Abstract
Apolipoprotein E (apoE) is a 34-kDa glycoprotein secreted from various cells including hepatocytes and macrophages and plays an important role in remnant lipoprotein clearance, immune responses, Alzheimer disease, and atherosclerosis. Cellular apoE and plasma apoE exist as multiple glycosylated and sialylated glycoforms with plasma apoE being less glycosylated/sialylated than cell-derived apoE. Some of the glycan structures on plasma apoE are characterized; however, the more complicated structures on plasma and cellular/secreted apoE remain unidentified. We investigated glycosylation and sialylation of cellular and secreted apoE from primary human macrophages by one- and two-dimensional gel electrophoresis and mass spectrometry. Our results identify eight different glycoforms with (HexNAc)2-Hex2-(NeuAc)2 being the most complex glycan detected on Thr194 in both cellular and secreted apoE. Four additional glycans were identified on apoE(283–299), and using β-elimination/alkylation by methylamine in vitro, we identified Ser290 as a novel site of glycan attachment. Comparison of plasma and cellular/secreted apoE from the same donor confirmed that cell-derived apoE is more extensively sialylated than plasma apoE. Given the importance of the C terminus of apoE in regulating apoE solubility, stability, and lipid binding, these results may have important implications for our understanding of apoE biochemistry.
Apolipoprotein E (apoE)1 is a 34-kDa glycosylated apolipoprotein of 299 amino acids. ApoE is synthesized and secreted by most cells including hepatocytes, smooth muscle cells, neuronal cells, and macrophages (1–3) and demonstrates extraordinary functional diversity. It has important roles in remnant lipoprotein clearance, the immune response, Alzheimer disease, cell proliferation, and lymphocyte activation (4, 5). More recent studies suggest that elevated plasma apoE precedes elevation of C-reactive protein and confers increased risk of cardiovascular death in the elderly (6). Proteomics-based approaches have identified elevated high density lipoprotein (HDL)-apoE as being associated with coronary disease (7). In contrast, macrophage-specific expression of apoE protects against atherosclerosis in mice (8, 9). The mechanisms by which macrophage apoE is antiatherogenic may include stimulating the removal of excess cholesterol from macrophage foam cells as well as anti-inflammatory, antiproliferative, and immunomodulatory properties (4, 5, 10–12). An accurate understanding of the structure of apoE secreted from macrophages is important for our understanding of its properties and its role in the atherosclerotic process.
Structural studies on apoE have provided important insights into its biological properties (13). Crystallography has demonstrated that the N-terminal domain is structured in a globular four-helix bundle with the helices orientated in an antiparallel alignment (14). The structure of the C terminus has not been resolved by crystallography, but circular dichroism spectroscopy indicates it to be highly α-helical (14). Recently, NMR studies of monomeric, full-length human apoE indicated that the C-terminal domain in the intact protein adopts a more defined structure than it does as an isolated fragment (15). Lipid binding occurs at the C terminus (residues 244–272), resulting in unfolding of the molecule into a helical hairpin with the binding region for the low density lipoprotein (LDL) receptor contained within the N terminus at its apex (16).
Mucin-type O-glycosylation is a particularly common, complex, and important post-translational modification of secreted and cell surface glycoproteins (17, 18) that is difficult to accurately characterize; however, several recent reports have facilitated analysis (19, 20). Cellular apoE and plasma apoE exist as multiple glycoforms, which vary in charge because of variable sialylation. The initial analysis of the carbohydrate content of plasma very low density lipoprotein (VLDL)-apoE by colorimetric methods and gas chromatography demonstrated that the major unmodified hexose in apoE was galactose and that N-acetylglucosamine, N-acetylgalactosamine, and sialic acid were present (21, 22). Two-dimensional gel electrophoresis (2-DE) identified up to six sialylated apoE (Es) glycoforms in cells for any given genotype and fewer sialylated glycoforms in plasma (22). ApoE does not contain the consensus sequence (NX(T/S/C)) required for N-linked glycans, and carbohydrate residues are attached to apoE via an O-linkage to residue Thr194 (23–25). More recent studies using 2-DE and MALDI-TOF/TOF (23) confirmed previous results and identified five glycosylated glycoforms of apoE in plasma VLDL with the most complex sugar structures containing two sialic acid residues (HexNAc-Hex-NeuAc-NeuAc). There were more negatively charged glycoforms present on 2-DE than were distinguished by MALDI-TOF/TOF, raising the possibility that complex structures containing more than two sialic acid residues may be inherently unstable during MS analysis. Importantly, this recent study did not analyze apoE glycoforms in, or secreted from, cells.
The purpose of this study was to undertake the first detailed characterization of the glycan structures of apoE from primary human macrophages by 1-DE, 2-DE, and mass spectrometry. We found that cellular and secreted apoE in human macrophages has at least eight different glycoforms with (HexNAc)2-Hex2-(NeuAc)2 being the most complex glycan identified. We extend previous studies by the identification of a novel site of glycan attachment on Ser290 near the functionally important apoE C terminus in addition to glycosylation of Thr194 and show that a major glycoform is present in each of the spots separated by 2-DE.
EXPERIMENTAL PROCEDURES
Chemicals
Protein A-Sepharose™ LC-4B was purchased from GE Healthcare. Goat anti-apolipoprotein E polyclonal antibodies to human apoE were obtained from Chemicon International Inc. ZOOM strips (5 × 70 mm, pH 4–7), carrier ampholyte pH 4–7, thiourea, urea, CHAPS, ultrapure dithiothreitol (DTT), ultrapure agarose, DNase I, SilverQuest™ silver staining kit, and SimplyBlue™ SafeStain were purchased from Invitrogen. Biotinylated Maackia amurensis lectin II (MAA), biotinylated Sambucus nigra bark lectin (SNA), and horseradish peroxidase (HRP)-avidin D were purchased from Vector Laboratories. α-(2→3,6,8,9)-Neuraminidase, α-(2→3)-neuraminidase, BSA, and RNase A were supplied by Sigma. LDL, acetylated LDL (AcLDL), and lipoprotein-deficient serum were prepared as described (23).
Isolation and Culture of Human Monocyte-derived Macrophages (HMDMs)
Human monocytes were isolated from white cell buffy coat concentrates from healthy donors using density gradient centrifugation after layering on Ficoll-Paque Plus (GE Healthcare). Purified monocytes were differentiated in 6-well Primaria plates (BD Biosciences) by culturing in RPMI 1640 medium containing 50 units/ml penicillin G, 50 μg/ml streptomycin, 2 mm l-glutamine, 10% heat-inactivated human serum, and 25 ng/ml macrophage colony-stimulating factor (PreproTech) for 3 days followed by culturing in the same medium without macrophage colony-stimulating factor for up to 7 days. After differentiation, the cells were washed and enriched with cholesterol by incubation in RPMI 1640 medium including 10% lipoprotein-deficient serum and 50 μg/ml acetylated LDL for 2 days. After enrichment, the cultures were washed twice with prewarmed RPMI 1640 medium and incubated in RPMI 1640 medium for between 1 and 24 h. At the indicated time points, the cells and medium samples were harvested. Cells were lysed using radioimmune precipitation assay buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 0.1% SDS, 1% Triton X-100, 0.5% deoxycholate, and protease inhibitors). White cell buffy coat concentrates and human serum were supplied by the New South Wales Red Cross blood transfusion service, Sydney, Australia. Donors were genotyped for apoE by the laboratory of Prof. D. Sullivan, Royal Prince Alfred Hospital, Sydney, Australia, by restriction enzyme analysis (24).
Isolation and Culture of Human Monocytes and Preparation of Human Plasma Proteins
Blood samples in EDTA-containing tubes were obtained from a healthy volunteer with an apoE3/3 genotype. Monocytes were isolated as described above. After density gradient centrifugation, plasma supernatant was collected. Total plasma proteins were prepared as described (25). Briefly, 12 μl of plasma was mixed with 20 μl of a 10% SDS and 2.3% DTT solution boiled at 95 °C for 5 min. The sample was diluted to 500 μl with rehydration buffer (9 m urea, 2 m thiourea, 4% CHAPS, and trace bromophenol blue). 30 μl of the sample was separated by 2-DE, and apoE was detected by Western blot.
Immunoprecipitation
To isolate apoE from cholesterol-enriched HMDMs, cell lysates and medium were immunoprecipitated using a goat antibody to human apoE and protein A-Sepharose. 1.2 mg of cell lysates and medium samples was precleared for 30 min by the addition of 50 μl of protein A-Sepharose, then mixed with 5 μl of goat anti-apoE antibody, and incubated for 1 h with rotation. After 1 h, 50 μl of protein A-Sepharose was added, and the samples were incubated for another 1 h with rotation. Beads were spun down and washed five times with radioimmune precipitation assay buffer. ApoE was eluted using rehydration buffer.
One-dimensional Electrophoresis
To detect apoE protein bands in HMDMs, 9 mg of cell lysates and corresponding medium samples were immunoprecipitated, eluted in 150 μl of sample buffer (50 mm Tris-HCl, pH 6.8, 100 mm DTT, 2% SDS, 0.1% bromphenol blue, and 10% glycerol), and separated by Tris-glycine SDS-PAGE using 10% polyacrylamide gels. ApoE was detected by Coomassie staining.
