Abstract
The use of electron transfer dissociation (ETD) fragmentation for analysis of peptides eluting in liquid chromatography tandem mass spectrometry experiments is increasingly common and can allow identification of many peptides and proteins in complex mixtures. Peptide identification is performed through the use of search engines that attempt to match spectra to peptides from proteins in a database. However, software for the analysis of ETD fragmentation data is currently less developed than equivalent algorithms for the analysis of the more ubiquitous collision-induced dissociation fragmentation spectra. In this study, a new scoring system was developed for analysis of peptide ETD fragmentation data that varies the ion type weighting depending on the precursor ion charge state and peptide sequence. This new scoring regime was applied to the analysis of data from previously published results where four search engines (Mascot, Open Mass Spectrometry Search Algorithm (OMSSA), Spectrum Mill, and X!Tandem) were compared (Kandasamy, K., Pandey, A., and Molina, H. (2009) Evaluation of several MS/MS search algorithms for analysis of spectra derived from electron transfer dissociation experiments. Anal. Chem. 81, 7170–7180). Protein Prospector identified 80% more spectra at a 1% false discovery rate than the most successful alternative searching engine in this previous publication. These results suggest that other search engines would benefit from the application of similar rules.
The recently developed fragmentation approach of electron transfer dissociation (ETD)1 has become a genuine alternative to the more ubiquitous collision-induced dissociation (CID) for high throughput and high sensitivity proteomic analysis (1–3). ETD (4) and the related fragmentation process electron capture dissociation (ECD) (5) have been demonstrated to have particular advantages for the analysis of large peptides and small proteins (6–8) as well as the analysis of peptides bearing labile post-translational modifications (9–11). The results achieved through ETD and ECD analysis have been shown to be highly complementary to those obtained through CID fragmentation analysis, both through increasing confidence in particular identifications of peptides and also by allowing identification of extra components in complex mixtures (10, 12, 13). As CID and ETD can be sequentially or alternatively performed on precursor ions in the same mass spectrometric run, it is expected that the combined use of these two fragmentation analysis techniques will become increasingly common to enable more comprehensive sample analysis.
Software for analysis of CID spectra is significantly more advanced than that for ECD/ETD data. This is partly because the behavior of peptides under CID fragmentation is better characterized and understood so software has been developed that is better able to predict the fragment ions expected. The fragment ion types observed in ETD and ECD are largely known (5, 14, 15), but information about the frequency and peak intensities of the different ion types observed is less well documented.
We recently performed a study to characterize how frequently the different fragment ion types are detected in ETD spectra when analyzing complex digest mixtures produced by proteolytic enzymes or chemical cleavage reagents of different sequence specificity (16). These results were analyzed with respect to precursor charge state and location of basic residues, which were both shown to be significant factors in controlling the fragment ion types observed. The results showed that ETD spectra of doubly charged precursor ions produced very different fragment ions depending on the location of a basic residue in the sequence.
Based on this statistical analysis of ETD data from a diverse range of peptides (16), in the present study, a new scoring system was developed and implemented in the search engine Batch-Tag within Protein Prospector that adjusts the weighting for different fragment ion types based on the precursor charge state and the presence of basic amino acid residues at either peptide terminus. The results using this new scoring system were compared with the previous generation of Batch-Tag, which used ion score weightings based on the average frequency of observation of different fragment types in ETD spectra of tryptic peptides and used the same scoring irrespective of precursor charge and sequence. The performance of this new scoring was also compared with those reported by other search engines using results previously published from a large standard data set (17). The new scoring system allowed identification of significantly more spectra than achieved with the previous scoring system. It also assigned 80% more spectra than the most successful of the compared search engines when using the same false discovery rate threshold.
EXPERIMENTAL PROCEDURES
Samples
All data analyzed in this study were derived from samples that have been described in previous publications (16, 17). Briefly, the samples used to create the data for comparison of search engine performance with different enzymes were in-solution digests of a mouse synaptosomal preparation and a nuclear preparation from a mouse stem cell line. These samples were analyzed by LC-MSMS using an LTQ-Orbitrap (Thermo) where precursor ions were measured in the Orbitrap and fragments were measured in the linear ion trap. Sequential CID and ETD (with supplemental activation) spectra were acquired of each precursor, but only the ETD spectra were used for this study. The raw data and peak lists for these data can be downloaded from Tranche, https://proteomecommons.org/tranche/, using the following hash: sNYKSLLfYxWfpTyf3qpB1ACy2HEwKgbudgpasiiIzSOI9BsM+Fm6ZNBx683DeGnVIrWBTHhyN1Gy8hfjb93LxlCjswYAAAAAAAAc/A==.
The samples for the search engine comparison were a tryptic digest of a standard protein mixture (Universal Proteomics Standard UPS1 from Sigma), a set of Lys-C phosphopeptides enriched from either HEK293T cells or the p196 human pancreatic cancer cell line, and strong cation exchange fractions from a tryptic digest of p196 cells. These data were acquired by LC-MSMS in an Agilent 6340 three-dimensional ion trap using supplemental activation.
