Abstract
The estrogen receptors (ERs) are ligand-dependent transcription factors that activate transcription by binding to estrogen response elements. Estrogen-mediated effects are tissue- and cell type-specific, determined by the cofactor recruitment to the ERs among other factors. To understand these differences in estrogen action, it is important to identify the various compositions of the ER complexes (ER receptosomes). In this report, we describe a fast and efficient method for the isolation of the ERα receptosome for proteomics analysis. Using immobilized estrogen response element on a Sepharose column in combination with two-dimensional electrophoresis and MALDI-TOF MS, significant amounts of proteins could be isolated and identified. Differences in ERα complex composition with the ER ligands 17β-estradiol, 4-hydroxytamoxifen, and ICI-182,780 could also be observed. Thus, this approach provides an easy and relevant way of identifying ERα cofactor and transcription factor recruitment under different conditions.
The steroid hormone 17β-estradiol (E2)1 plays important roles in numerous physiological processes such as growth, differentiation, function of the male and female reproductive systems, and maintenance of bone mass (1). In addition, E2 is thought to have protective effects in cardiovascular and neurodegenerative diseases (2, 3). However, E2 also represents a risk factor for the development of breast and endometrial cancers. The complex E2 signaling pathways giving rise to the diverse effects of E2 are mainly mediated through the estrogen receptors (ERs). The ERs belong to the superfamily of nuclear receptor (NR) transcription factors. Upon binding of ligand (e.g. E2), the ERs change conformation, leading to dimerization and recruitment of cofactors. This complex binds with very high affinity to estrogen response elements (EREs) in promoter regions of E2-responsive genes, which in turn leads to gene transcription. Two subtypes of estrogen receptors exist, ERα and ERβ, and although they share high sequence homology and E2 affinities and both bind to ERE sequences, they have radically different, sometimes opposing, effects in different tissues (4). In general, ERα is thought to promote proliferation, whereas ERβ is thought to promote differentiation (5). Furthermore, effects of various ligands appear to differ between different tissues, both normal and pathological. For example, the general ER antagonist 4-hydroxytamoxifen (4-OHT) displays antagonistic effects on ERα in some tissues and is thus used as a potent therapy for estrogen-dependent breast cancers, whereas in other tissues, it displays agonistic effects (6). To add to the complexity, many breast cancers develop resistance toward 4-OHT via unknown mechanisms (6, 7). The molecular mechanisms underlying the effects of ERs in different tissues upon ligand exposure are far from understood. However, the release of some and recruitment of other coregulatory proteins by the ERs is thought to play a major role. Hence, the investigation of the composition of the DNA-bound ER protein complex (the ER receptosome) is of great importance.
Several attempts to isolate nuclear receptor complexes have been made, and much useful information has been generated regarding complex compositions (8–13). However, the methods used often rely on laborious prefractionations and enormous amounts of starting material, making these methods inconvenient for efficient analysis of nuclear receptor complexes. In addition, ligand effects are either not addressed or cannot be addressed using previous methods. As mentioned above, upon agonist ligand exposure, the active ER complexes bind to the ERE sequence regions with exceptionally high affinity (Kd of 0.5 nm) (14, 15). This implies that ER receptosomes, at least in their active form, need to be isolated bound to DNA. Because stringent purification procedures often include several steps and thus take time, many relevant proteins are lost when using techniques like classical tandem affinity purification tagging or immunoprecipitations, yielding too low amounts of isolated proteins for efficient MS identification. Thus, a method for fast, stringent, and reproducible nuclear receptor protein complex isolation and identification would greatly benefit the understanding of nuclear receptor function and endocrine signaling pathways.
Here we report an effective method for the isolation of DNA-bound ERα complexes in the presence of the ligands E2, 4-OHT, and ICI-182,780. The method used relies on a single step stringent purification of proteins bound to a 9×ERE oligonucleotide immobilized on Sepharose beads. Although DNA affinity chromatography of ERα has been reported previously (16), we expanded the method for proteomics analysis of ligand-bound ERα complexes. This method was very efficient in isolating protein complexes bound to the ERE oligonucleotide and showed a high preference for immobilizing ERα. Surprisingly, low amounts of starting material yielded enough purified proteins for MS analysis. Hence, this technique proved to be a fast and efficient method for ERα complex isolation that can be used for studying ligand-specific effects on the ER receptosome.
EXPERIMENTAL PROCEDURES
Construction of ERE Columns
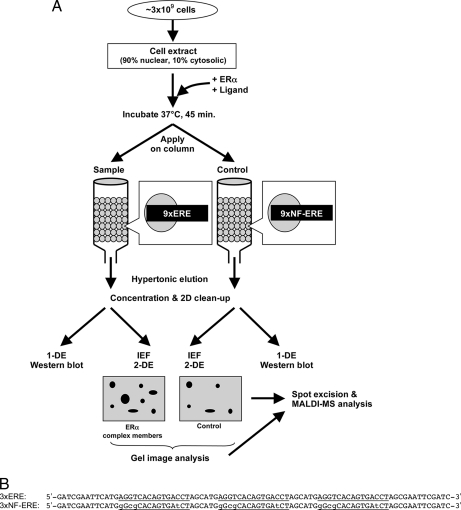
80 pmol each of the oligonucleotides representing either a functional 3×ERE (ERE-5′-2, “GATCGAATTCATGAGGTCACAGTGACCTAGCATGAGGTCACAGTGACCTAGCATGAGGTCACAGTGACCTAGCGAATTCGATC”, ERE-3′-2, “GATCGAATTCGCTAGGTCACTGTGACCTCATGCTAGGTCACTGTGACCTCATGCTAGGTCACTGTGACCTCATGAATTCGATC”) or a non-functional (NF; control) 3×ERE (ERE-5′-Mut, “GATCGAATTCATGGGCGCACAGTGATCTAGCATGGGCGCACAGTGATCTAGCATGGGCGCACAGTGATCTAGCGAATTCGATC”; ERE-3′-Mut, “GATCGAATTCGCTAGATCACTGTGCGCCCATGCTAGATCACTGTGCGCCCATGCTAGATCACTGTGCGCCCATGATTCGATC”), all containing flanking EcoRI restriction sites, were annealed and digested with EcoRI. The digested double-stranded 3×ERE oligonucleotides were then allowed to ligate with each other for 20 min at room temperature and then ligated into an EcoRI-digested and dephosphorylated pCDNA3 vector (Invitrogen) at +14 °C overnight. The vector constructs were then propagated in XL1(blue) bacteria, extracted, purified, and sequenced. A construct with the 3 × 3×ERE (9×ERE) sequence and another one with the 3 × 3×NF-ERE (9×NF-ERE) sequence were used as templates for PCR amplification using 5 nmol of each of the primers (pCDNA3-FW-ERE, “ATAGGGAGACCCAAGCTTGG”; pCDNA3-Rev-ERE, “GACACTATAGAATAGGGCC”), yielding 202-bp oligonucleotides. After the PCR that was carried out for 47 cycles, the oligonucleotides were sodium acetate/ethanol-precipitated, dried, and resuspended in 1 ml of coupling buffer (0.2 m NaHCO3, 0.5 m NaCl, pH 8.3). The amino groups of the nucleotide bases were used for linking the EREs to a HiTrap NHS column (GE Healthcare). Although it is common to use 5′-aminated sequences for this linking reaction to the Sepharose, we noticed that the efficiency of the reaction was equal or better when using non-5′-aminated nucleotides, utilizing the amino groups of the nucleotide bases to form the amide bond to the NHS-Sepharose (data not shown). Approximately 2 nmol of 9×ERE sequences were immobilized on the column following the protocol of the column supplier (GE Healthcare), yielding a theoretical binding capacity of 18 nmol of ERα protein complexes, supposedly sufficient for proper identification. The coupling was stopped, and the columns were washed by flushing with alternating volumes of 0.5 m ethanolamine, 0.5 m NaCl, pH 8.3 and 0.1 m sodium acetate, 0.5 m NaCl, pH 4. Unless directly used, the deactivated columns could be stored in 10% ethanol at +4 °C.