Two-dimensional Electrophoresis
To detect individual apoE glycoforms, 40 μl of immunoprecipitated apoE was subjected to 2-DE. For the first dimension, isoelectric focusing was performed with a ZOOM IPGRunner system (Invitrogen) using 7-cm, pH 4–7 strips at 2000 V-h at room temperature. Samples were then reduced in 1× NuPAGE sample reducing agent for 15 min and alkylated with 125 mm iodoacetamide for 15 min after which second-dimension SDS-PAGE was performed using NuPAGE Novex 4–12% Bis-Tris ZOOM gels. After electrophoresis, the gels were fixed, and protein spots were visualized using a SilverQuest silver staining kit (Invitrogen) and SimplyBlue SafeStain (Invitrogen) for mass spectrometry analysis. Preliminary experiments confirmed that this separation clearly distinguished apoE 2, 3, and 4 glycoforms as described (26), consistent with calculated pI values of 5.65 (apoE3), 5.81 (apoE4), and 5.52 (apoE2) (ExPASy Compute pI/Mw tool). For all experiments described herein, apoE3/3 donor macrophages were used exclusively.
In-gel Digestion
Protein spots were excised from one- or two-dimensional gels and destained to remove Coomassie stain by incubation with 100 mm NH4HCO3 in CH3CN for 1 h. Reduction and alkylation were then performed to maximize digestion efficiency. For reduction, gel spots were incubated with 10 mm DTT in 20 mm NH4HCO3 at 37 °C for 1 h, and this was followed by incubation with 25 mm iodoacetamide in 20 mm NH4HCO3 at 37 °C for 1 h for alkylation. Gel spots were dehydrated with 100 μl of 100% CH3CN for 10 min. Sequencing grade trypsin (Promega) was added to the dehydrated gel spots at 10 ng/μl in 15 mm NH4HCO3, and samples were incubated overnight at 37 °C. Digested peptides were extracted from gel spots with 1% formic acid for 10 min followed by incubation with 100% CH3CN for another 10 min. Peptide mixtures were evaporated in a SpeedVac for 1 h and finally dissolved in 0.05% heptafluorobutyric acid and 1% formic acid.
Mass Spectrometry
Q-Tof Ultima
Digest peptides were separated by nano-LC using a CapLC and autosampler system (Waters). Samples (5 μl) were concentrated and desalted onto a micro-C18 precolumn (500 μm × 2 mm; Michrom Bioresources, Auburn, CA) with H2O:CH3CN (98:2, 0.05% heptafluorobutyric acid) at 15 μl/min. After a 4-min wash, the precolumn was switched (Valco 10-port valve, Dionex) in line with a fritless nanocolumn (75 μm × ∼10 cm) containing C18 medium (5 μm, 200 Å; Magic Bioresources, Michrom) manufactured according to Gatlin et al. (27). Peptides were eluted using a linear gradient of H2O:CH3CN (98:2, 0.1% formic acid) to H2O:CH3CN (65:35, 0.1% formic acid) at ∼300 nl/min over 30 or 60 min using a CapLC system (Waters). The precolumn was connected via a fused silica capillary (25 μm × 10 cm) to a low volume tee (Upchurch Scientific) where high voltage (2400 V) was applied, and the column tip was positioned ∼1 cm from the Z-spray inlet of a Q-Tof Ultima API hybrid tandem mass spectrometer (Micromass, Manchester, UK). Positive ions were generated by electrospray, and the Q-Tof Ultima was operated in data-dependent acquisition (DDA) mode. A TOF MS survey scan was acquired (m/z 350–1700; 1 s), and the two largest multiply charged ions (counts >20) were sequentially selected by Q1 for MS/MS analysis. Argon was used as the collision gas, and an optimum collision energy was chosen (based on charge state and mass). Tandem mass spectra were accumulated for up to 2 s (m/z 50–2000). Peak lists were generated by MassLynx (version 4.0 SP4, Micromass) using the Mass Measure program and submitted to the database search program Mascot (version 2.2, Matrix Science, London, UK). Search parameters were as follows: precursor and product ion tolerances were ±0.25 and 0.2 Da, respectively; Met(O) and carboxyamidomethyl-Cys were specified as variable modifications; enzyme specificity was trypsin; one missed cleavage was possible; and the non-redundant protein database from NCBI (March 2008) was searched.
LTQ-FT Ultra
Digest peptides were separated by nano-LC using an Ultimate 3000 HPLC and autosampler system (Dionex, Amsterdam, Netherlands) as described above. Peptides were eluted using a linear gradient of H2O:CH3CN (98:2, 0.1% formic acid) to H2O:CH3CN (64:36, 0.1% formic acid) at 350 nl/min over 30 or 60 min. High voltage (1800 V) was applied to a low volume tee (Upchurch Scientific), and the column tip was positioned ∼0.5 cm from the heated capillary (T = 200 °C) of an LTQ-FT Ultra (Thermo Electron, Bremen, Germany) mass spectrometer. Positive ions were generated by electrospray, and the LTQ-FT Ultra was operated in DDA mode.
A survey scan (m/z 350–1750) was acquired in the FT ICR cell (resolution = 100,000 at m/z 400 with an accumulation target of 1,000,000 ions). Up to six of the most abundant ions (>3000 counts) with charge states >2+ were sequentially isolated and fragmented within the linear ion trap using collisionally induced dissociation with a normalized collision energy of 25 V, activation q of 0.25, and activation time of 30 ms at a target value of 30,000 ions. m/z values selected for MS/MS were dynamically excluded for 30 s. Peak lists were generated using Mascot Daemon/extract_msn (Matrix Science, Thermo) using the default parameters and submitted to the database search program Mascot (version 2.2, Matrix Science). General search parameters were as follows: precursor tolerance was 4 ppm, product ion tolerance was ±0.4 Da, Met(O) and Cys carboxyamidomethylation were specified as variable modifications, enzyme specificity was trypsin, one missed cleavage was possible, and the non-redundant protein database from NCBI or Swiss-Prot (March 2008) was searched. Additional searches with variable modifications N-acetylation, Cys-sulfenic acid, Cys-sulfonic acid, deamidation, and phosphorylation were performed. All peptides assigned by Mascot had Mowse scores >20, and spectra of glycopeptides were interpreted and validated manually. Extracted ion chromatograms (XICs) were derived from either MS or MS/MS spectra from the calculated monoisotopic mass of each ion (±m/z 0.1) using the QualBrowser in XCalibur (version 2.07), and abundances were calculated from the area of each extracted ion.
β-Elimination and Alkylation of Glycopeptides
Peptide digests were treated with NH2CH3 vapor as described (20). Briefly, a portion of the digests (∼15%) was dried (SpeedVac), tubes were placed in scintillation vials together with a microcentrifuge tube containing NH2CH3 (50 μl), and the vial was flushed with N2. After capping, the vials were left at 70 °C for 60 min. Peptides were solubilized and analyzed by nano-LC MS/MS using the Q-Tof Ultima as described above.
Lectin Blot Analysis and Neuraminidase Assay
Lectin blot analysis was performed as described (28). This is based on the differing relative affinities for α-(2→6) and α-(2→3) linkages of SNA and MAA, respectively (29, 30). In short, after 2-DE and protein transfer, nitrocellulose membranes were incubated with 2% periodate-oxidized BSA buffer containing 0.1% Tween 20 in PBS (BSA-Tween buffer) at 40 °C for 1 h. Membranes were subsequently incubated for 1 h with biotinylated lectins in BSA-Tween buffer containing 0.5 μg/ml SNA or 1 μg/ml MAA. After five 5-min washes in PBS containing 0.5% Tween 20, the membranes were incubated with 1 μg/ml HRP-avidin D for 1 h, washed five times with PBS containing 0.5% Tween 20, and revealed by ECL. For neuraminidase cleavage of sialic acid residues, HMDM cell lysates and medium samples were treated with α-(2→3,6,8,9)-neuraminidase or α-(2→3)-neuraminidase in 50 mm sodium phosphate, pH 5.0 at 37 °C for 3 h.
Analysis of Ser290 on Lipid-bound ApoE
A high resolution EPR structure of lipid-bound apoE was recently published, and the Protein Data Bank file was a kind gift from Prof K. Weisgraber (31). Analysis of the glycosylation site on Ser290 was performed using Discovery Studio 2.0 (Accelrys Software Inc.) on this structure.
RESULTS
Identification of O-Glycosylated Peptides of ApoE Using Q-TOF MS
To analyze the sugar structures of apoE glycoforms, cellular and secreted apoE was immunoprecipitated and separated by 1-DE (Fig. 1, inset). After Coomassie staining, 34-kDa apoE bands were excised, destained, and treated with trypsin, and peptides were analyzed by nano-LC data-dependent tandem mass spectrometry. Total ion chromatograms (TICs) and XICs derived from all MS/MS spectra using m/z 204.08, 292.09, and 366.13 for glycan oxonium ions (32) were obtained from Q-Tof Ultima mass analysis (not shown). These were used to indicate likely glycopeptides from which MS/MS spectra were manually interpreted. Recently, HexNAc- Hex oxonium ions (calculated mass [M + H]+ = 366.13) were identified in the sugar structures of apoE (25).
