their tentative sequences were confirmed as above with the respective synthetic peptide (supplemental Figs. 2–9). Thus, these results indicate that a total of nine HRSV ligands were endogenously processed and presented in the infected cell line.
Fig. 1.
Identification of the N 100–109 ligand in infected cell extracts by mass spectrometry. MS/MS fragmentation spectra obtained from quadrupole ion trap mass spectrometry of the ion peaks at m/z 614.3 (upper panel) and m/z 409.9 (lower panel) from the HRSV-infected B27-C1R cell extract are shown. The vertical axis represents the relative abundance of the parental ion and each fragmentation ion detected. The horizontal axis corresponds to the m/z region in which significant daughter ions were detected. Ions generated in the fragmentation are detailed, and the sequence deduced from the indicated fragments is shown in the upper right box of each panel.
Identification of viral HLA ligands by immunoproteomics analysis is still very limited. A previous study identified 12 viral ligands presented by HLA-A*0201 by differential stable isotope labeling of large amounts of vaccinia virus Ankara-infected cells (41). In another study with a similar approach but 10-fold fewer infected cells, only one HRSV ligand was identified for each HLA-A*0201 or -B*0702 class I molecule (22). In our current report in which the amount of cells used was similar to that in the vaccinia virus study and the peptide pools were compared directly without any labeling, nine endogenously processed HRSV HLA ligands were found. Thus, our current report shows a similar number of HLA ligands between viruses that differ 13-fold in their respective proteome sizes. Therefore, large scale immunoproteomics could allow for the systematic identification of the array of viral HLA ligands.
HRSV-specific Ligands Efficiently Bind to B*2705 Molecule
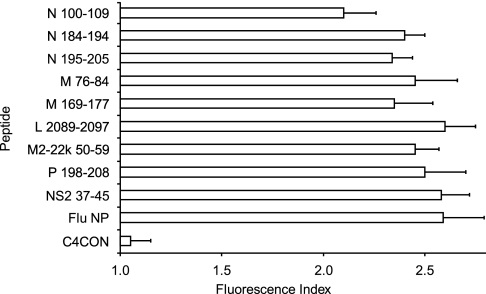
The classical anchor motifs for HLA-B*2705 binding, Arg at position 2 (P2) and basic or aliphatic C-terminal residues (SYFPEITHI database (4)), were present in all detected viral ligands (Table I). To confirm that HLA-B*2705 is the MHC class I molecule that presents these ligands, MHC-peptide complex stability assays were performed using TAP-deficient RMA-S cells transfected with the HLA-B*2705 molecule. The nine HRSV synthetic peptides induced similar numbers of HLA-peptide surface complexes to a well known HLA-B*2705 epitope from the influenza virus (Fig. 2). In addition, the relative MHC class I affinity was determined for all HRSV peptides. These peptides bound to HLA-B*2705 class I molecules with EC50 values in the range commonly found among natural ligands (Table II). These data indicate that all ligands detected in HRSV-infected cells were endogenously presented in association with the B*2705 molecule.
Fig. 2.
HLA stabilization assay of HRSV synthetic ligands. The stability of HLA-B*2705-peptide complexes on the surface of RMA-S transfectant cells was measured by flow cytometry. The indicated peptides were used at 200 μm. The C4CON (38) and Flu NP (37) peptides were used as negative and positive controls, respectively. The results, calculated as fluorescence index (see “Experimental Procedures”), are the mean (bars) ± S.D. (error bars) of three to four independent experiments.
Table II. Summary of binding of HLA-B27 ligands of HRSV.
| Peptidea | Sequence | EC50 ± S.D.b |
|---|---|---|
| μm | ||
| N 100–109 | HRQDINGKEM | 26 ± 5 |
| N 184–194 | RRANNVLKNEM | 10 ± 1 |
| N 195–205 | KRYKGLLPKDI | 5 ± 2 |
| M 76–84 | SRSALLAQM | 9 ± 2 |
| M 169–177 | VRNKDLNTL | 12 ± 5 |
| L 2089–2097 | GRNEVFSNK | 18 ± 3 |
| M2–22k 150–159 | KRLPADVLKK | 9 ± 2 |
| P 198–208 | LRNEESEKMAK | 14 ± 3 |
| NS2 37–45 | HRFIYLINH | 11 ± 4 |
| Flu NP | SRYWAIRTR | 8 ± 2 |
a All peptides were derived from the sequence of the Long strain of HRSV except for Flu NP 383–391 (37), which was used as positive control for HLA-B27 binding.
b Data are means of three to five independent experiments and are expressed as EC50 ± S.D. (see “Experimental Procedures”).
