Abstract
Obtaining accurate protein profiles from homogeneous cell populations in heterogeneous tissues can enhance the capability to discover protein biomarkers. In this context, methodologies to access specific cellular populations and analyze their proteome with exquisite sensitivity have to be selected. We report here the results of an investigation using a combination of laser microdissection and accurate mass and time tag proteomics. The study was aimed at the precise determination of proteome alterations in intrahepatic cholangiocarcinoma ICC, a markedly heterogeneous tumor. This cancer, which is difficult to diagnose and carries a very poor prognosis, has shown an unexplained increase in incidence over the last few years. Among a pool of 574 identified proteins, we were able to report on altered abundance patterns affecting 39 proteins conforming to a variety of potential tumorigenic pathways. The reliability of the proteomics results was confirmed by Western blot and immunohistochemistry on matched samples. Most of the proteins displaying perturbed abundances had not yet been described in the setting of ICC. These include proteins involved in cell mobility and actin cytoskeleton remodeling, which may participate in the epithelial to mesenchymal transition, a process invoked in migration and invasion of cancer cells. The biological relevance of these findings was explored using a tissue microarray. An increased abundance of vimentin was thus detected in 70% of ICC and none of the controls. These results suggest that vimentin could play a role in the aggressiveness of ICC and provide a basis for the serious outcome of this cancer.
Cholangiocarcinoma (CCA),1 which arises from the hepatic bile ducts, is the primary cancer accounting for ∼10–15% of all hepatobiliary malignancies. CCA is categorized by the International Classification of Diseases for Oncology into intrahepatic and extrahepatic forms, the latter including perihilar, hilar, and distal bile duct CCA. The global incidence rate of intrahepatic cholangiocarcinoma (ICC) has increased by 2–6% during recent decades, whereas the annual incidence rate of the more common, extrahepatic form has remained relatively stable (1, 2). The diagnosis of ICC remains particularly challenging because the disease can mimic metastasis to the liver or hepatocellular carcinoma (HCC). The only potentially curative treatment options available at present are surgical. Unfortunately, the majority of patients are diagnosed at an advanced, unresectable stage because of the initially silent clinical characteristics of this malignancy. The prognosis of ICC is therefore devastating with survival of less than 24 months following diagnosis. Although several risk factors have been reported as contributing to the development of the disease, most cases of ICC occur in the absence of known etiological factors (3–5). As a consequence, considerable efforts have been made to identify reliable markers to enable the early detection of biliary cancers and provide new insights into the pathogenesis of this deadly disease (6). Recent studies have focused on the cytokines and growth factors (7, 8) produced by CCA cells as well as on the proteomics analysis of serum and bile (9, 10). Follow-up studies are ongoing to determine the sensitivity and specificity of the markers that have emerged from these investigations.
Tumor tissue is undeniably the most appropriate resource to investigate tumor-specific signals. Indeed, the determination of alterations to the protein profiles of ICC may offer opportunities to identify tumor-specific markers. To date, proteomics studies performed using tumor tissues have mainly tried to identify proteins with a differential expression between HCC and ICC to prevent the misclassification of these pathologies (11, 12). Furthermore, these studies, performed on total liver homogenates, were not appropriate to detect proteins with altered expression in tumorous cholangiocytes. Indeed, ICC tumor cells are essentially embedded in an abundant stroma containing inflammatory cells and fibroblasts, which may impair the detection of proteins displaying tumor-specific expression patterns (13). In summary, because no proteomics studies have so far been performed on isolated cholangiocytes, any proteome alterations in the setting of ICC remain a matter of speculation.
Laser microdissection (LM) has emerged as a suitable tool to selectively extract cells of interest from their natural environment. This technology has been used extensively to profile global gene expression in purified tumor cells (14–16). It has also been used successfully in experiments designed to discover protein biomarkers through mass spectrometry-based proteomics studies (17–19). However, working with such small samples challenges conventional proteomics techniques in terms of sensitivity and the precision of quantitation.
One particularly efficient method for proteome analysis is the so-called accurate mass and time (AMT) tag approach where high performance liquid chromatography (HPLC) and high resolution Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS or FTMS) work in synergy to achieve a broad proteome coverage even for small sample amounts (20, 21). A recent report demonstrated the significant advantage of the AMT tag method over more conventional proteomics technologies in enabling significantly broader proteome coverage using the minute protein quantities available from tissue microdissection (22).
In the present study, we used an AMT tag strategy to determine the differential protein profile between tumorous and non-tumorous microdissected cholangiocytes to define specific proteome alterations of human ICC. Among proteins with altered expression in ICC, we detected a panel of proteins conforming to potential tumorigenic pathways that could be candidates as therapeutic targets and tumor markers for this lethal disease.
EXPERIMENTAL PROCEDURES
Patients and Tissue Samples
Liver specimens were obtained from the “Centre de Ressources Biologiques Paris-Sud,” Paris-Sud University, Villejuif, France. Access to this material was in agreement with French legislation. Tissue samples were collected at the time of surgical resection with the prior informed consent of patients. The local research ethics committee specifically approved this procedure. Detailed clinical data regarding these subjects are summarized in Table I. Tumorous and adjacent non-tumorous hepatic tissues from four patients with intrahepatic cholangiocarcinoma and five perihilar samples containing large bile duct sections from control subjects without ICC were obtained within 30 min of surgical resection. These samples were immediately frozen in liquid nitrogen and stored at −80 °C until use. The tumors were graded by a pathologist by means of microscopic examination.
Table I. Clinical data of subjects included in mass spectrometry analysis.
F, female; M, male; N/A, not applicable to the controls.
| Identification | Gender | Age | Liver lesion | Tumor size | Histological type |
|---|---|---|---|---|---|
| yr | cm | ||||
| Tumors | |||||
| CA | F | 39 | Intrahepatic cholangiocarcinoma | 7 | Glandular structure |
| CB | F | 62 | Intrahepatic cholangiocarcinoma | 6 | Mixed architecture |
| CC | M | 77 | Intrahepatic cholangiocarcinoma | 12 | Glandular structure |
| CD | M | 55 | Intrahepatic cholangiocarcinoma | 14 | Compact architecture |
| Controls | |||||
| TB | M | 48 | Cirrhosis | N/A | N/A |
| TC | M | 53 | Cirrhosis | N/A | N/A |
| TD | F | 45 | Subacute hepatitis | N/A | N/A |
| TE | F | 61 | Cirrhosis | N/A | N/A |
| TF | M | 53 | Hepatocellular carcinoma | N/A | N/A |
Laser Microdissection
The liver sections were prepared and treated as described previously (23). LM was performed using a PALM Microbeam system (Carl Zeiss Inc.) (see supplemental Fig. 1). From each liver sample, ∼100,000 cells corresponding to 10 mm2 were microdissected and catapulted directly into the cap of a microcentrifuge tube containing 10 μl of 10% SDS. Cell lysates were stored at −80 °C until use. Based on a careful review, each section was estimated to contain ≥90% of the desired cells.
