Abstract
The versatile metabolism of the green alga Chlamydomonas reinhardtii is reflected in its complex response to anaerobic conditions. The anaerobic response is also remarkable in the context of renewable energy because C. reinhardtii is able to produce hydrogen under anaerobic conditions. To identify proteins involved during anaerobic acclimation as well as to localize proteins and pathways to the powerhouses of the cell, chloroplasts and mitochondria from C. reinhardtii in aerobic and anaerobic (induced by 8 h of argon bubbling) conditions were isolated and analyzed using comparative proteomics. A total of 2315 proteins were identified. Further analysis based on spectral counting clearly localized 606 of these proteins to the chloroplast, including many proteins of the fermentative metabolism. Comparative quantitative analyses were performed with the chloroplast-localized proteins using stable isotopic labeling of amino acids ([13C6]arginine/[12C6]arginine in an arginine auxotrophic strain). The quantitative data confirmed proteins previously characterized as induced at the transcript level as well as identified several new proteins of unknown function induced under anaerobic conditions. These proteins of unknown function provide new candidates for further investigation, which could bring insights for the engineering of hydrogen-producing alga strains.
Due to an urgent demand for clean energy for the future, there has been an increased interest in research regarding Chlamydomonas reinhardtii in the context of renewable energy. Among the numerous possibilities for clean energy, hydrogen is considered to be one of the most attractive because its combustion produces zero carbon emission (1). C. reinhardtii is a promising organism for renewable energy because it is able to produce hydrogen as a photosynthetic product (1–3). This is possible because C. reinhardtii possesses one of the most efficient [Fe-Fe]-hydrogenases that is induced under anaerobic conditions and sulfur starvation (4, 5).
There has been an array of studies that have investigated C. reinhardtii under anaerobic conditions and provided valuable insights into the metabolic changes undertaken by the cell to acclimate to an anaerobic condition. Despite the wide range of knowledge regarding C. reinhardtii and anaerobiosis, many of the studies have been based on transcript or metabolite levels (6–10). To expand the current knowledge on the subject, we investigated the chloroplast and mitochondrial proteomes of C. reinhardtii under anaerobiosis.
It is now well established that under anaerobic conditions C. reinhardtii induces a wide range of fermentative pyruvate-dependent metabolic pathways (11–13). The induction of these pathways has been confirmed at the transcript level for dark anaerobic and sulfur-depleted anaerobic conditions (7, 8, 10) as well as through the increase in fermentative products such as formate, ethanol, and acetate (6, 9). Despite the identification of these induced proteins of the fermentative metabolism, there have been little biochemical data to support the localization for some of the proteins (7, 14). Although discovering induced proteins is crucial for the understanding of the anaerobic response, it is equally important to understand the localization of these proteins to engineer a strain that potentially produces higher amounts of hydrogen.
In this study, we aimed to localize currently known key proteins involved in the anaerobic response to within or outside of the chloroplast as well as to identify proteins that are significantly induced under anaerobiosis through quantitative proteomics. Qualitative and semiquantitative analyses of isolated chloroplasts and mitochondria from aerobic and anaerobic C. reinhardtii cultures allowed for the identification and localization of proteins, including a handful of fermentative proteins. We identified 606 proteins highly likely to be chloroplast-localized that well supplement the recently published significant list of mitochondrial proteins by Atteia et al. (15) as well as aspects of the chloroplast proteome already characterized (16–21).
We further analyzed the identified chloroplast proteins by means of quantitative proteomics, which allowed for identification of proteins that are induced under anaerobiosis. These consist of the proteins previously characterized to be highly expressed under anaerobiosis, including those that are co-induced under anaerobic and copper-deficient conditions. Additionally, induced proteins of particular interest are those of unknown function, some of which are part of the GreenCut proteins (22), making them favorable candidates for further analyses.
EXPERIMENTAL PROCEDURES
Strains and Cultures
The arginine auxotrophic C. reinhardtii strain CC424 mt− was used for all experiments. Cells were grown under standard conditions (23) or supplemented with isotopically labeled l-[13C6]arginine as described in Naumann et al. (23) and grown under 50 microeinsteins·m−2·s−1 light. Isotopically labeled cultures were maintained in standard, aerobic conditions and cultivated to a cell density of 3–4 × 106 cells/ml. Unlabeled cell cultures were also grown to a cell density of 3–4 × 106 cells/ml followed by anaerobic induction by bubbling with argon for 8 h under 80 microeinsteins·m−2·s−1 light.
Formate Measurements
Formate levels were measured with a test kit (catalog number 10979732035) from R-Biopharm AG, Darmstadt, Germany, following the supplier's instructions.
Isolation of Chloroplasts and Mitochondria
Chloroplasts were isolated as described by Naumann et al. (23). Mitochondria were isolated as described by Eriksson et al. (24) with a few modifications as described by Busch et al. (25).
Protein Analysis, Immunodetection, and LC-MS/MS Analysis of Proteins
Protein analysis and immunodetection were performed as described in Naumann et al. (23) and Hippler et al. (26). Antibodies against CoxIIB were purchased from AgriSera, and antibodies for TEF7 (against peptide sequence EEIYIGFVKEEGFGS) were purchased from Eurogentec.
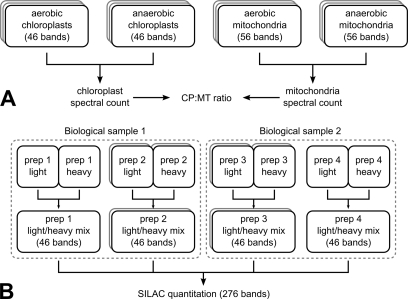
Samples for mass spectrometric analyses were prepared as in Naumann et al. (23). Samples fractionated by SDS-PAGE were excised into 46 bands for the chloroplast samples and 56 bands for the mitochondrial samples and digested tryptically. A schematic diagram in Fig. 1 demonstrates the number of preparations and measurements that were performed in this study. LC-MS/MS analyses were performed as described in Naumann et al. (23) with the following modifications: the nano-LC was performed on an Ultimate 3000 system (Dionex, Sunnyvale, CA) with the solvent gradients described in Stauber et al. (19) and Naumann et al. (23). An HD-05 μPrecolumn holder from Dionex (order number 6720.0012) was used for trapping (4 min), and a 3-μm Atlantis® (part number 186002197) column from Waters was used for peptide separation (56 min). An LTQ Orbitrap XL (Thermo, Bremen, Germany) mass spectrometer was used with FT Programs 2.0.7, Xcalibur 2.0.7, and LTQ Orbitrap XL MS 2.4 SP1. Peptides were measured in the FTMS (mass range, 375–2000 m/z; resolution, 60,000) and selected for fragmentation in the ion trap mass spectrometer (CID collision energy, 35 V) with a 2-Da isolation width using the “Big 5” method (which selects the five most abundant precursor ions detected in the full scan), requiring a minimum precursor charge state of 2. Automatic gain control was used, and dynamic exclusion was enabled for 90 s.
Fig. 1.
Schematic overview of identification and SILAC quantitation MS/MS measurements. A, chloroplast (CP) and mitochondrial (MT) samples were cut into 46 and 56 bands, respectively. Both isolations were performed for aerobic and anaerobic cells with a total of three replicates, resulting in a total band count of 612. B, for SILAC quantitation, four preparations (prep 1–4) stemming from two biological samples were prepared and measured once or twice, resulting in a total of six SILAC measurements with 46 bands each.
Protein Identification from Mass Spectrometric Data
For the MS2 identification of peptides, OMSSA1 (version 2.1.4) (27) was used with a target/decoy approach (28). The maximum number of missed cleavages allowed was set to 2. No modifications were used except a variable heavy [13C6]arginine modification for the SILAC runs. Mass tolerances were set to 0.02 Da for the precursor ions and 0.5 Da for the product ions. The JGI Chlamydomonas gene model database v3.1 and NCBI databases BK000554.2 and NC_001638.1 were merged in a resulting database containing 15001 protein sequences that was used for the database search, and a decoy protein was generated for each of these proteins by sequence reversal. For each set of bands from one sample, an adaptive E-value threshold was determined in such a way that the estimated false positive rate (FPR) was 1% or lower, derived from the following formula: FPR = 2·ndecoys/(ntargets + ndecoys). In addition, the peptides were filtered through a 5-ppm mass accuracy requirement.
As an alternative to peptide-mass spectrum matching via a protein database search algorithm, de novo prediction (using PEAKS (29)) was used together with Genomic Peptide Finder (GPF) (16) to yield additional peptide identifications or to confirm peptide identifications that originated from the protein database search. GPF is a tool that takes de novo predicted amino acid sequences and aligns them to the genomic DNA sequence in an error-tolerant way that compensates for de novo sequencing errors. In addition, GPF allows for intron splits to occur within peptide sequence matches. Recently, GPF has been redesigned to allow for intron splits to occur within a single nucleotide triplet.2 In addition, GPF is now much faster due to the use of an indexing strategy that, although requiring a correctly predicted sequence of three amino acids within one exon, achieves a 300-fold speed increase compared with the previous GPF. In addition, the N- and C-terminal masses of the correctly predicted amino acid trimer must be correct within a certain mass accuracy. Finally, all GPF alignments are filtered in such a way that a correctly predicted amino acid pentamer must be present in the final (possibly spliced) alignment.
Combining putative GPF peptides with a target/decoy approach for the protein database posed a challenge because it is not known how many of the putative GPF peptides are already false positives, which means that if decoys were added to an unknown mixture of true and false positives, a correct target FPR could not be estimated. This problem was solved in the following way: GPF peptides and gene model proteins were combined into a single mixed database, and decoys were only created for gene model proteins. Then OMSSA was run on this combined database to yield comparable E-values, and the target FPR estimation was performed on the target and decoy entries from the protein database alone. The resulting E-value threshold was then applied to all identified peptides, and all remaining decoy identifications were discarded. Because of this setup, we were able to identify putative GPF peptides at a predefined estimated target FPR.
From a total of 12,216 model peptides, 49.5% of the peptides could be independently identified by PEAKS/GPF. A protein was considered for localization only if it was identified with at least two distinct peptides or at least one GPF-confirmed peptide having at least two spectral counts because the PEAKS/GPF identification can be regarded as an independent verification of the database identification (16). In our chloroplast data set, 142 proteins were additionally included in the chloroplast proteome after GPF confirmation.
Identifying Possible Cleavage Sites for Chloroplast Transit Peptides
To check whether a transit peptide consensus sequence can be predicted from the mass spectrometric data, OMSSA was used to search for semitryptic peptides. The parameters used for the search were the same as described above except that enzymatic cleavage was set to “semitryptic.” Due to the tryptic digestion of the protein mixtures, we would expect fully tryptic peptides in the sample. However, if a semitryptic peptide can be identified in the sample and the non-tryptic cleavage site is located at the N terminus, we can assume that the cleavage might have happened not due to the tryptic digestion but due to a transit peptide having been cleaved off the protein. From all significantly identified, proteotypic semitryptic peptides, those that were not part of the experimentally deduced chloroplast proteome or not previously annotated as chloroplast proteins were discarded. In addition, we required a non-tryptic cleavage site at the N terminus of the semitryptic peptide and that the resulting transit peptide is no longer than 100 amino acids. Finally, all deduced transit peptides that consist of a single methionine residue were discarded. Whenever multiple semitryptic peptides could be identified with the same protein, the one yielding the shortest transit peptide was selected. The 20 amino acids surrounding the putative transit peptide cleavage site were recorded and converted into a WebLogo (see Fig. 5B).
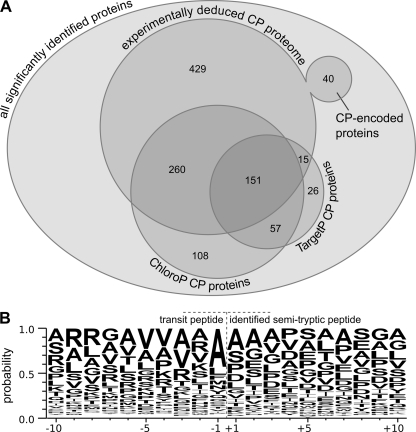
Fig. 5.