Cell Culturing
The human mammary carcinoma cell line MCF-7 (ATCC) was cultured in Dulbecco's modified Eagle's medium containing 1 mg/ml d-glucose and supplemented with 0.3 mg/ml l-glutamine and 10% FCS (Invitrogen). To increase E2 sensitivity and endogenous ERα expression and suppress E2-mediated complex formation (17), the medium was exchanged to medium without phenol red supplemented with 0.3 mg/ml l-glutamine (Invitrogen) and 5% dextran-charcoal-treated FCS (Hyclone) 48 h prior to harvest. All cell culturing was performed at 37 °C at 5% CO2 in a humidified incubator, and the medium was changed every 2–3 days.
Isolation of ERα Complexes
Approximately 3 × 109 MCF-7 cells were used for each purification procedure. The cells were washed twice in 1× PBS and then collected in 40 ml of ice-cold cell collection buffer (200 mm Tris-HCl, 10 mm DTT, pH 9.4). The cells were sedimented, resuspended in 600 μl of a hypotonic buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1× protease inhibitor mixture), homogenized with 20 strokes using a Dounce glass-glass homogenizer (B-pestle), and sedimented at 4000 × g for 15 min at +4 °C. The cytosol (supernatant) was saved, and the pellet was resuspended in 500 μl of 200 mm Tris-HCl, pH 7.4, 1.5 mm MgCl2, 25% (v/v) glycerol, 1× protease inhibitor mixture. 10% (v/v) of the cytosolic fraction and 10 μg of salmon sperm DNA (Sigma) were added to this nuclear extract, and the sample was sonicated in a +4 °C waterbath for 15 min. Thereafter, 20 μg of recombinant ERα (Invitrogen) and 100 nm ligand (E2, 4-OHT, or ICI-182,780; all from Sigma) were added to the extract and incubated at 37 °C for 45 min. Insoluble cell debris were sedimented, and the supernatant was split into two samples, one applied to a 1-ml HiTrap NHS column (GE Healthcare) with covalently immobilized 9×ERE sequences and the other applied to a 1-ml HiTrap NHS column with immobilized 9×NF-ERE sequences. The flow rate was set to 0.2 ml/min using 10 mm Tris-HCl, pH 7.4 as mobile phase, and the samples were reapplied twice. The columns were washed with 10 mm NaCl, 10 mm Tris-HCl, pH 7.4 for 10 column volumes at 0.5 ml/min and eluted with 1 m NaCl, 10 mm Tris-HCl, pH 7.4 at 0.5 ml/min. After ∼4 ml, all proteins were eluted and concentrated to 250 μl using Amicon ultraconcentration filter devices (Millipore). Thereafter, protein concentration was measured. All steps were performed at +4 °C.
Western Blot Analysis
15% of the volume of concentrated eluted proteins (∼40 μg; except for control sample, which had a lower protein concentration, ∼15 μg) was denatured, separated on a 4–20% gradient Tris-glycine one-dimensional gel (Invitrogen) by SDS-PAGE, and electrotransferred onto a nitrocellulose membrane (GE Healthcare). The membrane was blocked using 10% (w/v) fat-free milk powder in 1× PBS supplemented with 0.1% (v/v) Tween 20 (PBS-T). Primary antibodies were rabbit anti-human ERα (sc-543, HC-20, Santa Cruz Biotechnology) diluted 1:1000, chicken anti-human ERβ antibody (ab14021, Abcam) diluted 1:700, mouse anti-human p300 (RW128, Millipore) diluted 1:300, mouse anti-human RNA polymerase II (Pol-II) (610985, BD Biosciences) diluted 1:300, mouse anti-human histone deacetylase 3 (HDAC3) (sc-17795, B-12, Santa Cruz Biotechnology) diluted 1:300, mouse anti-human Hsp70 (sc-24, W27, Santa Cruz Biotechnology) diluted 1:2000, rabbit anti-human splicing factor proline/glutamine-rich (SFPQ) (ab38148, Abcam) diluted 1:600, goat anti-human 100-kDa coactivator SND1 (sc-34753, C-17, Santa Cruz Biotechnology) diluted 1:200, and goat anti-human acidic nuclear phosphoprotein 32 family member 3 (PHAPI2) (sc-68219, G-12, Santa Cruz Biotechnology) diluted 1:200. Secondary antibodies were sheep anti-mouse HRP-linked IgG (NA931, GE Healthcare) diluted 1:5000, donkey anti-rabbit HRP-linked IgG (NA934, GE Healthcare) diluted 1:5000, donkey anti-goat HRP-linked IgG (sc-2020, Santa Cruz Biotechnology) diluted 1:2000, and rabbit anti-chicken HRP-linked IgY (A-9046, Sigma) diluted 1:2000. All antibodies were diluted in PBS-T supplemented with 1% milk (w/v). The immunoblotted proteins were detected by enhanced chemoluminescence (ECL kit, GE Healthcare) and exposure to a light-sensitive charge-coupled device camera (ChemiDocXRS, Bio-Rad).
Two-dimensional Electrophoresis (2-DE)
All eluted and concentrated proteins (with the exception of the aliquot for 1-DE and Western blot analysis) were purified from salts by acetone precipitation using a 2-D Cleanup kit (Bio-Rad). The precipitate was resuspended in 7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 50 mm DTT, 0.5% pH 3–11 non-linear ampholyte, 0.002% (w/v) bromphenol blue and allowed to rehydrate an 18-cm non-linear pH 3–11 IPG strip (GE Healthcare) for 12 h. IEF was then carried out at +20 °C at 500 V for 1000 V-h, 1000 V for 2000 V-h, and 8000 V for 40,000 V-h (maximum 50 μA/strip) using an IPGphor (GE Healthcare). The strips were then equilibrated for the second dimension by soaking them first in equilibration buffer (6 m urea, 30% (v/v) glycerol, 5% (w/v) SDS, 50 mm Tris-HCl, pH 8.8, 0.002% (w/v) bromphenol blue) containing 1% DTT and then in equilibration buffer containing 5% iodoacetamide (IAA; Sigma) for 15 min each. For the second dimension, 1-mm-thick plastic-backed slab gels (T = 12.5%) (GE Healthcare) were used. The IPG strips were applied onto the second dimension by embedding them in 0.5% (w/v) agarose in electrophoresis running buffer (25 mm Tris, 192 mm glycine, 0.1% (w/v) SDS). Thereafter, the second dimension electrophoresis was carried out with the Ettan DALT six system (GE Healthcare) at 2.5 watts/gel for 1 h and then at 20 watts/gel until the bromphenol dye front reached the bottom of the gel. The gels were washed 3 × 10 min with H2O, then stained with Coomassie Rapid Stain (G Biosciences) for 1 h, and destained with H2O overnight. The 2-DE gels were scanned using an image scanner and analyzed using the PDQuest Advanced 8.0.1 two-dimensional gel analysis software (Bio-Rad).