Mass Spectrometry Analysis
Sample Preparation
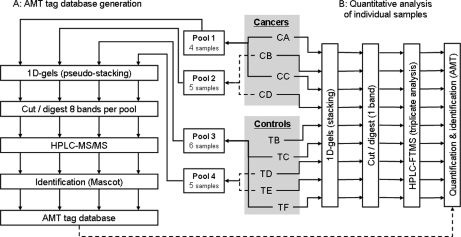
For AMT tag database generation, LC-MS/MS analyses have to be performed on fractionated samples to achieve decent proteome coverage. For this purpose, four to six samples were pooled per pair of patients with or without ICC (see Fig. 1A). Thirty microliters of lysis buffer (50 mm Tris, pH 6.8, 2% SDS, 0.35 m β-mercaptoethanol, and 4 m urea) were added to each pool, and the samples were deposited on a one-dimensional SDS-PAGE gel before being subjected to a short separation (pseudostacking) to prevent the sample from being distributed over the whole gel lane. After Coomassie Blue staining to reveal proteins, each pseudostacking band was cut into eight slices, and the corresponding fractions were digested in gel with trypsin (Promega, Charbonnières-les-bains, France) as described previously (24). An estimated 0.8–1.2 μg of total protein was injected for each gel fraction. All fractions were analyzed with LC-MS/MS in triplicate, and the resulting peptide identifications were used to compile an AMT tag database.
Fig. 1.
Study design. Cholangiocytes microdissected from control livers (TB–TF) and from intrahepatic cholangiocarcinomas (CA–CD) were pooled prior to sample fractionation and LC-MS/MS analysis to generate an AMT tag database (A). Individual samples were subsequently analyzed by LC-FTMS to determine differential alterations of their proteome (B).
For quantitative studies, protein extracts from individual patient biopsies were deposited on a one-dimensional SDS-PAGE gel and separated in the stacking mode (see Fig. 1B). The stacked gel band associated with each patient was then excised and digested in gel as described above. The resulting peptide mixtures were analyzed in triplicate using LC-FTMS. Peptides from ∼20,000 cells were injected for each LC-FTMS analysis.
Nano-HPLC-MS and MS/MS Analysis
All proteomics analyses were performed using the 7-tesla hybrid linear ion trap Fourier transform ion cyclotron resonance mass spectrometer LTQ-FT Ultra (Thermo Electron, Bremen, Germany) coupled to an Ultimate 3000 (LC Packings, Amsterdam, Netherlands) nano-HPLC system mounted with an LC Packings PepMap nanocolumn (75-μm inner diameter, 15 cm long, 3-μm particles, 100-Å pores). The chromatographic separation conditions were as described previously (25).
AMT Tag Database Generation (MS/MS Mode)
The FTMS detector was used for the survey scan within the m/z range of 400–1850 at a resolution setting of 50,000 (25). MS/MS was performed in the linear ion trap in parallel with high resolution acquisition of the MS signal on the FT detector. Dynamic exclusion prevented the acquisition of multiple MS/MS spectra for a given precursor ion (mass tolerance, ±2 ppm; repeat MS/MS acquisition, two times within 30 s; exclusion duration, 180 s; exclusion list size, 500 entries.)
MS/MS data were processed using Mascot Distiller (Matrix Science) to produce peak lists (e.g. .mgf files) that were subsequently submitted to the Mascot v.2.2.03 (Matrix Science) search engine for database searches against the Swiss-Prot-TrEMBL database (compiled from Swiss-Prot release v54.8 and TrEMBL release v37.8) using the taxonomy “Homo sapiens (human)” (72,036 protein sequences). Mascot search parameters are listed in supplemental Table 1. A target-decoy database search was performed as described previously (26) to estimate false positive rates. Identifications were validated automatically (filters, Rank = 1 and score > identity threshold at p value <0.05) and compiled in an MS identification database using the IRMa homemade software (27). The MS identification database was then converted into an AMT tag database (Microsoft Access format) by grouping the multiple occurrences of each identified peptide. To reduce false positives, we opted for the removal of peptides with length ≤6 amino acids. In addition, only proteins covered by at least two peptides were considered for inclusion in the database with the exception of proteins covered by a single peptide (length, >6) with a Mascot score greater than or equal to 50. Peptides spectrum matches for these peptides have been compiled and are provided as supplemental Fig. 2.
Quantitative MS Acquisitions (MS Mode)
All quantitative MS data were obtained under the FTMS detection mode on an m/z range of 400–1850 and resolution of 50,000. We took advantage of the parallel acquisition capability of the LTQ-FT instrument to acquire a single MS/MS spectrum during each survey scan. This allowed us to further populate the AMT tag database without compromising the MS data acquisition rate.
MS data were processed using the Decon-2LS/VIPER open source software suite provided by Pacific Northwest National Laboratory. Individual MS spectra were deisotoped using Decon-2LS. The VIPER software then ensured LC-MS feature detection, retention time alignment, and feature identification in the AMT tag database as described previously (28) We opted to further filter the list of identifications by keeping only matches with a spatially localized confidence score (i.e. SLIC score) (29) higher than 0.9.
Statistical Analysis
To extract lists of peptides that were differentially abundant in the two cholangiocytes populations, we applied a statistical analysis to the data based on the spectral index (SpI) approach proposed by Heinecke and co-workers (30). The SpI method was developed in the context of label-free quantification using spectral counting techniques (31). It was necessary to adapt it slightly to fit the needs of our AMT-based label-free strategy, which is reliant on peptide signal extraction. To achieve this, we replaced protein spectral counts by individual peptide abundances, and instead of defining the SpI for each identified protein, we defined an abundance index (AbI) for each identified peptide. Thus, the AbI was defined as
 |
where Ā and B̄ is the average abundance of a peptide in population A respectively B, NAD and NBD are the numbers of patients in each population where the peptide was detected, and NAT and NBT are the total numbers of samples in each population. The distribution of observed AbI for all the detected peptides was then compared with the AbI distribution obtained by random permutation of the data sets across the two populations to estimate the variance. Permutations thus allowed the establishment of a confidence interval to assess differential abundances. It is worth mentioning here that, although the AbI method was designed to handle missing values, it is only applicable to peptides that have been detected at least once in each population. All calculations were performed using JMP v.7.0.1 software (SAS Institute Inc.).