Among the nine HRSV peptides discovered by the proteomics analysis, four (matrix 76–84-, matrix 169–177-, polymerase-, and non-structural protein 2-derived peptides) were nonamers. In addition, two (matrix 2-22k and nucleoprotein 100–109 ligands) were decamers, and the other three (phosphoprotein, nucleoprotein 184–194, and nucleoprotein 195–205) were undecamers. No correlation was found between the length of the peptides and their affinity for the HLA-B*2705 molecule (Table II). Of the 239 B*2705-bound cellular ligands previously sequenced (SYFPEITHI database), 4% were octamers, 65% were nonamers or decamers, and 11% were undecamers or longer peptides with both HLA-B*2705 anchor motifs. Thus, this HLA molecule could easily accommodate bulged peptides. The results shown here (Table I) indicate that bulged viral ligands could be presented by the B*2705 molecule.
Internal Protein, but Not F Protein, Ligands Are Presented by HLA-B*2705 in HRSV-infected Cells
Eleven proteins are encoded by the HRSV genome (8). In general, the larger proteins in this proteome contain more Arg residues with the exception of the G protein (Table III) and thus are candidates for the source of HLA-B27-restricted epitopes. All nine identified HLA-B*2705 ligands are derived from viral proteins with a significant Arg content in their sequences with the one exception. The F protein is the second largest protein in the HRSV proteome and is also the second most Arg-enriched protein of HRSV, containing 18 Arg residues (Table III). The epitope prediction for HLA-B*2705 binding using various web software tools showed HLA-B27 peptide scores similar to those for the binding of other HRSV proteins with HLA-B*2705 ligands (data not shown). In addition to the fusion protein F, the HRSV envelope contains two other transmembrane surface glycoproteins, the attachment protein G and the small hydrophobic (SH) protein (8). Both G and SH proteins contain only two Arg in their amino acid sequences; these residues are needed to anchor peptides to the HLA-B27 molecule. Thus, the lack of HLA-B27 ligands in these envelope proteins could be expected. In summary, the absence of F and, to a lesser extent, G and SH protein ligands indicates that HLA-B*2705 exclusively sampled internal HRSV viral proteins. This lack of envelope protein ligands was also recently described for influenza epitopes restricted by the HLA-B*0702 allele (42). Therefore, for some pairs of HLA and virus, only internal ligands are found associated to class I molecules.
Table III. Distribution of Arg content and HLA-B27 ligands in HRSV proteome.
| Long strain proteina | Number of Argb | Percentage of Argc | Number of residuesd | Percentage of proteomee | Ligandsf |
|---|---|---|---|---|---|
| L | 81 | 51.6 | 2165 | 49.2 | 1 |
| F | 18 | 11.5 | 573 | 13.0 | 0 |
| N | 16 | 10.2 | 391 | 8.9 | 3 |
| P | 11 | 7.0 | 219 | 5.0 | 1 |
| M2-22k | 11 | 7.0 | 195 | 4.4 | 1 |
| M | 6 | 3.8 | 256 | 5.8 | 2 |
| NS2 | 6 | 3.8 | 124 | 2.8 | 1 |
| M2-2 | 3 | 1.9 | 90 | 2.0 | 0 |
| G | 2 | 1.3 | 186 | 4.2 | 0 |
| SH | 2 | 1.3 | 65 | 1.5 | 0 |
| NS1 | 1 | 0.6 | 138 | 3.1 | 0 |
a The abbreviations used are: L, polymerase; F, fusion protein; N, nucleoprotein; P, phosphoprotein; M2-22k, matrix protein 22k; M, matrix protein; NS2, non-structural protein 2; M2-2, matrix protein 2; G, attachment protein; SH, small hydrophobic protein; and NS1, non-structural protein 1.
b Number of Arg in the viral proteome.
c Percentage of Arg included in the viral proteome .
d Total number of residues.
e Number of residues of each protein/number of total residues as a percentage.
f Number of HLA-B27 ligands in HRSV.