Analyses of organelle targeting prediction and transit peptide cleavage specificity using chloroplast-localized proteins. A, from a total of 2315 significantly identified proteins (each of which was identified with at least two distinct peptides or at least one GPF-confirmed peptide at an estimated peptide FPR of 1%), 895 proteins could be localized to the chloroplast (CP). 40 of these proteins are chloroplast-encoded, and the remaining 855 proteins are encoded in the nucleus. The majority of the proteins that were localized to the chloroplast by the localization prediction tools ChloroP and TargetP overlap with the experimentally deduced chloroplast proteome. B, the WebLogo of the identified semitryptic peptides shows an increased probability for alanine at the C terminus of the chloroplast transit peptide as well as the trend that the charge of the 10 amino acids before the cleavage site of the transit peptide is overall positive, whereas the charge of the 10 amino acids after the cleavage site is overall negative. A total of 111 significantly identified semitryptic transit peptides were used along with the amino acid residues surrounding each of the putative transit peptide cleavage sites to generate the WebLogo.
Automated Protein Quantitation from MS Data Using qTrace
For each band, the list of significantly identified peptides and the MS1 scans acquired were passed to the novel open source SILAC quantitation tool qTrace,3 which identifies peaks in every MS1 scan and checks whether the isotope envelope patterns of the previously identified peptides are present. For the peak determination, local maxima were determined. For each of these points, a set of three points consisting of the local maximum plus the two adjacent points was used to fit a Gaussian curve through the peak, given that the signal to noise ratio was at least 2. The center and the area under the Gaussian curve were then used as the peak m/z value and intensity.
For every identified peptide, a target isotope envelope pattern was created, consisting of three isotope peaks for the unlabeled peptide (A, A + 1, and A + 2) and three isotope peaks for the labeled peptide (A*, A* + 1, and A* + 2). In addition to these six peaks, the absence of the A − 1 peak was required to ensure that the A peak was not actually the A + n peak of another isotope envelope. Because proline can be synthesized from arginine, the quantitation results of proline-containing peptides can be biased toward the unlabeled sister peptide due to the fact that an unknown amount of the proline residues has been labeled as well. To accommodate for this, an additional set of three isotope peaks was required for every proline residue, covering all possibilities from one to all proline residues being labeled.
The mass accuracy used for checking for peak presence was set to 5 ppm. To test for peak absence, a mass accuracy of 30 ppm was used to increase the confidence that the A − 1 peak is really absent, even if there were small deviations in mass accuracy. Three peaks were regarded for every isotope envelope, and isotope envelopes were created for charge states 2 and 3.
The raw output of qTrace was a list of peptide quantitation events, which were then filtered using the following steps.
Most abundant band selection. From the MS2 identification results, the most abundant band was determined for each protein. Subsequently, all quantitation events that occurred in other bands except for the most abundant band (±1 band) were discarded.
MS2 coupling. All quantitation events for which no MS2 identification event in the same band was available within 60 s of the full scan were discarded.
Normalization. To accommodate for inaccuracies in the sample preparation process, the quantitation results were normalized to a small set of proteins expected to remain unaffected by the different treatment of the samples (Lhca3, PsaD, Lhca1, and PsaB).
During the course of MS1 full scans, two sister peptides with a certain charge state appear at some point, and their abundances increase and then decrease again during the course of elution until the precursor signals have disappeared. It can be noticed that for some peptide pairs with very different abundances the elution of the less abundant peptide starts later and ends earlier than the elution of the more abundant peptide. Also, the elution profile of one peptide might be shifted by a small amount due to elution artifacts. To accommodate for these effects, the ratios were not determined on a per scan basis but were calculated for every elution profile by grouping the individual quantitation events by their combination of peptide, band, and charge state (PBC combination). Adding the unlabeled and labeled amounts within each PBC group, a ratio was determined for each of these groups. The protein ratio was then calculated as the mean value of the individual PBC group ratios.
For those proteins that could only be quantified in a single PBC combination, the ratios of the individual scans were used to determine a mean and S.D. In general, all quantitation results with a relative S.D. of more than 0.6 were discarded.
A subset of the protein quantitation results were determined by using a targeted accurate mass/time tag (AMT) strategy. This subset contains all proteins that were only found in a single state plus a couple of proteins of interest that could not be quantified using the high throughput quantitation pipeline, which used MS2 coupling to confirm full-scan quantitation events.
For the AMT quantitation, the MS2 coupling step was replaced by a peptide retention time filter. Because of this, all proteotypic peptides that were identified in all MS/MS runs, including non-SILAC runs, could be used for the AMT quantitation, leading to potentially more raw quantitation events. To assess the correctness of a raw AMT quantitation event, the full-scan retention time was required to be within 1 min of a previously determined average peptide retention time that was determined from various SILAC and non-SILAC MS/MS runs. To accommodate for slight variations in the HPLC elution profiles, retention time offsets were determined for every individual MS/MS run by the following procedure: the retention time alignment was performed using all peptides that were identified in more than one band. For each of these peptides, the retention time of the first identification was determined for every band. After that, the average retention time for a peptide in all bands was subtracted from the actual retention time of that peptide in a certain band, yielding per peptide retention time alignments for every band. To reflect the retention time shift for a whole band, the median of the peptide retention time differences of a band was determined and later used for correcting the retention times of the raw AMT quantitation results.
RT-PCR
The Promega ImPromII Reverse Transcription system was used for RT-PCR. Sampling, RNA extraction, and cDNA synthesis were performed following the supplier's manual. A total of 1.5 ml of cells at a density of 3–4 × 106 cells/ml were spun down and resuspended per 1 ml of TRI Reagent. Anaerobiosis was induced as described above, and duplicate samples (2 × 1 ml of TRI Reagent) were taken for each anaerobic and aerobic sample. After the RNA drying step, the duplicate samples were merged by resuspending the RNA to a total volume of 20 μl. The following primers were used: hyd1, same primers were used as for the hydA1 by Kruse et al. (30); cgld22 forward primer (F), AAGTCGAGCAGCAGCTCT; cgld22 reverse primer (R), AGGTTGCCATCAAACACATCC; cpld51 (F), TGTGCCTGGCATCTGGTC; cpld51 R, CCCAGCTCCTTCCAGAAG; tef5 F, CTGGAGATCTACCCGGTCAA; tef5 R, GGTCCATGCCCTCAAAGTACA; tef7 F, AAGCAGGTGAAGGTGTCGT; tef7 R, GGAGTCCTTCTCCTTAATGACC; JGI v3.1: 183968 F, AAGGCCGATGGTGAGTACGC; JGI v3.1: 183968 R, AAGACAGAGCGTCCTGCAGG; JGI v3.1: 114525 F, TTCTTCCACAAGGACGGCAT; JGI v3.1: 114525 R, GGCTTGACGTGGATCCACTT; JGI v3.1: 190196 F, AAGAGCACAGTTGCCATC; JGI v3.1: 190196 R, CCGATCAGCTGCAGGATA; tub1 F, TGCCTGCAGGGCTTCCAGG; tub1 R, GGGATCCACTCGACGAAGTA.
RESULTS
Confirmation of Anaerobic Conditions with Induction of hyd1 and Formate Production
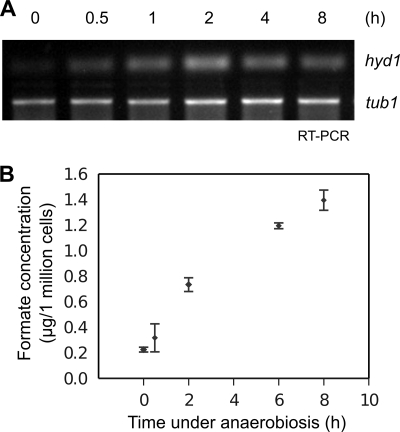
To analyze the changes in the chloroplast and mitochondria proteomes under anaerobic conditions, hydrogenase 1 transcript (hyd1) and formate production levels were investigated to confirm anaerobic conditions after 8 h of argon bubbling. In addition to hydrogen production, formate is a known by-product under anaerobic conditions (31). Fig. 2A shows a fast and continuous hyd1 expression level under anaerobiosis. This is in line with the nearly 7-fold increase in formate levels after 8 h of argon bubbling (Fig. 2B).
Fig. 2.
Hyd1 transcript levels and formate production increase under anaerobiosis. A, Hyd1 transcript levels in C. reinhardtii increased under anaerobic conditions as seen by RT-PCR of samples taken at 0, 0.5, 2, 4, and 8 h of argon gas bubbling. B, formate accumulated under continuous anaerobic conditions. Cell samples in the light were measured for formate levels after 0, 0.5, 2, 6, and 8 h of argon gas bubbling. Error bars indicate the standard deviation between measurements from two biological samples.
Isolated Organelles from Aerobically and Anaerobically Grown Cells Show Marginal Amounts of Contamination
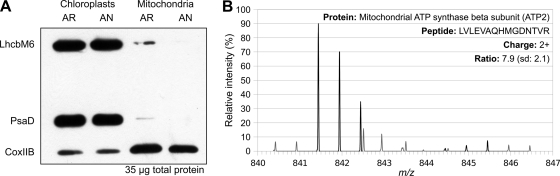
Immunoblot analysis of isolated mitochondria and chloroplasts was performed against chloroplast (LhcbM6 and PsaD) and mitochondrial (CoxIIB) marker proteins to investigate the purity of the isolated samples from aerobic and anaerobic cultures. The immunoblot shown in Fig. 3A demonstrates a low, but equal amount of mitochondrial protein contamination in the aerobic and anaerobic chloroplast samples as well as some minor contamination from chloroplast proteins in the mitochondrial samples. Presence of mitochondrial protein in the chloroplast sample could be detected in MS/MS-SILAC analysis of mitochondria mixed with labeled chloroplasts as seen in Fig. 3B where the mitochondrial ATP synthase β subunit (ATP2) was also detected in the heavy isotope form (see below and Table IV). The ratio of ATP2 present in the mitochondrial sample to ATP2 present in the chloroplast sample is 10.9 (S.D. 3.5), consistent with the 10–15% mitochondria contamination level in the Chloroplasts intuited from the immunoblots.
Fig. 3.
Immunoblot and MS/MS-SILAC analyses of chloroplast and mitochondrial samples show low contamination levels. A, immunoblot analysis of 35 μg of total protein from chloroplast and mitochondrial samples from AR and AN conditions. Detection using antibodies against chloroplast (LhcbM6 and PsaD) and mitochondrial (CoxIIB) marker proteins shows low, but constant contamination of mitochondrial proteins in the chloroplast samples. On the other hand, the mitochondrial samples contain significantly less chloroplast protein contamination. B, MS/MS-SILAC analysis of mitochondrial samples mixed with labeled chloroplasts reveals detection of mitochondrial proteins in the heavy isotope range. For the ATP2, the ratio of mitochondria to chloroplasts is 10.895 (S.D. 3.545). A spectrum for a peptide belonging to this protein is shown.
Table IV. Quantitation of mitochondrial samples against labeled chloroplast samples provides information on sample contamination levels and further confirms protein localization.
SILAC was performed on AN (induced by 8 h of argon bubbling) and AR C. reinhardtii mitochondrial samples against labeled chloroplast samples (CP, chloroplast; MT, mitochondria; PX, peroxisome). 30 μg of protein from each of the respective mitochondria samples were loaded together with 30 μg of protein of labeled chloroplast sample and separated by SDS-PAGE; bands were digested with trypsin; and the peptides were measured by the LTQ Orbitrap XL hybrid FTMS, identified using SEQUEST and OMSSA, and quantified using the MS/MS-SILAC method with the quantitation program qTrace. Candidate chloroplast-localized proteins from this study are indicated with an asterisk (*). Proteins localized from this data set alone without additional localization confirmation from Atteia et al. (15) are marked with a dagger (†).
Qualitative Proteomics Combined with Spectral Counting Led to Identification of Chloroplast Core Proteome
The SDS-PAGE-fractionated proteins from aerobic and anaerobic chloroplast and mitochondrial samples were excised, digested in-gel with trypsin, and measured separately using the LTQ Orbitrap XL hybrid FTMS. Peptides and corresponding proteins were identified and quantitatively analyzed using spectral count as an indicator for protein abundance (32). Three independent runs were measured for each condition (aerobic chloroplasts, anaerobic chloroplasts, aerobic mitochondria, and anaerobic mitochondria) stemming from a total of eight biological samples. 2315 proteins were identified from 612 measured samples at an estimated FPR of 1% with at least two distinct peptides identified by OMSSA (27) or with one peptide identified independently by combining de novo amino acid sequencing using PEAKS (29) as well as GPF (16) and OMSSA (Table I and supplemental Table 2). Of these 2315 proteins, 1800 proteins were identified from the chloroplast samples, and 1498 proteins were identified from the mitochondrial samples. Some proteins were identified in both the chloroplast and mitochondrial samples due to the co-isolation of mitochondrial proteins in the chloroplast samples and vice versa in addition to those proteins that may be co-localized to both organelles.