Immunochemical Analysis
Western Blot Analysis
For Western blot analysis, samples from the batches of microdissected cholangiocytes were run on SDS-PAGE and transferred onto nitrocellulose membranes as described previously (23). The membranes were then blocked with PBS, 5% skimmed milk, and 0.1% Tween 20 for 2 h at room temperature before being incubated overnight with appropriate dilutions of antibodies raised against vimentin (Dako, V9 clone), carbonic anhydrase II (Santa Cruz Biotechnology), and profilin-1 (Cell Signaling Technology). The primary antibody was detected using appropriate horseradish peroxidase-coupled secondary antibodies and the ECL Plus kit (GE Healthcare) for signal visualization.
Construction of Tissue Microarray
The hepatic tumors selected for inclusion in a tissue microarray (TMA) comprised 23 ICC, 17 hilar cholangiocarcinoma, 19 HCC, and 22 non-tumorous liver samples (see supplemental Table 2). All cases were reviewed by a pathologist. Intrahepatic cholangiocarcinoma cases were categorized as trabecular-tubular (19 cases) or invasive papillary (four cases) carcinomas. Before arraying, sections from each tissue block were stained with hematoxylin and eosin to define morphologically representative areas of the tumor. Three tissue cylinders with a diameter of 1 mm were then punched from carefully selected, morphologically representative regions of each donor tissue block and deposited in a recipient block using a tissue arrayer (MTAI Beecher Instruments). Four-micrometer-thick sections of this array block were stained with hematoxylin and eosin to enable histological verification of the adequacy of the arrayed tumor tissues.
Immunohistochemical Analysis
Immunohistochemical analysis was performed using formalin-fixed and paraffin-embedded tissue specimens that were matched with the samples used for LM. Deparaffinized 4-μm-thick sections were treated with primary antibodies against vimentin and carbonic anhydrase II overnight at 4 °C after appropriate antigen retrieval treatment. Primary antibody/antigen binding was detected using the Vectastain ABC system (Vector Laboratories). Nuclei were counterstained with hematoxylin. Labeling specificity was determined by omitting the primary antibody during the experiment. Using the same procedure, labeling of microarrayed tissue sections by the anti-vimentin antibody was carefully analyzed by a pathologist and was considered to be negative when fewer than 5% of the cells of interest were immunostained. Statistical significance was calculated using a two-tailed Fisher's exact test in StatEL software (Ad Science) with p < 0.01 for statistical significance.
RESULTS
A major feature of ICC is the presence of an important stromal reaction in the tumor. Thus, to obtain protein abundance profiles that were relevant to ICC tumor cells, we opted for the laser microdissection technique to collect enriched populations of cholangiocytes (see supplemental Fig. 1). ICC tumor cells were isolated from the livers of four patients. Biliary cells dissected from large bile ducts present in the perihilar sections of the livers of five patients without ICC or any biliary tract pathology were used as controls.
Proteomics Study Using AMT Tag Strategy
Generation of AMT Tag Database
The first step in the AMT tag methodology consists of generating a database of peptides that can be identified in the sample under study. To achieve this, we analyzed peptide mixtures resulting from digestion of the various fractions obtained from pools of microdissected cells separated on one-dimensional gels (see Fig. 1A). Following LC-MS/MS analysis and a Mascot search, peptide identifications were compiled in a so-called AMT tag database. Only proteins identified by at least two peptides were considered for inclusion except when a single peptide had a Mascot score higher than 50. In total, 21,902 peptide identifications were validated and grouped by sequence, molecular weight, and average retention time. The median Mascot score for the peptides that passed our filtering criteria for inclusion in the AMT database was 50.56, and 90% of the peptides considered scored higher than 33.27. The resulting AMT tag database thus contained 2,499 distinct peptides indicative of 4,508 protein accessions. When shotgun approaches are used for protein identification, a certain degree of redundancy can be observed in the list of proteins identified. Indeed, several proteins in our database, and particularly isoforms or sequence variants, were covered by the same subsets of peptides. To reduce this redundancy in the protein list, proteins characterized by the same subset of peptides were grouped into so-called protein groups (32). Supplemental Table 3 lists the 574 protein groups thus identified and the individual proteins within these groups. This list can be considered as the minimal list of proteins required to account for all the peptides identified. Supplemental Table 6 lists the peptides identified with their sequence, molecular weight, modifications and corresponding protein group. In fact, the average peptide coverage for the 574 proteins incorporated in the AMT database was of the order of five peptides per protein.
The subcellular localization of identified proteins was determined using Ingenuity Pathway Analysis software (Ingenuity Systems). As shown in supplemental Fig. 3, whereas the majority (67.4%) of the proteins identified were from the cytoplasm, notable proportions (18.4 and 6.5%) arose from the nucleus and plasma membrane, respectively. Overall, most subcellular compartments were represented in the AMT database, suggesting that our approach enabled access to a broad cross-section of cellular proteins.
Label-free Quantitative LC-FTMS Data Evaluation
After compiling the AMT tag database, we then considered the quantitative data obtained by LC-FTMS analysis of individual microdissected samples (see Fig. 1B). Following data acquisition, several tests were performed to assess their quality. This was achieved by evaluating the raw signals in all analyses using dedicated software and by calculating correlations between the peptide abundances obtained by different acquisitions. The Chaorder software (33) was used as a first means of data evaluation. This algorithm, based on a score that measures the similarity between pairs of mass spectra, enables an assessment of reproducibility and similarity between LC-MS experiments. For this purpose, Chaorder computes a pseudodistance between acquisitions based solely on the raw signal. According to Prakash et al. (33), a distance between two points between 0 and 0.2 represents levels of reproducibility usually seen in repeat experiments with the exact same experimental setup. Fig. 2A shows the Chaorder plot obtained by comparing the LC-MS acquisitions collected during our experiment. Given the randomized order in which analyses were performed, the limited scattering of replicate acquisitions of the same sample was indicative of an absence of significant drift in the analytical system. Data consistency was further evaluated using pairwise correlations of peptide abundances between acquisitions. The higher the Pearson coefficient, the more alike the two data sets were. When plotting the correlation matrix of all experiments (see Fig. 2B), it became clear that triplicate experiments on the same subject always resulted in strongly correlated abundances (r = 0.94 ± 0.04).