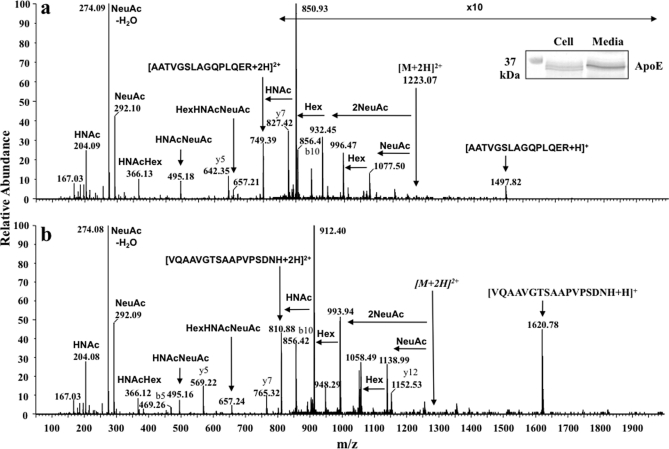
Fig. 1.
Q-Tof Ultima MS/MS spectra of protonated O-glycosylated peptides of cellular apoE. Immunoprecipitated apoE was separated by 1-DE, Coomassie-stained bands (Mr ∼34,000) were excised and digested with trypsin, and peptides were separated and analyzed by nano-LC data-dependent tandem MS. The inset shows Coomassie-stained cellular and 24-h secreted apoE. [M + 2H]2+ and [M + 3H]3+ precursor ions of two different glycopeptides were observed after analysis of cellular apoE. a, abundant glycan-specific ions from CID of the [M + 2H]2+ ion of apoE(192–206)-HexNAc-Hex-(NeuAc)2. b, abundant glycan-specific ions from CID of the [M + 3H]3+ ion of apoE(283–299)-HexNAc-Hex-(NeuAc)2 were observed (for ease of interpretation, fragment ions are shown with respect to a double charged precursor in both spectra). Fragment ions and glycan oxonium ions at m/z 204.08, 292.09, and 366.13 are labeled in each spectrum. Similar spectra were obtained with secreted apoE (not shown). HNAc, HexNAc.
Cellular apoE and secreted apoE showed very similar XIC profiles, and two different potential glycosylated peptides were readily identified (not shown). An MS/MS spectrum of the [M + 2H]2+ precursor ion m/z 1223.07 (single charged m/z 2445.12) was obtained. This was identified as modified AATVGSLAGQPLGER (apoE(192–206)) and is consistent with glycosylation of Thr194 (25). The delta mass between 2445.12 and 1497.80 is consistent with HexNAc-Hex-(NeuAc)2 glycosylation of this peptide (ExPASy GlycoMod tool). Four double charged glycan fragment ions (m/z 850.93, 932.45, 996.47, and 1077.50) were detected in the spectrum derived from this protonated peptide, and the compositions of sugar structures of these fragments are described (Fig. 1a and legend).
Unexpectedly, we detected a second putative glycopeptide from the XIC of the glycan oxonium ions. The MS/MS spectrum of precursor ion m/z 1284.57 is shown in Fig. 1b. MS/MS analysis of [M + 2H]2+ precursor ions showed b- and y-type ions consistent with the sequence VQAAVGTSAAPVPSDNH of the C-terminal tryptic peptide of apoE, apoE(283–299). The delta mass between 2568.14 and 1620.80 is also consistent with HexNAc-Hex-(NeuAc)2 glycosylation. Similar to the spectrum of protonated apoE(192–206)-HexNAc-Hex-(NeuAc)2, four differently glycosylated double charged fragment ions (m/z 912.40, 993.94, 1058.49, and 1139.49) were detected, and the structures of these peaks, described in Fig. 1, confirm HexNAc-Hex-(NeuAc)2 glycosylation. As apoE(283–299) (VQAAVGTSAAPVPSDNH) does not contain the consensus sequence (NX(T/S/C)) required for N-linked glycans and N-linked glycosylation occurs commonly with a (GlcNAc)2-Man3 core structure, it was concluded that this peptide also contains O-linked glycans potentially on Thr289, Ser290, or Ser296. Although the two glycosylated peptides were well separated, identical small m/z peaks (m/z 204.08, 274.08, 292.09, 366.13, 495.16, and 657.24) were present in both MS/MS spectra, consistent with known glycan oxonium ions (32) and facilitating their detection within the mixture. Some signals corresponding to the m/z of oxonium ions were also detected in the XICs. The fragmentation spectra did not contain ions corresponding to the m/z of protonated apoE(192–206) or apoE(283–299) or typical glycan fragmentations and were therefore likely from unknown contaminating substances.
Identification of O-Glycosylated Peptides of ApoE Using LTQ-FT MS
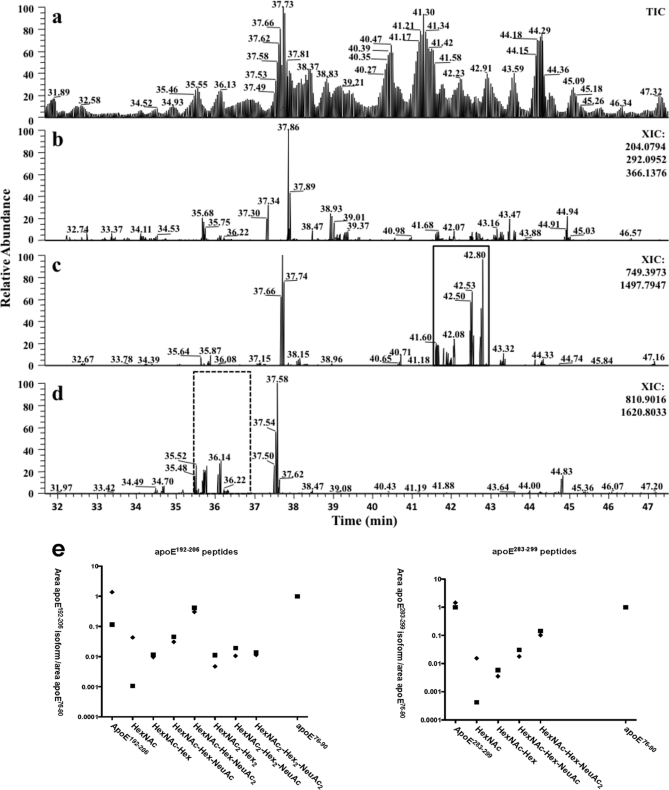
To further confirm the Q-Tof Ultima MS/MS results and detect additional glycopeptides, we performed similar experiments and analysis using a more sensitive LTQ-FT Ultra mass spectrometer. In these data-dependent experiments, the linear ion trap was used to collect MS/MS spectra, and the ICR cell was used for MS. Fig. 2 shows the partial TIC (Fig. 2a) and XICs (MS2 spectra) (Fig. 2, b–d) of separated apoE protonated peptides. To identify specific glycopeptides observed from the Q-TOF experiments and other glycopeptides, the XICs were extracted for the oxonium ions m/z 204.08 + 292.09 + 366.12 (Fig. 2b), m/z 749.40 + 1497.80 ([M + 2H]2+ and [M + H]+) of apoE(192–206) (Fig. 2c), and 810.90 + 1620.80 ([M + 2H]2+ and [M + H]+) of apoE(283–299) (Fig. 2d). Despite recording these spectra in the linear ion trap with the concomitant loss of low mass ions and mass accuracy of approximately ±m/z 0.4, many abundant peaks characteristic of oxonium ion or apoE glycopeptides were observed, and the glycosylation patterns were characterized (Table I and Fig. 3).
Fig. 2.
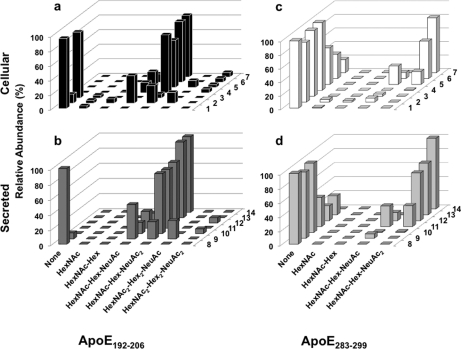
LTQ-FT TIC and XICs of apoE. Immunoprecipitated apoE was separated by 1-DE, Coomassie-stained bands were excised and digested with trypsin, and peptides were separated and analyzed by nano-LC data-dependent tandem MS. a, TIC of protonated apoE tryptic peptides. b, XIC obtained by adding glycan oxonium ion masses 204.08 (HexNAc), 292.09 (NeuAc), and 366.12 (HexNAc-Hex) from all MS/MS spectra. Abundant signals were observed despite the obligatory low mass cutoff of ion trap mass spectrometers. c, XIC for apoE(192–206) obtained by adding the expected precursor ion masses (m/z 749.40 [M + 2H]2+ and 1497.80 [M + H]+) from all MS/MS spectra. The variously O-glycosylated apoE(192–206) peptides are only detected in the solid square area. d, XIC for apoE(283–299) obtained by adding expected precursor ion masses (m/z 810.90 [M + 2H]2+ and 1620.80 [M + H]+) from all MS/MS spectra. The variously O-glycosylated apoE(283–299) peptides are only present in the dotted square area. e, scatter plots depicting proportional glycan content of the indicated glycosylation/sialylation structures on apoE(192–206) and apoE(283–299) peptides in cellular (♦) and secreted (■) apoE normalized to unmodified apoE(76–90) peptide. Data were obtained by automatic peak area detection from double and triple charged ion XICs (see text for details), and to bring out the large differences in normalized abundance, the y axis is in log scale.
Table I. LTQ-FT MS analysis of O-glycosylated apoE peptides isolated from macrophages.