Table I. 2315 total proteins were identified from chloroplast and mitochondrial samples.
Proteins were measured using the LTQ Orbitrap XL hybrid FTMS and identified with the requirement of at least two unique peptides identified by OMSSA or one peptide identified independently using OMSSA and GPF. The number of proteins identified in each condition (aerobic chloroplasts, anaerobic chloroplasts, aerobic mitochondria, and anaerobic mitochondria) is shown. These results are based on three independent runs for each condition, representing eight biological samples.
| Total proteins identified (2315) | |||
|---|---|---|---|
| Chloroplast samples (1800) |
Mitochondrial samples (1498) |
||
| Aerobic | Anaerobic | Aerobic | Anaerobic |
| 1760 | 1536 | 1406 | 1364 |
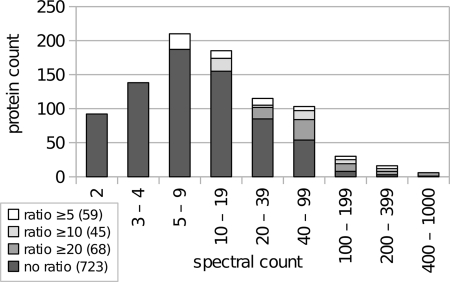
Comparison of total spectral counts between chloroplast and mitochondrial samples allows for the localization of proteins to within or outside of the chloroplast. The spectral count calculation was carried out as follows: peptide spectral counts were determined by counting the number of OMSSA peptide-spectrum matches, and based on this information, protein spectral counts were determined by adding the peptide spectral counts of all peptides that unambiguously identified a protein; peptides that appeared in multiple proteins were discarded. Of the total proteins identified with the criteria mentioned above, proteins with a ratio of chloroplast to mitochondrial spectral counts of at least 5 were localized to the chloroplast. Low abundance proteins were localized to the chloroplast only if identified solely in the chloroplast runs with at least two peptides or two spectral counts in the case of GPF-recognized peptides and none in the mitochondrial runs. This resulted in a total of 895 chloroplast-localized proteins (Fig. 4; see supplemental Table 1 for a complete list). Fig. 4 shows the distribution of spectral counts for the 895 experimentally chloroplast-localized proteins. The histogram reveals that 665 proteins have more than four spectral counts, 92 proteins have two, and another 138 proteins have three to four spectral counts. If the minimum spectral count requirement is raised to five instead of two, the number of predicted chloroplast-localized proteins drops to 665 or 606 proteins, given a minimum chloroplast/mitochondrial ratio of 5 or 10, respectively. This indicates that the C. reinhardtii chloroplast proteome data set with 606 proteins is very robust. For further analysis, however, we decided to also consider the set with 895 proteins to also have low abundance proteins such as ACK1 and HYD1 with two and four spectral counts, respectively, included. However, we consider the 606 proteins resulting from a minimum of five spectral counts with a chloroplast/mitochondrial spectral count ratio of 10 to be safe chloroplast-localized proteins. The remaining 289 proteins that were included when the requirement was set at a minimum spectral count of 2 and a chloroplast/mitochondrial spectral count ratio of 5 will be referred to as candidate chloroplast-localized proteins. The chloroplast proteins belonging to this group of 289 proteins will henceforth be indicated with an asterisk (*).
Fig. 4.
Distribution of spectral counts for 895 chloroplast-localized proteins. Proteins were measured using the LTQ Orbitrap XL hybrid FTMS and identified using OMSSA and GPF. The histogram demonstrates that the majority of the proteins were identified in the range of 5–19 spectra. In addition, it shows how the size of the experimentally deduced chloroplast proteome is affected with increasing stringency of protein inclusion requirements. Requiring at least five spectra for every protein and a ratio of at least 10 still results in a chloroplast proteome of 606 proteins.
The question may arise whether cytosolic contamination is higher in the chloroplast samples as compared with the mitochondrial samples, which could result in false localization of cytosolic proteins to the chloroplast. Within the 2315 total identified proteins, 62 cytosolic ribosomal protein subunits were found. However, when the ratio of chloroplast to mitochondrial spectral count is examined for these 62 proteins, the ratio ranges from 0 to 1.3 with a mean ratio at 0.48. This implicates that the mitochondrial samples have a higher abundance of cytosolic proteins and, seeing that none of these 62 identified cytosolic ribosomal subunit proteins made it into the chloroplast proteome list, even with the lowest stringency, one could expect low amounts of false localization of cytosolic proteins to the chloroplast.
Of the 895 proteins, 855 are nucleus-encoded, and 40 are chloroplast-encoded (supplemental Table 1). We tested organelle target prediction tools such as ChloroP (33) or TargetP (34) (Fig. 5A) and asked how many of the 855 nucleus-encoded experimentally chloroplast-localized proteins were targeted to the chloroplast. ChloroP predicted 411 proteins to possess a chloroplast transit sequence, whereas TargetP only sorted 166 proteins into the chloroplast (Fig. 5A and Table II) and 357 proteins into the mitochondria (supplemental Table 1). This suggests that TargetP has a bias and sorts C. reinhardtii chloroplast proteins predominantly to the mitochondria. In this respect, for C. reinhardtii proteins, ChloroP seems to be a better tool for the prediction of chloroplast localization, although it seems to have a large false negative rate. It is of note that of the 166 proteins predicted by TargetP 151 polypeptides were also recognized by ChloroP. In addition, ChloroP predicted another 165 and TargetP predicted 83 proteins to be chloroplast-localized that are not included in the experimentally determined chloroplast proteome (Fig. 5A). Interestingly, 72 proteins were predicted by TargetP to be secretory pathway proteins (see Table II and supplemental Table 1). Among these proteins, 26 proteins possessed a signal peptide signature, including proteins such as ATPG (chloroplast ATP synthase subunit II precursor), an aspartic proteinase, and HSP70G, suggesting that trafficking of proteins into the chloroplast may also involve vesicular transport via the secretory pathway as described for vascular plants (35).
Table II. Summary of chloroplast-localized proteins analyzed using organelle targeting programs and BLAST algorithm.
From a total of 895 chloroplast-localized proteins, 279 proteins were also localized to the chloroplast by PPDB. The 855 nucleus-encoded, chloroplast-localized proteins were processed by the localization prediction tools ChloroP and TargetP, yielding an overlap of 411 proteins for ChloroP and 166 proteins for TargetP. CP, chloroplast. Numbers in parentheses refer to the safe CP proteome.
| Description | Candidate + safe CP proteome | Safe CP proteome |
|---|---|---|
| Total CP proteome size | 895 | 606 |
| With a chloroplast/mitochondria ratio ≥5 (10) | 172 | 113 |
| With no mitochondria identifications but at least 2 (5) chloroplast identifications | 723 | 493 |
| Nucleus-encoded | 855 | 570 |
| Chloroplast-encoded | 40 | 36 |
| PPDB chloroplast-localized proteins | 279 | 219 |
| Comparison with localization prediction tools (regarding the 855 (570) nucleus-encoded proteins) | ||
| ChloroP chloroplast-localized proteins | 411 | 308 |
| TargetP chloroplast-localized proteins | 166 | 119 |
| TargetP mitochondria-localized proteins | 357 | 263 |
| TargetP secretory pathway-localized proteins | 72 | 40 |
| With signal peptide | 26 | 19 |
| BLAST results for the 855 (570) nucleus-encoded, chloroplast-localized proteins | ||
| Best non-Chlamydomonas hit in V. carteri | 789 | 534 |
| Results for best non-Chlamydomonas and non-Volvox best hit | ||
| Photosynthesis | ||
| Photosynthetic | 623 | 432 |
| Not photosynthetic | 122 | 76 |
| Unknown | 110 | 62 |
| Kingdom | ||
| Plantae | 553 | 382 |
| unknown | 110 | 62 |
| Bacteria | 87 | 61 |
| Animalia | 42 | 26 |
| Other | 63 | 39 |
| Algae-specific proteins | 68 | 42 |
To further define a putative cleavage site motif within chloroplast transit peptides of C. reinhardtii, the mass spectrometric data for the nucleus-encoded, chloroplast-localized proteins were searched for the occurrence of semitryptically digested peptides that harbor a non-tryptic cleavage site within the first 100 N-terminal amino acids as deduced from their nuclear genes. In total, 111 semitryptic peptides were found (supplemental Table 4 and supplemental Fig. 1). Manual inspection of chloroplast-imported proteins with experimentally known cleavage sites revealed that for sequences such as Lhca1 and Lhca8 (36), plastocyanin (37), PsbP (38), and PsbQ (39) transit peptide cleavage sites were correctly determined. For PsbQ, where the mature protein would start with LTPVDLFDDR rather than TPVDLFDDR (39), high similarity was also observed, although the two sequences differ by one amino acid. The 20 amino acids surrounding the putative transit peptide cleavage site were recorded and converted into a WebLogo (40) (Fig. 5B). The WebLogo shows only a weak pattern except that the probability for an alanine residue at the C terminus of the transit peptide is around 40%. In addition, it should be of note that the charge of the 10 amino acids before the cleavage site of the transit peptide is overall positive, whereas the charge of the 10 amino acids after the cleavage site is overall negative. These patterns might be useful to improve prediction programs that assess chloroplast targeting for C. reinhardtii proteins with unknown location.
It should be of note that the semitryptic peptide survey also revealed putative regulatory proteolytic cleavages. For LhcbM3, for example, a putative transit peptide with a length of 31 amino acids has been recorded, suggesting that the mature protein would start with the amino acid sequence APASSGIEFY. However, protein phosphorylation of Thr residue 24 or 27 of LhcbM3 has been reported (19), indicating that a longer phosphorylated and a shorter non-phosphorylated form of LhcbM3 exist as suggested previously (19).
To evaluate the C. reinhardtii chloroplast proteome further, BLAST searches were performed on the 855 nucleus-encoded, chloroplast-localized proteins. Of these proteins, 789 proteins had the best non-Chlamydomonas hit to Volvox carteri protein sequences, confirming the close relation between these two organisms. For 623 proteins, the non-Chlamydomonas and non-Volvox best hit was to an ortholog from a photosynthetic organism. Of the remaining nucleus-encoded, chloroplast-localized proteins, the non-Volvox best hit was to 122 proteins for sequences from non-photosynthetic organisms, and 110 proteins were deduced from DNA sequences stemming from environmental samples (Table II and supplemental Table 1). It is of note that from our chloroplast proteome list 137 proteins are GreenCut proteins of which 98 belong to the 606 safe chloroplast-localized proteins, and the remaining 39 GreenCut proteins stem from the candidate chloroplast-localized proteins. Interestingly, a number of proteins localized to the chloroplast in C. reinhardtii have similarities to non-photosynthetic organisms. This suggests that the C. reinhardtii chloroplast is a site for metabolic activities that are not necessarily directly linked with photosynthesis and carbon fixation in addition to demonstrating the large array of metabolic processes that allow C. reinhardtii to live in non-photosynthetic conditions. In total, 279 of the 895 putative chloroplast proteins were predicted to be chloroplast-localized by PPDB, the Plant Proteome Database for Arabidopsis thaliana and maize (41, 42), and accordingly have orthologs in vascular plants (see Table II and supplemental Table 1). It is of note that about 553 C. reinhardtii proteins possess vascular plant orthologs, indicating that in addition to the known PPDB proteins novel chloroplast vascular plant proteins can be predicted. Furthermore, 47 algae-specific chloroplast proteins could be determined in this study (see Table II) of which 39 are proteins of unknown function, suggesting that many Chlamydomonas-specific metabolic aspects are not yet fully understood.