MALDI-TOF Analysis
All visible spots on the two-dimensional gels were excised with a 1.5-mm-inner diameter needle using the ExQuest spot-picking robot (Bio-Rad). The gel pieces were washed three times with water and then dehydrated with 50 and 100% acetonitrile for 10 min each. Disulfide bonds were reduced by incubating the gel pieces in 1% (w/v) DTT in 100 mm NH4HCO3, pH 8 for 30 min at 50 °C. The gel pieces were then washed three times with water and again dehydrated with acetonitrile, and thereafter, the cysteines were alkylated with 2% IAA (Sigma) in 100 mm NH4HCO3, pH 8 for 30 min at room temperature. The IAA was removed by washing three times with water, and the gel pieces were dehydrated with acetonitrile. The acetonitrile supernatant was removed after 10 min, and the gel pieces were air-dried. The proteins were in-gel digested by rehydrating the gel pieces with 19 μl of trypsin solution (10 ng/μl trypsin (porcine, Promega) in 50 mm NH4HCO3, pH 8) and incubating at 37 °C overnight. The tryptic digests were acidified by addition of 1 μl of 10% TFA (Sigma), and the peptides were concentrated and desalted using μ-C18 ZipTip microcolumns (Millipore). Peptides were eluted with 1 μl of 50% acetonitrile onto a MALDI target plate. The solvent was evaporated, and the spots were covered with 0.2 μl of α-cyano-4-hydroxycinnamic acid matrix solution (Agilent Technologies).
The MALDI-TOF/TOF mass spectrometer (Ultraflex 2, Bruker Daltonics) was calibrated with peptide calibration standard 2 (Bruker Daltonics), and mass spectra were acquired in the positive ion mode at an acceleration voltage of 25 kV and pulsed ion extraction time of 80 ns. On average, 300 shots were combined for each spectrum that were annotated using the FlexAnalysis 2.2 software (Bruker Daltonics) and searched against the NCBI non-redundant database (NCBInr, July 9, 2009, containing 9,775,507 sequences of which 224,015 originated from human protein sequences) using the MASCOT search engine (version 2.1, Matrix Science) with the restriction to human proteins. The search parameters used were: peptide tolerance, 120 ppm; fixed modification, carbamidomethylation; variable modification, methionine oxidation; enzyme, trypsin; allow one missed cleavage. Molecular weight search (MOWSE) scores higher than 70 were considered significant (at >95% confidence level). Peptide mass fingerprint (PMF) spectra were internally calibrated using the trypsin autolysis peptides with monoisotopic masses (m/z 842.51, 1045.56, and 2211.10).
RESULTS
Isolation of Liganded ERα Complexes on 9×ERE-Sepharose
The purification of ERα complexes has been hampered by the low amounts of receptor in most cells. Additionally, activated receptors bind with high affinity to EREs and have to be eluted under conditions that are not compatible with complex purification (e.g. high salt). We therefore used a Sepharose column containing immobilized EREs and let complex formation occur by addition of cell extracts, ligands, and recombinant unliganded ERα. To exclude false positive identifications, we repeated this procedure in parallel on NF-EREs that do not bind ERα. Additionally, to diminish the effect of in-gel variations, staining was always performed in parallel between the ligand treatments. The basic flow scheme of the method used for the isolation of the ERE-bound ERα is depicted in Fig. 1A.
Fig. 1.
Strategy for isolation and identification of DNA-bound ERα complex. A, 90% nuclear and 10% cytosolic extract from ∼3 × 109 MCF-7 cells was used for protein complex isolation. 100 nm ligand and 20 μg of recombinant human ERα were added to the cell extract. The extract was incubated at 37 °C for 45 min and split into two samples, one applied to a column containing 9×ERE-Sepharose and the other applied to a column containing 9×NF-ERE (control) run in parallel. Elution was performed with 1 m NaCl, 10 mm Tris-HCl, pH 7.4, and the eluted proteins were concentrated and desalted. The proteins were then separated both by 1-DE (e.g. for Western blot analysis) and by IEF-2-DE for better separation. Spots were compared between gels using advanced two-dimensional gel analysis software and then excised for MALDI MS identification. B, sequences immobilized onto the HiTrap NHS column: functional trimers of 3×ERE or non-functional trimers of 3×NF-ERE. The ERE sites are underlined, and the lowercase letters indicate exchanged bases in the NF-ERE.
Because many cofactors are situated in the cytoplasm under basal conditions, we decided to include the cytoplasm in the sample. However, using whole cell extracts yielded heavy contamination with structural proteins and chaperones; hence, a ratio of 90% nuclear and 10% cytosolic extract from ∼3 × 109 MCF-7 cells (a fast growing, E2-dependent human mammary carcinoma cell line) was used as the working sample applied on the column. The cell extracts were treated with 100 nm ligand, either E2 or the ER antagonists ICI-182,780 and 4-OHT. In addition, 20 μg of recombinant human ERα were added to the extract for quantifiable reproducibility and to avoid ERα from being the limiting factor for complex formation. An incubation step was included to enable the formation of complexes, and then the extracts were divided into two samples, one added to a column containing 2 nmol of 9×ERE and the other added to a column containing 2 nmol of 9×NF-ERE oligonucleotides composed of a sequence that has no affinity for ERα (Fig. 1B) (18–20). The latter will be referred to as control sample. It is important to process all samples in parallel to avoid variations. Elution was performed using the mild conditions of a hypertonic buffer to disrupt DNA-protein and protein-protein interactions.
Analysis of Endogenous ER Content and Identification of Known ERα Interactors
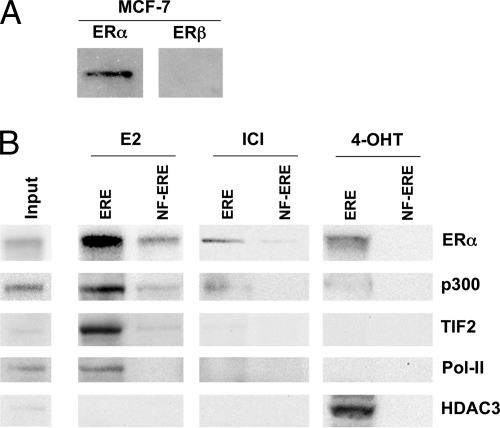
To ensure that the MCF-7 cells do contain endogenous amounts of ERα and thus represent a relevant biological system for ERα complex formation, we performed Western blot analysis on cell extracts. Indeed, we could detect significant amounts of ERα (Fig. 2A). In addition, no ERβ was detected in this cell line, suggesting that ER ligands act solely through the ERα subtype in this system. To evaluate our procedure, we used Western blot to detect ERα and known ERα complex members in the eluted fractions. We could detect enrichment not only of ERα itself but also of different known classical transcriptional regulators and Pol-II (21) (Fig. 2B). ERα was found in all ERE eluates although mostly in those from E2- and 4-OHT-treated extracts, suggesting that ERα binds to the ERE regardless of ligand type. Small traces of receptor were found in the E2 control sample purified on NF-ERE, representing its background binding to DNA or to proteins that bind DNA. The classical NR coactivator and acetyltransferase p300 (22) was seen enriched only in the eluate from E2-treated extracts. The transcription intermediary factor 2 (TIF2), a protein belonging to the p160 family of NR coactivators (23, 24), was enriched in the eluate from E2-treated extracts. HDAC3 (25) could only be observed in the eluates from 4-OHT-treated extracts, whereas Pol-II, the effector of NR-mediated transcription, was more enriched in E2 eluates. The discrepancy between E2 and 4-OHT ligand-induced receptosomes supports the common notion that different ligands induce a different ER conformation, thus attracting different cofactors (21, 26). Enrichment of these cofactors showed that the ERα complex isolation was specific and effective.