The 34-kDa band of apoE was excised after 1-DE (see legend to Fig 3) and subjected to LTQ-FT MS/MS. Separate analyses were undertaken for cell lysate apoE and apoE secreted into media. Peptide sequence AATVGSLAGQPLQER corresponds to apoE(192–206), and peptide sequence VQAAVGTSAAPVPSDNH corresponds to apoE(283–299). XICs are included in supplemental Fig. 1.
| Glycan | Peptide AATVGSLAGQPLQER (mass, 1496.7947) |
Peptide VQAAVGTSAAPVPSDNH (mass, 1619.7903) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular apoE |
Secreted apoE |
Cellular apoE |
Secreted apoE |
|||||||||
| Observed mass (0 charge) | Theoretic mass | Mass difference | Observed mass (0 charge) | Theoretic mass | Mass difference | Observed mass (0 charge) | Theoretic mass | Mass difference | Observed mass (0 charge) | Theoretic mass | Mass difference | |
| ppm | ppm | ppm | ppm | |||||||||
| None | 1496.79 | 1496.79 | −1.710 | 1496.79 | 1496.79 | −0.895 | 1619.79 | 1619.79 | −1.222 | 1619.79 | 1619.79 | −0.926 |
| HexNAc | 1699.88 | 1699.87 | 1.182 | 1822.87 | 1822.87 | 2.299 | 1822.88 | 1822.87 | 3.286 | |||
| Hex-HexNAc | 1861.93 | 1861.93 | −0.091 | 1861.93 | 1861.93 | 0.038 | 1984.93 | 1984.93 | −0.408 | 1984.92 | 1984.93 | −1.728 |
| Hex-HexNAc-NeuAc | 2153.03 | 2153.03 | −0.980 | 2153.02 | 2153.03 | −1.770 | 2276.02 | 2276.02 | −0.822 | 2276.02 | 2276.02 | 0.053 |
| Hex-HexNAc-(NeuAc)2 | 2444.12 | 2444.12 | 0.745 | 2444.12 | 2444.12 | −0.777 | 2567.11 | 2567.12 | −1.843 | 2567.12 | 2567.12 | −1.375 |
| Hex2-(HexNAc)2 | 2227.07 | 2227.07 | −1.976 | 2227.06 | 2227.07 | −2.483 | ||||||
| Hex2-(HexNAc)2-NeuAc | 2518.17 | 2518.17 | −0.036 | 2518.17 | 2518.17 | 1.346 | ||||||
| Hex2-(HexNAc)2-(NeuAc)2 | 2809.26 | 2809.26 | −1.289 | 2809.26 | 2809.26 | −0.295 | ||||||
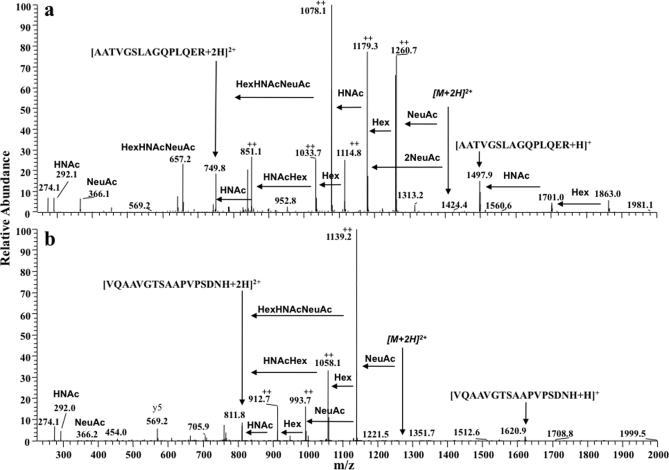
Fig. 3.
Ion trap MS/MS spectra of two distinct O-glycosylated peptides of apoE. Spectra of the most highly glycosylated peptides identified from each apoE glycosylation site are shown. Structures were determined from ion trap spectra even though the mass accuracy was ±m/z 0.4 and fragment ion charge states could not be accurately resolved. a, MS/MS analysis of an [M + 3H]3+ ion (m/z 937.08) present in cellular apoE. Glycan fragment ions (with respect to a double charged precursor) and losses are annotated, and the spectrum indicates glycopeptide apoE(192–206)-(HexNAc)2-Hex2-(NeuAc)2. b, ion trap MS/MS analysis of the [M + 3H]3+ ion (m/z 856.67) present in cellular apoE. The spectrum is of protonated apoE(283–299)-HexNAc-Hex-(NeuAc)2 and shows the same fragment ions observed with the spectrum acquired using the Q-Tof Ultima (Fig. 1b). The fragment ions indicated by ++ are assumed to be double charged. HNAc, HexNAc.
Glycosylated apoE(192–206) peptides were detected between the retention times 41.65 and 42.91 min (Fig. 2c and Table I), and glycosylated apoE(283–299) peptides were detected between 35.63 and 36.52 min (Fig. 2d and Table I). Seven differently glycosylated peptide MS/MS spectra of apoE(192–206) and four different glycosylated peptide spectra of the apoE(283–299) precursor peptide were detected and manually interpreted (Table I). Many glycopeptides were present in apoE from cell lysates and from apoE secreted into medium, and both demonstrated at least one asialylated sugar residue. ApoE(192–206) peptides contained larger glycan structures than apoE(283–299) peptides, but for both, the maximum number of sialic acid residues detected per glycan was 2.
The precursor and product ion measurements in MS/MS spectra from the Q-Tof Ultima were all within ∼50 ppm of the calculated values, and with a resolution of ∼10,000, each product charge state was accurately determined, allowing confident assignment of glycan fragmentation patterns (Fig. 1). Fig. 3 shows MS/MS spectra of the most extensively glycosylated peptides identified from the LTQ-FT. The two glycoforms detected were triple charged ions, and these were readily resolved in the MS scan. An [M + 3H]3+ precursor ion, m/z 937.08 (±2 ppm, single charged, m/z 2810.28), was identified for apoE(192–206) (Fig. 3a); the delta mass between 2810.28 and 1497.80 corresponds to apoE(192–206)- (HexNAc)2-Hex2-(NeuAc)2, and the glycosylation pattern was consistent with the observed fragment ions (Fig. 3a). An [M + 3H]3+ precursor ion, m/z 856.67 (±2 ppm, single charged, m/z 2568.14), was identified for apoE(283–299) (Fig. 3b) and was consistent with apoE(283–299)-HexNAc-Hex-(NeuAc)2 (Fig. 3b). Although MS/MS spectra measured within the LTQ were without isotopic resolution, many abundant glycosylated peptide fragment ions and oxonium ions were detected in both MS/MS spectra, allowing confident prediction of the glycan structure. The determined compositions of sugar structures of these fragment ions are described in Fig. 3.
The relative abundances of each ionized glycoform and unmodified peptides were determined by calculating monoisotopic XICs using the summed area of the 2+ and 3+ charge states (±m/z 0.1) of potential precursor ions from all LTQ-FT MS spectra (supplemental Fig. 1). Separation of each of the glycoforms was observed with no apparent co-elution that could be attributed to in-source fragmentation or detectable loss of sialic acid (supplemental Fig. 1). No estimates of the relative amounts of each form within the samples can be made because of potential differences in ionization efficiencies from these types of analyses, but reliable comparisons of the relative amounts between samples can be made because any differences in ionization remains the same in each measurement and glycoform. A scatter plot was generated using the area of the double and triple charged ions of apoE(76–90) as a control. We assumed this procedure would normalize the variable amount of apoE in each extract. For each peptide, the relative abundance of each glycan was similar in secreted and cellular apoE with the exception of HexNAc, which was more abundant in cellular than secreted apoE (Fig. 2e). Interestingly, the apoE(192–206) peptide showed proportionately more glycosylation than the apoE(283–299) peptide, suggesting preferential glycosylation of the apoE(192–206). The figure also shows that more complex sugars detected in apoE(192–206) peptides were not detected in apoE(283–299) peptides and that the relative abundance of sugars such as HexNAc-Hex-NeuAc and HexNAc-Hex-(NeuAc)2 was greater in apoE(192–206) peptides than in apoE(283–299) peptides.
Analysis of ApoE Glycoforms Synthesized in HMDMs Using 2-DE
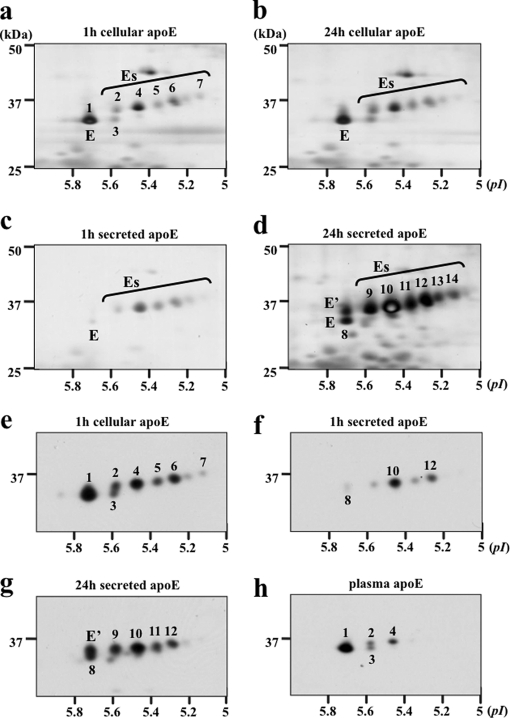
Cholesterol-enriched HMDMs were washed and incubated in serum-free medium, and media and cells were collected at 1 and 24 h. ApoE was immunoprecipitated from lysates and from culture media, separated by 2-DE (IP-2-DE), and detected by silver staining and Western blotting (Fig. 4). Cellular apoE and secreted apoE were separated within a pI range of 5.1–5.8. One major asialylated apoE (E) glycoform (also designated glycoform 1) was detected, and at least seven Es glycoforms were detected in cellular and medium samples (Fig. 4). Glycoforms detected in cell lysates and in culture medium were sequentially numbered 1–14 for later MS/MS analysis. On the basis of apparent molecular weight and pI, cellular glycoforms 1, 2, 4, 5, 6, and 7 appeared to correspond to secreted glycoforms 8, 9, 10, 11, 12, and 14.