Spectral Counting Allows for Localization of Key Proteins Involved in Anaerobic Response
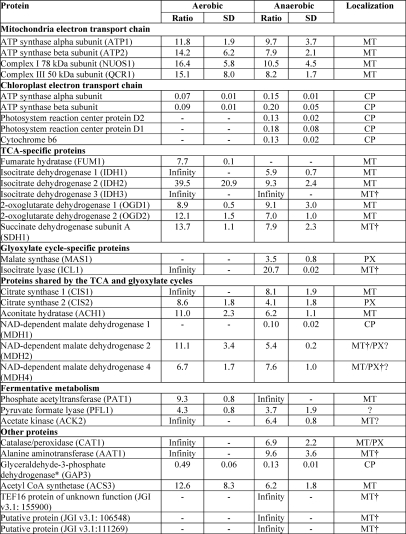
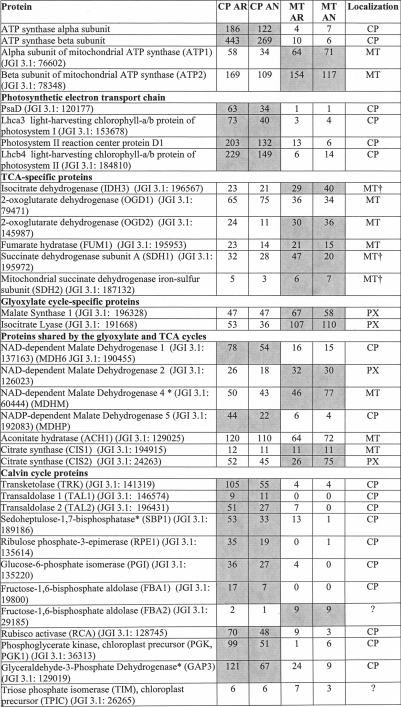
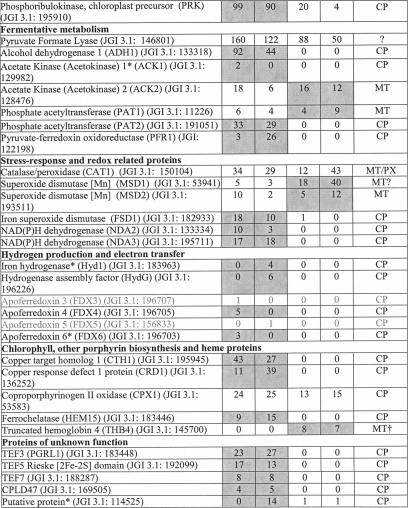
To localize C. reinhardtii metabolic processes to the chloroplast, the putative chloroplast proteome was searched for proteins of interest, and the proteins were grouped according to the corresponding metabolic pathways (Table III) with a special focus on metabolic processes involved in the anaerobic response of C. reinhardtii. Spectral count values for these proteins provide information for the identification of metabolic pathways that are localized to within or outside of the chloroplast. As expected, proteins involved in the tricarboxylic acid cycle and glyoxylate cycle appear to be largely localized outside of the chloroplast (15). The majority of the tricarboxylic acid cycle proteins have been characterized as mitochondrial. However, spectral count data suggest exceptions to this notion, implying that some proteins may not exclusively be involved in these metabolic pathways. The numerous malate dehydrogenases (MDHs) found in C. reinhardtii show these tendencies. MDH2 (JGI v3.1: 126023) was localized to the peroxisome, and MDH4 (JGI v3.1: 60444) was found to be mitochondrial (15). The higher spectral counts for both proteins in the mitochondrial data further support localization outside of the chloroplast. SILAC quantitation of isolated mitochondria from aerobic and anaerobic conditions mixed with labeled chloroplasts at equal protein levels could not provide further evidence for mitochondrial or peroxisomal localization (see below and Table IV). The spectral counts for the MDH1 (JGI v3.1: 190455) and MDH5* (JGI v3.1: 192083) predict these proteins as chloroplast-localized.
Table III. Spectral counts provide insights to protein localization and abundance.
A comparison of spectral counts for chloroplast and mitochondrial samples taken from AR and AN conditions measured using the LTQ Orbitrap XL hybrid FTMS and identified using OMSSA and GPF is shown. The total spectral count from chloroplasts was compared with the total spectral count from mitochondria to determine protein localization. Proteins identified with at least five spectral counts with at least 10-fold more spectral counts in the chloroplast samples were assigned as safe chloroplast-localized proteins. Proteins belonging to the candidate chloroplast proteins, stemming from proteins identified with a minimum of two spectral counts and at least 5-fold more spectral counts in the chloroplast samples are indicated with an asterisk (*). Mitochondrial proteins were assigned based on data presented in Atteia et al. (15) except for those indicated with a dagger (†), which were localized solely from this experimental data. CP, chloroplast; MT, mitochondria; PX, peroxisome. Localization of the protein based on the spectral count to CP or MT has been indicated by the gray shading. Proteins identified with only one spectral count and therefore not included in the identified protein list are printed in gray.
It is no surprise that the Calvin cycle proteins are localized to the chloroplast (43). However, an interesting exception is the fructose-1,6-bisphosphate aldolases. Isoform 1 is clearly localized to the chloroplast as the protein was never detected in the mitochondrial samples. However, isoform 2 was detected in both anaerobic and aerobic mitochondria more abundantly than in the chloroplasts, suggesting possible localization outside of the chloroplast.
Of particular interest under anaerobic condition are proteins of the fermentative metabolism (44). Proteins such as the alcohol dehydrogenase 1 (ADH1), phosphate acetyltransferase 2 (PAT2), acetate kinase 1* (ACK1), and pyruvate-ferredoxin oxidoreductase 1 (PFR1) are only identified in the chloroplast samples. On the other hand, acetate kinase 2 (ACK2) and phosphate acetyltransferase 1 (PAT1) are more abundant in mitochondrial samples. Once again, some proteins are less clear-cut, which may suggest dual targeting such as the pyruvate-formate lyase 1 (PFL1).
Proteins involved in chlorophyll biosynthesis were also largely identified in the chloroplast samples. The overlap between proteins induced under copper deficiency and anaerobic conditions has been described previously (45, 46). The spectral count data further confirm this as seen from the higher abundance of the copper response defect 1 protein (CRD1) in the anaerobic condition, whereas this was not observed for the copper target homolog 1 (CTH1). In addition, several proteins involved in heme biosynthesis were detected in low abundance. The truncated hemoglobin, THB4, is of particular interest because it is annotated in JGI v4.0 to be chloroplast-localized; however, it was only identified in mitochondrial runs.
In addition, proteins involved in hydrogenase assembly and hydrogen production were also identified, despite the low abundance, solely in the anaerobic sample. Several isoforms of ferredoxins (FDXs) were also identified solely in the chloroplast samples (FDX3, FDX4, FDX5, and FDX6*). FDX3 and FDX5 were identified only with one spectral count and therefore are not included in our chloroplast proteome list. However, the chloroplast localization of these ferredoxins, along with all of the other above mentioned ferredoxins, have already been demonstrated in recent findings by Jacobs et al. (47) and Terauchi et al. (47). Of the four ferredoxins identified in the chloroplast, only FDX5 was detected under anaerobic conditions. This again is in line with the likely variable expression of FDXs under differing conditions (47, 48).
It is of note that in contrast to A. thaliana an NAD(P) transhydrogenase is present in the C. reinhardtii chloroplast, pointing to important differences in chloroplast redox metabolism between C. reinhardtii and vascular plants. In respect to chloroplast redox metabolism, the chloroplast localization of the type II NADH dehydrogenase NDA2 (49, 50) could be confirmed. In addition, the spectral count data indicate the chloroplast localization of another putative NADH dehydrogenase, NDA3 (JGI v3.1: 195711). However, neither of the two proteins seem to be induced under oxygen deprivation. A complete list of the 2315 proteins identified along with the spectral count information can be found in supplemental Table 2.
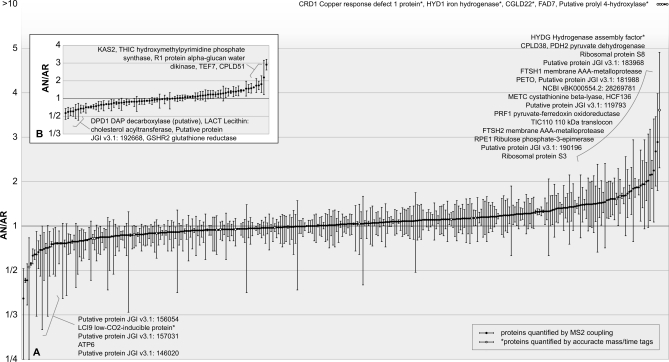
SILAC Quantitation of Chloroplast-localized Proteins Using Quantitation Program qTrace Shows Novel Proteins Induced under Anaerobic Conditions
Proteins localized in the chloroplast were quantified using the SILAC method and the quantitation program qTrace. Chloroplasts were isolated from anaerobic cultures and compared with chloroplasts from aerobic cultures labeled with a heavy isotope arginine (23). A total of 425 chloroplast proteins were quantified of which 345 proteins were quantified with multiple peptide/band/charge combinations (Fig. 6A), and 80 proteins were quantified with one peptide in a single band (Fig. 6B). 275 quantitations stem from two separate biological samples for each condition (see supplemental Table 3 for a complete list of the 425 proteins quantified). A vast majority of these proteins hover at an anaerobic/aerobic (AN/AR) ratio of 1, suggesting that after 8 h of anaerobic conditions the cells do not pursue a dramatic alteration of the proteome but rather appear to use an array of metabolic pathways and biological processes to cope with the various stresses and limitations that may accompany anaerobiosis.
Fig. 6.
Relative abundance of 425 anaerobic to aerobic Chlamydomonas chloroplast proteins quantified using quantitation program qTrace. Anaerobic chloroplasts were isolated after 8 h of argon bubbling, loaded onto the gel (the amount was based on 5 μg of chlorophyll) along with 5 μg of chlorophyll from the labeled chloroplast sample, and separated by SDS-PAGE. The bands were cut out and digested with trypsin, and the resulting peptides were measured by the LTQ Orbitrap XL hybrid FTMS followed by identification using OMSSA. Subsequently, the arginine-containing peptides were selected and quantified using qTrace. Quantitation events were confirmed by MS2 coupling (black dots) or by an AMT approach (white dots). Full scans from six independent runs stemming from two biological samples for each condition were used for quantitation, and the AN/AR ratio for each quantified protein is shown. A, 345 proteins were quantified with multiple peptide/band/charge combinations. B, 80 proteins were quantified with one peptide/band/charge combination. Error bars indicate standard deviations between different combinations of peptide, band and charge state for graph A and standard deviations between individual scans for graph B.
Consistent with proteins described previously as induced under anaerobic conditions (11, 12, 44, 51) and the spectral count data described above, the quantitation results show up-regulation (1.5-fold or more induced) of ADH1 and PFR1, both chloroplast-localized proteins involved in fermentative metabolism. ADH1 has an AN/AR ratio of 1.5 (S.D. 0.6), and PFR1 has a ratio of 1.9 (S.D. 0.01). As expected, HYD1* was detected exclusively in the anaerobic sample, whereas the hydrogenase assembly factor, HYDG, was detected at an AN/AR ratio of 3.6 (S.D. 1.3). HYDG may be present even under aerobic conditions to allow quick assembly of the hydrogenase under a sudden shift to anaerobiosis.
Aside from the hydrogenase and PFR1, other ferredoxin-interacting proteins were induced. A ferredoxin-sulfite reductase (SIR1) was induced with an AN/AR ratio of 1.6 (S.D. 0.02); SIR1 receives electrons from FDX to reduce sulfite to sulfide (52). In addition, the cystathionine β-lyase (METC) involved in cysteine and methionine metabolism, working downstream of SIR1, was quantified and was also induced with an AN/AR ratio of 1.9 (S.D. 0.5). Aside from SIR1, a protein related to the phycocyanobilin-ferredoxin oxidoreductase (PCYA; JGI v3.1: 156256), which also appears to interact with FDX, was induced with a ratio of 1.5 (S.D. 0.1). It has been shown that the PCYA from Thermosynechococcus elongatus interacts with FDX (53).
CRD1 was also largely induced under anaerobiosis with an AN/AR ratio of 15.9 (S.D. 5.4), further supporting evidence for a link between the response to anaerobiosis and copper deficiency (45, 46). The paralog of CRD1, CTH1 (54), was only slightly decreased under anaerobic condition with a ratio of 0.7 (S.D. 0.2). Additionally, coproporphyrinogen III oxidase (CPX1) was induced at a ratio of 2.3 (S.D. 0.5). This protein was not included in the 895 chloroplast-localized proteins because the chloroplast to mitochondrial spectral count ratio of 1.8 was too low to fit the criteria for chloroplast localization, which required at least a ratio of 5. This protein is also described to be induced under copper deficiency as well as under anaerobic condition observed at the transcript level (8). Along the same lines, uroporphyrinogen decarboxylase (UROD2), which acts upstream of CPX1 by producing coproporphyrinogen from uroporphyrinogen, was induced under anaerobic condition with a ratio of 1.6 (S.D. 0.3). Other proteins involved in chlorophyll biosynthesis were not induced. Magnesium chelatase subunit D (CHLD) and subunit H (CHLH1) have a ratio of 1.4 (S.D. 0.3) and 0.8 (S.D. 0.3), respectively. In addition, GUN4, which was found to interact with CHLH1 to activate magnesium chelatase in A. thaliana and cyanobacteria (55, 56), showed a similar ratio of 1.3 (S.D. 0.04).