Peak List Generation
Descriptions of peak list generation methods were described in previous publications (16, 17). For the search engine comparison data set, the headers of the peak lists in the files were edited slightly for ease of searching within Batch-Tag, but the lists of masses were identical to the previous study.
Search Parameters for Enzyme Comparison Data
Data were searched against a version of the UniProtKB database (combination of Swiss-Prot and TrEMBL) downloaded on July 7, 2009 to which was concatenated a sequence-randomized version of the database. Only rodent entries were considered, leading to consideration of 187,236 database entries. Data were searched allowing a precursor mass tolerance of ±15 ppm and a fragment mass tolerance of ±0.6 Da. Cysteines were assumed to be carbamidomethylated, and the following other modifications were considered: acetyl (protein N terminus), acetyl + oxidation (protein N-terminal Met), Gln → pyro-Glu (N-terminal Gln), Met loss (protein N-terminal Met), Met loss + acetyl (protein N-terminal Met), and oxidation (Met). Different enzyme cleavage specificities were selected depending on the enzyme (or chemical agent) used for cleavage, but in each case, up to one missed cleavage was permitted.
False discovery rate (FDR) thresholds were calculated by importing results into Excel, sorting results by expectation value and then charge, and then reporting the number of hits to the normal part of the database at the point at which the relevant FDR threshold was reached, assuming the frequency of randomly matching peptides to the normal and random database components is the same. The estimated FDR = 2 × Ndecoy hits/Ntotal IDs, but the decoy hits are known so they can be removed to give a list with an estimated Ndecoy hits random assignments to the normal database sequences.
Search Parameters for Search Engine Comparison Data
Search parameters were as close as possible to the previous publication (17). Data were searched against the human subset of RefSeq from September 14, 2009 (the previously published search engine data were queried against the same database and subset but from March 5, 2007). This database contained a total of 37,878 protein entries. Separate searches were performed against a sequence-randomized version of this database in the same way as the previous publication (17). A precursor mass error tolerance of ±2.5 Da and fragment mass tolerance of ±0.7 Da were considered. All cysteines were assumed to be carbamidomethylated. The variable modifications considered were oxidation (Met) and phosphorylation (Ser/Thr/Tyr) with up to five modifications per peptide. Data were searched with the appropriate enzyme cleavage specificity (either trypsin or Lys-C) allowing for up to three missed cleavages.
A minimum peptide and protein score of 12 was used, and a minimum discriminant score threshold of −0.8 was used. FDR thresholds were calculated by combining the results from all searches against normal and randomized databases in Excel, sorting results by expectation value and then charge, and then calculating the estimated FDR in the same way as described for the enzyme comparison data above.
Differences between Batch-Tag Versions 5.3 and 5.4
The scoring and expectation value calculation for version 5.3 have been described previously (18, 19). Briefly, the scores listed in supplemental Table 1 for each ion matched are summed together to give a score for the spectrum to peptide match; e.g. if four observed masses correspond to z· ions and the score for each z· ion is 3.2, then the spectrum to peptide match scores 4 × 3.2 = 12.8. The scores for all peptide matches in the database with the correct precursor mass of each spectrum are stored, and then the probability of a particular score being part of this distribution of scores (where at best all but one are random matches) is calculated by a linear tail fit to a survival plot of this distribution.
Four changes were made in version 5.4. First, neutral loss peaks from charge-reduced species are removed prior to database searching in a way similar to that proposed by other researchers (20). For 1+ charge-reduced peaks, all masses within 60 Da were removed. For multiply charged charge-reduced peaks, peaks corresponding to the following mass differences were removed: −60, −59, −58, −45, −44, −43, −29, −28, −17, −16, −2, −1, and +1 Da. These losses were found to be the most common peaks observed when analyzing more than 10,000 spectra in house (some of these are second isotope peaks). Second, doubly charged fragment ions are now considered in ETD data (for precursors of charge state 3+ or higher). Third, new scoring was introduced that gives different fragment ion type weighting depending on the precursor ion charge state and presence of basic residues at the peptide termini (scoring is listed in supplemental Table 1). Finally, the calculation of the number of precursor ions considered when converting a probability into an expectation value was altered such that PTM positional isomers counted as a single precursor for this calculation (Protein Prospector still considers all the different site assignments; the change is only in the expectation value calculation). This prevents a significant degradation in expectation value calculation performance when a large number of modifications per peptide is permitted, which is an increasing problem if larger peptides are analyzed with multiple modification types considered.