Fig. 4.
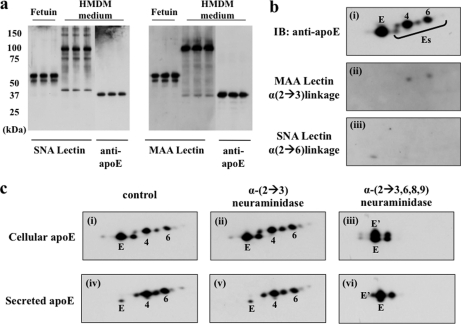
Two-dimensional gel electrophoresis of apoE glycoforms in HMDMs. HMDMs from two independent donors (a–d, donor 1; e–h, donor 2) were cholesterol-enriched by incubation for 2 days with 50 μg/ml AcLDL, washed, and incubated in serum-free and BSA-free RPMI 1640 medium for up to 24 h. At the indicated times, apoE was immunoprecipitated from cell lysates (a, b, and e) and media (c, d, f, and g), separated by 2-DE, and detected by silver staining (a–d) or Western blot (e–h). The distribution of cellular apoE glycoforms did not change between 1 (a) and 24 h (b), whereas there was an increase in the abundance of glycoforms E and E′ relative to Es glycoforms between 1 and 24 h in the medium (c versus d and f versus g). Western blots show that plasma apoE (h) comprises less charged apoE glycoforms than secreted (g) or cellular apoE (e) from the same donor. On the basis of mass and pI, the following glycoforms appear to be the same: 1, E, and 8; 2 and 9; 4 and 10; 5 and 11; 6 and 12; and 7 and 14.
Between 1 and 24 h, cellular apoE glycoforms did not change (Fig. 4, a versus b), whereas apoE secreted into medium changed significantly over this time with increased abundance of E and E′ relative to Es glycoforms (Fig. 4, c versus d). E′ was more prominent in medium than in cells (Fig. 4, b versus d), whereas glycoform 3 was more prominent in cells than in medium (Fig. 4, a versus c). Similar results were obtained with silver staining (Fig. 4, a–d) and Western blot (Fig. 4, e–g) using macrophages from two independent donors. Consistent with previous studies (22, 33–35), apoE in cell lysates and cell culture media were more extensively sialylated than was plasma apoE even when the cells and plasma were from the same donor (Fig. 4, e and g versus h).
Although 2-DE had indicated the presence of up to seven sialic acid residues per molecule of apoE, 1-DE and MS/MS detected only up to two sialic acid residues per glycan. To investigate in more detail the identity of each glycoform and whether the apparent differences in charge of apoE glycoforms identified by 2-DE corresponded to differences in sialylation and/or glycosylation, immunoprecipitated cellular apoE and secreted apoE were separated by 2-DE, destained, and digested, and DDA MS analysis using the LTQ-FT was performed (Table II). As observed after MS analysis of 1-DE, the same two distinct glycosylated peptides were detected in many of the apoE spots with the same glycosylation patterns observed. In earlier studies, glycoform 1 was identified previously as asialylated and non-glycosylated (25, 36). Our analysis confirmed the absence of sialic acid residues but identified HexNAc in glycoform 1 as well as a non-glycosylated peptide, suggesting that this spot is a mixture of non-sialylated, glycosylated, and non-glycosylated apoE. Glycoform 3 had a similar molecular weight but was more highly charged than glycoform 1, suggesting either failure to detect sialic acid residues due to glycan instability or an alternative source of negative charge to explain its pI relative to glycoform 1. Mascot searches were repeated including phosphorylation, deamidation, and Cys-sulfonic acid variable modifications as these are hypothesized to change pI values. The Mascot search result of glycoform 3 (supplemental Fig. 2) shows a peptide containing Cys-sulfonic acid, and this peptide was absent from the search of glycoform 1, indicating that the likely cause of the pI shift of glycoform 3 was conversion of Cys to Cys-sulfonic acid (supplemental Fig. 2). Glycoforms of increasing charge (e.g. glycoforms 2, 4, 5, 6, and 7) demonstrated increasing glycan complexity but not beyond that of (HexNAc)2-Hex2-(NeuAc)2 (Table II), suggesting the presence of multiple sites of glycosylation on apoE or some unidentified post-translational modifications to explain their difference in charge. The presence of multiple glycans per spot may also be due to instability during sample preparation or LC/MS analysis (Table II). Analysis of the relative proportion, determined from XICs, of individual apoE(192–206) and apoE(283–299) glycopeptides in each separated 2-DE spot demonstrated similar glycan distribution patterns in cellular and secreted apoE for each peptide (Fig. 5). For all 2-DE glycoforms of apoE, the relative proportion of glycosylated apoE(192–206) peptides appeared greater than the proportion of glycosylated apoE(283–299) peptides (Fig. 5), and the more highly glycosylated/sialylated forms of apoE occurred at higher pI values.
Table II. LTQ-FT MS analysis of glycosylated apoE peptides from apoE glycoforms isolated by 2-DE.
| 2-DE assignment | Glycan | Peptide AATVGSLAGQPLQER (mass, 1496.79) |
Peptide VQAAVGTSAAPVPSDNH (mass, 1619.8) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Retention time | Mass (0 charge) | Theoretic mass | Mass difference | Retention time | Mass (0 charge) | Theoretic mass | Mass difference | ||
| ppm | ppm | ||||||||
| 1 | HexNAc | 34.38 | 1699.88 | 1699.87 | 1.818 | 30.63 | 1822.87 | 1822.87 | 1.053 |
| 2 | Hex-HexNAc-NeuAc | 27.51 | 2276.02 | 2276.02 | −0.909 | ||||
| Hex2-(HexNAc)2-NeuAc | 31.07 | 2518.16 | 2518.17 | −3.157 | |||||
| 3 | Hex-HexNAc-NeuAc | 27.53 | 2276.02 | 2276.02 | −1.424 | ||||
| Hex-HexNAc-(NeuAc)2 | 31.09 | 2444.12 | 2444.12 | −1.342 | |||||
| 4 | Hex-HexNAc-(NeuAc)2 | 29.36 | 2567.12 | 2567.121 | −1.223 | ||||
| Hex2-(HexNAc)2-(NeuAc)2 | 33.4 | 2809.25 | 2809.26 | −2.100 | |||||
| 5 | Hex-HexNAc-(NeuAc)2 | 26.99 | 2567.12 | 2567.12 | −0.721 | ||||
| Hex2-(HexNAc)2-(NeuAc)2 | 30.87 | 2809.24 | 2809.26 | −5.614 | |||||
| 6 | Hex-HexNAc-(NeuAc)2 | 29.35 | 2567.12 | 2567.12 | −1.024 | ||||
| Hex2-(HexNAc)2-(NeuAc)2 | 33.42 | 2809.25 | 2809.26 | −3.083 | |||||
| 7 | Hex-HexNAc-(NeuAc)2 | 29.36 | 2567.11 | 2567.12 | −2.555 | ||||
| Hex2-(HexNAc)2-(NeuAc)2 | 33.41 | 2809.25 | 2809.26 | −3.189 | |||||
| 8 | None | 35.29 | 1496.80 | 1496.80 | 1.243 | 31.32 | 1619.79 | 1619.79 | 0.407 |
| 9 | Hex-HexNAc-NeuAc | 29.99 | 2276.02 | 2276.02 | 0.211 | ||||
| Hex2-(HexNAc)2-NeuAc | 33.6 | 2518.16 | 2518.17 | −2.144 | |||||
| 10 | Hex-HexNAc-(NeuAc)2 | 29.36 | 2567.11 | 2567.12 | −2.006 | ||||
| Hex2-(HexNAc)2-(NeuAc)2 | 33.41 | 2809.25 | 2809.26 | −3.798 | |||||
| 11 | Hex-HexNAc-(NeuAc)2 | 33.79 | 2444.12 | 2444.12 | −1.428 | 29.49 | 2567.12 | 2567.12 | −0.802 |
| 12 | Hex-HexNAc-(NeuAc)2 | 29.42 | 2567.12 | 2567.12 | 0.016 | ||||
| Hex2-(HexNAc)2-(NeuAc)2 | 33.37 | 2809.26 | 2809.26 | −1.951 | |||||
| 13 | Hex-HexNAc-(NeuAc)2 | 33.72 | 2444.12 | 2444.12 | −1.943 | 29.37 | 2567.12 | 2567.12 | −0.464 |
| 14 | Hex-HexNAc-(NeuAc)2 | 33.68 | 2444.13 | 2444.12 | 1.052 | 29.39 | 2567.11 | 2567.12 | −1.971 |
Fig. 5.
Relative proportion of glycan residues on apoE(192–206) and apoE(283–299) peptides of cellular and secreted apoE glycoforms separated by 2-DE. Immunoprecipitated cellular apoE and secreted apoE were separated by 2-DE, apoE glycoform spots were excised after Coomassie staining and destained, and XICs were obtained. Proportional glycan content on cellular (a) and secreted (b) apoE(192–206) peptide and cellular (c) and secreted (d) apoE(283–299) peptide was determined for each glycoform and is presented as a percentage of total residues detected within each 2-DE glycoform spot.