In addition, proteins involved in photosystem II degradation and assembly were induced. The two membrane AAA metalloproteases, FTSH1 and -2, which are involved in photosystem II degradation (57), were induced along with a putative photosystem assembly factor, HCF136 (58–61). FTSH1 has an AN/AR ratio of 2.2 (S.D. 1.0), FTSH2 has a ratio of 1.8 (S.D. 1.1), and HCF136 has a ratio of 1.9 (S.D. 0.8).
Another protein that was more abundant in the anaerobic sample is ribulose-phosphate 3-epimerase (RPE1) with a ratio of 1.8 (S.D. 0.5), and the rubisco activase was slightly induced with a ratio of 1.4 (S.D. 0.3). Contrarily, numerous other proteins involved in the Calvin cycle were quantified but were not found to be induced. Rubisco, sedoheptulose-1,7-bisphosphatase*, phosphoglucose isomerase, fructose-1,6-bisphosphate aldolase, transketolase, transaldolase, phosphoglycerate kinase, and glyceraldehyde-3-phosphate dehydrogenase* all had AN/AR ratios close to 1 in the range from 0.7 to 1.2. Aside from proteins involved in carbon fixation, proteins involved in fatty acid biosynthesis were identified: 3-ketoacyl-acyl carrier protein synthase* (KAS2) was induced at an AN/AR ratio of 2.9 (S.D. 0.3); FAD7, a chloroplast glycerolipid ω-3-fatty acid desaturase, was induced; and a putative protein, 157545 (JGI v3.1), annotated as being related to lipocalin, a protein involved in the transport of hydrophobic molecules, including lipids, was induced at a ratio of 1.6 (S.D. 0.4).
A number of ribosomal polypeptides were increased at the protein level. The plastid ribosomal proteins L21 and L31 have an AN/AR ratio of 1.5 (S.D. 0.3) and 1.7 (S.D. 0.2), respectively. Ribosomal proteins S3 and S8 have a ratio of 1.7 (S.D. 0.5) and 2.3 (S.D. 1.2), respectively. Although these ribosomal proteins were induced, this was not characteristic for all ribosomal proteins because the majority of the ribosomal proteins quantified were not induced. These were plastid ribosomal proteins L1, L2, L6, L9, L13, L14, L15, L16, L19, L23, L28, S1, S2, S4, S5, S6, S9, S15, S18, and S16 and plastid-specific ribosomal proteins 1 and 3. The ratios for these proteins ranged from 0.7 to 1.4. Protein expression is known to be controlled largely through the regulation of translation (62, 63) and the chloroplast-unique ribosomal structures found especially on the small subunit potentially play an important role in the translation regulation (64). These control mechanisms may explain the varying levels of the ribosomal subunits between the two conditions.
23 putative proteins were induced under anaerobic conditions. 19 putative proteins were identified in the results stemming from multiple peptide/band/charge combinations of which 13 have an AN/AR ratio in the range of 1.5–2.9, whereas two were exclusively detected in the anaerobic sample. A total of four induced putative proteins were identified in the results stemming from a single peptide/band/charge combination with ratios ranging from 1.6 to 1.7. Among these 23 putative proteins, TEF5, CGLD22, CPLD38, and CPLD51* (JGI v3.1: 192099, 102133, 121963, and 120574) are part of the GreenCut proteins, making these candidates of particular interest (22). TEF5 is annotated to have a Rieske [2Fe-2S] domain, CPLD51* is required for the covalent heme binding to cytochrome b6f during assembly, and CGLD22 is annotated as being similar to the ATP synthase 1 protein. In addition, TEF5 and a putative prolyl 4-hydroxylase* (JGI v3.1: 114525) are annotated as having oxidoreductase activity. Proteins with oxidoreductase activity could possibly be important in the context of anaerobiosis due to the consequent accumulation of reducing equivalents (65). Other interesting candidates include a protein with weak similarity to PsbP (JGI v3.1: 183968) and a membrane protein, YCF78 (NCBI BK000554.2: 28269781). Three proteins have a calcium-binding EF-hand (JGI v3.1: 154066, 179211*, and 183554).
To assess the source of deviation within the individual PBC quantitation results, the 20 most divergently quantified proteins were examined. In more than 50% of the cases, the high deviation was either due to single outlier peptides, which might be the result of an unrecognized modification or a protein appearing in multiple isoforms, or due to band smear in which the sister peptides seem to have been distributed to slightly different bands, possibly due to varying running characteristics of the SDS-PAGE. Only a minor fraction of these variations seem to stem from variations in the biological samples or the preparations. It should be noted, however, that these variations are reflected in the relative standard deviation and can thus be considered during interpretation of the results.
It is of note that the semiquantitative spectral counts also show differences in abundance for proteins differentially expressed to a large degree between aerobic and anaerobic conditions. This can be observed, for example, with the PFR1 where there were three spectral counts under aerobic conditions and 26 under anaerobic conditions. In addition, CRD1, HYD1*, HYDG, and putative prolyl 4-hydroxylase* (JGI v3.1: 114525) all suggest induction from the higher spectral counts under anaerobic conditions. In this way, for the highly induced proteins, induction is already evident from the semiquantitative spectral count data, which support the SILAC results. However, the slight adjustments of the chloroplast proteome in response to the anaerobiosis could only be revealed through direct quantitation using SILAC.
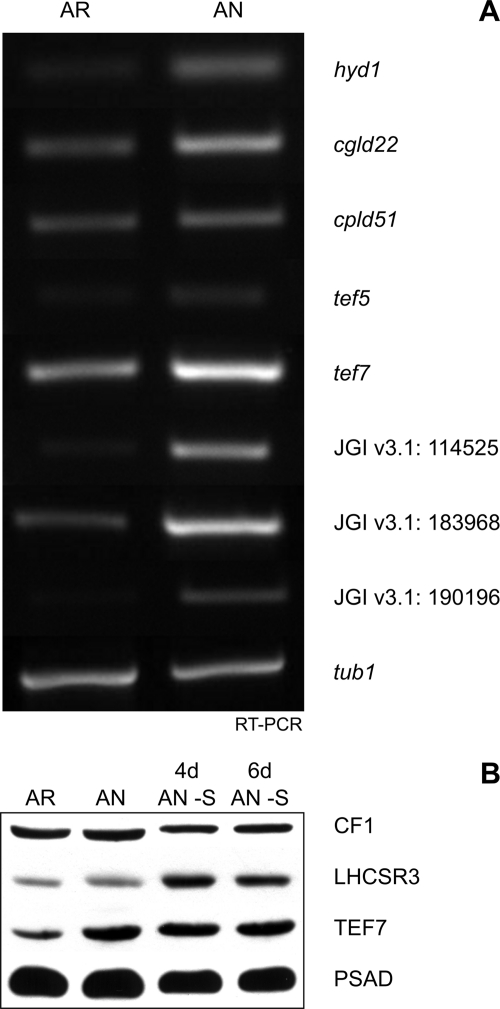
Selected Putative Proteins Show Induction Also at Transcript Level
RT-PCR performed on selected putative proteins observed to be induced in the proteomics data were also found to be induced, at varying degrees, at the transcript level (Fig. 7A). Candidates with especially high induction at the transcript level were hyd1* and genes encoded by gene models JGI v3.1: 114525*, 183968, and 190196*. High expression for hyd1* and JGI v3.1: 114525 encoding for a putative prolyl 4-hydroxylase* were expected because both were only found at the protein level under anaerobic conditions. In contrast, CGLD22 (JGI v3.1: 102133) was also only found under anaerobic conditions; however, at the transcript level, it was only slightly induced. The moderate induction that was seen for tef5, tef7, and cpld51* (JGI v3.1: 192099, 188287, and 120574) is reflected in their slight increase at the protein level under anaerobic conditions with values of 1.5 (S.D. 0.2), 1.7 (S.D. 0.2), and 1.7 (S.D. 0.4), respectively. TEF7 induction was also seen under sulfur-deprived anaerobic conditions as seen from immunoblot analysis (Fig. 7B). Interestingly, transcript levels of genes JGI v3.1: 190196* encoding for a putative signal peptide peptidase and JGI v3.1: 183968 encoding for a PsbP-like protein that were found to be significantly higher under anaerobic versus aerobic conditions were simply 1.8- and 2.2-fold induced at the protein level by oxygen deprivation. Although the quantitative RT-PCR data are confirmative for the quantitative proteomics data, differences may reflect another level of regulation due to post-transcriptional mechanisms.
Fig. 7.
RT-PCR analyses of selected proteins of unknown function found to be induced under anaerobic conditions from the SILAC quantitative data show varying inductions at transcript level, whereas immunoblot detection of CF1 β subunit of chloroplast ATP synthase, LHCSR3, TEF7, and PsaD shows consistencies with SILAC data. A, Cgld22, cpld51*, and tef5 are GreenCut proteins (21). Cgld22 (JGI v3.1: 102133) is a protein similar to ATP synthase I, cpld51* (JGI v3.1: 120574) is a protein required for cytochrome b6f assembly, and tef5 (JGI v3.1: 192099) has a Rieske [2Fe-2S] domain. Tef7 is a Chlamydomonas-specific protein of unknown function. Protein 114525 is a prolyl 4-hydroxylase*, protein 183968 has a weak homology to PsbP, and protein 190196* is annotated as a signal peptide peptidase-like protein. TEF5 and protein 114525* have been annotated as having oxidoreductase activity. B, immunoblot analysis of 5 μg of chlorophyll of AR, AN, 4-day sulfur-deprived anaerobic cultures (4d AN −S), and 6-day sulfur-deprived anaerobic cultures (6d AN −S) using αCF1, αLHCSR3, αTEF7, and αPsaD antibodies. The immunoblot demonstrates induction of TEF7 under all anaerobic conditions, whereas LHCSR3 is induced only under sulfur-deprived anaerobic conditions.
SILAC Quantitation of Mitochondrial Proteins against Labeled Chloroplasts Further Confirms Localization and Level of Organelle Contamination
Isolated mitochondria from aerobic and anaerobic conditions were mixed based on equal amounts of protein with labeled chloroplasts and quantified using the SILAC method and qTrace. Because of the presence of mitochondrial proteins in the chloroplast sample and vice versa (Fig. 3), highly abundant mitochondrial and chloroplast proteins can be quantified. The resulting ratios provide further localization confirmation (Table IV). Abundant mitochondrial proteins, such as the components of the electron transport chain, quantified in this manner have a ratio ranging from around 8 to 16. Contrarily, proteins of the chloroplast electron transport chain have a ratio lower than 0.2. The ability for SILAC quantitation between these two samples shows the extent of contamination, which appears to be in the range of 10–15% (see also Fig. 3). Contamination levels appear to be slightly lower in the aerobic conditions, resulting in generally higher ratios for mitochondrial proteins as seen when the mitochondrial electron transport chain ratios are compared. Quantitation of other proteins allowed for confirmation of mitochondrially localized proteins due to derived ratios similar to those of the electron transport chain proteins or simply due to the fact that a ratio could not be obtained because the protein was absent in the chloroplast samples. This phenomenon was observed for many of the tricarboxylic acid cycle-specific proteins. Interestingly, the glyoxylate cycle-specific malate synthase 1 (MAS1) and isocitrate lyase 1 (ICL1) show varying ratios. MAS1 has a ratio of 3.5 (S.D. 0.8) for the anaerobic sample. Contrarily, the ICL1 was detected only in the mitochondria under aerobic conditions and had a significantly higher ratio of 20.7 (S.D. 0.02) under anaerobic conditions. Previous research has localized both proteins to the peroxisome (15). However, quantitative data suggest different localization patterns with MAS1 being possibly localized in the peroxisome and ICL1 being mitochondrial because the ratio of the latter is comparable to those of the mitochondrial electron transport chain proteins.
In line with previous research as well as the spectral count results presented above, the proteins involved in both the tricarboxylic acid and the glyoxylate cycles show inconsistent ratios, suggesting variation in localization or dual localization to peroxisomes and mitochondria for selected proteins. The two isoforms for citrate synthase (CIS1 and -2) have been previously localized to mitochondria and the peroxisome, respectively. The quantitative data show a difference between these isoforms. CIS1 was only found in the mitochondria for the aerobic sample and has a ratio of 8.1 (S.D. 1.9) for the anaerobic sample. On the other hand, CIS2 has a lower ratio in both conditions, 8.6 (S.D. 1.8) under aerobic conditions and 4.1 (S.D. 1.8) under anaerobic conditions. The slightly lower ratios seem to be consistent with proteins characterized to be peroxisomal as proposed for MAS1.