RESULTS
In a previous study, we acquired ETD spectra of peptides produced using the enzymes trypsin, endoprotease Lys-C, endoprotease Lys-N, and the chemical cleavage agent CNBr (16). These data were acquired using an LTQ-Orbitrap where precursor ions were measured with high resolution and mass accuracy in the Orbitrap and ETD fragments were measured at low resolution but high sensitivity in the LTQ linear ion trap. These spectra were then analyzed to determine how frequently the different fragment ion types were observed in each data set. Data were also analyzed with respect to the precursor ion charge state. Noticeable differences were observed between the frequency of occurrence of different fragment ion types in doubly charged spectra compared with those from higher charge state precursors. There were also dramatic differences between ion type frequencies observed when different cleavage agents were used for peptide production.
It was predicted that utilizing this information should allow development of improved algorithms for performing database searching to identify peptides from ETD fragmentation data. Hence, a new scoring system was implemented in Batch-Tag in Protein Prospector that uses different weightings based on two parameters: 1) precursor ion charge state and 2) presence of basic residues at the N terminus, C terminus, both termini, or neither terminus. These weightings are reported in supplemental Table 1.
Effect of Implementation of Sequence- and Charge-dependent Scoring
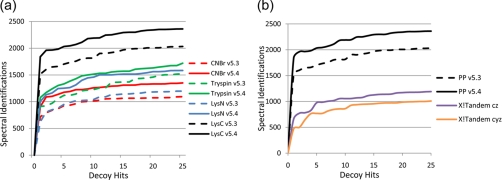
To test the effect of this new scoring system, the spectra from the peptides formed by different protein cleavage agents were reanalyzed. Results were compared between two scoring systems, one that used score weightings that were derived from the average frequency of observation of different ion types in ETD spectra of tryptic peptides of all precursor charges and peptide sequences (Protein Prospector version 5.3) and the sequence- and charge state-dependent scoring system documented in supplemental Table 1 (Protein Prospector version 5.4). Data were searched against a database with random sequences concatenated onto the normal database to allow estimation of peptide FDRs (21). All peptide identifications were considered independent; i.e. there was no adjustment of scoring based on the presence of other peptides from the same protein. Fig. 1a shows receiver operating characteristic plots for the different enzymes and scoring systems, and Table I reports the numbers of spectra identified at an estimated peptide FDR threshold of 1%.
Fig. 1.
Receiver operating characteristic plots showing numbers of spectral identifications for given number of matches to decoy part of database. a, results for all enzyme data. For each enzyme/chemical cleavage, the results for Protein Prospector (PP) version 5.3 are represented by a dashed line, whereas results for the newer version 5.4 are represented by a solid line. b, results for Lys-C data when searched using two versions of Protein Prospector and using X!Tandem allowing for c and z· ion (cz) or c, y, and z· ions (cyz).
Table I. Identifications at 1% threshold when analyzing data produced using a range of enzymatic or chemical cleavages.
Results are presented from Protein Prospector versions 5.3 and 5.4, the latter of which uses new sequence- and charge state-dependent scoring.
| FDR | Lys-N | Lys-C | CNBr | Trypsin | |
|---|---|---|---|---|---|
| % | |||||
| v5.3 | 1 | 1091 | 2007 | 1027 | 1370 |
| v5.4 | 1 | 1516 | 2359 | 1295 | 1570 |
For all enzymes, a significant increase in the number of spectra identified was observed. At the 1% FDR threshold, the increased percentage of spectra identified was 18% for Lys-C data, 15% for spectra of tryptic peptides, 26% for CNBr products, and 39% for Lys-N peptide spectra. The improvement for the Lys-N data stands out but is not particularly surprising as the weightings for the Protein Prospector version 5.3 were trained on, and hence optimized for, data from tryptic peptides as these are the most common type of peptide being analyzed in proteomic studies. Hence, the previous weighting favored peptides with basic C-terminal residues. It was shown in the fragment ion statistics results that the ratio of N-terminal to C-terminal ions observed in Lys-N digests, especially for spectra derived from doubly charged precursors, differs most dramatically from those observed from tryptic peptide spectra (16), statistically confirming the ad hoc observations of other researchers (22). Thus, it was expected that the original scoring would perform suboptimally with Lys-N peptide fragmentation data. Nevertheless, the new scoring yielded a dramatic improvement in the number of spectra reliably identified.
For comparison with another search engine, the Lys-C data set was searched using the latest version of X!Tandem (version 2009.10.01). The data were searched with two different settings, either allowing only for c and z· ions or allowing for c, y, and z· ions. The results are shown in Fig. 1b. It can be seen that X!Tandem performed better when not considering y ions, but even this search only identified roughly half the number of spectra compared with the improved version of Protein Prospector.