Most of HLA-B*2705 Ligands Are Not Conserved between HRSV Antigenic Subgroups
HRSV exists as a single serotype but has two antigenic subgroups, A and B (8). Most proteins are highly conserved between these subgroups (88–96% amino acid identity). Greater differences are found in M2-2, G, and SH proteins (61–71% amino acid identity). Within subgroups, the percentage of nucleotide and amino acid identity between viruses is much higher for all proteins (97–100% amino acid identity). In addition, mutations that enable escape from host immunity do not appear to accumulate with time (8). Thus, it would be reasonable to infer the identity of HLA-B*2705-restricted ligands between strains of different subgroups. Therefore, amino acid sequence comparison was performed for the ligands identified in the Long strain with two or three representative strains of subgroups A and B, respectively. Table IV shows that two-thirds of the ligands detected in the Long strain are mutated in all of the subgroup B strains sequenced. N 195–205 and M 76–84 ligands had mutations in T cell receptor-interacting residues, whereas N 100–109 and N 184–194 were mutated at the N- and C-terminal anchor residues, respectively. The sequences of the B subgroup present alterations in the P2 anchor motif as well as a change in the sequence of both NS2 37–45 and M 169–177 ligands. Only P 198–208, M2-22k 150–159, and L 2089–2097 peptides were conserved in all strains studied. Two ligands were not conserved across subgroup A strains: Arg was changed to Lys in the NS2 37–45 ligand of the A2 strain, and a Val → Leu substitution was detected in both S2 and A2 strains. In addition, a substitution at position 80 is the only change in the M protein sequence of the Long and A2 strains. Similarly, only residues 8, 26, and 38 were altered in the NS2 protein across different strains of this subgroup. In summary, HLA-B*2705 mainly binds non-conserved peptides of the Long strain of HRSV compared with other strains in accordance with the percentage of amino acid identity of proteins of antigenic subgroups of this virus. In addition to evidence that CD8+ T cells are partially functionally inactivated in the respiratory syncytial virus-infected respiratory tract in model mice (43), the surface expression of non-conserved HRSV ligands could explain the mechanism of reinfection with different HRSV antigenic subgroups (44).
Table IV. Conservation of HLA-B27 viral ligands in several HRSV strains.
| Straina | N 195–205 | M 76–84 | N 100–109 | N 184–194 | NS2 37–45 | M 169–177 | P 198–208 | M2–22k 150–159 | L 2089–2097 |
|---|---|---|---|---|---|---|---|---|---|
| Long | KRYKGLLPKDI | SRSALLAQM | HRQDINGKEM | RRANNVLKNEM | HRFIYLINH | VRNKDLNTL | LRNEESEKMAK | KRLPADVLKK | GRNEVFSNK |
| S2 | ----------- | ----V---- | ---------- | ----------- | --------- | --------- | ----------- | ---------- | --------- |
| A2 | ----------- | ----V---- | ---------- | ----------- | -K------- | --------- | ----------- | ---------- | --------- |
| 9320 | ------I---- | ----V---- | Y--------- | ----------I | -K------N | -K-----S- | ----------- | ---------- | --------- |
| 18537 | ------I---- | ----V---- | Y--------- | ----------I | -K------N | -K-----S- | ----------- | ---------- | --------- |
| B1 | ------I---- | ----V---- | Y--------- | ----------I | -K------N | -K-----S- | ----------- | ---------- | --------- |
a Long, S2, and A2 strains are representative of HRSV subtype A, whereas 9320, 18537, and B1 are HRSV subtype B strains.
Significant Fraction of Viral Proteome Was Monitored in Association to HLA-B*2705 Class I Molecule
The proteome of the Long strain of HRSV contains 4402 residues; 157 of them are Arg. Thus, the nine detected ligands represent 2% of the proteome and 6% of the possible B*2705-restricted ligands (Table V). In other RNA viruses with similar proteome sizes and Arg content, about 6–13 B*2705-restricted epitopes were identified (summarized in the Immune Epitope Database (45)). As shown in Table V, the percentages of “immune coverage” are similar in these RNA viruses: around 2–4% of the total proteome and 2–7% of the maximum number of potential HLA-B*2705 ligands. If these data are typical for all HLA class I molecules, the cellular immune response would monitor 14–25% of the proteome of these viral pathogens in heterozygous HLA-A, -B, and -C infected individuals. In a study similar to our report, 12 ligands of vaccinia virus restricted by HLA-A*0201 and three restricted by HLA-B*0702 were identified (41). Namely, around eight ligands were identified per HLA studied, very close to the nine detected in our report. As both viruses have a similar ligand/HLA ratio but very different proteome sizes, although different studies are not always comparable, these data suggest that the cellular immune pressure could be higher in some small viruses.