Fig. 2.
Western blot analysis of ERα and ERβ levels and identification of known ERα interactors. A, analysis of endogenous levels of ERα and ERβ in MCF-7 cell extracts (90% nuclear, 10% cytosolic). B, 15% of the volume of eluted ERE-interacting proteins from extracts treated with 100 nm E2, ICI-182,780, or 4-OHT. The proteins were separated by one-dimensional SDS-PAGE, electrotransferred to a nitrocellulose membrane, and immunoblotted using antibodies directed against ERα, p300, TIF2, Pol-II, and HDAC3. Control samples were treated equally but separated on 9×NF-ERE. Input represents 40 μg of cell extract prior to column chromatography. ICI, ICI-182,780.
Identification of ERE-interacting Proteins by 2-DE and MALDI-MS
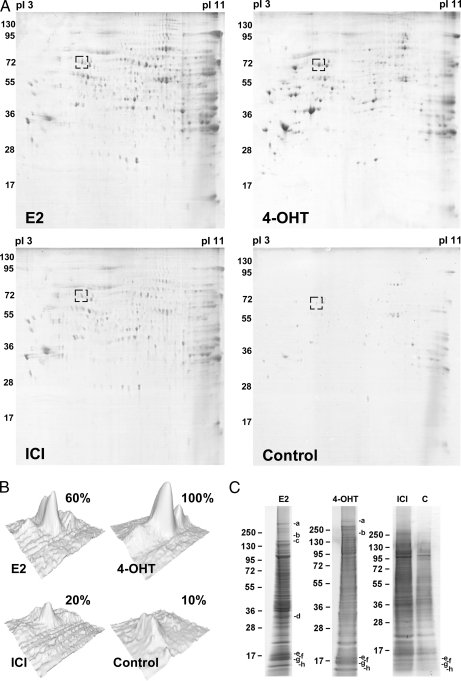
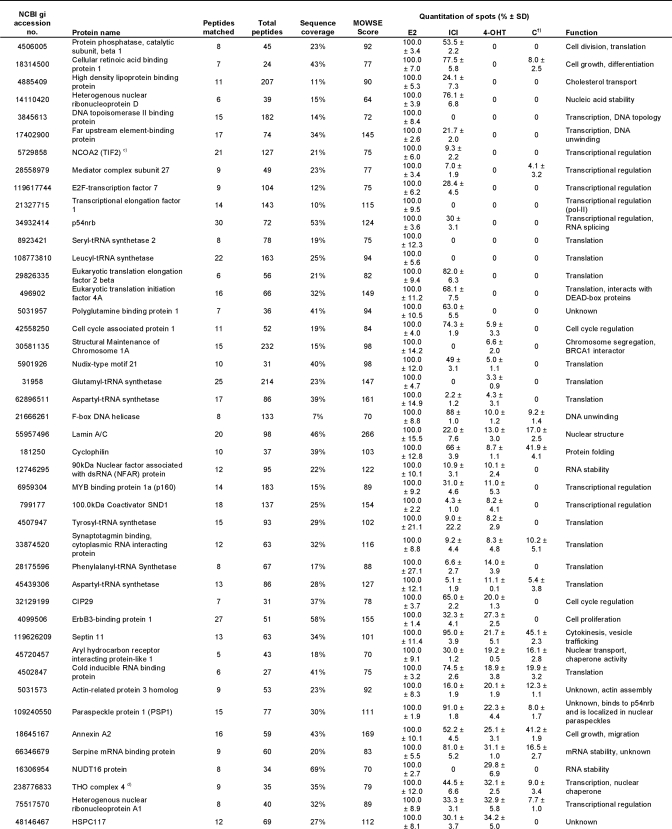
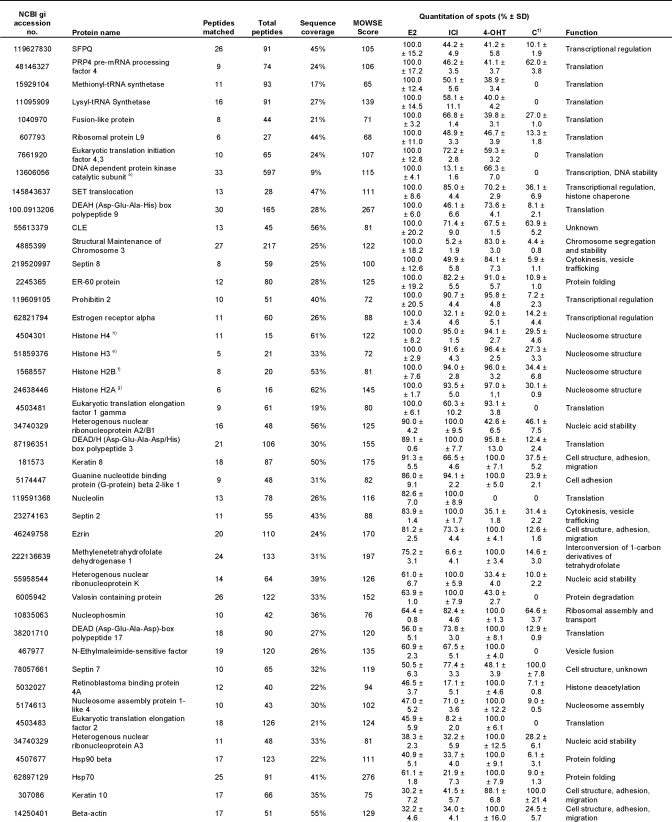
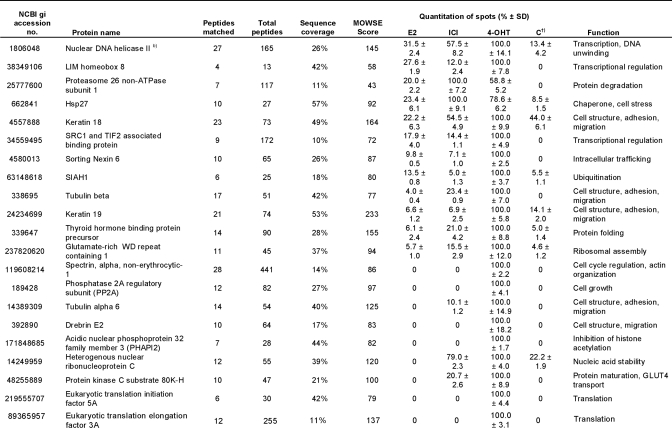
To separate and quantify the isolated proteins, a broad range 2-DE covering pI 3–11 and 10–150 kDa was performed. 240 distinct spots could be detected on the gel from E2-treated extracts, 206 spots could be detected from 4-OHT-treated extracts, 177 spots could be detected from ICI-182,780-treated extracts, and at most, 96 spots could be detected from each control sample (Fig. 3A). The identification of the spots was performed using MALDI-TOF MS and resulted in the total detection of 108 proteins. The identified proteins are summarized in Table I. Proteins matching a PMF spectrum with a MOWSE score of 70 or higher were considered significant. Hence, the protein spot identification was estimated to encompass around 80% of total spots, not counting spots of post-translationally modified isoforms. The intensity of each spot on the two-dimensional gel was quantified by measuring the Gaussian distribution of the spot in x, y, and z directions using the PDQuest Advanced gel image analysis software and compared between the gels. This is summarized as percentage of the highest intensity in Table I. For simplicity, control sample here is represented by the NF-ERE eluate from the E2 treatment. Fig. 3B shows a typical comparison between the same protein spot (Hsp70) on the different gels. However, not all proteins could be detected by 2-DE alone, probably due to solubility problems, large protein mass, difficulties for some proteins to enter the second dimension, and/or protein degradation. In this case, one-dimensional gels were run as complements to the two-dimensional gels by which eight additional proteins could be identified (Fig. 3C and Table I). Among these were the histones H2A, H2B, H3, and H4, which could suggest that nucleosomal structures actually can be formed on the immobilized DNA or that the histones are attracted to co-purify with the ERα complexes by specific interactions with receptosomal proteins. The latter hypothesis is supported by lower amounts of histones detected in the control samples.