Characterization of Sialic Acid Modification in ApoE Glycoforms
To identify the types of sialic acid linkage of apoE, lectin blots were performed using MAA (which detects terminal α-(2→3)-sialic acid linkage) and SNA (which detects terminal α-(2→6)-sialic acid linkage) (37). Secreted apoE was used for 1-DE analysis in preference to cellular apoE because preliminary studies demonstrated extensive cross-reactivity of lectins with a number of cell proteins in cell lysates, causing smearing (not shown). Fetuin protein, which has both α-(2→3)- and α-(2→6)-sialic acid linkages, was stained by both MAA and SNA. Several bands were detected by each lectin in secreted apoE samples (Fig. 6). Comparison of the SNA lectin blot (middle three lanes) and apoE Western blot (far right lanes) demonstrated that the bands identified in the SNA lectin blot do not correspond to apoE. In contrast, a 34-kDa band corresponding to apoE was detected by MAA lectin blot.
Fig. 6.
Contribution of sialic acid linkages to apoE glycoforms: lectin blots and effect of neuraminidase treatment. 24-h HMDM-conditioned medium (a) was separated by 10% SDS-PAGE, specific sialic acid linkages in secreted apoE were detected using SNA (left panel) and MAA (right panel) lectin blots (lanes 1–6), and apoE protein was detected by Western blot (lanes 7–9 in both panels). Fetuin was used as a standard (lanes 1–3) for both α-(2→3)- and α-(2→6)-sialic acid linkage. HMDM cellular apoE (b) was immunoprecipitated, separated by 2-DE, and stained for apoE protein by Western blot (i), MAA (ii), and SNA (iii). c, 24-h HMDM cell lysate and medium were incubated with α-(2→3)-neuraminidase or α-(2→3,6,8,9)-neuraminidase for 3 h at 37 °C, separated by 2-DE, and detected by silver stain: i, cellular apoE control; ii, cellular apoE after α-(2→3)-neuraminidase treatment; iii, cellular apoE after α-(2→3,6,8,9)-neuraminidase; iv, macrophage-secreted apoE control; v, macrophage-secreted apoE after α-(2→3)-neuraminidase treatment; vi, macrophage-secreted apoE after α-(2→3,6,8,9)-neuraminidase treatment. Note that glycoform E′ is generated after treatment of cellular or secreted apoE with α-(2→3,6,8,9)-neuraminidase. IB, immunoblot.
To confirm that cellular apoE also demonstrated α-(2→3)-sialic acid linkage, IP-2-DE lectin blots were performed on cellular IP-2-DE apoE to eliminate smearing (Fig. 6b). MAA weakly stained two Es glycoforms in cell lysates corresponding to glycoforms 2 and 4. However, there were only minor changes in apoE glycoform distribution after treatment with α-(2→3)-neuraminidase and substantial elimination of Es glycoforms after α-(2→3,6,8,9)-neuraminidase treatment, suggesting that linkages other than α-(2→3) were present (Fig. 6c). The loss of glycoforms 4–7 in cell lysates and glycoforms 10–14 from medium samples after neuraminidase treatment confirmed the presence of sialic acid residues in cell and secreted apoE. Interestingly, E′ was generated after treating apoE from cell lysates and media with α-(2→3,6,8,9)-neuraminidase, suggesting that E′ arises from the desialylation of Es glycoforms.
Determination of Location of Second Site of O-Glycosylation in ApoE(283–299)
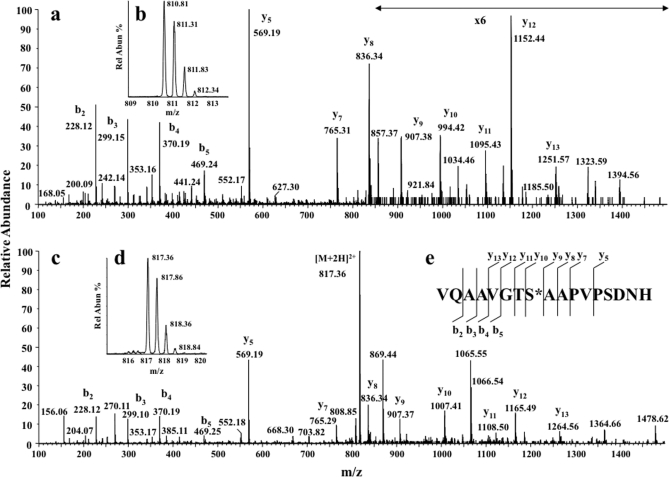
The newly identified glycosylated tryptic peptide (apoE(283–299)) has three potential O-linked glycosylation sites (two Ser and one Thr in VQAAVGTSAAPVPSDNH). To identify which amino acid represented the site of O-linked glycan attachment, peptide digests were treated with methylamine vapor, and under these conditions, glycopeptides underwent β-elimination of the glycan and alkylation, forming stable methylamine derivatives of either Ser or Thr with a mass addition of m/z 13 (20).
After nano-LC and Q-Tof Ultima DDA MS analysis, Mascot database searches were performed including variable modifications Ser-methylamine and Thr-methylamine together with the standard search parameters. Both Thr194 residue, which is present in the apoE(192–206) glycopeptide, and Ser290 residue, which is present in apoE(283–299), were detected as modified by addition of methylamine. The Mascot search assignment was confirmed by manually interpreting the MS/MS spectra of the two modified peptides. The MS/MS spectra of apoE(192–206) peptide exposed to methylamine identified modification of Thr194 only (supplemental Fig. 3). Fig. 7 shows the MS/MS spectrum of double charged ions of apoE(283–299) (Fig. 7a) and methylamine-derivatized apoE(283–299) (Fig. 7b). Comparison of the y-ion series of product ions present in Fig. 7, a and b, showed a mass difference of m/z 13 in y-ions 13, 12, 11, and 10. The mass of the ions in both spectra (i.e. y-ions 9, 8, 7, and 6) were identical after Ser290, thereby confirming that the methylamine-modified residue and the major glycosylation site was Ser290 within apoE(283–299). A similar analysis of the fragmentation spectrum containing Thr194 confirmed that this was the major glycosylated amino acid in apoE(192–206) (supplemental Fig. 3).
Fig. 7.
Determination of location of second O-glycosylation site. Nano-LC MS spectra of the reaction products of apoE tryptic peptides after treatment with methylamine are shown. a, MS/MS of [M + 2H]2+ ion (m/z 810.85) of apoE(283–299) (inset b). c, MS/MS of the [M + 2H]2+ ion (m/z 817.35) of apoE(283–299) modified by methylamine after treatment with CH3NH2 and β-elimination of the O-glycan (inset d). e, sequence-specific b- and y-type ions identified in both spectra. Intense product ions indicate conversion of Ser290 to Ser290-methylamine, indicating that the O-glycosylation site was predominately on Ser290 (product ions containing Ser290 were shifted by +m/z 13). Spectra were recorded using a Q-Tof Ultima with precursor/production ion resolution ∼10,000 and a root mean square error of ∼50 ppm. Rel Abun, relative abundance.
DISCUSSION
We undertook the first detailed analysis of the glycosylation of apoE from primary human macrophages. Our results confirm previous studies identifying that apoE is extensively glycosylated and sialylated but extend these previous observations by demonstrating multiple glycosylation sites on apoE and identifying Ser290 on the apoE C terminus as a novel site of glycosylation. Given the importance of the C terminus of apoE in regulating apoE solubility, stability, and lipid binding, these results may have important implications for our understanding of apoE biochemistry.
Mucin-type O-glycosylations as occur on apoE are common, complex, and important post-translational protein modifications (17, 18) that are difficult to accurately characterize (19, 20). They are typically initiated in the Golgi with attachment of an N-acetylgalactosamine (GalNAc) residue to the side chain of an exposed Ser or Thr residue. Stepwise elongation generates up to eight core structures before further modification by processes such as sialylation. O-Linked sugars are frequently located in clusters in short regions of peptide chains containing repeating units of Ser, Thr, and Pro. Consistent with this, the peptide apoE(283–299) is rich in Ser and Thr and is the most highly predicted site for O-glycosylation in apoE (NetOGlyc 3.1 Server), supporting our identification of glycosylation on the apoE C terminus.
Our results confirm the complementary nature of MS and 2-DE approaches to identify apoE glycoforms. We identified at least seven consistently observed glycoforms of apoE in cells and in media by 2-DE, but analysis of these glycoforms by MS did not precisely distinguish between more highly charged and less charged glycoforms because multiple glycopeptides were detected in each 2D spot. Mancone et al. (25), who studied apoE associated with VLDL, also identified multiple glycoforms on 2-DE but reported no more than two sialic acid residues per glycoform. As variations in apoE charge on 2-DE were largely eliminated by treatment with neuraminidase, our data indicate that there may be up to six sialic acid residues per molecule of cell-derived apoE, and we propose that this variation relates to both the number of amino acid residues with O-linkage and variation in the number of sialic acid residues (between zero and two) on each O-linkage. Our data indicate that glycoform 3 differs from glycoform 1 as a result of conversion of Cys to Cys-sulfonic acid, which in other systems has been associated with both loss and gain of function (38). For the other spots, it is possible that there may be glycosylation sites on apoE in addition to both Thr194 and Ser290. Preliminary analysis of lower abundance ions and MS/MS spectra (data not shown) suggests the presence of glycosylated residues other than Ser290 within apoE(283–299), and candidates include Thr289 and Ser296. Other modifications of apoE may also contribute to differences in glycoform charge. ApoE can be phosphorylated by protein kinase CK2 in vitro on Ser296 (39). Mascot searches, however, did not identify other common modifications including phosphorylation of macrophage apoE in these studies.