Fig. 2.
Quantitative LC-FTMS data quality control. A, Chaorder plot for the 27 acquisitions (three for each sample) grouped by sample. B, Pearson pairwise correlation (R) matrix of peptide abundances measured in each acquisition sorted by sample.
Determination of Deregulated Proteins
Before statistical analysis, triplicate acquisitions of the same sample were combined by averaging the abundances in all runs for each peptide that had been detected at least in two of the three replicates. This procedure produced four cancer and five control data sets (see supplemental Table 4). During the quantitative study, we were able to detect and quantify ∼1,850 of the 2,499 previously sequenced peptides corresponding to 349 proteins identified by at least two peptides.
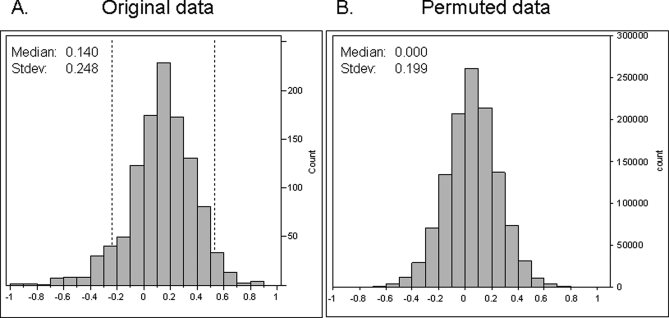
The nine data sets were subjected to statistical analysis based on the approach proposed by Heinecke and co-workers (30). This method combined the relative species abundance and the number of cases within a group for which the species was detected into a so-called AbI. Permutation analysis then enabled the establishment of a confidence interval to assess differential abundances. The resulting distributions of AbI obtained for all the detected peptides in the two populations are shown in Fig. 3. Peptides with AbI values falling outside the 95% confidence interval (−0.25 < AbI < 0.53), based on permuted data, were determined to be differentially abundant between tumorous and control cholangiocytes (p < 0.05). Peptides were then grouped into proteins, and only proteins for which at least one proteotypic peptide was detected with differential abundance between the two cholangiocyte populations were retained. This process led to a list of 82 peptides (associated with 23 proteins) with decreased abundance in tumorous cholangiocytes and 42 peptides (corresponding to 16 proteins) with higher abundance in these cells (see Table II).
Fig. 3.
Distributions of AbI for 1121 peptides detected in both populations for original data (A) and for permuted data (B). The permutation allowed an estimation of the variance of the AbI measure under the assumption that all samples were equivalent. The dashed lines in A delimit the 95% confidence interval (−0.24 < AbI < 0.53), which is equal to the median AbI (0.14) of the original data ±1.96 times the standard deviation (Stdev) (0.199) of the permuted data, a region where no change in abundance between the two populations was observed.
Table II. Identification of deregulated proteins in tumorous cholangiocytes.
| Accession number | Protein name | Gene name | -Fold change cancer/normal | Sequence coverage | Unique peptides/proteotypic peptides | Protein score | Postulated biological processa |
|---|---|---|---|---|---|---|---|
| % | |||||||
| Proteins having peptides detected with increased abundance in tumorous samples | |||||||
| VIME_HUMAN | Vimentin | VIM | 9.26 | 68.5 | 39/34 | 2,496 | Cell motion |
| LDHB_HUMAN | l-Lactate dehydrogenase B chain | LDHB | 7.72 | 37.4 | 11/10 | 808 | Monocarboxylic acid metabolic process |
| Q5JP53_HUMAN | Tubulin β polypeptide | TUBB | 4.76 | 33.8 | 16/3 | 1,098 | Cell motion |
| Q1WM23_HUMAN | Nucleoside-diphosphate kinase | NME1 | 3.66 | 32.6 | 8/8 | 431 | Regulation of cell proliferation, metastatic process |
| LDHA_HUMAN | l-Lactate dehydrogenase A chain | LDHA | 3.45 | 34.0 | 10/9 | 620 | Anaerobic glycolysis |
| S10AB_HUMAN | Protein S100-A11 (calgizzarin) | S100A11 | 3.01 | 43.8 | 5/5 | 293 | Signal transduction, regulation of cell proliferation |
| ALDOA_HUMAN | Fructose-bisphosphate aldolase A | ALDOA | 2.28 | 31.0 | 10/10 | 531 | Actin cytoskeleton organization, hexose metabolic process |
| KPYM_HUMAN | Pyruvate kinase M2 | PKM2 | 2.17 | 45.2 | 20/20 | 1,410 | Hexose metabolic process |
| A8K220_HUMAN | Peptidyl-prolyl cis-trans isomerase (cyclophilin) | PPIA | 2.16 | 50.3 | 11/11 | 811 | Protein folding |
| K1C18_HUMAN | Keratin, type I cytoskeletal 18 | KRT18 | 2.12 | 64.9 | 33/32 | 2,571 | Cell morphogenesis |
| PROF1_HUMAN | Profilin-1 | PFN1 | 1.97 | 55.0 | 8/8 | 457 | Actin cytoskeleton organization |
| Q6FGI1_HUMAN | Transgelin-2 | TAGLN2 | 1.86 | 33.2 | 6/6 | 355 | Actin cytoskeleton regulation |
| PTMA_HUMAN | Prothymosin α | PTMA | 1.86 | 21.6 | 2/2 | 126 | Transcription |
| COF1_HUMAN | Cofilin-1 | CFL1 | 1.70 | 50.0 | 6/6 | 433 | Actin cytoskeleton organization |
| MDHC_HUMAN | Malate dehydrogenase, cytoplasmic | MDH1 | 1.68 | 29.9 | 8/8 | 517 | Carboxylic acid metabolic process |
| HS90B_HUMAN | Heat shock protein HSP 90-β | HSP90AB 1 | 1.47 | 15.5 | 11/5 | 739 | Molecular chaperone, protein folding |
| Proteins having peptides detected with decreased abundance in tumorous samples | |||||||
| AGR2_HUMAN | Anterior gradient protein 2 | AGR2 | 0.09 | 22.3 | 4/4 | 208 | Unknown |
| CAH2_HUMAN | Carbonic anhydrase II | CA2 | 0.09 | 25.4 | 6/6 | 353 | Response to osmotic stress |
| ANX13_HUMAN | Annexin A13 | ANXA13 | 0.23 | 33.9 | 9/9 | 480 | Cell differentiation |
| A8K008_HUMAN | cDNA FLJ78387 | 0.26 | 11.2 | 4/4 | 237 | Unknown | |
| A0A5C9_HUMAN | IGLV2-14 protein | IGLV2-14 | 0.31 | 12.4 | 2/2 | 107 | Unknown |