Fig. 3.
1-DE and 2-DE separation of ERE-interacting proteins. A, Coomassie staining of proteins isolated on 9×ERE-Sepharose and separated by 2-DE (pI 3–11, 10–150 kDa) from extracts treated with 100 nm E2, ICI-182,780 (ICI), or 4-OHT. The protein pattern of each complex is a representation of at least three independent two-dimensional gels. The box represents the Hsp70 spot and its quantification by comparing its peak intensity in the different two-dimensional gels (B). C, 1-DE separation of 15% of the volume of eluted ERE-interacting proteins from 100 nm E2, ICI-182,780, and 4-OHT treatments as well as control sample (E2-treated and isolated on 9×NF-ERE column). The indicated bands (a–h) correspond to the proteins listed in Table I. Control samples were treated equally but separated on 9×NF-ERE. For simplicity, only the E2 control sample is shown (4-OHT and ICI-182,780 control samples are available as supplemental Fig. S1).
Table I. Isolated proteins bound to the ERE-speharose upon different ligand treatments.
†) Control sample represent E2-treated extracts separated on NF-ERE.
a)-h): Identified on 1D gel.
Interestingly, apart from some of the known ERα cofactors, many novel interactors were identified of which a large part is involved in not only transcription but also in, among others, cell structure, translation, and nucleic acid stability. The general ligand specificities (if any) toward functional categories of proteins identified are summarized in Table II. Studying Table I in detail, it is apparent that several of the proteins eluted from the columns are contaminants such as abundant structural proteins (annexin A2, septins, β-actin, and keratins), which are also abundant in the control sample. Several protein families were overrepresented in the eluates from E2- and 4-OHT-treated extracts. These include the heterogeneous nuclear ribonucleoproteins (hnRNPs) of which the function of many is still unknown. Another group belongs to the DEA(D/H) box family of RNA helicases that have a conserved Asp-Glu-Ala-(Asp/His) motif and are multifunctional proteins that, in addition to RNA unwinding, are involved in RNA processing, export, and translation. A third group of abundant eluted proteins belong to eukaryotic translation initiation and elongation factors. These proteins are members of the translation complexes involved in initiation of translation and stabilizing elongation of mRNA and participate in polypeptide synthesis by attracting tRNAs (27, 28). The eukaryotic elongation factors and eukaryotic translation initiation factors are basically not found in the control samples, suggesting that these factors bind indirectly (or nonspecifically) to the large protein complexes assembled on the ERE. An interesting group of eluted proteins is associated with mRNA splicing: paraspeckle protein 1 (PSP1), SFPQ, and the p54nrb splicing factor. At least the latter two proteins form heterodimers and have been shown, in addition to their RNA splicing ability, to have several other functions, including transcriptional regulation of NRs (29–31). SF2 on the other hand is known to interact with at least hnRNP A1 and PP2A (both found in Table I) as part of a larger translation/splicing complex (32, 33). Most of these proteins were mainly found in eluates from E2-treated extracts, but some were also in ICI-182,780 and 4-OHT eluates.
Table II. Summary of ligand specificity versus function of proteins identified bound to ERE-ER.
| Category | No. proteins found | Ligand specificitya |
|---|---|---|
| Cell cycle regulation | 7 | E2 |
| Cell structure, adhesion, migration | 12 | 4-OHT |
| Cytokinesis, vesicle trafficking | 4 | No specificity |
| Histone deacetylation | 2 | 4-OHT |
| Nucleic acid stability | 14 | No specificity |
| Nucleosome structure | 4 | No specificity |
| Protein degradation | 2 | E2/ICIb |
| Protein folding | 7 | E2/4-OHT |
| Transcriptional regulation | 20 | E2 |
| Translation | 31 | E2 |
| Other | 7 | No specificity |
a Cutoff of 2-fold enrichment between ligand treatments used.
b ICI-182,780.
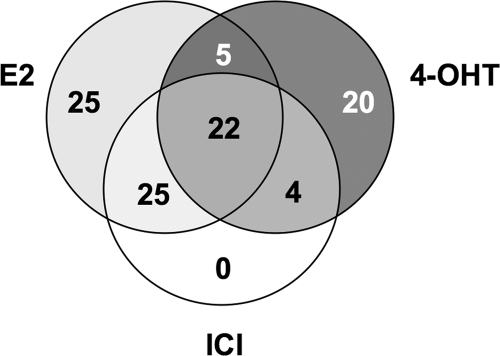
The overlap of proteins found in the E2-, ICI-182,780-, and 4-OHT-liganded complexes are summarized in Fig. 4. Interestingly, using the cutoff of 2-fold enrichment, no unique proteins could be detected in the ICI-182,780 complex, suggesting the effect of this ligand to be mediated differently than by ERα-ERE binding.
Fig. 4.
Venn diagram showing overlap of proteins found in E2-, ICI-182,780-, and 4-OHT-induced ERα complexes. Of 108 proteins identified, 25 proteins were found enriched at the ERE under E2 treatment and 20 were enriched under 4-OHT treatment, whereas no significant enrichment could be observed under ICI-182,780 (ICI) treatment. Additionally, seven proteins were found in all eluates, including control sample (not shown here). A 2-fold enrichment cutoff was set to discriminate between the treatments.