The functional consequences of apoE sialylation are likely to be complex. ApoE was secreted normally from UDP-galactose/UDP-N-acetylgalactosamine 4-epimerase-deficient ldlD CHO cells expressing a reversible defect in protein O-glycosylation (36, 40), implying that glycosylation is not essential for secretion. However, sialylated apoE was reportedly released from the plasma membrane of macrophages more efficiently than apoE that was not sialylated (41). Desialylation of apoE markedly decreased binding to HDL (42), which may be critical for facilitating HDL-mediated recycling of apoE and removal of cellular cholesterol. Protein solubility and stability can be markedly enhanced by glycosylation (43–45) with a linear dependence on the number of carbohydrate residues attached to the protein (46). In the case of apoE, we expect that C-terminal glycosylation may protect against self-association, spontaneous aggregation, and fibril formation as occurs in Alzheimer disease and atherosclerotic plaques (47, 48). Ser290 is situated just after the last C-terminal α-helix at the start of an unstructured C-terminal tail. Therefore, glycosylation of Ser290 is unlikely to interfere with the interhelical interactions (supplemental Fig. 4). More likely, the presence of highly negatively charged sialic acid residues on Ser290 could modify interactions between apoE and phospholipid head groups or affect interactions between apoE molecules.
All sialylation and glycosylation of plasma VLDL-derived apoE have been attributed to O-linkage to residue Thr194 (36), which resides within the peptide AATVGSLAGQPLQER (apoE(192–206)). In our studies, secreted apoE(192–206) peptides were more commonly glycosylated and contained larger glycan structures than apoE(283–299) peptides. As previous studies found that mutagenesis of Thr194 precluded apoE glycosylation (36), we propose that Thr194 provides an initial and necessary site for apoE glycosylation and that this is followed by glycosylation of other sites on the C terminus. Such a stepwise pathway has been described for MUC1 (49).
Although it has been postulated that apoE undergoes postsecretory desialylation by plasma neuraminidases (50), VLDL-apoE reinjected in humans was not desialylated (51), and the mechanism by which apoE becomes desialylated is unknown. Our 2-DE analysis indicates that the glycoform distribution of secreted apoE changes over time with less sialylated forms (E and E′) appearing in medium during longer incubations. This suggests either that macrophages desialylate apoE extracellularly or that sialic acid-poor apoE is more slowly secreted and accumulated in the medium than is sialylated apoE. Desialylation appears the more likely mechanism as the glycoform E′ is generated by treatment of cellular or secreted apoE with neuraminidase. Our studies suggest the possibility that neuraminidase activity at the macrophage surface or in adjacent extracellular space acts on secreted apoE to generate lower molecular weight glycoforms after secretion. Importantly, recent studies have described endogenous sialidases Neu 1 and Neu 3 in human macrophages (52). The ability of cells other than macrophages, such as hepatocytes, to similarly generate sialic acid-poor apoE requires further investigation.
Sialylation most commonly occurs through α-(2→3) and α-(2→6) linkages to galactose. Our data based on lectin blotting indicate that macrophage-derived apoE has some terminal α-(2→3)-sialic acid linkages. However, the almost complete elimination of ES glycoforms with α-(2→3,6,8,9)-neuraminidase and weak effect of α-(2→3)-neuraminidase strongly suggest that other sialic acid linkages such as α-(2→6), α-(2→8), and α-(2→9) are likely to be present. Some α-(2→6)-sialic acid linkages may be below the limit of detection by SNA lectin blot especially if these are non-terminal sialic acid residues (29). The site specificity of O-mucins is tissue-specific as the GalNAc-transferases have tissue-specific expression patterns. It will therefore be interesting to compare the sialic acid linkages of macrophage-secreted, hepatocyte-secreted, and plasma apoE as differences may allow robust distinction between hepatocyte-derived, lipoprotein-derived, and macrophage-derived apoE in various tissues such as the arterial wall.
Supplementary Material
Acknowledgments
Mass spectrometric results were obtained at the Bioanalytical Mass Spectrometry Facility within the Analytical Centre of the University of New South Wales. This work was undertaken using, in part, infrastructure provided by New South Wales Government co-investment in the National Collaborative Research Infrastructure Scheme. Subsidized access to this facility is gratefully acknowledged.
* This work was supported by National Health and Medical Research Council of Australia Project Grant 455251 and Program Grant 482800. C-terminal sialidation of either T289 or S290 of apoE was also identified using independent techniques by Nilsson et al Nature Methods, 2009, 6, 809–811.
 This article contains supplemental Figs. 1–4.
This article contains supplemental Figs. 1–4.
1 The abbreviations used are:
- apoE
- apolipoprotein E
- LTQ
- linear ion trap quadrupole
- 1-DE
- one-dimensional gel electrophoresis
- 2-DE
- two-dimensional gel electrophoresis
- IP
- immunoprecipitation
- MAA
- M. amurensis lectin II
- SNA
- S. nigra bark lectin
- HMDM
- human monocyte-derived macrophage
- E
- asialylated apoE
- Es
- sialylated apoE
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- DDA
- data-dependent acquisition
- XIC
- extracted ion chromatogram
- TIC
- total ion chromatogram
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Mahley R. W., Rall S. C., Jr. (2000) Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 1, 507–537 [DOI] [PubMed] [Google Scholar]
- 2.Greenow K., Pearce N. J., Ramji D. P. (2005) The key role of apolipoprotein E in atherosclerosis. J. Mol. Med. 83, 329–342 [DOI] [PubMed] [Google Scholar]
- 3.Kockx M., Jessup W., Kritharides L. (2008) Regulation of endogenous apolipoprotein E secretion by macrophages. Arterioscler. Thromb. Vasc. Biol. 28, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 4.Kothapalli D., Fuki I., Ali K., Stewart S. A., Zhao L., Yahil R., Kwiatkowski D., Hawthorne E. A., FitzGerald G. A., Phillips M. C., Lund-Katz S., Puré E., Rader D. J., Assoian R. K. (2004) Antimitogenic effects of HDL and APOE mediated by Cox-2-dependent IP activation. J. Clin. Investig. 113, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Elzen P., Garg S., León L., Brigl M., Leadbetter E. A., Gumperz J. E., Dascher C. C., Cheng T. Y., Sacks F. M., Illarionov P. A., Besra G. S., Kent S. C., Moody D. B., Brenner M. B. (2005) Apolipoprotein-mediated pathways of lipid antigen presentation. Nature 437, 906–910 [DOI] [PubMed] [Google Scholar]
- 6.Mooijaart S. P., Berbée J. F., van Heemst D., Havekes L. M., de Craen A. J., Slagboom P. E., Rensen P. C., Westendorp R. G. (2006) ApoE plasma levels and risk of cardiovascular mortality in old age. PLoS Med. 3, e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., Chea H., Knopp R. H., Brunzell J., Geary R., Chait A., Zhao X. Q., Elkon K., Marcovina S., Ridker P., Oram J. F., Heinecke J. W. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Investig. 117, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linton M. F., Atkinson J. B., Fazio S. (1995) Prevention of atherosclerosis in apolipoprotein E-deficient mice by bone marrow transplantation. Science 267, 1034–1037 [DOI] [PubMed] [Google Scholar]
- 9.Bellosta S., Mahley R. W., Sanan D. A., Murata J., Newland D. L., Taylor J. M., Pitas R. E. (1995) Macrophage-specific expression of human apolipoprotein E reduces atherosclerosis in hypercholesterolemic apolipoprotein E-null mice. J. Clin. Investig. 96, 2170–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazzone T. (1996) Apolipoprotein E secretion by macrophages: its potential physiological functions. Curr. Opin. Lipidol. 7, 303–307 [DOI] [PubMed] [Google Scholar]
- 11.Kelly M. E., Clay M. A., Mistry M. J., Hsieh-Li H. M., Harmony J. A. (1994) Apolipoprotein E inhibition of proliferation of mitogen-activated T lymphocytes: production of interleukin 2 with reduced biological activity. Cell. Immunol. 159, 124–139 [DOI] [PubMed] [Google Scholar]
- 12.Ali K., Middleton M., Puré E., Rader D. J. (2005) Apolipoprotein E suppresses the type I inflammatory response in vivo. Circ. Res. 97, 922–927 [DOI] [PubMed] [Google Scholar]
- 13.Hatters D. M., Peters-Libeu C. A., Weisgraber K. H. (2006) Apolipoprotein E structure: insights into function. Trends Biochem. Sci. 31, 445–454 [DOI] [PubMed] [Google Scholar]