| K1C19_HUMAN | Keratin, type I cytoskeletal 19 | KRT19 | 0.32 | 59.3 | 32/21 | 2,330 | Actin cytoskeleton organization |
| EST1_HUMAN | Liver carboxylesterase 1 | CES1 | 0.32 | 34.0 | 16/16 | 1,091 | Metabolic process |
| LEG4_HUMAN | Galectin-4 | LGALS4 | 0.35 | 12.7 | 3/3 | 187 | Cell adhesion |
| HBA_HUMAN | Hemoglobin subunit α | HBA1 | 0.39 | 39.4 | 6/6 | 328 | Oxygen transport |
| Q06AH7_HUMAN | Transferrin | TF | 0.45 | 5.7 | 4/4 | 172 | Iron ion transport |
| AT1A1_HUMAN | Sodium/potassium-transporting ATPase subunit α1 | ATP1A1 | 0.49 | 25.1 | 19/19 | 1,244 | Cellular ion homeostasis, carcinoma cell motility |
| ANXA4_HUMAN | Annexin A4 | ANXA4 | 0.49 | 56.4 | 23/23 | 1,885 | Signal transduction, antiapoptosis |
| ERLN2_HUMAN | Erlin-2 | ERLIN2 | 0.50 | 10.6 | 3/3 | 180 | Unknown |
| ALBU_HUMAN | Serum albumin | ALB | 0.51 | 56.5 | 39/39 | 2,418 | Transport |
| A4GX73_HUMAN | Hemoglobin subunit β | HBB | 0.51 | 75.5 | 9/4 | 678 | Oxygen transport |
| A4D1PA_HUMAN | Aldo-keto reductase family 1, member B10 | AKR1B10 | 0.55 | 13.6 | 4/4 | 271 | Cellular lipid metabolic process |
| TALDO_HUMAN | Transaldolase | TALDO1 | 0.56 | 14.8 | 5/5 | 213 | Carbohydrate metabolic process |
| A0A5E5_HUMAN | IGKC protein | IGKC | 0.57 | 24.2 | 4/4 | 304 | Immune response |
| AK1C1_HUMAN | Aldo-keto reductase family 1 member C1 | AKR1C1 | 0.62 | 14.6 | 3/3 | 116 | Steroid metabolic process |
| HSP71_HUMAN | Heat shock 70-kDa protein 1 | HSPA1A | 0.63 | 27.5 | 15/4 | 975 | Ubiquitin proteasome pathway, protein folding |
| PDIA3_HUMAN | Protein-disulfide isomerase A3 | PDIA3 | 0.68 | 26.3 | 11/11 | 646 | Protein folding |
| ANXA2_HUMAN | Annexin A2 | ANXA2 | 0.79 | 53.1 | 17/17 | 1,189 | Actin cytoskeleton organization |
| A1A5C4_HUMAN | RRBP1 protein | RRBP1 | 0.80 | 10.6 | 9/9 | 427 | Translation |
a Postulated biological process was determined by gene ontology and bibliographic features.
With respect to functional considerations, most proteins identified with an altered expression in tumorous cells could be grouped in two categories. A first pool of proteins was associated with metabolic pathways (glycolysis, neoglucogenesis, or fatty acid metabolism), and a second pool consisted of proteins involved in cell structure and/or motility.
It is worth noting that only peptides detected at least once in each population (ICC and controls) are amenable to this type of statistical analysis. As a matter of fact, several peptides were detected only in one of the two populations. When assigning these peptides to proteins, it turned out that several proteins exclusively covered by population-specific peptides were involved in a biological process relevant to carcinogenesis (see supplemental Table 5). Among them, the up-regulation of Mac 2-binding protein (LGALS3BP), a metastasis-related protein C (34), had been reported in various cancers including biliary tract carcinoma (35). Likewise, IQGAP2 had shown reduced levels in cancers, and the targeted disruption of the murine IQGAP2 gene resulted in the development of HCC (36, 37).
Confirmation of Proteomics Findings
Western Blot and Immunohistochemistry on Matched Samples
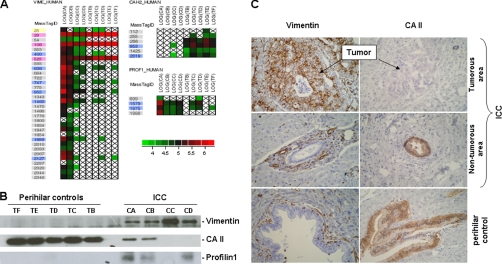
To verify the reliability of the data obtained during this study, the expression of vimentin, profilin-1, and carbonic anhydrase II (CAII) was assessed using a Western blot in microdissected cholangiocytes from the patients and controls included in the proteomics analysis. As determined during mass spectrometry analysis (Fig. 4A), the expression of vimentin and profilin-1 was stronger in ICC samples, whereas the immunolabeling of CAII was reduced in these samples when compared with the controls (Fig. 4B).
Fig. 4.
Expression analysis of vimentin, CAII, and profilin-1 in intrahepatic cholangiocarcinoma. A, cell plot of the log of peptide abundances in cancer and control samples. Outline colors correspond to peptides returned as significant by the statistical analysis (blue), peptides shared with other protein groups (pink), peptides not returned as significant by the statistical analysis (yellow), and population-specific peptides (gray). B, immunodetection of vimentin, CAII, and profilin-1. Western blots carried out as described under “Experimental Procedures” were performed using microdissected tumorous and non-tumorous cholangiocytes from subjects included in the study. C, immunohistochemical analysis of vimentin and CAII. Immunohistochemistry was performed as described under “Experimental Procedures.” Data are from a representative experiment including the tumorous and non-tumorous areas of the liver of patient CA and a control liver, TB. Vimentin is expressed in tumorous cholangiocytes from ICC (top left) and negative in non-tumorous cholangiocytes of peripheral (middle left) and perihilar (bottom left) bile ducts. Internal controls (vascular smooth muscle cells and inflammatory cells) are positive. Conversely, CAII is negative in tumorous cholangiocytes from ICC (top right) and expressed in non-tumorous cholangiocytes of peripheral (middle right) and perihilar (bottom right) bile ducts.