Table V. Proteome size, Arg content, and HLA-B27-restricted epitope/ligands in several RNA viruses.
| Virusa | Proteome |
Arg |
Epitopesf | ||
|---|---|---|---|---|---|
| Residuesb | %c | Numberd | %e | ||
| HRSV | 4402 | 2 | 157 | 6 | 9 |
| Influenza | 4112 | 2 | 284 | 2 | 6 |
| HIV | 3151 | 4 | 184 | 7 | 13 |
a The sequences used were Long strain (HRSV), A/Puerto Rico/8/34 strain (influenza), and human immunodeficiency virus (HIV) clade B consensus (Los Alamos National Laboratory, National Institutes of Health).
b Total number of residues.
c Percentage of proteome included in the detected epitopes/ligands.
d Number of Arg in each viral proteome.
e Number of epitopes or ligands/number of total Arg as a percentage.
f Number of ligands in HRSV.
In summary, the present report demonstrates that the endogenous processing of HRSV proteins generates multiple different peptidic species that are bound to the HLA-B27 molecule in infected cells. These ligands, identified by mass spectrometry of intricate cellular extracts, induce high numbers of HLA-peptide complexes, and their respective MHC class I affinities are similar to those of other natural HLA-B27 ligands. All nine peptides are derived from internal proteins of the infective virus, and the sequences of most of the peptides are not conserved between the two HRSV antigenic subgroups described above. Lastly, these nine ligands represent a significant fraction of the proteome of this virus monitored by the same MHC class I allele.
Supplementary Material
Acknowledgments
We thank Dr. J. A. López de Castro (Centro de Biología Molecular Severo Ochoa, Madrid, Spain) for the cell lines.
* This work was supported by grants from the Programa Ramón y Cajal and the Fondo de Investigaciones Sanitarias de la Seguridad Social (to D. L.) and Israel Science Foundation Grant 916/05 (to A. A.).
 This article contains supplemental Figs. 1–9.
This article contains supplemental Figs. 1–9.
2 S. Infantes, E. Lorente, E. Barnea, I. Beer, J. J. Cragnolini, R. García, F. Lasala, M. Jiménez, A. Admon, and D. López, manuscript in preparation.
1 The abbreviations used are:
- HLA
- human leukocyte antigen
- Ab
- antibody
- B27-C1R
- HMy2.C1R transfected with HLA-B*2705
- CTL
- cytotoxic T lymphocyte
- EC50
- half-maximal effective concentration
- HRSV
- human respiratory syncytial virus
- MHC
- major histocompatibility complex
- TAP
- transporter associated to antigen processing
- μLC
- microcapillary LC
- SH
- small hydrophobic.
REFERENCES
- 1.Shastri N., Schwab S., Serwold T. (2002) Producing nature's gene-chips: the generation of peptides for display by MHC class I molecules. Annu. Rev. Immunol. 20, 463–493 [DOI] [PubMed] [Google Scholar]
- 2.Del-Val M., López D. (2002) Multiple proteases process viral antigens for presentation by MHC class I molecules to CD8+ T lymphocytes. Mol. Immunol. 39, 235–247 [DOI] [PubMed] [Google Scholar]
- 3.Parker K. C., Bednarek M. A., Coligan J. E. (1994) Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152, 163–175 [PubMed] [Google Scholar]
- 4.Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanoviæ S. (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 [DOI] [PubMed] [Google Scholar]
- 5.Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. (1987) Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329, 506–512 [DOI] [PubMed] [Google Scholar]
- 6.Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. (1987) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329, 512–518 [DOI] [PubMed] [Google Scholar]
- 7.York I. A., Goldberg A. L., Mo X. Y., Rock K. L. (1999) Proteolysis and class I major histocompatibility complex antigen presentation. Immunol. Rev. 172, 49–66 [DOI] [PubMed] [Google Scholar]
- 8.Collins P. L., Chanock R. M., Murphy B. R. (2007) Respiratory syncytial virus, in Fields Virology (Knipe D. M., Howley P. M. eds) 5th Ed., Lippincott Williams and Wilkins, Philadelphia: 1443–1486 [Google Scholar]
- 9.Hall C. B. (2001) Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 344, 1917–1928 [DOI] [PubMed] [Google Scholar]