- 14.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 15.Zhang Y., Vasudevan S., Sojitrawala R., Zhao W., Cui C., Xu C., Fan D., Newhouse Y., Balestra R., Jerome W. G., Weisgraber K., Li Q., Wang J. (2007) A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry 46, 10722–10732 [DOI] [PubMed] [Google Scholar]
- 16.Peters-Libeu C. A., Newhouse Y., Hatters D. M., Weisgraber K. H. (2006) Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J. Biol. Chem. 281, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 17.Van den Steen P., Rudd P. M., Dwek R. A., Opdenakker G. (1998) Concepts and principles of O-linked glycosylation. Crit. Rev. Biochem. Mol. Biol. 33, 151–208 [DOI] [PubMed] [Google Scholar]
- 18.Hanisch F. G. (2001) O-Glycosylation of the mucin type. Biol. Chem. 382, 143–149 [DOI] [PubMed] [Google Scholar]
- 19.Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. (2002) Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791–804 [DOI] [PubMed] [Google Scholar]
- 20.Mirgorodskaya E., Hassan H., Clausen H., Roepstorff P. (2001) Mass spectrometric determination of O-glycosylation sites using beta-elimination and partial acid hydrolysis. Anal. Chem. 73, 1263–1269 [DOI] [PubMed] [Google Scholar]
- 21.Jain R. S., Quarfordt S. H. (1979) The carbohydrate content of apolipoprotein E from human very low density lipoproteins. Life Sci. 25, 1315–1323 [DOI] [PubMed] [Google Scholar]
- 22.Zannis V. I., vanderSpek J., Silverman D. (1986) Intracellular modifications of human apolipoprotein E. J. Biol. Chem. 261, 13415–13421 [PubMed] [Google Scholar]
- 23.Kritharides L., Jessup W., Mander E. L., Dean R. T. (1995) Apolipoprotein A-I-mediated efflux of sterols from oxidized LDL-loaded macrophages. Arterioscler. Thromb. Vasc. Biol. 15, 276–289 [DOI] [PubMed] [Google Scholar]
- 24.Kontula K., Aalto-Setälä K., Kuusi T., Hämäläinen L., Syvänen A. C. (1990) Apolipoprotein E polymorphism determined by restriction enzyme analysis of DNA amplified by polymerase chain reaction: convenient alternative to phenotyping by isoelectric focusing. Clin. Chem. 36, 2087–2092 [PubMed] [Google Scholar]
- 25.Mancone C., Amicone L., Fimia G. M., Bravo E., Piacentini M., Tripodi M., Alonzi T. (2007) Proteomic analysis of human very low-density lipoprotein by two-dimensional gel electrophoresis and MALDI-TOF/TOF. Proteomics 7, 143–154 [DOI] [PubMed] [Google Scholar]
- 26.Zannis V. I., Breslow J. L., Utermann G., Mahley R. W., Weisgraber K. H., Havel R. J., Goldstein J. L., Brown M. S., Schonfeld G., Hazzard W. R., Blum C. (1982) Proposed nomenclature of apoE isoproteins, apoE genotypes, and phenotypes. J. Lipid Res. 23, 911–914 [PubMed] [Google Scholar]
- 27.Gatlin C. L., Kleemann G. R., Hays L. G., Link A. J., Yates J. R., 3rd (1998) Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263, 93–101 [DOI] [PubMed] [Google Scholar]
- 28.Glass W. F., 2nd, Briggs R. C., Hnilica L. S. (1981) Use of lectins for detection of electrophoretically separated glycoproteins transferred onto nitrocellulose sheets. Anal. Biochem. 115, 219–224 [DOI] [PubMed] [Google Scholar]
- 29.Shibuya N., Goldstein I. J., Broekaert W. F., Nsimba-Lubaki M., Peeters B., Peumans W. J. (1987) The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2–6)Gal/GalNAc sequence. J. Biol. Chem. 262, 1596–1601 [PubMed] [Google Scholar]
- 30.Wang W. C., Cummings R. D. (1988) The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked alpha-2,3 to penultimate galactose residues. J. Biol. Chem. 263, 4576–4585 [PubMed] [Google Scholar]
- 31.Hatters D. M., Voss J. C., Budamagunta M. S., Newhouse Y. N., Weisgraber K. H. (2009) Insight on the molecular envelope of lipid-bound apolipoprotein E from electron paramagnetic resonance spectroscopy. J. Mol. Biol. 386, 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carr S. A., Huddleston M. J., Bean M. F. (1993) Selective identification and differentiation of N- and O-linked oligosaccharides in glycoproteins by liquid chromatography-mass spectrometry. Protein Sci. 2, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zannis V. I., Breslow J. L., SanGiacomo T. R., Aden D. P., Knowles B. B. (1981) Characterization of the major apolipoproteins secreted by two human hepatoma cell lines. Biochemistry 20, 7089–7096 [DOI] [PubMed] [Google Scholar]
- 34.Reardon C. A., Lau Y. F., Paik Y. K., Weisgraber K. H., Mahley R. W., Taylor J. M. (1986) Expression of the human apolipoprotein E gene in cultured mammalian cells. J. Biol. Chem. 261, 9858–9864 [PubMed] [Google Scholar]
- 35.Basu S. K., Brown M. S., Ho Y. K., Havel R. J., Goldstein J. L. (1981) Mouse macrophages synthesize and secrete a protein resembling apolipoprotein E. Proc. Natl. Acad. Sci. U.S.A. 78, 7545–7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wernette-Hammond M. E., Lauer S. J., Corsini A., Walker D., Taylor J. M., Rall S. C., Jr. (1989) Glycosylation of human apolipoprotein E. The carbohydrate attachment site is threonine 194. J. Biol. Chem. 264, 9094–9101 [PubMed] [Google Scholar]
- 37.Knibbs R. N., Goldstein I. J., Ratcliffe R. M., Shibuya N. (1991) Characterization of the carbohydrate binding specificity of the leukoagglutinating lectin from Maackia amurensis. Comparison with other sialic acid-specific lectins. J. Biol. Chem. 266, 83–88 [PubMed] [Google Scholar]
- 38.Lim J. C., Choi H. I., Park Y. S., Nam H. W., Woo H. A., Kwon K. S., Kim Y. S., Rhee S. G., Kim K., Chae H. Z. (2008) Irreversible oxidation of active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 283, 28873–28880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raftery M., Campbell R., Glaros E. N., Rye K. A., Halliday G. M., Jessup W., Garner B. (2005) Phosphorylation of apolipoprotein-E at an atypical protein kinase CK2 PSD/E site in vitro. Biochemistry 44, 7346–7353 [DOI] [PubMed] [Google Scholar]
- 40.Zanni E. E., Kouvatsi A., Hadzopoulou-Cladaras M., Krieger M., Zannis V. I. (1989) Expression of ApoE gene in Chinese hamster cells with a reversible defect in O-glycosylation. Glycosylation is not required for apoE secretion. J. Biol. Chem. 264, 9137–9140 [PubMed] [Google Scholar]
- 41.Zhao Y., Mazzone T. (2000) Transport and processing of endogenously synthesized ApoE on the macrophage cell surface. J. Biol. Chem. 275, 4759–4765 [DOI] [PubMed] [Google Scholar]
- 42.Marmillot P., Rao M. N., Liu Q. H., Lakshman M. R. (1999) Desialylation of human apolipoprotein E decreases its binding to human high-density lipoprotein and its ability to deliver esterified cholesterol to the liver. Metabolism 48, 1184–1192 [DOI] [PubMed] [Google Scholar]
- 43.Paulson J. C. (1989) Glycoproteins: what are the sugar chains for? Trends Biochem. Sci. 14, 272–276 [DOI] [PubMed] [Google Scholar]
- 44.Ioannou Y. A., Zeidner K. M., Grace M. E., Desnick R. J. (1998) Human alpha-galactosidase A: glycosylation site 3 is essential for enzyme solubility. Biochem. J. 332, 789–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Høiberg-Nielsen R., Fuglsang C. C., Arleth L., Westh P. (2006) Interrelationships of glycosylation and aggregation kinetics for Peniophora lycii phytase. Biochemistry 45, 5057–5066 [DOI] [PubMed] [Google Scholar]
- 46.Tams J. W., Vind J., Welinder K. G. (1999) Adapting protein solubility by glycosylation. N-glycosylation mutants of Coprinus cinereus peroxidase in salt and organic solutions. Biochim. Biophys. Acta 1432, 214–221 [DOI] [PubMed] [Google Scholar]
- 47.Howlett G. J., Moore K. J. (2006) Untangling the role of amyloid in atherosclerosis. Curr. Opin. Lipidol. 17, 541–547 [DOI] [PubMed] [Google Scholar]
- 48.Westerlund J. A., Weisgraber K. H. (1993) Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 268, 15745–15750 [PubMed] [Google Scholar]
- 49.Sihlbom C., van Dijk Härd I., Lidell M. E., Noll T., Hansson G. C., Bäckström M. (2009) Localisation of O-glycans in MUC1 glycoproteins using electron-capture dissociation fragmentation mass spectrometry. Glycobiology 19, 375–381 [DOI] [PubMed] [Google Scholar]
- 50.Zannis V. I., McPherson J., Goldberger G., Karathanasis S. K., Breslow J. L. (1984) Synthesis, intracellular processing, and signal peptide of human apolipoprotein E. J. Biol. Chem. 259, 5495–5499 [PubMed] [Google Scholar]
- 51.Ghiselli G., Beigel Y., Soma M., Gotto A. M., Jr. (1986) Plasma catabolism of human apolipoprotein E isoproteins: lack of conversion of the doubly sialylated form to the asialo form in plasma. Metabolism 35, 399–403 [DOI] [PubMed] [Google Scholar]
- 52.Stamatos N. M., Liang F., Nan X., Landry K., Cross A. S., Wang L. X., Pshezhetsky A. V. (2005) Differential expression of endogenous sialidases of human monocytes during cellular differentiation into macrophages. FEBS J. 272, 2545–2556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.