These results were further corroborated when altered abundance patterns for vimentin and CAII were detected in cholangiocytes using immunohistochemistry (Fig. 4C). Tissue sections from formalin-fixed and paraffin-embedded tissue specimens matched with the samples used for the microdissection of cholangiocytes were covered by this analysis. CAII was only detected in biliary ducts from control livers and non-tumorous areas of the ICC, whereas anti-CAII antibody did not label tumorous cholangiocytes. Conversely, the anti-vimentin antibody labeled vascular smooth muscle, Kupffer, and inflammatory cells in the analyzed tissue sections. However, vimentin was not detected in cholangiocytes lining normal biliary ducts from the control liver or non-tumorous area of the cholangiocarcinoma but was revealed in tumorous intrahepatic cholangiocytes. Similar results were obtained with the other tissue specimens (data not shown). Taken together, these data established the reliability of our data and the robustness of our strategy regarding the identification of proteins displaying altered expression in the setting of ICC.
Vimentin Expression in Primary Liver Cancers
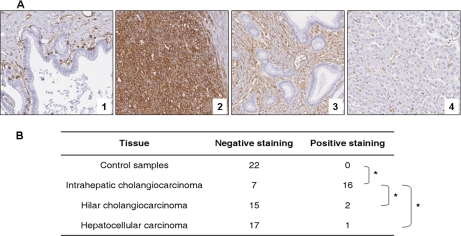
To further validate the overabundance of vimentin in ICC, the expression of this protein was examined using a tissue microarray comprising samples from a series of patients with primary liver cancers including ICC, hilar CCA, and HCC. Briefly, we confirmed, in a larger series of liver samples, that vimentin was not detected in non-tumorous cholangiocytes (zero of 22) but was expressed in the tumor cells of most (16 of 23) ICC samples included in the tissue microarray. On the other hand, only two of 17 hilar CCA and one of 18 HCC samples showed positive staining for vimentin in tumor cells. The specificity of vimentin expression in ICC was established by statistical analysis using Fisher's exact test (see Fig. 5).
Fig. 5.
Vimentin expression in liver lesions by immunohistochemical staining using tissue microarray. A, representative cores of perihilar bile duct (1), intrahepatic cholangiocarcinoma (2), hilar cholangiocarcinoma (3), and hepatocellular carcinoma (4). Immunohistochemical staining was performed as described under “Experimental Procedures.” Vimentin is negative in normal cholangiocytes from perihilar bile duct, whereas inflammatory cells stained as positive internal controls (1). Vimentin is diffusely positive in the tumorous cholangiocytes from ICC (2) and totally negative in the tumorous cholangiocytes from hilar cholangiocarcinoma (3) and tumorous hepatocytes from HCC (4). B, vimentin expression in 23 intrahepatic cholangiocarcinoma, 17 hilar cholangiocarcinoma, 18 hepatocellular carcinoma, and 22 controls patients (seven perihilar areas from liver without ICC, four normal livers from amyloid neuropathy, and 11 non-tumoral counterparts of ICC). Staining was considered negative when less than 5% of the cells of interest were immunostained. *, p < 0.001 (two-tailed Fisher's exact text).
DISCUSSION
To our knowledge, this is the first study to investigate the proteome alterations that occur in ICC while taking into account the important cellular heterogeneity of this tumor. Our strategy combining LM to select cholangiocytes and an AMT tag proteomics methodology appropriate for the analysis of small cellular samples was devised to overcome this challenge. And indeed, this strategy provided strong evidence of specific alterations to glycolysis and organization of the cytoskeleton, two protein networks involved in carcinogenesis.
ICC is a devastating cancer with an increasing incidence worldwide. For this reason, studies have been carried out in recent years to clarify the mechanisms underlying the development of this cancer with the aim of developing novel therapeutic strategies. These investigations provided evidences that ICC was associated with genetic alterations (38–40) and impaired protein expression that could contribute to the proliferation of tumorous cholangiocytes (41, 42). Until now, global genome and proteome alterations in ICC have been the subject of little investigation. In particular, proteome impairments in ICC had only been explored during one study using liver homogenates (43). Indeed, the accurate determination of proteome alterations in ICC is a particularly challenging task because in most tumors cholangiocytes are embedded in an abundant stroma that may mask cholangiocyte signals. Therefore, to obtain accurate insights into proteome alterations in ICC, we used LM to selectively extract cholangiocytes from liver specimens.
Another important prerequisite to this study was the choice of a proteomics technique appropriate for the analysis of small samples. An AMT tag strategy was adopted because this method had been shown to significantly improve proteome coverage for samples originating from microdissected breast carcinoma cells (22). To exploit each specimen to the maximum, we used microdissected cholangiocytes both for the generation of the AMT tag database and the quantitative study. The resulting AMT database contained 2,499 distinct peptides belonging to 574 proteins; 460 of these were characterized by at least two peptides.
The first step in quantitative data processing consisted of assessing data quality using two independent methods: Chaorder and pairwise correlations. As can be seen from the Chaorder plot (Fig. 2A) and the pairwise correlation matrix (Fig. 2B), acquisitions corresponding to the same patient displayed less dispersion and better correlations than analyses of unrelated samples even though the run order was randomized. Moreover, acquisitions corresponding to the controls tended to display better homogeneity than those of cancer patients. The Chaorder plot and the correlations between replicates thus confirmed that our data were consistent with sample classification and that no outlier run was present.
We then proceeded to extract the list of peptides that were differentially abundant between the two populations using an AbI methodology. The observed AbI distribution was skewed toward positive values (see Fig. 3A), suggesting that the abundance of peptides in tumorous samples was slightly higher than in the control samples. Taking account of this observation, we therefore considered the median AbI value (0.140) as the reference point at which there was no difference between cancer and control samples. Statistical processing and the subsequent filtering of proteins for which no proteotypic peptides were found to be deregulated led to a list of 23 proteins with decreased abundance in ICC and 16 proteins exhibiting higher abundance in tumor cells (see Table II). We believe that the rigorous criteria chosen to perform this proteomics study warrant a high level of confidence in the final list of deregulated proteins. Indeed, using alternative methods (Western blot and immunohistochemistry), the altered expression of some of them (vimentin, profilin-1, and CAII) could be confirmed in tumorous cholangiocytes (see Fig. 4).