- 10.Shay D. K., Holman R. C., Roosevelt G. E., Clarke M. J., Anderson L. J. (2001) Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979–1997. J. Infect. Dis. 183, 16–22 [DOI] [PubMed] [Google Scholar]
- 11.Thompson W. W., Shay D. K., Weintraub E., Brammer L., Cox N., Anderson L. J., Fukuda K. (2003) Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289, 179–186 [DOI] [PubMed] [Google Scholar]
- 12.Wendt C. H., Hertz M. I. (1995) Respiratory syncytial virus and parainfluenza virus infections in the immunocompromised host. Semin. Respir. Infect. 10, 224–231 [PubMed] [Google Scholar]
- 13.Ison M. G., Hayden F. G. (2002) Viral infections in immunocompromised patients: what's new with respiratory viruses? Curr. Opin. Infect. Dis. 15, 355–367 [DOI] [PubMed] [Google Scholar]
- 14.Han L. L., Alexander J. P., Anderson L. J. (1999) Respiratory syncytial virus pneumonia among the elderly: an assessment of disease burden. J. Infect. Dis. 179, 25–30 [DOI] [PubMed] [Google Scholar]
- 15.Falsey A. R., Hennessey P. A., Formica M. A., Cox C., Walsh E. E. (2005) Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352, 1749–1759 [DOI] [PubMed] [Google Scholar]
- 16.Anderson L. J., Heilman C. A. (1995) Protective and disease-enhancing immune responses to respiratory syncytial virus. J. Infect. Dis. 171, 1–7 [DOI] [PubMed] [Google Scholar]
- 17.Brandenburg A. H., de Waal L., Timmerman H. H., Hoogerhout P., de Swart R. L., Osterhaus A. D. (2000) HLA class I-restricted cytotoxic T-cell epitopes of the respiratory syncytial virus fusion protein. J. Virol. 74, 10240–10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rock M. T., Crowe J. E., Jr. (2003) Identification of a novel human leucocyte antigen-A*01-restricted cytotoxic T-lymphocyte epitope in the respiratory syncytial virus fusion protein. Immunology 108, 474–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venter M., Rock M., Puren A. J., Tiemessen C. T., Crowe J. E., Jr. (2003) Respiratory syncytial virus nucleoprotein-specific cytotoxic T-cell epitopes in a South African population of diverse HLA types are conserved in circulating field strains. J. Virol. 77, 7319–7329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidema J., de Bree G. J., De Graaff P. M., van Maren W. W., Hoogerhout P., Out T. A., Kimpen J. L., van Bleek G. M. (2004) Human CD8(+) T cell responses against five newly identified respiratory syncytial virus-derived epitopes. J. Gen. Virol. 85, 2365–2374 [DOI] [PubMed] [Google Scholar]
- 21.Terrosi C., Di Genova G., Savellini G. G., Correale P., Blardi P., Cusi M. G. (2007) Immunological characterization of respiratory syncytial virus N protein epitopes recognized by human cytotoxic T lymphocytes. Viral Immunol. 20, 399–406 [DOI] [PubMed] [Google Scholar]
- 22.Meiring H. D., Soethout E. C., Poelen M. C., Mooibroek D., Hoogerbrugge R., Timmermans H., Boog C. J., Heck A. J., de Jong A. P., van Els C. A. (2006) Stable isotope tagging of epitopes: a highly selective strategy for the identification of major histocompatibility complex class I-associated peptides induced upon viral infection. Mol. Cell. Proteomics 5, 902–913 [DOI] [PubMed] [Google Scholar]
- 23.Calvo V., Rojo S., López D., Galocha B., López de Castro J. A. (1990) Structure and diversity of HLA-B27-specific T cell epitopes. Analysis with site-directed mutants mimicking HLA-B27 subtype polymorphism. J. Immunol. 144, 4038–4045 [PubMed] [Google Scholar]
- 24.Storkus W. J., Howell D. N., Salter R. D., Dawson J. R., Cresswell P. (1987) NK susceptibility varies inversely with target cell class I HLA antigen expression. J. Immunol. 138, 1657–1659 [PubMed] [Google Scholar]
- 25.Zemmour J., Little A. M., Schendel D. J., Parham P. (1992) The HLA-A,B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J. Immunol. 148, 1941–1948 [PubMed] [Google Scholar]
- 26.Ljunggren H. G., Kärre K. (1985) Host resistance directed selectively against H-2-deficient lymphoma variants. Analysis of the mechanism. J. Exp. Med. 162, 1745–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villadangos J. A., Galocha B., López de Castro J. A. (1994) Unusual topology of an HLA-B27 allospecific T cell epitope lacking peptide specificity. J. Immunol. 152, 2317–2323 [PubMed] [Google Scholar]