Confirmation of MALDI-MS Analysis
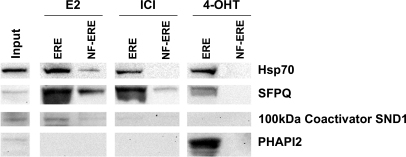
Four proteins identified by MALDI MS in eluted fractions were selected for Western blot confirmation (Fig. 5): two proteins found to a larger or smaller extent in all fractions, Hsp70 and SFPQ; one protein found predominantly in the eluate from E2-treated extracts, 100-kDa coactivator SND1; and one protein only found in the eluate from 4-OHT-treated extract, PHAPI2 (also known as pp32). The Western blot analysis confirmed the two-dimensional gel image analysis of these proteins and the MALDI MS identification.
Fig. 5.
Western blot analysis of MS-identified ERE-interacting proteins. 15% of the volume of eluted ERE-interacting proteins from extracts treated with 100 nm E2, ICI-182,780 (ICI), and 4-OHT was analyzed. The proteins were separated by one-dimensional SDS-PAGE, electrotransferred to a nitrocellulose membrane, and immunoblotted using antibodies against Hsp70, SFPQ, 100-kDa coactivator SND1, and PHAPI2. Control samples were treated equally but separated on 9×NF-ERE. Input represents 40 μg of cell extract prior to column chromatography.
DISCUSSION
It is clear that cofactors are responsible for mediating balanced and accurate transcriptional effects of NRs. Understanding the cofactor complex composition of NRs (the receptosomes) in different tissues, on different promoter elements, and upon different ligand binding would thus be of great importance in deciphering the complex signaling of the NR-mediated transcription. To date, several attempts have been made to isolate nuclear receptor complexes either whole or in smaller parts (8–13). However, several difficulties have limited the isolation and proteomics analyses of NR complexes of which the largest have been the low amounts of endogenous NRs and the compromise between stringency and efficiency of the methods applied. In addition, the strong association of liganded NRs to DNA has also posed a problem for efficient complex isolation (15). Recently, Schultz-Norton et al. (13, 34) demonstrated a method for the isolation of DNA-bound ERα complex separated on an agarose gel. Although their technique is very straightforward, we could not reproduce their results stringently enough using MCF-7 cells and the ligands E2, ICI-182,780, and 4-OHT. Instead, the efficiency of this method was hampered by co-purification of large amounts of chaperones and structural proteins (probably due to co-migration of numerous other protein complexes with the ERα complex in the gel), making it difficult to distinguish false positives from true positives. In this report, we present a different approach, using the high affinity of liganded ERα toward ERE sequences for its receptor complex isolation from MCF-7 cells in a one-step column chromatography procedure. This mammary carcinoma cell line has high endogenous levels of ERα (no ERβ) and is referred to as an E2-sensitive cell line, thus representing a relevant biological system for ERα complex isolation. Although we used the MCF-7 cell line exclusively, this method could be adapted to other cell lines or even tissue samples. The method would also be applicable to other NRs that have affinity toward a specific DNA sequence. Encouragingly, the method presented here uses relatively low amounts of starting material (approximately 3 × 109 cells) and is fast and efficient in isolating significant amounts of ERα complexes. Previous attempts in NR complex isolations have required enormous amounts of starting material as is common in many proteomics analyses of low abundance proteins, making every experiment extremely laborious. Our method isolates anything bound to the ERE sequence, not only the ERα complex but also many factors associated with the DNA. Hence, it is important to include a good control for false positive interactors (background binding). Here we used an NF-ERE sequence to identify any proteins that might associate to the DNA itself in vitro. In addition to various factors associated with nucleic acid stability, mRNA processing, translation, and cell structure, we also detected small amounts of ERα bound to the NF-ERE. The binding of ERα to NF-ER could imply a weak (probably unspecific and tethered) binding of ERα to this sequence, and we cannot rule out that some of the identified proteins in the control sample are false negatives. In this sense, it is important to analyze the relative amounts of each protein spot between the gels and draw conclusions on specificity based on this. This method is thus compatible with relative quantitative analyses such as two-dimensional DIGE. Another interesting benefit of this method is the limitless modification of the DNA sequence immobilized on the beads. Any classical promoter site and variations thereof may be used, making this method an easy way of studying the effect of promoter region heterogeneity (20, 35), in combination with different ligands, on cofactor recruitment. A drawback of this method was apparent heavy contamination with structural and chaperone proteins (e.g. keratins, β-actin, Hsp70, and Hsp90) when using whole cell extracts of MCF-7 cells. To reduce this contamination, we used starting material consisting of a 9:1 ratio between nuclear and cytosolic extracts. Although this adds to the artificiality, we reasoned that this would not affect the method per se. In fact, it is very hard to mimic the complex from an in vivo environment with compartmentalization of different proteins that are affected by E2 signaling, especially when isolating protein complexes. In this sense, one needs to determine the parameters (e.g. choice of cell type, cellular extract, ligand exposure, and DNA sequence) for the purification beforehand based on known facts and previous experiences.
The identified proteins were all detected using Coomassie staining, which has a good stoichiometric linear range and sensitivity down to 0.5 pmol of protein. Using a more sensitive staining dye (e.g. SYPRO Ruby or silver staining) for detection of some very low abundance proteins proved insufficient for significant MALDI MS identification. However, using a combination of Western blot and MALDI MS, we could detect several classical cofactors such as Pol-II, p300, and TIF2, suggesting this method to work and that scaling up the sample size would likely identify even very low abundance proteins efficiently. Interestingly, not only transcriptional regulatory proteins were found, but also many proteins associated with translation, mRNA processing, cell structure, and cell cycle regulation were isolated. Some overrepresented proteins belong to the hnRNP and DEA(D/H) box multifunctional protein families. Although the function of many of these proteins is still not completely understood, it has been demonstrated that hnRNPs can bind specifically to DNA and RNA (36, 37) and also to EREs (38), modulating ER transcriptional activity. Members of the DEA(D/H) box family have also been recognized as cofactors of ERα-mediated transcription (39, 40). It is not known whether these proteins are components of the ERα complex or are associated directly or indirectly with the ERE, and future experiments to characterize these interactions would be of great interest. Our data possibly propose the ERα complex to be much larger than previously thought, probably due to dynamic interactions with surrounding structural and nucleotide-binding proteins. For example, the histones H2A, H2B, H3, and H4 were found in all eluates, which could suggest that the immobilized ERE sequences (202 bp) are large enough for nucleosome assembly. Lower amounts of all histones were found in the control eluate, suggesting that they have some affinity to the ERα complex or specificity to functional EREs. It is known that ERα-mediated gene transcription is associated with histone acetylation and chromatin remodeling (41), a process that is mediated by histone acetyltransferase cofactors such as p300. On the contrary, histone deacetylation by corepressors such as HDACs inhibits ERα transcription. It is still debated whether and how a physical interaction between the ER transcriptional complex and nucleosomes would be of functional significance, although an interaction between histone acetyltransferases and other cofactors (e.g. ATP-dependent chromatin remodelers such as Brahma-related gene 1) could lead to such interactions (42, 43).