Comparison of Results with Other Search Engines
Encouraged by these results, a more in-depth investigation of how Protein Prospector performance compared with alternative software for analyzing peptide ETD data was undertaken. A study has been published comparing the results of four different software search engines, Mascot (version 2.2) (23), OMSSA (version 2.1.0) (24), Spectrum Mill (version 3.03.078) (Agilent), and X!Tandem (version 2007.04.01.1) (25), on the combination of three different data sets of ETD spectra (17). These data sets included spectra produced from both trypsin and endoprotease Lys-C enzymatic cleavages. Strong cation exchange fractions and phosphopeptide data were also analyzed in the study. Hence, these data represent different types of peptides in large enough numbers to allow assessment of software performance when analyzing sequences with different characteristics.
Search parameters used by Protein Prospector were matched to those in the previous study, and FDR thresholds were determined by separate searches of target and decoy databases to match the approach used in the previously published analysis of these data. Many measures of search engine performance were reported for the different search engines in the previously published analysis (17). Table II reports the performance metrics of Protein Prospector version 5.3 and the new scoring system in version 5.4 at a 1% FDR threshold and compares these values with the previously generated results from the other search engines when analyzing the sum of all of the data. All of the spectral identifications by Protein Prospector are reported in supplemental Table 2, and supplemental Table 3 reports the spectral identification overlap between Protein Prospector version 5.4 and the other search engines. The numbers of spectra reported for Spectrum Mill and X!Tandem differ slightly from the previously published values (17) but are the values we derived when analyzing their results (in the supplemental table to their paper).
Table II. Acceptance criteria corresponding to 1% FDR threshold for all search engines and corresponding number of spectra identified using these parameters.
The values for search engines other than Protein Prospector (PP) were derived from reanalysis of searches performed by Kandasamy et al. (17).
| OMSSA | Mascot | Spectrum Mill | X!Tandem | PP v 5.3 | PP v 5.4 | |
|---|---|---|---|---|---|---|
| All data, 1% FDR threshold | 3.7 × 10−3 | 41.2 | 10.1, 10.3, 12.5, 12.5 | −2.797 | 0.075 | 0.028 |
| Total spectral identifications at 1% FDR threshold | 4,491 | 5,529 | 7,779 | 4,997 | 9,589 | 14,028 |
These results show that the scoring system based on ion frequency independent of charge state and sequence (Protein Prospector version 5.3) fared well against the other search engines, identifying 23% more spectra than the most successful alternative software. The results from the newer scoring system (Protein Prospector version 5.4) represent an even bigger improvement with an additional 46% more reported identifications at the 1% FDR threshold. Thus, the sequence- and charge state-dependent scoring assigned 80–212% more spectra than other search engines at this common threshold.
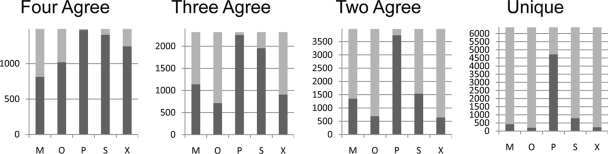
The overlap between confident spectral identifications assigned by the new scoring compared with the other four search engines is summarized in Table III, and Fig. 2 plots the membership of different search engines in each level of agreement among the search engines. These results show that a total of 15,985 spectra were assigned a confident result by one or more search engines, but for only 1815 of these did all the search engines confidently assign the same spectrum. In the previous comparison of the four search engine results, it was shown that about 45% of all the identifications were reported by only one search engine (17). The inclusion of Protein Prospector results significantly reduced this number. The majority of the single search engine matches are now to Protein Prospector. Excluding these matches, the inclusion of Protein Prospector reduced the single search engine matches from 45% to about 10% (although it should be noted that results here are reported at a 1% FDR threshold, whereas a 5% FDR threshold was used in the previously published study (17)). This shows that Protein Prospector results overlap with many of the unique results from other search engines.
Table III. Overlap of spectral identifications between Protein Prospector version 5.4 and the four compared search engines at 1% FDR threshold.
Values for search engines other than Protein Prospector were derived from filtering of the results created by Kandasamy et al. (17). M, Mascot; O, OMSSA; P, Protein Prospector; S, Spectrum Mill; X, X!Tandem.
| No. search engines that identified spectrum | Total | Search engine combination | No. spectra | Search engine combination | No. spectra | Search engine combination | No. spectra | Search engine combination | No. spectra |
|---|---|---|---|---|---|---|---|---|---|
| All 5 | 1815 | ||||||||
| Only 4 | 1489 | O, P, S, X | 676 | M, P, S, X | 469 | M, O, P, S | 246 | M, O, P, X | 82 |
| M, O, S, X | 16 | ||||||||
| Only 3 | 2320 | M, P, S | 924 | P, S, X | 559 | O, P, S | 409 | O, P, X | 211 |
| M, P, X | 105 | M, O, P | 48 | M, O, S | 32 | M, S, X | 17 | ||
| O, S, X | 13 | M, O, X | 2 | ||||||
| Only 2 | 3976 | P, S | 1344 | M, P | 1195 | O, P | 613 | P, X | 585 |
| M, S | 122 | S, X | 36 | O, S | 36 | M, O | 24 | ||
| O, X | 16 | M, X | 5 | ||||||
| Only 1 | 6385 | P | 4724 | S | 790 | M | 427 | X | 239 |
| O | 205 |
Fig. 2.