- 28.Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., Ziegler A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14, 9–20 [DOI] [PubMed] [Google Scholar]
- 29.Ellis S. A., Taylor C., McMichael A. (1982) Recognition of HLA-B27 and related antigen by a monoclonal antibody. Hum. Immunol. 5, 49–59 [DOI] [PubMed] [Google Scholar]
- 30.Cragnolini J. J., López de Castro J. A. (2008) Identification of endogenously presented peptides from Chlamydia trachomatis with high homology to human proteins and to a natural self-ligand of HLA-B27. Mol. Cell. Proteomics 7, 170–180 [DOI] [PubMed] [Google Scholar]
- 31.Cragnolini J. J., Garcia-Medel N., López de Castro J. A. (2009) Endogenous processing and presentation of T-cell Epitopes from chlamydia trachomatis with relevance in HLA-B27-associated reactive arthritis. Mol. Cell. Proteomics 8, 1850–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 33.Beer I., Barnea E., Ziv T., Admon A. (2004) Improving large-scale proteomics by clustering of mass spectrometry data. Proteomics 4, 950–960 [DOI] [PubMed] [Google Scholar]
- 34.Eng J., McCormack A., Yates J. (2009) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 35.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 36.López D., Calero O., Jiménez M., García-Calvo M., Del Val M. (2006) Antigen processing of a short viral antigen by proteasomes. J. Biol. Chem. 281, 30315–30318 [DOI] [PubMed] [Google Scholar]
- 37.Wang M., Lamberth K., Harndahl M., Røder G., Stryhn A., Larsen M. V., Nielsen M., Lundegaard C., Tang S. T., Dziegiel M. H., Rosenkvist J., Pedersen A. E., Buus S., Claesson M. H., Lund O. (2007) CTL epitopes for influenza A including the H5N1 bird flu; genome-, pathogen-, and HLA-wide screening. Vaccine 25, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 38.Fan Q. R., Garboczi D. N., Winter C. C., Wagtmann N., Long E. O., Wiley D. C. (1996) Direct binding of a soluble natural killer cell inhibitory receptor to a soluble human leukocyte antigen-CW4 class I major histocompatibility complex molecule. Proc. Natl. Acad. Sci. U.S.A. 93, 7178–7183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.López D., Samino Y., Koszinowski U. H., Del Val M. (2001) HIV envelope protein inhibits MHC class I presentation of a cytomegalovirus protective epitope. J. Immunol. 167, 4238–4244 [DOI] [PubMed] [Google Scholar]
- 40.Bangham C. R., McMichael A. J. (1986) Specific human cytotoxic T cells recognize B-cell lines persistently infected with respiratory syncytial virus. Proc. Natl. Acad. Sci. U.S.A. 83, 9183–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer V. S., Kastenmuller W., Gasteiger G., Franz-Wachtel M., Lamkemeyer T., Rammensee H. G., Stevanovic S., Sigurdardottir D., Drexler I. (2008) Long-term immunity against actual poxviral HLA ligands as identified by differential stable isotope labeling. J. Immunol. 181, 6371–6383 [DOI] [PubMed] [Google Scholar]
- 42.Wahl A., Schafer F., Bardet W., Buchli R., Air G. M., Hildebrand W. H. (2009) HLA class I molecules consistently present internal influenza epitopes. Proc. Natl. Acad. Sci. U.S.A. 106, 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang J., Braciale T. J. (2002) Respiratory syncytial virus infection suppresses lung CD8+ T-cell effector activity and peripheral CD8+ T-cell memory in the respiratory tract. Nat. Med. 8, 54–60 [DOI] [PubMed] [Google Scholar]
- 44.Mufson M. A., Belshe R. B., Orvell C., Norrby E. (1987) Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J. Clin. Microbiol. 25, 1535–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters B., Sidney J., Bourne P., Bui H. H., Buus S., Doh G., Fleri W., Kronenberg M., Kubo R., Lund O., Nemazee D., Ponomarenko J. V., Sathiamurthy M., Schoenberger S., Stewart S., Surko P., Way S., Wilson S., Sette A. (2005) The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 3, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.