Three proteins of the fermentative metabolism could be quantified: PAT1, PFL1, and ACK2. From the ratios, PAT1 and ACK2 are mitochondrial as already suggested (7). PAT1 has a ratio of 9.3 (S.D. 0.8) for aerobic mitochondria and was present only in the anaerobic mitochondria. ACK2 was found only in the aerobic mitochondria and has a ratio of 6.4 (0.8) under anaerobic conditions. A slightly lower ratio for ACK2 under anaerobic conditions may indicate differential expression or localization of the protein under anaerobic conditions. A lowered ratio solely under anaerobic conditions implies that the protein is no longer as abundant in the mitochondria but provides no indication whether this protein is down-regulated overall. This can also explain other proteins with slightly inconsistent ratios between the aerobic and anaerobic conditions as seen for ACH1, MDH2, MDH4, ACK1, and CAT1.
The localization of PFL1 is not as clear-cut as inferred previously (14). In both conditions, the ratios were significantly lower: 4.3 (S.D. 0.8) in aerobic conditions and 3.7 (S.D. 1.9) in anaerobic conditions. This suggests that PFL1 is not exclusively localized to the mitochondria. However, it is of note that PFL1 was not induced under anaerobic conditions either in chloroplast or in mitochondrial samples. Quantitative data of PFL1 from the chloroplast sample resulted in a ratio of 1.2 (S.D. 0.8) (data not shown). Despite the high abundance of the protein (260 scans), the ratios determined by qTrace varied greatly as seen from the large standard deviation, which could be due to the variable mitochondrial contamination levels of the chloroplast samples. Whole cells were quantified to get accurate quantitative data for PFL1. On the whole cell level, PFL1 was found with a ratio of 0.6 (S.D. 0.4, 58 scans, seven peptide/band/charge combinations; data not shown), supporting the notion that PFL1 is not induced during the first hours after the onset of anaerobic conditions. Despite the high abundance of PFL1, the PFL-activating enzyme was not identified. However, the abundance of PFL1 even in anaerobic conditions suggests the protein to be regulated by activation rather than via an increase in protein abundance because an increase in the formate level can be observed after the onset of anaerobic conditions (Fig. 2B). In addition to the PFL1, catalase (CAT1) appears also to be dually localized (15) and was also quantified on the whole cell level (data not shown). CAT1 appears to be somewhat induced at a ratio of 1.7 (S.D. 0.7, 14 scans, four peptide/band/charge accounts). Induction of CAT1 is consistent with previous findings (7). A lower ratio of CAT1 in the mitochondrial samples in relation to chloroplast samples under anaerobic conditions suggests differential localization in which CAT1 is induced outside of the mitochondria, probably in the peroxisome, under anaerobic conditions.
DISCUSSION
Qualitative and quantitative analyses of chloroplast and mitochondrial samples derived from aerobically and anaerobically grown C. reinhardtii cells provided new insights into the chloroplast proteome and the anaerobic response of C. reinhardtii. Taking advantage of differential spectral counting, a safe chloroplast proteome of 606 proteins was identified in addition to 289 candidate chloroplast proteins. This experimentally determined set of chloroplast proteins contains numerous algae-specific chloroplast proteins and proteins with orthologs in vascular plants not identified in previous chloroplast proteome studies (66–69).
Of particular interest are proteins that have not been identified in the chloroplast before and are candidate genes involved in metabolic pathways important for anaerobic acclimation. Novel algae-specific chloroplast proteins that were induced under low oxygen (at least 1.5-fold) and may therefore have functions in the anaerobic response are proteins 154066, 172018, 181988, 188054*, 188281, and TEF7 (JGI v3.1). Protein 182514 is algae- and cyanobacteria-specific, and protein 183968, which show similarities to PsbP, also is largely algal in addition to having similarities in higher plants. Furthermore, proteins such as CGLD22 (JGI v3.1: 102133), CPLD38 (JGI v3.1: 121963), CPLD51* (JGI v3.1: 120574), 119793, and 114525* (JGI v3.1), which share highest similarity with vascular plant proteins, were up-regulated under oxygen deprivation and localized to the chloroplast.
Spectral Count Analyses to Localize Identified Proteins
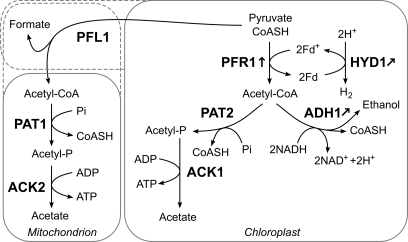
Spectral count data allowed further insights on the localization of proteins involved in fermentative metabolism. The localizations of ACK1, ADH1, PAT2, pyruvate decarboxylase (PDC), and PFR1 have been speculated although not biochemically confirmed (7, 14). The spectral count data confirmed the proteins that are highly likely to be localized to the chloroplast. Proteins with strong evidence for chloroplast localization are ACK1*, ADH1, PAT2, and PFR1 (Fig. 8). These proteins were identified with spectral counts in the chloroplast samples exclusively. ACK1*, PAT2, and PFR1 are in accordance with the localization posed by Mus et al. (7). On the other hand, ADH1 has been proposed to be cytosolic, but our data strongly suggest ADH1 to be localized to the chloroplast as we identified the protein with a total of 104 spectra in the chloroplast and none in mitochondrial samples. This further suggests that the reason Atteia et al. (15) were unable to detect the ADH1 in the proteomics analysis of mitochondrial samples is due to the proteins being localized in the chloroplast and not due to the absence of the protein in physiological conditions as seen from the presence of the protein under aerobic conditions in rather high abundance in our data. Interestingly, despite the high abundance of ADH1, PDC was identified with one peptide without GPF confirmation in the mitochondrial sample (data not shown) and therefore was not included in the data set of identified proteins. Our identification of PDC with one peptide does not provide strong localization insights; however, the identification is in line with the previous suggestion that PDC is a cytosolic protein (7). The extremely low abundance of PDC and the separate localization of the ADH1 suggest that ADH1 largely acts downstream of PFL1 or PFR1 rather than downstream of PDC under these conditions.
Fig. 8.
Localization of major fermentative pathways identified in this study. Localization via spectral count analysis performed in this study showed several proteins involved in the fermentative pathways in C. reinhardtii to be localized to the chloroplast. Confirmed chloroplast-localized enzymes are PFR1, PAT2, ACK1, and ADH1. PAT1 and ACK2 have been localized to the mitochondrion by Atteia et al. (15), and our spectral count was in accordance with these findings as these proteins were not localized to the chloroplast. The localization of PFL1 is still unclear from our data, which may be due to previously suggested dual localization of the protein (14, 15). Fd, ferredoxin.
Our spectral count data further confirm the observed dual localization of PFL1, inferred by the high abundance of the protein in both chloroplast and mitochondrial samples (14). In addition, Atteia et al. (15) have characterized the CAT1 as being localized both to mitochondria and the peroxisome. Our spectral count data also provide no clear-cut localization insights as the protein was highly abundant in both chloroplast and mitochondrial samples.
Looking at the proteins involved in the fermentative metabolism and spectral count-derived localization, it appears that pyruvate metabolism involving PFL1 and/or PFR1 with ADH1 leading to the production of ethanol takes place in the chloroplast along with another branch involving PFL1 and/or PFR1, PAT2, and ACK1* leading to acetate production. In parallel, the same biochemical pathway catalyzed by PFL1, PAT1, and ACK2 appears to take place outside of the chloroplast (Fig. 8). The presence of fermentative metabolism proteins in various cellular compartments reflects the ability for C. reinhardtii to cope with various metabolic demands brought about by a range of environmental conditions. Dually localized proteins, parallel pathways, and pathways that appear to cross organelle boundaries also suggest close communication and collaboration between the organelles.
It is likely that the cell utilizes the pathway involving ADH1 in the chloroplast due to the resulting oxidation of two NADH molecules per acetyl-CoA consumption. During anaerobiosis, reducing equivalents accumulate faster than they can be consumed (65), leading to the obvious benefits of C. reinhardtii favoring pyruvate metabolism that helps to rebalance the NAD(P)H/NAD(P)+ ratio. Suppressing ADH1 activity may be a straightforward approach to increase reducing equivalents in the chloroplast and force more hydrogenase activity in an attempt to compensate for the decreased NADH consumption. Identification of the HYD1* and HYDG in anaerobic chloroplasts further confirms hydrogenase localization and assembly in the chloroplast (70, 71) as well as points to its relevance for the regulation of the chloroplast redox poise for anaerobic acclimation. Further investigation is necessary to confirm whether electron transfer to hydrogenase is catalyzed solely by FDX1 and to understand the role of the anaerobically induced FDX5 (this study and Refs. 47 and 48).
Comparative Quantitative Proteomics
From our SILAC-MS/MS quantitation results of chloroplast-localized proteins, we detected the HYD1* exclusively in the anaerobic chloroplast samples. This along with induction of ADH1 and PFR1 suggests that the characterized fermentative metabolism and hydrogen production dominate under anaerobic conditions. Induction of ADH1 under anaerobiosis is in agreement with the real time PCR analyses performed by Mus et al. (7). In addition, numerous studies have reported the induction of PFL1 at the transcript level (14, 72). However, this was not found to be the case from the protein level in the chloroplast.
Aside from fermentative metabolism, many microorganisms reduce sulfate to hydrogen sulfide under anaerobic conditions with the use of 6 reducing equivalents stemming from FDX reducing the sulfite reductase (73). Quantitation showed induction of SIR1 also in C. reinhardtii under anaerobic conditions. Mus et al. also found this protein to be induced at the transcript level under dark anaerobiosis (7). With accumulating reducing equivalents, induction of SIR1 may allow an increased rate of FDX oxidation, allowing the cell to utilize FDXs for sulfur reduction and assimilation. In the presence of sulfur, its reduction could act in competition to hydrogen production. The induction of METC under anaerobic conditions is also an indicator that SIR1 acts in the direction to oxidize FDX as METC acts downstream of the reduced SIR1. The induction of PCYA is also in line with this strategy. It is conceivable that if these FDX-oxidizing pathways were inhibited more electrons could be passed to the hydrogenase for hydrogen production.
CRD1 was also found to be highly induced. CRD1, a protein involved in chlorophyll biosynthesis under copper-deficient conditions, is also induced under anaerobiosis (45). Aside from CRD1, other proteins induced under copper deficiency are also induced under anaerobiosis (46). CPX1 is another protein that falls in this category observed in our data. The overlap between response to copper deficiency and anaerobiosis has been characterized but not fully understood (74). It has been proposed, as observed also in Saccharomyces cerevisiae, that under anaerobiosis the oxygen-dependent CPX1 becomes the rate-limiting step in heme synthesis (75). To compensate for this limitation, the cell responds by producing more of the protein. Quinn et al. (74) suggest that the same is true for CRD1 under anaerobiosis. Induction of CRD1 and CPX1 at the protein level from our data further supports this notion.
Nguyen et al. (8) used sulfur deficiency to induce anaerobiosis. Although there are similarities between the transcriptome data obtained by Nguyen et al. (8) and our proteomics data, there are also large differences, which may be due to the different conditions in which anaerobiosis was induced. Contradiction in the results may separate sulfur starvation-specific responses from the anaerobic response. From the sulfur deficiency data, it is clear that the expression of stress-related light-harvesting lhcsr genes are largely up-regulated (8, 10). In contrast, LHCSR proteins are clearly not induced after 8 h of argon bubbling, pointing to the fact that stress-related responses that require efficient thermal dissipation of light energy (76) observable in sulfur deficiency are absent in argon-induced anaerobic conditions. Immunoblot analysis of LHCSR3 protein between aerobic, anaerobic, and sulfur-deprived anaerobic conditions showed induction of LHCSR3 only under the sulfur-deprived anaerobic conditions (Fig. 7B). This suggests that the observed induction of lhcsr3 at the transcript level is also consistent at the protein level but appears to be an effect seen largely under the sulfur-depleted anaerobic condition. In addition, Nguyen et al. (8) observed the LhcbM9 protein to be induced. We were unable to quantify a unique LhcbM9 peptide. However, quantification of peptide YRELELIHAR shared by all LhcbM proteins showed a general AN/AR ratio of 1.2 (S.D. 0.4). This is based on 10 independent quantitation events. Single unique peptides for LhcbM3 and LhcbM5 were successfully quantified, resulting in a ratio of 0.8 (S.D. 0.09) and 0.6 (S.D. 0.1), suggesting, if anything, a minor decrease in these two proteins. The slightly higher ratio of the general LhcbM peptide compared with the ratios of LhcbM3 and LhcbM5 suggests variations in LhcbM protein expression.