Bar charts plotting overlap in spectral identifications for each search engine at each level of agreement between search engine results. The total height of the plot is the number of spectra matched at each level of agreement, and the dark bar is the number of spectra at a given level of agreement for which the particular search engine is one of the members. M, Mascot; O, OMSSA; P, Protein Prospector; S, Spectrum Mill; X, X!Tandem.
Fig. 2 shows that Mascot was the most common search engine to miss out when all other search engines agreed. However, as one examines lower levels of overlap, OMSSA and X!Tandem were the least consensual. There were very few instances at any level where multiple search engines agreed on an interpretation and Protein Prospector did not return the same result. Protein Prospector also dominated the set of single search engine results.
Influence of Charge State on Results
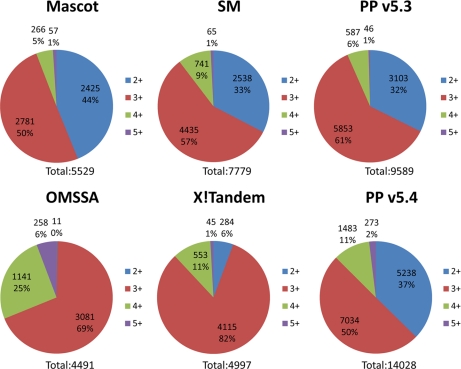
The difference between the performances of the other search engines was shown in the previously published analysis to be heavily due to the ability of the different softwares to identify spectra from doubly charged precursor ions (17). Fig. 3 presents pie charts showing the numbers and percentages of identifications at each charge state for the two versions of Protein Prospector and each of the other search engines. Identifications of spectra from doubly charged precursors make up roughly 35% of the results from both versions of Protein Prospector, which shows a good agreement with the results reported by Spectrum Mill, the most successful of the compared search engines. However, the newer scoring system assigned more than twice as many quadruply charged precursors and dramatically more 5+ precursor ion spectra than version 5.3.
Fig. 3.
Total numbers and distribution of spectra identified to precursors of different charge states by search engines OMSSA, Mascot, Spectrum Mill (SM), X!Tandem, and two versions of Protein Prospector (PP). The values for search engines other than Protein Prospector were derived from filtering of the results created by Kandasamy et al. (17). For each charge state, the total number and percentage of identifications are indicated.
The increased number of higher charge state precursors identified raises the question of whether the reliability of results for different charge states is the same; indeed, separate thresholds are reported for Spectrum Mill results because of this recognized concern. Table IV reports E-value thresholds for 1% FDR estimates for the two versions of Protein Prospector when the thresholds were separately calculated by charge state. These values show that the older scoring system exhibited little bias in terms of reporting precursors of different charge states with differing reliability, but the introduction of charge state-dependent scoring required lower expectation value thresholds to be applied for higher charge state precursor ions to maintain a given FDR threshold. This indicates that the application of a global expectation value threshold in the newer version of Protein Prospector caused some reliable doubly charged assignments to not be reported (referred to as false negatives in database searching terms) but also led to some 4+ and 5+ precursors of lower reliability being confidently reported (potential false positives). Hence, charge state-dependent thresholds were applied to all of the Protein Prospector results, and Table V compares the number of spectra identified at each charge state when using the global 1% FDR threshold and when using the charge state-specific FDR thresholds. These results show that the more lenient threshold for doubly charged precursors more than compensates for the more conservative thresholds for quadruply and quintuply charged precursors to lead to a total of 14,346 reported spectral matches. This value represents 318 identifications more than the number reported using the global FDR acceptance criterion.
Table IV. Expectation value thresholds corresponding to 1% FDR calculated for results from precursors with different charge states for Protein Prospector (PP) versions 5.3 and 5.4.
| Charge |
||||
|---|---|---|---|---|
| 2+ | 3+ | 4+ | 5+ | |
| PP v 5.3 | 0.037 | 0.096 | 0.083 | 0.082 |
| PP v 5.4 | 0.036 | 0.031 | 0.025 | 0.011 |
Table V. Comparison of number of spectra identified at each precursor charge state when using global or charge state-specific 1% FDR thresholds for Protein Prospector version 5.4.
| Charge | Global 1% FDR | Charge state-dependent 1% FDR |
|---|---|---|
| 2+ | 5,238 | 5,553 |
| 3+ | 7,034 | 7,127 |
| 4+ | 1,483 | 1,457 |
| 5+ | 273 | 209 |
| Total | 14,028 | 14,346 |
Analysis of Phosphopeptide Spectra
Table VI reports the number of phosphopeptides identified in the total data set. Two sets of values are included for Protein Prospector results, those using the global 1% FDR E-value threshold listed in Table II and also a 1% FDR threshold calculated based solely on peptides reported as being phosphorylated. To obtain this second value, all the results were filtered to only list phosphorylated peptides matched to the normal or random database, and then appropriate expectation values for a 1% FDR threshold were estimated from this data subset. Also listed are numbers of phosphopeptides reported by other search engines using the global 1% FDR thresholds listed in Table II. Using the global thresholds, Protein Prospector version 5.4 reported twice as many identifications as any other search engine. However, we believe the values reported using these global thresholds are inaccurate for PTM results, so they are only reported for the purpose of comparison with the other search engine values.