We found that ERα binds to the ERE after treatment, not only with E2 but also with 4-OHT and, to a lesser extent, ICI-182,780, two ER antagonists. The issue of the ERα complex affinity for the ERE under various ligand treatments is still debated, although the common notion is that the effect of the ligand is mediated via different cofactor affinities to ERα rather than via ERE binding. 4-OHT induces a conformation that attracts corepressors to the ERE, rendering the complex inactive, and deacetylates the histones. However, in some tissues, 4-OHT attracts other cofactors to activate gene transcription at estrogen-regulated promoters (44). The molecular action of ICI-182,780, on the other hand, seems to down-regulate ER protein levels by degrading the receptor (45, 46) rather than to inhibit ER-ERE binding. This could explain why we could not detect significant enrichment of any unique proteins at the ERE after ICI-182,780 treatment (Fig. 4). However, we found two novel proteins involved in proteasomal degradation to associate with the ERα complex mainly following ICI-182,780 treatment: valosin-containing protein and proteasome 26 non-ATPase subunit 1. The 4-OHT-induced ERα conformation was found to attract several histone deacetylases (or inhibitors of acetylation) such as PHAPI2 (pp32) and retinoblastoma protein binding members 4A and 7. The PHAPI2 and retinoblastoma proteins have been found to associate in large complexes to NR-regulated promoters inhibiting gene transcription (29, 47). A protein mainly isolated from the E2 treatment was the SET nuclear oncogene. This protein is a member of the SET complex of multifunctional histone chaperones of which PHAPI2 is also a member and has been shown to have different transcription modulating activities on different genes (47, 48) and to enhance transcription by interacting with the NR coactivator cAMP-response element-binding protein (CREB)-binding protein (CBP) (49). Furthermore, the members of the pre-mRNA processing machinery, SFPQ, p54nrb, and PSP1, found mostly in the eluates from E2 treatment, have also been shown to be associated with PHAPI2 and appear to modulate NR-mediated transcription in different ways (29, 30). Noteworthy is that ERα liganded with 4-OHT appears more prone to interact with tubulins and the heat shock proteins Hsp70 and Hsp90. Similar associations have been seen previously for NRs outside the nucleus and might be involved in the degradation of cytoplasmic ERα (50, 51).
In conclusion, we have developed an effective method for isolating the liganded ERα complex as it is assembled on the ERE. In addition to several classical cofactors, we identified proteins associated with e.g. chromatin remodeling, nucleic acid processing, cell structure, and transcriptional and translational regulation to interact directly or indirectly with ERα. This may result in a transcriptional megacomplex of various factors in an intricate network of interactions that delivers a measured, cell- and promoter-specific response to ligand stimulation (52, 53). The functional characterization of the identified proteins will be an exciting task for the future.
Supplementary Material
Acknowledgments
We thank Dr. Joëlle Rüegg for critically reading the manuscript.
* This work was supported by the Swedish Cancer Fund and the Inga-Britt and Arne Lundberg's Forskningsstiftelse.
 This article contains supplemental Table S1 and Fig. S1.
This article contains supplemental Table S1 and Fig. S1.
1 The abbreviations used are:
- E2
- 17β-estradiol
- 2-DE
- two-dimensional electrophoresis
- 1-DE
- one-dimensional electrophoresis
- 4-OHT
- 4-hydroxytamoxifen
- ER
- estrogen receptor
- ERE
- estrogen response element
- HDAC3
- histone deacetylase 3
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- HRP
- horseradish peroxidase
- Hsp
- heat shock protein
- IAA
- iodoacetamide
- MOWSE
- molecular weight search
- NF
- non-functional
- NHS
- N-hydroxysuccinimide
- NR
- nuclear receptor
- PHAPI2
- acidic nuclear phosphoprotein 32 family member 3
- PMF
- peptide mass fingerprint
- Pol-II
- RNA polymerase II
- PSP1
- paraspeckle protein 1
- SFPQ
- splicing factor proline/glutamine-rich
- TIF2
- transcription intermediary factor 2.
REFERENCES
- 1.Nilsson S., Gustafsson J. A. (2002) Estrogen receptor action. Crit. Rev. Eukaryot. Gene. Expr. 12, 237–257 [DOI] [PubMed] [Google Scholar]
- 2.Behl C. (2002) Estrogen can protect neurons: modes of action. J. Steroid. Biochem. Mol. Biol. 83, 195–197 [DOI] [PubMed] [Google Scholar]
- 3.Farhat M. Y., Lavigne M. C., Ramwell P. W. (1996) The vascular protective effects of estrogen. FASEB J. 10, 615–624 [PubMed] [Google Scholar]
- 4.Nilsson S., Mäkelä S., Treuter E., Tujague M., Thomsen J., Andersson G., Enmark E., Pettersson K., Warner M., Gustafsson J. A. (2001) Mechanisms of estrogen action. Physiol. Rev. 81, 1535–1565 [DOI] [PubMed] [Google Scholar]
- 5.Matthews J., Gustafsson J. A. (2003) Estrogen signaling: a subtle balance between ER alpha and ER beta. Mol. Interv. 3, 281–292 [DOI] [PubMed] [Google Scholar]
- 6.Dorssers L. C., Van der Flier S., Brinkman A., van Agthoven T., Veldscholte J., Berns E. M., Klijn J. G., Beex L. V., Foekens J. A. (2001) Tamoxifen resistance in breast cancer: elucidating mechanisms. Drugs 61, 1721–1733 [DOI] [PubMed] [Google Scholar]
- 7.Girault I., Bièche I., Lidereau R. (2006) Role of estrogen receptor alpha transcriptional coregulators in tamoxifen resistance in breast cancer. Maturitas 54, 342–351 [DOI] [PubMed] [Google Scholar]
- 8.Goodson M. L., Farboud B., Privalsky M. L. (2007) An improved high throughput protein-protein interaction assay for nuclear hormone receptors. Nucl. Recept. Signal. 5, e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedman E., Widén C., Asadi A., Dinnetz I., Schröder W. P., Gustafsson J. A., Wikström A. C. (2006) Proteomic identification of glucocorticoid receptor interacting proteins. Proteomics 6, 3114–3126 [DOI] [PubMed] [Google Scholar]
- 10.Hosohata K., Li P., Hosohata Y., Qin J., Roeder R. G., Wang Z. (2003) Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription. Mol. Cell. Biol. 23, 7019–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung S. Y., Malovannaya A., Wei J., O'Malley B. W., Qin J. (2005) Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol. Endocrinol. 19, 2451–2465 [DOI] [PubMed] [Google Scholar]
- 12.Malik S., Roeder R. G. (2003) Isolation and functional characterization of the TRAP/mediator complex. Methods Enzymol. 364, 257–284 [DOI] [PubMed] [Google Scholar]
- 13.Schultz-Norton J. R., Ziegler Y. S., Likhite V. S., Yates J. R., Nardulli A. M. (2008) Isolation of novel coregulatory protein networks associated with DNA-bound estrogen receptor alpha. BMC Mol. Biol. 9, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murdoch F. E., Gorski J. (1991) The role of ligand in estrogen receptor regulation of gene expression. Mol. Cell. Endocrinol. 78, C103–C108 [DOI] [PubMed] [Google Scholar]
- 15.Weinberg A. L., Carter D., Ahonen M., Alarid E. T., Murdoch F. E., Fritsch M. K. (2007) The DNA binding domain of estrogen receptor alpha is required for high-affinity nuclear interaction induced by estradiol. Biochemistry 46, 8933–8942 [DOI] [PubMed] [Google Scholar]