Many of the altered proteins in tumorous cholangiocytes were involved in glycolysis and regulation of the cytoskeleton. As observed previously in kidney and pancreatic cancers (44, 45), we detected an increased abundance of proteins from the glycolytic pathway in ICC. In addition to M2-PK, we were able to identify fructose-bisphosphate aldolase A and lactate dehydrogenase A. These findings indicate, as suggested many years ago by Warburg (46) for other tumor cells, that glycolysis could also be altered in tumorous cholangiocytes. It has recently been shown that aerobic glycolysis, a distinctive feature of tumor cells, requires the overexpression of M2-PK and provides a selective growth advantage for these cells (47). We therefore hypothesize that an overabundance of this isoform may participate in the development of ICC.
We also noted an altered protein network involved in cytoskeleton organization and cell motility. In particular, we noticed a striking increase in the abundance of vimentin, an intermediate filament protein. This protein, expressed in a number carcinomas, has been shown to correlate with the invasiveness and poor prognosis of these tumors (48–52). We also demonstrated the impaired expression of a group of proteins implicated in the equilibrium between globular and filamentous actin. This process, altered during cell transformation (53, 54), is involved in cell motility, which is notably increased during invasion and metastasis (55, 56). We found an increase in the abundance of actin-binding proteins (cofilin-1, profilin-1, and transgelin-2). Cofilin-1 had been reported as being overexpressed in gastrointestinal endocrine tumors, and the aggressiveness of these tumors had been linked with the expression of this protein (57). As for profilin-1 and transgelin-2, a contrasting expression pattern had been reported in breast and colon cancers (58, 59). The functional significance of these differences needs to be clarified, particularly because these proteins had been described as tumor suppressors (60, 61).
We also observed an overabundance in ICC of S100A11 (calgizzarin), a protein involved in the Ca2+ signaling network and in the regulation of cell growth and motility (62, 63). Such alterations had also been reported in pancreatic (64) and colon cancers where disease progression correlated with the abundance of this protein in tumor tissues (65, 66).
On the other hand, annexins exhibited lower abundance in our ICC samples compared with the controls. Increased expression of annexins had been reported in different types of carcinoma, e.g. colorectal, bladder, pancreatic, hepatocellular, and renal carcinomas (67–71). However, a reduced expression of annexin A2, also observed in head and neck squamous cell and prostate carcinomas (72, 73) and osteosarcoma (74), had been linked to the aggressive behavior and metastatic potential of these tumors.
Similarly, as reported in renal carcinoma (75), we observed that the catalytic subunit α1 of Na/K-ATPase was under-represented in ICC samples. Na/K-ATPase had been reported as regulating carcinoma cell motility (76) and had been shown to be involved in tight junction formation during the biogenesis of polarized cells (77). It has thus been speculated (75) that reduced Na/K-ATPase activity might be implicated in the loss of cell-cell adhesion.
Cancer cells attain the migratory and invasive capacities required for metastasis by undergoing a phenotypic conversion referred to as the epithelial to mesenchymal transition (EMT) (78). The expression of TGFβ, a potent inducer of EMT (79), is increased in many human cancers (80) including ICC (81, 82). Interestingly, the protein alterations found in tumorous cholangiocytes (present study) displayed striking similarities with those reported in the A549 human lung adenocarcinoma cell line (83) during TGFβ-induced EMT. Specifically, similar impairments of vimentin, M2-PK, cofilin-1, transgelin-2, Na/K-ATPase, and S100A11 expression were observed in both cases, suggesting that the protein profile of tumorous cholangiocytes fits with that of epithelial cells displaying a transition to the mesenchymal phenotype.
Vimentin is a mesenchymal marker that had been reported to be overexpressed in a number of cancers, its expression associated with EMT (78). During the present study, we were able to confirm using tissue microarrays that the overexpression of vimentin was a typical feature of ICC because 16 of 23 (∼70%) of the tumors studied expressed this protein. Further experiments will be necessary to correlate this finding with EMT. If confirmed, however, this hypothesis might provide a basis for the serious outcome of this cancer. It is worth noting that all invasive papillary tumors included in the TMA were negative for vimentin (see supplemental Table 2 and supplemental Fig. 4), suggesting that these tumors could represent a subtype of ICC. Studies on a larger patient population (with trabecular and papillary cases) are underway to assess the generality of these findings. Interestingly, an increased abundance of vimentin was not found in all types of CCA: it was only detected in two of the 17 tumors analyzed from patients with hilar CCA. As previously shown by Guedj et al. (84), this tends to confirm that hilar CCA and ICC are distinct entities displaying specific protein abundance profiles. Similarly, vimentin expression was not found to be a common feature in HCC: it was only expressed in one of the 18 tumor tissue samples analyzed. Overall, these findings suggest, as reported in others cancers (44–48), that vimentin could play a role in aggressiveness of ICC and provide a basis for the serious outcome of this cancer.
CONCLUSION
Until now, because of the important cellular heterogeneity of ICC, the specific protein profile of these tumors could not be adequately determined. In the present study, we selected an appropriate strategy based on LM and AMT tag proteomics to access specific cellular populations and analyze their proteome with high sensitivity. We were thus able to describe a panel of 39 proteins showing altered abundance profiles in tumorous versus healthy cholangiocytes. Most of the proteins displaying perturbed abundances had not yet been described in the setting of ICC. Interestingly, a majority of deregulated proteins were involved in glycolysis and cytoskeleton plasticity/cell motility, pathways that had been reported to be implicated in carcinogenesis. Although further experiments are required to characterize the consequences of these deregulations, we anticipate that these data may open the way to novel therapeutic targets or diagnostic tools. In particular, achieving a specific diagnosis of ICC among other primary liver cancers is often problematic. Results from our tissue microarray experiment suggest that vimentin, in combination with other markers, might prove useful as an indicator of particular subtypes of ICC. This will be the subject of a follow-up study on a larger number of patients with ICC, hilar CCA, and HCC.
Supplementary Material
Acknowledgments
We gratefully acknowledge Prof. Valerie Paradis for tissue procurement. We thank Christophe Bruley and Véronique Dupierris for assistance with setting up the AMT tag database, Sabine Brugière for assistance with the analytical setup, and Virginie Hossard for TMA construction. Laser microdissection was performed with technical assistance from Sylvie Dumont at the laser microdissection core facility directed by Dominique Wendum at IFR65, Saint Antoine, Paris, France. The Decon-2LS and VIPER software used for data processing were provided by the W. R. Wiley Environmental Molecular Science Laboratory, a national scientific user facility sponsored by the United States Department of Energy's Office of Biological and Environmental Research located at the Pacific Northwest National Laboratory. Pacific Northwest National Laboratory is operated by the Battelle Memorial Institute for the United States Department of Energy under Contract DE-AC05-76RL0 1830.