Table VI. Number of phosphopeptides reported by different software at 1% FDR threshold.
For Protein Prospector (PP), two values are reported, one value using a global threshold as acceptance criteria and one value using thresholds calculated using only phosphopeptide identifications.
| FDR | OMSSA | Mascot | Spectrum Mill | X!Tandem | PP v 5.4 |
|---|---|---|---|---|---|
| Global 1% FDR | 996 | 562 | 916 | 549 | 1839 |
| 1% FDR for reported phosphopeptides | 1480 |
The two methods used for calculating FDR values reported significantly different numbers of phosphopeptide spectra, but we believe the lower values are more accurate estimations of the number of phosphopeptides reported at a 1% FDR. The difference between the values results because these searches considered significantly more potential phosphopeptides than unmodified peptides. This causes random matches to phosphopeptides to be more common than to unmodified peptides. This phenomenon is described in more detail under “Discussion.”
DISCUSSION
Because of the relatively recent availability of ETD on commercial instruments, the software for analyzing this type of data is still being developed. In this study, the results of a systematic analysis of how often different fragment ion types are observed in ETD data were used to develop a charge state- and sequence-dependent scoring system in the program Batch-Tag, which is part of the Protein Prospector software package.
The new results, when compared with results obtained using a scoring system that used average ion frequencies independent of peptide sequence and precursor charge, showed improved performance on data derived from all four different protein cleavage specificities examined. The most significant improvement was observed with data derived from peptides formed by cleavage with the endoprotease Lys-N. Researchers who promote the suitability of endoprotease Lys-N in combination with ETD for peptide analyses have commented that one current drawback to their approach is that search engines are not optimized for analysis of such spectra (22). The new scoring system presented is now tailored for use with different enzymes, so it should provide sensitive results for different cleavage options. With the optimized scoring, among the data sets used for testing the software performance, the endoprotease Lys-C digestion was comfortably the most effective preparation for subsequent peptide identification using ETD fragmentation.
A thorough comparison of the performance of four different search engines when analyzing ETD data acquired in a three-dimensional ion trap has been published previously (17). The same data sets now were analyzed using the two generations of scoring systems of Batch-Tag to evaluate their performance in comparison with these other tools. The results showed that both scoring systems outperformed other software with the new scoring (version 5.4) identifying 46% more spectra than version 5.3 at a 1% peptide FDR threshold. Conceptually, comparable levels of improvement may be obtainable with other search engines through the application of sequence- and/or charge state-dependent adaptations to their scoring systems.
The improvements in the search engine comparison were significantly higher than those observed in the comparison of the two scoring systems using spectra from peptides produced by different enzymes or chemical cleavages. We predict that the cause for this disparity is that the enzyme cleavage comparison data were acquired in an LTQ-Orbitrap where precursors were measured at high mass accuracy. In contrast, the published software comparison data set was acquired with low resolution and low mass accuracy precursor ion mass measurement where precursor charge state determination was not known prior to database searching. Hence, the search engines had to consider many more precursors for each spectrum with the latter data set because of both the wider precursor mass tolerance and the consideration of several potential charge states. This makes it much more difficult to discriminate between correct and random results with the second data set, meaning that there was more potential for improvement in the analysis of these data.
The overlap between search engine results was investigated. The numbers in the present study differ slightly from those published in the previous analysis due to our only querying whether the spectra were assigned and not differentiating between alternative modification site assignments (17). For only about 11% of reported identifications did all search engines agree on a given spectrum, a surprisingly low number but similar to the observations when the four search engines were compared previously (17). However, a marked reduction in the number of spectra identified by only one search engine was observed, indicating that the new scoring in Protein Prospector assigns many of the spectra that were previously only identified by one search engine. It should be noted that these values do not simply represent whether search engines reported the same peptide result but also require the assignment to meet the 1% FDR threshold; the overlap in peptide assignments is actually much greater than these values indicate. For example, of the sixteen assignments that were reported by all search engines except Protein Prospector, the same peptide was also reported by Protein Prospector in 14 of the cases but did not meet the 1% FDR acceptance criterion.