Nguyen et al. (8) observed a decrease in proteins involved in amino acid synthesis, and Timmins et al. (9) observed a decrease in amino acid abundance. Our spectral count and quantitative data showed no marked decrease of proteins in this category. One example is the aspartate-semialdehyde dehydrogenase (ASSD) (JGI v3.1: 148810), which was decreased in the transcriptome data but was not found to be the case in our SILAC-MS/MS data with a ratio of 1.4 (S.D. 0.5).
Along the same lines, the transcriptome data showed strong suppression of the Calvin cycle probably also due to sulfur deficiency stress response, whereas our data showed no change with the exception of selective proteins being induced. RPE1 was significantly induced along with a slight induction of rubisco activase. RPE1 is involved in the conversion of d-ribulose 5-phosphate to d-xylulose 5-phosphate, which takes part in the Calvin cycle and the non-oxidative part of the pentose phosphate pathway. The Calvin cycle, sometimes also referred to as the reductive pentose phosphate cycle, leads to the consumption of reducing equivalents. The marked differences in the expression levels of proteins involved in the Calvin cycle under sulfur-depleted and sulfur-replete anaerobic conditions might be also explained by a key difference in the redox state of the cell. Under sulfur starvation, the cells face oxidative stress (10) due to limitations in photosynthetic electron transfer in the presence of an oxygenic atmosphere, whereas under anaerobiosis in sulfur-replete conditions, the cells face the accumulation of reducing equivalents because respiratory electron transfer is restricted due to oxygen deprivation (65). In this way, it appears as though under sulfur-replete anaerobic conditions the Calvin cycle is utilized to consume some of the accumulating reducing equivalents to balance the NAD(P)H/NAD(P)+ ratio. It has also been suggested that the RPE1 is likely to be situated in the metabolic network that allows for the rate control of the non-oxidative part of the pentose shunt pathway, which could explain the induction of this protein under reducing anaerobic conditions (77). In addition, the induction of the α-glucan, water dikinase R1 protein and α-amylase, which are both involved in starch degradation (78), might suggest a higher turnover rate of starch production and degradation under anaerobic conditions.
Contrarily, under sulfur-deficient anaerobic conditions, Nguyen et al. (8) reported an increase in 6-phosphogluconate dehydrogenase, an enzyme involved in the oxidative pentose phosphate pathway, coupled with a decrease in proteins involved in the reductive Calvin cycle. However, under sulfur-replete anaerobic conditions, no induction of proteins involved in the oxidative pentose phosphate cycle and glycolysis was observed.
The putative proteins found to be induced in the SILAC-MS/MS quantitative data shed light to new interesting candidates for further investigation. One interesting candidate highly induced under anaerobiosis is the putative prolyl 4-hydroxylase* (JGI v3.1: 114525), which was also observed by Mus et al. (7) to be induced at the transcript level under dark anaerobiosis as well as by Nguyen et al. (8) at the transcript level under anaerobiosis induced by sulfur starvation (listed as contig 15.82.2.11). This putative prolyl 4-hydroxylase* was localized to the chloroplast by the spectral count data and was exclusively identified in the anaerobic sample. We also found the protein to be induced at the transcript level (Fig. 7A). Mus et al. (7) proposed a possible role of the prolyl hydroxylases to be acting as oxygen sensors that initiate the metabolic switch to aerobic respiration when oxygen becomes available again. Although not a concern under the dark anaerobic conditions investigated by Mus et al. (7), under anaerobic conditions in the light implemented in this study, prolyl hydroxylases could not only act to sense the return of aerobic conditions but could also act as scavengers for minimal oxygen present in the chloroplast produced from the photosynthetic electron transport chain. Under anaerobic conditions, minimal oxygen in the chloroplast can cause harm due to an abundance of reducing equivalents, which can in turn lead to the production of ROS. Oxygen produced from photosynthesis under anaerobiosis induced by argon bubbling may be too scarce or may diffuse out of the cell too quickly to support aerobic respiration, but the production of ROS could be instantaneous under a highly reducing environment. Production of ROS could be reduced if the prolyl hydroxylases could directly consume the produced oxygen. This can in turn act hand in hand with CAT1, which was slightly induced in the whole cell quantitative results. Although CAT1 was not localized to the chloroplast, it may have a role in preventing and eliminating ROS in other cellular compartments. Up-regulation of a putative lipocalin protein (JGI v3.1: 157545) may also act to control ROS damage. It has been shown in A. thaliana that the chloroplast lipocalins are involved in the protection of thylakoid membrane lipids from ROS by preventing lipid peroxidation (79). The induction of KAS2* and FAD7 involved in fatty acid biosynthesis may also be a sign for prepared defense against potentially increased lipid damage, permitting rapid lipid replenishment in the case of lipid peroxidation in the thylakoid membrane.
Proteins that allow for the cell to control the redox state and prevent the formation of and the damage from ROS would be of importance for the cell to survive under anaerobic conditions in the light. Proteins with oxidoreductase activities such as the putative prolyl hydroxylase and TEF5 are interesting new targets because of their possible role in regulating/balancing the NAD(P)H/NAD(P)+ ratio. Compromising proteins that may consume the accumulating reducing equivalents may lead to more electrons being funneled to the hydrogenase for increased hydrogen production. Other interesting putative proteins are those grouped in the GreenCut proteins (22): TEF5, CPLD38, CPLD51*, and CGLD22. GreenCut proteins are naturally of interest because of their likely importance in relation to photosynthesis. Considering the interaction of ferredoxin and hydrogenase with the photosynthetic electron transport chain, these induced putative GreenCut proteins are also of great interest in the context of anaerobiosis and hydrogen production. TEF5 with its putative Rieske [2Fe-2S] domain is a promising candidate gene product that might be involved in light-driven electron transfer processes related to oxygen deprivation. The induction of CPLD51*, a factor involved in covalent heme binding to cytochrome b6f during assembly (80), might implicate that other heme-containing proteins are induced under low oxygen because the cytochrome b6f itself remains rather constant. This is further supported by the observed induction of CPX1 and UROD2, proteins involved in heme biosynthesis. A function cannot be suggested for algae-specific chloroplast proteins such as (JGI v3.1) 172018, 181988, 188054*, 188281, and TEF7 along with algae- and cyanobacteria-specific protein 182514 that possess no annotated known protein domain and were induced under oxygen deprivation. However, because hydrogen production by means of hydrogenases is also unique to green algae and cyanobacteria, these proteins may be related to such functions. Immunoblot analysis using αTEF7 antibody confirmed the induction of TEF7 under anaerobic conditions (Fig. 7B). In addition, TEF7 also showed an induction under sulfur-deficient anaerobic conditions and may play an important role in acclimation to anaerobic stress.
Qualitative and quantitative proteomics of chloroplast and mitochondrial samples stemming from aerobic and anaerobic C. reinhardtii cultures allowed for the localization of 606 proteins (with an additional 289 candidates) to the chloroplast, including many proteins involved in fermentative metabolism. Further SILAC quantitative analyses of chloroplast-localized proteins resulted in the identification of proteins induced under anaerobic condition. These included HYD1*, HYDG, and key proteins in the fermentative metabolism such as PFR1 and ADH1. The majority of the proteins were mildly induced, demonstrating that the anaerobic response comprises an array of metabolic changes, likely allowing for versatile and fast acclimation to anaerobiosis. In addition to these insights, quantitative data provided new targets for further research, including proteins with oxidoreductase and ROS-protective activities as well as many proteins of unknown functions that were induced under anaerobiosis. Many of these proteins were also found to be up-regulated at the transcript level. This discovery-driven proteomics approach along with the quantitation program qTrace allowed for fast and effective protein identification and quantitation, leading to the discovery of novel targets for further hypothesis-driven projects.
Acknowledgments
We are grateful to Susan Hawat for measuring mass spectrometric data during the early stages of the project, Ingrid Hirschmann for providing quantitative mass spectrometric data from whole cells, and Irina Tolstygina for providing sulfur-deprived cell samples.
 This article contains supplemental Tables 1–4 and Fig. 1.
This article contains supplemental Tables 1–4 and Fig. 1.
2 M. Specht, M. Stanke, and M. Hippler, manuscript in preparation.
3 M. Specht, M. Terashima, B. Naumann, and M. Hippler, manuscript in preparation.
1 The abbreviations used are:
- OMSSA
- Open Mass Spectrometry Search Algorithm
- ACK
- acetate kinase
- ADH1
- alcohol dehydrogenase 1
- AMT
- accurate mass/time tag
- ATP2
- mitochondrial ATP synthase β subunit
- CAT1
- catalase
- CHL
- magnesium chelatase subunit
- CIS
- citrate synthase
- CPX1
- coproporphyrinogen III oxidase
- CoxIIB
- cytochrome c oxidase subunit II
- CRD1
- copper response defect 1 protein
- CTH1
- copper target homolog 1
- FAD7
- glycerolipid ω-3-fatty-acid desaturase
- FDX
- ferredoxin
- FPR
- false positive rate
- FTSH
- membrane AAA metalloproteases
- GPF
- Genomic Peptide Finder
- HYD1
- Fe-hydrogenase
- ICL1
- isocitrate lyase 1
- KAS2
- 3-ketoacyl-acyl carrier protein synthase
- Lhca
- light-harvesting chlorophyll a/b protein of photosystem I
- LhcbM
- light-harvesting chlorophyll a/b protein of photosystem II
- MAS1
- malate synthase 1
- MDH
- malate dehydrogenase
- NDA
- NADH dehydrogenase
- METC
- cystathionine β-lyase
- PAT
- phosphate acetyltransferase
- PCYA
- phycocyanobilin-ferredoxin oxidoreductase
- PDC
- pyruvate decarboxylase
- PFL
- pyruvate-formate lyase
- PFR1
- pyruvate-ferredoxin oxidoreductase 1
- PsaB
- photosystem I protein subunit PsaB
- PsaD
- photosystem I protein subunit PsaD
- PsbB
- photosystem II protein subunit PsbB
- PsbP
- photosystem II protein subunit PsbP
- PsbQ
- photosystem II protein subunit PsbQ
- ROS
- reactive oxygen species
- RPE1
- ribulose-phosphate 3-epimerase
- SIR1
- ferredoxin-sulfite reductase
- UROD2
- uroporphyrinogen decarboxylase
- SILAC
- stable isotope labeling with amino acids in cell culture
- JGI
- Joint Genome Institute
- NCBI
- National Center for Biotechnology Information
- PBC
- peptide, band, and charge state
- F
- forward primer
- R
- reverse primer
- PPDB
- Plant Proteome Database
- AN
- anaerobic
- AR
- aerobic
- BLAST
- Basic Local Alignment Search Tool
- FTMS
- FT mass spectrometer
- HYDG
- Hydrogenase assembly factor
- LHCSR
- Stress-related chlorophyll a/b binding protein
- THIC
- Hydroxymethylpyrimidine phosphate synthase
- LACT
- Lecithin:cholesterol acyltransferase
- PETO
- Cytochrome b6f complex subunit V.