- 16.Landel C. C., Kushner P. J., Greene G. L. (1995) Estrogen receptor accessory proteins: effects on receptor-DNA interactions. Environ. Health Perspect. 103, Suppl. 7, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santen R. J., Song R. X., Masamura S., Yue W., Fan P., Sogon T., Hayashi S., Nakachi K., Eguchi H. (2008) Adaptation to estradiol deprivation causes up-regulation of growth factor pathways and hypersensitivity to estradiol in breast cancer cells. Adv. Exp. Med. Biol. 630, 19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driscoll M. D., Sathya G., Muyan M., Klinge C. M., Hilf R., Bambara R. A. (1998) Sequence requirements for estrogen receptor binding to estrogen response elements. J. Biol. Chem. 273, 29321–29330 [DOI] [PubMed] [Google Scholar]
- 19.Gruber C. J., Gruber D. M., Gruber I. M., Wieser F., Huber J. C. (2004) Anatomy of the estrogen response element. Trends Endocrinol. Metab. 15, 73–78 [DOI] [PubMed] [Google Scholar]
- 20.Yi P., Driscoll M. D., Huang J., Bhagat S., Hilf R., Bambara R. A., Muyan M. (2002) The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ER alpha and ER beta. Mol. Endocrinol. 16, 674–693 [DOI] [PubMed] [Google Scholar]
- 21.Edwards D. P. (2000) The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors. J. Mammary Gland Biol. Neoplasia 5, 307–324 [DOI] [PubMed] [Google Scholar]
- 22.Voegel J. J., Heine M. J., Tini M., Vivat V., Chambon P., Gronemeyer H. (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong H., Kohli K., Garabedian M. J., Stallcup M. R. (1997) GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 17, 2735–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voegel J. J., Heine M. J., Zechel C., Chambon P., Gronemeyer H. (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15, 3667–3675 [PMC free article] [PubMed] [Google Scholar]
- 25.Wolffe A. P. (1997) Transcriptional control. Sinful repression. Nature 387, 16–17 [DOI] [PubMed] [Google Scholar]
- 26.Glass C. K., Rosenfeld M. G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 27.Brant-Zawadzki P. B., Schmid D. I., Jiang H., Weyrich A. S., Zimmerman G. A., Kraiss L. W. (2007) Translational control in endothelial cells. J. Vasc. Surg. 45, Suppl. A, A8–A14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I., Hellen C. U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 98, 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adegbola O., Pasternack G. R. (2005) A pp32-retinoblastoma protein complex modulates androgen receptor-mediated transcription and associates with components of the splicing machinery. Biochem. Biophys. Res. Commun. 334, 702–708 [DOI] [PubMed] [Google Scholar]
- 30.Dong X., Yu C., Shynlova O., Challis J. R., Rennie P. S., Lye S. J. (2009) p54nrb is a transcriptional corepressor of the progesterone receptor that modulates transcription of the labor-associated gene, connexin 43 (Gja1). Mol. Endocrinol. 23, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishitani K., Yoshida T., Kitagawa H., Ohta H., Nozawa S., Kato S. (2003) p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 306, 660–665 [DOI] [PubMed] [Google Scholar]
- 32.Black D. L. (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 33.Michlewski G., Sanford J. R., Cáceres J. F. (2008) The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 30, 179–189 [DOI] [PubMed] [Google Scholar]
- 34.Schultz-Norton J. R., Ziegler Y. S., Likhite V. S., Nardulli A. M. (2009) Isolation of proteins associated with the DNA-bound estrogen receptor alpha. Methods Mol. Biol. 590, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anolik J. H., Klinge C. M., Brolly C. L., Bambara R. A., Hilf R. (1996) Stability of the ligand-estrogen receptor interaction depends on estrogen response element flanking sequences and cellular factors. J. Steroid Biochem. Mol. Biol. 59, 413–429 [DOI] [PubMed] [Google Scholar]
- 36.Bomsztyk K., Van Seuningen I., Suzuki H., Denisenko O., Ostrowski J. (1997) Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 403, 113–115 [DOI] [PubMed] [Google Scholar]
- 37.Matunis M. J., Michael W. M., Dreyfuss G. (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12, 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., Hu B., Gacad M. A., Adams J. S. (1998) Cloning and expression of a novel dominant-negative-acting estrogen response element-binding protein in the heterogeneous nuclear ribonucleoprotein family. J. Biol. Chem. 273, 31352–31357 [DOI] [PubMed] [Google Scholar]
- 39.Endoh H., Maruyama K., Masuhiro Y., Kobayashi Y., Goto M., Tai H., Yanagisawa J., Metzger D., Hashimoto S., Kato S. (1999) Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol. Cell. Biol. 19, 5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Wilson B. J., Bates G. J., Nicol S. M., Gregory D. J., Perkins N. D., Fuller-Pace F. V. (2004) The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol. Biol. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng A. S., Jin V. X., Fan M., Smith L. T., Liyanarachchi S., Yan P. S., Leu Y. W., Chan M. W., Plass C., Nephew K. P., Davuluri R. V., Huang T. H. (2006) Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol. Cell 21, 393–404 [DOI] [PubMed] [Google Scholar]
- 42.Green K. A., Carroll J. S. (2007) Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat. Rev. Cancer 7, 713–722 [DOI] [PubMed] [Google Scholar]
- 43.Narlikar G. J., Fan H. Y., Kingston R. E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487 [DOI] [PubMed] [Google Scholar]
- 44.Shang Y., Brown M. (2002) Molecular determinants for the tissue specificity of SERMs. Science 295, 2465–2468 [DOI] [PubMed] [Google Scholar]
- 45.Dauvois S., Danielian P. S., White R., Parker M. G. (1992) Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl. Acad. Sci. U.S.A. 89, 4037–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakeling A. E. (2000) Similarities and distinctions in the mode of action of different classes of antioestrogens. Endocr. Relat. Cancer 7, 17–28 [DOI] [PubMed] [Google Scholar]
- 47.Kutney S. N., Hong R., Macfarlan T., Chakravarti D. (2004) A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression. J. Biol. Chem. 279, 30850–30855 [DOI] [PubMed] [Google Scholar]
- 48.Gamble M. J., Erdjument-Bromage H., Tempst P., Freedman L. P., Fisher R. P. (2005) The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol. Cell. Biol. 25, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karetsou Z., Martic G., Sflomos G., Papamarcaki T. (2005) The histone chaperone SET/TAF-Ibeta interacts functionally with the CREB-binding protein. Biochem. Biophys. Res. Commun. 335, 322–327 [DOI] [PubMed] [Google Scholar]
- 50.Azuma K., Horie K., Inoue S., Ouchi Y., Sakai R. (2004) Analysis of estrogen receptor alpha signaling complex at the plasma membrane. FEBS Lett. 577, 339–344 [DOI] [PubMed] [Google Scholar]
- 51.Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 52.Kadonaga J. T. (2004) Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116, 247–257 [DOI] [PubMed] [Google Scholar]
- 53.Ptashne M., Gann A. (1997) Transcriptional activation by recruitment. Nature 386, 569–577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.