A bias was discovered with the new scoring on the search engine comparison data set that gave peptide identifications to higher charge state precursors slightly elevated confidence when a global expectation value threshold was applied. This is a phenomenon that is apparently also present in results from Spectrum Mill as it uses different acceptance thresholds for each charge state. It is also a well recognized situation in CID spectral identifications returned by the search engine Sequest (26). It may also be present in other search engine ETD results but to our knowledge has not been evaluated. We believe this bias was mainly caused by the inability to determine the precursor ion charge state for spectra in this data set. A higher charge state precursor matches to a significantly longer peptide and will generally score higher than the best match to a lower charge state interpretation of the same spectrum because of the higher number of potential fragment ions possible. We investigated whether this same bias existed in the enzyme comparison data set where the precursor ions were measured in the Orbitrap and did not observe a noticeable bias.
Finally, the performance of Batch-Tag at identifying phosphopeptides was evaluated. Using the same global FDR threshold criteria for comparison with the other search engines led to the new version of Protein Prospector reporting roughly twice as many identifications as any compared search engine. However, we then demonstrated that this method of FDR estimation provides a poor measure of reliability for phosphopeptide identifications when the majority of peptides present are not modified. In database searches where post-translational modifications are considered, all of a particular amino acid(s) are considered as either being unmodified or modified with each possibility given equal likelihood. Because of the potential combinations of modified residues within a peptide, when allowing for a modification of common amino acids such as serine and threonine, the number of modified peptides considered is dramatically higher than unmodified peptides. Permitting phosphorylation in these database searches increased the average number of precursor ions (with or without modifications) considered per spectrum from roughly 30,000 (when phosphorylation was not considered) to around 260,000. Hence, nearly 90% of all peptide sequences considered are phosphorylated. Assuming an equal likelihood of matching all peptides at random, one would predict that nearly 90% of the random matches to the normal database are to phosphopeptides.
By filtering the results to list only those that are reported as phosphorylated in the normal and random database searches and then determining the FDR threshold, a much more accurate estimate of the number of phosphopeptides reliably identified could be derived. This is a filtering step that should be used in all analyses that consider modifications that lead to a significant increase in the database search space if the majority of the peptides present are not modified. It is important to note that even for the results with the stricter acceptance criteria we are not claiming that the reported phosphorylation sites are necessarily correct, merely that the peptide sequence and modification state are reliable. Either manual verification or the use of a secondary program to evaluate site assignments would be necessary prior to publication of modification sites (21).
Comparison of different database search engines always produces biases introduced as a result of the different parameters and processing of results by each search engine. Also, attempts to use as similar parameters as possible for each software invariably lead to compromises in the performance of each program. The parameters used in this search engine comparison were not sensible options for searching the majority of the data. For example, the consideration of up to five phosphorylations per peptide when the majority of the data were from samples that were not expected to contain many phosphopeptides led to elevated numbers of false matches to phosphopeptides. If all the data other than the phosphopeptide-enriched data set had been searched without considering phosphorylation a higher number of spectra would have been identified (even though the few phosphopeptides in these other data sets would no longer have been reported).
Permitting five modifications per peptide leads to the consideration of nearly 10 times as many precursors compared with a search not considering phosphorylation. We have already highlighted that PTM site assignments returned by search engines are not as reliable as the reporting of peptide with PTM state. Many of the considered precursors are the same peptide with the same modification state but are PTM positional isomers. The new version of Protein Prospector counts these variants as one precursor for the expectation value calculation but still considers all of the modification combinations for spectral assignment. If the search engine reduced the effort in trying to assign the sites reliably and focused more on just identifying the peptide and PTM state it would be possible to noticeably accelerate search times with minimal effect on the reliability of peptide results. To demonstrate this theory, the data were re-searched restricting the number of PTM permutations per peptide to 64 on the phosphopeptide data set in this study. Adding this threshold reduced the duration of the search to one-eighth of the original time and led to the loss of only three phosphopeptide identifications. Combining results from a search like this with software for determining PTM site assignments and reliabilities (21) would significantly reduce data analysis times while having minimal impact on results.
All of the results presented here were analyzed under the assumption that peptide identifications are independent of each other. In reality, a peptide is more likely to be matched to a protein that has already been identified in the sample, and most search engines have different strategies for converting peptide results into protein assignments that may include reporting peptides below a given threshold if they are from proteins already confidently identified. This conversion of peptide identifications to protein identifications and assessment of the reliability of this process were not addressed in this study.
CONCLUSIONS
The implementation of a scoring system adapted to different precursor charge states and peptide sequences for analysis of ETD data led to a significant increase in the number of spectra identified in a variety of different data sets. The new scoring appeared to be particularly beneficial for peptides produced by digestion with endoprotease Lys-N. It also proved to have a larger effect on the number of components identified in data that were acquired with poor mass accuracy and resolution measurement of precursor ions than equivalent fragmentation data where the precursor ions were measured in an LTQ-Orbitrap. Problems with the use of global FDR estimation approaches when there are different peptide populations within the data set were highlighted, particularly the issues when reporting phosphopeptide results (or other analyses that consider multiple modifications per peptide).