REFERENCES
- 1.Hemschemeier A., Melis A., Happe T. (2010) Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynth. Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaffron H., Rubin J. (1942) Fermentative and photochemical production of hydrogen in algae. J. Gen. Physiol. 26, 219–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melis A. (2007) Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae). Planta 226, 1075–1086 [DOI] [PubMed] [Google Scholar]
- 4.Fouchard S., Hemschemeier A., Caruana A., Pruvost J., Legrand J., Happe T., Peltier G., Cournac L. (2005) Autotrophic and mixotrophic hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. Appl. Environ. Microbiol. 71, 6199–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Happe T., Hemschemeier A., Winkler M., Kaminski A. (2002) Hydrogenases in green algae: do they save the algae's life and solve our energy problems? Trends Plant Sci. 7, 246–250 [DOI] [PubMed] [Google Scholar]
- 6.Klöck G., Kreuzberg K. (1991) Compartmented metabolite pools in protoplasts from the green alga Chlamydomonas reinhardtii: changes after transition from aerobiosis to anaerobiosis in the dark. Biochim. Biophys. Acta 1073, 410–415 [DOI] [PubMed] [Google Scholar]
- 7.Mus F., Dubini A., Seibert M., Posewitz M. C., Grossman A. R. (2007) Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282, 25475–25486 [DOI] [PubMed] [Google Scholar]
- 8.Nguyen A. V., Thomas-Hall S. R., Malnoë A., Timmins M., Mussgnug J. H., Rupprecht J., Kruse O., Hankamer B., Schenk P. M. (2008) Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell 7, 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timmins M., Zhou W., Rupprecht J., Lim L., Thomas-Hall S. R., Doebbe A., Kruse O., Hankamer B., Marx U. C., Smith S. M., Schenk P. M. (2009) The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur deprivation. J. Biol. Chem. 284, 23415–23425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Shrager J., Jain M., Chang C. W., Vallon O., Grossman A. R. (2004) Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot. Cell 3, 1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubini A., Mus F., Seibert M., Grossman A. R., Posewitz M. C. (2009) Flexibility in anaerobic metabolism as revealed in a mutant of Chlamydomonas reinhardtii lacking hydrogenase activity. J. Biol. Chem. 284, 7201–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman A. R., Croft M., Gladyshev V. N., Merchant S. S., Posewitz M. C., Prochnik S., Spalding M. H. (2007) Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10, 190–198 [DOI] [PubMed] [Google Scholar]
- 13.Hemschemeier A., Happe T. (2005) The exceptional photofermentative hydrogen metabolism of the green alga Chlamydomonas reinhardtii. Biochem. Soc. Trans. 33, 39–41 [DOI] [PubMed] [Google Scholar]
- 14.Atteia A., van Lis R., Gelius-Dietrich G., Adrait A., Garin J., Joyard J., Rolland N., Martin W. (2006) Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281, 9909–9918 [DOI] [PubMed] [Google Scholar]
- 15.Atteia A., Adrait A., Brugière S., Tardif M., van Lis R., Deusch O., Dagan T., Kuhn L., Gontero B., Martin W., Garin J., Joyard J., Rolland N. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26, 1533–1548 [DOI] [PubMed] [Google Scholar]
- 16.Allmer J., Naumann B., Markert C., Zhang M., Hippler M. (2006) Mass spectrometric genomic data mining: novel insights into bioenergetic pathways in Chlamydomonas reinhardtii. Proteomics 6, 6207–6220 [DOI] [PubMed] [Google Scholar]
- 17.Michelet L., Zaffagnini M., Vanacker H., Le Maréchal P., Marchand C., Schroda M., Lemaire S. D., Decottignies P. (2008) In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J. Biol. Chem. 283, 21571–21578 [DOI] [PubMed] [Google Scholar]
- 18.Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 19.Stauber E. J., Fink A., Markert C., Kruse O., Johanningmeier U., Hippler M. (2003) Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2, 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi K., Beligni M. V., Prieto S., Haynes P. A., McDonald W. H., Yates J. R., 3rd, Mayfield S. P. (2003) Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to th e70 S ribosome. J. Biol. Chem. 278, 33774–33785 [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi K., Prieto S., Beligni M. V., Haynes P. A., McDonald W. H., Yates J. R., 3rd, Mayfield S. P. (2002) Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell 14, 2957–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naumann B., Stauber E. J., Busch A., Sommer F., Hippler M. (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280, 20431–20441 [DOI] [PubMed] [Google Scholar]
- 24.Eriksson M., Gardestrom P., Samuelsson G. (1995) Isolation, purification, and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol. 107, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busch A., Rimbauld B., Naumann B., Rensch S., Hippler M. (2008) Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 55, 201–211 [DOI] [PubMed] [Google Scholar]
- 26.Hippler M., Klein J., Fink A., Allinger T., Hoerth P. (2001) Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 28, 595–606 [DOI] [PubMed] [Google Scholar]
- 27.Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 28.Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 29.Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 30.Kruse O., Rupprecht J., Bader K. P., Thomas-Hall S., Schenk P. M., Finazzi G., Hankamer B. (2005) Improved photobiological H2 production in engineered green algal cells. J. Biol. Chem. 280, 34170–34177 [DOI] [PubMed] [Google Scholar]
- 31.Kreuzberg K. (1984) Starch fermentation via a formate producing pathway in Chlamydomonas reinhardtii, Chlorogonium elongatum and Chlorella fusca. Physiol. Plant. 61, 87–94 [Google Scholar]
- 32.Wang G., Wu W. W., Zeng W., Chou C. L., Shen R. F. (2006) Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J. Proteome Res. 5, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 33.Emanuelsson O., Nielsen H., von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 35.Villarejo A., Burén S., Larsson S., Déjardin A., Monné M., Rudhe C., Karlsson J., Jansson S., Lerouge P., Rolland N., von Heijne G., Grebe M., Bako L., Samuelsson G. (2005) Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 36.Bassi R., Soen S. Y., Frank G., Zuber H., Rochaix J. D. (1992) Characterization of chlorophyll a/b proteins of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 267, 25714–25721 [PubMed] [Google Scholar]
- 37.Redinbo M. R., Yeates T. O., Merchant S. (1994) Plastocyanin: structural and functional analysis. J. Bioenerg. Biomembr. 26, 49–66 [DOI] [PubMed] [Google Scholar]
- 38.Mayfield S. P., Rahire M., Frank G., Zuber H., Rochaix J. D. (1987) Expression of the nuclear gene encoding oxygen-evolving enhancer protein 2 is required for high levels of photosynthetic oxygen evolution in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 84, 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayfield S. P., Schirmer-Rahire M., Frank G., Zuber H., Rochaix J. D. (1989) Analysis of the genes of the OEE1 and OEE3 proteins of the photosystem II complex from Chlamydomonas reinhardtii. Plant Mol. Biol. 12, 683–693 [DOI] [PubMed] [Google Scholar]
- 40.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friso G., Giacomelli L., Ytterberg A. J., Peltier J. B., Rudella A., Sun Q., Wijk K. J. (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Q., Zybailov B., Majeran W., Friso G., Olinares P. D., van Wijk K. J. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37, D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suss K. H., Prokhorenko I., Adler K. (1995) In situ association of Calvin cycle enzymes, ribulose-1,5-bisphosphate carboxylase/oxygenase activase, rerredoxin-NADP+ reductase, and nitrite reductase with thylakoid and pyrenoid membranes of Chlamydomonas reinhardtii chloroplasts as revealed by immunoelectron microscopy. Plant Physiol. 107, 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gfeller R. P., Gibbs M. (1984) Fermentative metabolism of Chlamydomonas reinhardtii: I. analysis of fermentative products from starch in dark and light. Plant Physiol. 75, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moseley J., Quinn J., Eriksson M., Merchant S. (2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinn J. M., Barraco P., Eriksson M., Merchant S. (2000) Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275, 6080–6089 [DOI] [PubMed] [Google Scholar]
- 47.Jacobs J., Pudollek S., Hemschemeier A., Happe T. (2009) A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett. 583, 325–329 [DOI] [PubMed] [Google Scholar]
- 48.Terauchi A. M., Lu S. F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J. N., Knaff D. B., Farmer P. J., Lemaire S. D., Hase T., Merchant S. S. (2009) Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 284, 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desplats C., Mus F., Cuiné S., Billon E., Cournac L., Peltier G. (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J. Biol. Chem. 284, 4148–4157 [DOI] [PubMed] [Google Scholar]
- 50.Jans F., Mignolet E., Houyoux P. A., Cardol P., Ghysels B., Cuiné S., Cournac L., Peltier G., Remacle C., Franck F. (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 105, 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hemschemeier A., Fouchard S., Cournac L., Peltier G., Happe T. (2008) Hydrogen production by Chlamydomonas reinhardtii: an elaborate interplay of electron sources and sinks. Planta 227, 397–407 [DOI] [PubMed] [Google Scholar]
- 52.Nakayama M., Akashi T., Hase T. (2000) Plant sulfite reductase: molecular structure, catalytic function and interaction with ferredoxin. J. Inorg. Biochem. 82, 27–32 [DOI] [PubMed] [Google Scholar]
- 53.Okada K. (2009) HO1 and PcyA proteins involved in phycobilin biosynthesis form a 1:2 complex with ferredoxin-1 required for photosynthesis. FEBS Lett. 583, 1251–1256 [DOI] [PubMed] [Google Scholar]
- 54.Moseley J. L., Page M. D., Alder N. P., Eriksson M., Quinn J., Soto F., Theg S. M., Hippler M., Merchant S. (2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larkin R. M., Alonso J. M., Ecker J. R., Chory J. (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906 [DOI] [PubMed] [Google Scholar]
- 56.Sobotka R., Dühring U., Komenda J., Peter E., Gardian Z., Tichy M., Grimm B., Wilde A. (2008) Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J. Biol. Chem. 283, 25794–25802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindahl M., Spetea C., Hundal T., Oppenheim A. B., Adam Z., Andersson B. (2000) The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komenda J., Nickelsen J., Tichý M., Prásil O., Eichacker L. A., Nixon P. J. (2008) The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 22390–22399 [DOI] [PubMed] [Google Scholar]
- 59.Meurer J., Plücken H., Kowallik K. V., Westhoff P. (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plücken H., Muller B., Grohmann D., Westhoff P., Eichacker L. A. (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532, 85–90 [DOI] [PubMed] [Google Scholar]
- 61.Shen G., Zhao J., Reimer S. K., Antonkine M. L., Cai Q., Weiland S. M., Golbeck J. H., Bryant D. A. (2002) Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 277, 20343–20354 [DOI] [PubMed] [Google Scholar]
- 62.Barkan A., Goldschmidt-Clermont M. (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572 [DOI] [PubMed] [Google Scholar]
- 63.Eberhard S., Drapier D., Wollman F. A. (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31, 149–160 [DOI] [PubMed] [Google Scholar]
- 64.Manuell A. L., Quispe J., Mayfield S. P. (2007) Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol. 5, e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein U., Betz A. (1978) Fermentative metabolism of hydrogen-evolving Chlamydomonas moewusii. Plant Physiol. 61, 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjölander K., Gruissem W., Baginsky S. (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362 [DOI] [PubMed] [Google Scholar]
- 67.Kleffmann T., von Zychlinski A., Russenberger D., Hirsch-Hoffmann M., Gehrig P., Gruissem W., Baginsky S. (2007) Proteome dynamics during plastid differentiation in rice. Plant Physiol. 143, 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peltier J. B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A. J., Rutschow H., van Wijk K. J. (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5, 114–133 [DOI] [PubMed] [Google Scholar]
- 69.Peltier J. B., Emanuelsson O., Kalume D. E., Ytterberg J., Friso G., Rudella A., Liberles D. A., Söderberg L., Roepstorff P., von Heijne G., van Wijk K. J. (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Happe T., Mosler B., Naber J. D. (1994) Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 222, 769–774 [DOI] [PubMed] [Google Scholar]
- 71.Happe T., Naber J. D. (1993) Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 214, 475–481 [DOI] [PubMed] [Google Scholar]
- 72.Hemschemeier A., Jacobs J., Happe T. (2008) Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot. Cell 7, 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knaff D. B., Hirasawa M. (1991) Ferredoxin-dependent chloroplast enzymes. Biochim. Biophys. Acta 1056, 93–125 [DOI] [PubMed] [Google Scholar]
- 74.Quinn J. M., Eriksson M., Moseley J. L., Merchant S. (2002) Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol. 128, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zagorec M., Labbe-Bois R. (1986) Negative control of yeast coproporphyrinogen oxidase synthesis by heme and oxygen. J. Biol. Chem. 261, 2506–2509 [PubMed] [Google Scholar]
- 76.Peers G., Truong T. B., Ostendorf E., Busch A., Elrad D., Grossman A. R., Hippler M., Niyogi K. K. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521 [DOI] [PubMed] [Google Scholar]
- 77.Karmali A., Drake A. F., Spencer N. (1983) Purification, properties and assay of D-ribulose 5-phosphate 3-epimerase from human erythrocytes. Biochem. J. 211, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reimann R., Ritte G., Steup M., Appenroth K. J. (2002) Association of alpha-amylase and the R1 protein with starch granules precedes the initiation of net starch degradation in turions of Spirodela polyrhiza. Physiol. Plant. 114, 2–12 [DOI] [PubMed] [Google Scholar]
- 79.Levesque-Tremblay G., Havaux M., Ouellet F. (2009) The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J. 60, 691–702 [DOI] [PubMed] [Google Scholar]
- 80.Kuras R., Saint-Marcoux D., Wollman F. A., de Vitry C. (2007) A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 9906–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]