The results assigned using the new scoring showed major improvements over other search engine performances. This is probably largely because other search engines have not yet been optimized for ETD data analysis. The results presented here will hopefully spur other softwares to also improve their performance. The software described here is freely available through the web at http://prospector.ucsf.edu.
Supplementary Material
Acknowledgments
We are very grateful to Henrik Molina for sharing the peak lists and analysis results from his study.
* This work was supported, in whole or in part, by National Institutes of Health Grant P41 RR001614 from the National Center for Research Resources.
 This article contains supplemental Tables 1–3.
This article contains supplemental Tables 1–3.
1 The abbreviations used are:
- ETD
- electron transfer dissociation
- ECD
- electron capture dissociation
- OMSSA
- Open Mass Spectrometry Search Algorithm
- FDR
- false discovery rate
- PTM
- post-translational modification.
REFERENCES
- 1.Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Good D. M., Wirtala M., McAlister G. C., Coon J. J. (2007) Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 3.Swaney D. L., McAlister G. C., Wirtala M., Schwartz J. C., Syka J. E., Coon J. J. (2007) Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 79, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zubarev R. A. (2004) Electron-capture dissociation tandem mass spectrometry. Curr. Opin. Biotechnol. 15, 12–16 [DOI] [PubMed] [Google Scholar]
- 6.Kjeldsen F., Haselmann K. F., Budnik B. A., Sørensen E. S., Zubarev R. A. (2003) Complete characterization of posttranslational modification sites in the bovine milk protein PP3 by tandem mass spectrometry with electron capture dissociation as the last stage. Anal. Chem. 75, 2355–2361 [DOI] [PubMed] [Google Scholar]
- 7.McLafferty F. W., Horn D. M., Breuker K., Ge Y., Lewis M. A., Cerda B., Zubarev R. A., Carpenter B. K. (2001) Electron capture dissociation of gaseous multiply charged ions by Fourier-transform ion cyclotron resonance. J. Am. Soc. Mass Spectrom. 12, 245–249 [DOI] [PubMed] [Google Scholar]
- 8.Coon J. J., Ueberheide B., Syka J. E., Dryhurst D. D., Ausio J., Shabanowitz J., Hunt D. F. (2005) Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 102, 9463–9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelleher N. L., Zubarev R. A., Bush K., Furie B., Furie B. C., McLafferty F. W., Walsh C. T. (1999) Localization of labile posttranslational modifications by electron capture dissociation: the case of gamma-carboxyglutamic acid. Anal. Chem. 71, 4250–4253 [DOI] [PubMed] [Google Scholar]
- 10.Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) O-Linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 12.Swaney D. L., McAlister G. C., Coon J. J. (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zubarev R. A., Zubarev A. R., Savitski M. M. (2008) Electron capture/transfer versus collisionally activated/induced dissociations: solo or duet? J. Am. Soc. Mass Spectrom. 19, 753–761 [DOI] [PubMed] [Google Scholar]
- 14.Cooper H. J., Hudgins R. R., Håkansson K., Marshall A. G. (2002) Characterization of amino acid side chain losses in electron capture dissociation. J. Am. Soc. Mass Spectrom. 13, 241–249 [DOI] [PubMed] [Google Scholar]
- 15.Fung Y. M., Chan T. W. (2005) Experimental and theoretical investigations of the loss of amino acid side chains in electron capture dissociation of model peptides. J. Am. Soc. Mass Spectrom. 16, 1523–1535 [DOI] [PubMed] [Google Scholar]
- 16.Chalkley R. J., Medzihradszky K. F., Lynn A. J., Baker P. R., Burlingame A. L. (2010) Statistical analysis of peptide electron transfer dissociation fragmentation mass spectrometry. Anal. Chem. 82, 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandasamy K., Pandey A., Molina H. (2009) Evaluation of several MS/MS search algorithms for analysis of spectra derived from electron transfer dissociation experiments. Anal. Chem. 81, 7170–7180 [DOI] [PubMed] [Google Scholar]
- 18.Chalkley R. J., Baker P. R., Huang L., Hansen K. C., Allen N. P., Rexach M., Burlingame A. L. (2005) Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer: II. new developments in Protein Prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol. Cell. Proteomics 4, 1194–1204 [DOI] [PubMed] [Google Scholar]
- 19.Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., Burlingame A. L. (2008) In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics 7, 2386–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Good D. M., Wenger C. D., McAlister G. C., Bai D. L., Hunt D. F., Coon J. J. (2009) Post-acquisition ETD spectral processing for increased peptide identifications. J. Am. Soc. Mass Spectrom. 20, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 22.Boersema P. J., Taouatas N., Altelaar A. F., Gouw J. W., Ross P. L., Pappin D. J., Heck A. J., Mohammed S. (2009) Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol. Cell. Proteomics 8, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 24.Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 25.Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 26.Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.