* This work was supported in part by Institut National du Cancer Grant PL027.
 This article contains supplemental Tables 1–6 and Figs. 1–4.
This article contains supplemental Tables 1–6 and Figs. 1–4.
1 The abbreviations used are:
- CCA
- cholangiocarcinoma
- AMT
- accurate mass and time
- EMT
- epithelial to mesenchymal transition
- HCC
- hepatocellular carcinoma
- ICC
- intrahepatic cholangiocarcinoma
- LM
- laser microdissection
- TMA
- tissue microarray
- SpI
- spectral index
- AbI
- abundance index
- CAII
- carbonic anhydrase II.
REFERENCES
- 1.Patel T. (2001) Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology 33, 1353–1357 [DOI] [PubMed] [Google Scholar]
- 2.Ustundag Y., Bayraktar Y. (2008) Cholangiocarcinoma: a compact review of the literature. World J. Gastroenterol. 14, 6458–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Menachem T. (2007) Risk factors for cholangiocarcinoma. Eur. J. Gastroenterol. Hepatol. 19, 615–617 [DOI] [PubMed] [Google Scholar]
- 4.Blechacz B. R., Gores G. J. (2008) Cholangiocarcinoma. Clin. Liver Dis. 12, 131–150, ix [DOI] [PubMed] [Google Scholar]
- 5.El-Serag H. B., Engels E. A., Landgren O., Chiao E., Henderson L., Amaratunge H. C., Giordano T. P. (2009) Risk of hepatobiliary and pancreatic cancers after hepatitis C virus infection: a population-based study of U.S. veterans. Hepatology 49, 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonney G. K., Craven R. A., Prasad R., Melcher A. F., Selby P. J., Banks R. E. (2008) Circulating markers of biliary malignancy: opportunities in proteomics? Lancet Oncol. 9, 149–158 [DOI] [PubMed] [Google Scholar]
- 7.Alpini G., Invernizzi P., Gaudio E., Venter J., Kopriva S., Bernuzzi F., Onori P., Franchitto A., Coufal M., Frampton G., Alvaro D., Lee S. P., Marzioni M., Benedetti A., DeMorrow S. (2008) Serotonin metabolism is dysregulated in cholangiocarcinoma, which has implications for tumor growth. Cancer Res. 68, 9184–9193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohira S., Itatsu K., Sasaki M., Harada K., Sato Y., Zen Y., Ishikawa A., Oda K., Nagasaka T., Nimura Y., Nakanuma Y. (2006) Local balance of transforming growth factor-beta1 secreted from cholangiocarcinoma cells and stromal-derived factor-1 secreted from stromal fibroblasts is a factor involved in invasion of cholangiocarcinoma. Pathol. Int. 56, 381–389 [DOI] [PubMed] [Google Scholar]
- 9.Alvaro D. (2009) Serum and bile biomarkers for cholangiocarcinoma. Curr. Opin. Gastroenterol. 25, 279–284 [DOI] [PubMed] [Google Scholar]
- 10.Farina A., Dumonceau J. M., Lescuyer P. (2009) Proteomic analysis of human bile and potential applications for cancer diagnosis. Expert Rev. Proteomics 6, 285–301 [DOI] [PubMed] [Google Scholar]
- 11.Nishino R., Honda M., Yamashita T., Takatori H., Minato H., Zen Y., Sasaki M., Takamura H., Horimoto K., Ohta T., Nakanuma Y., Kaneko S. (2008) Identification of novel candidate tumour marker genes for intrahepatic cholangiocarcinoma. J. Hepatol. 49, 207–216 [DOI] [PubMed] [Google Scholar]
- 12.Wang A. G., Yoon S. Y., Oh J. H., Jeon Y. J., Kim M., Kim J. M., Byun S. S., Yang J. O., Kim J. H., Kim D. G., Yeom Y. I., Yoo H. S., Kim Y. S., Kim N. S. (2006) Identification of intrahepatic cholangiocarcinoma related genes by comparison with normal liver tissues using expressed sequence tags. Biochem. Biophys. Res. Commun. 345, 1022–1032 [DOI] [PubMed] [Google Scholar]
- 13.Tietz P., de Groen P. C., Anderson N. L., Sims C., Esquer-Blasco R., Meheus L., Raymackers J., Dauwe M., LaRusso N. F. (1998) Cholangiocyte-specific rat liver proteins identified by establishment of a two-dimensional gel protein database. Electrophoresis 19, 3207–3212 [DOI] [PubMed] [Google Scholar]
- 14.Grützmann R., Pilarsky C., Ammerpohl O., Lüttges J., Böhme A., Sipos B., Foerder M., Alldinger I., Jahnke B., Schackert H. K., Kalthoff H., Kremer B., Klöppel G., Saeger H. D. (2004) Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia 6, 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luzzi V., Mahadevappa M., Raja R., Warrington J. A., Watson M. A. (2003) Accurate and reproducible gene expression profiles from laser capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J. Mol. Diagn. 5, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sriuranpong V., Mutirangura A., Gillespie J. W., Patel V., Amornphimoltham P., Molinolo A. A., Kerekhanjanarong V., Supanakorn S., Supiyaphun P., Rangdaeng S., Voravud N., Gutkind J. S. (2004) Global gene expression profile of nasopharyngeal carcinoma by laser capture microdissection and complementary DNA microarrays. Clin. Cancer Res. 10, 4944–4958 [DOI] [PubMed] [Google Scholar]
- 17.Li C., Hong Y., Tan Y. X., Zhou H., Ai J. H., Li S. J., Zhang L., Xia Q. C., Wu J. R., Wang H. Y., Zeng R. (2004) Accurate qualitative and quantitative proteomic analysis of clinical hepatocellular carcinoma using laser capture microdissection coupled with isotope-coded affinity tag and two-dimensional liquid chromatography mass spectrometry. Mol. Cell. Proteomics 3, 399–409 [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Rudnick P. A., Evans E. L., Li J., Zhuang Z., Devoe D. L., Lee C. S., Balgley B. M. (2005) Proteome analysis of microdissected tumor tissue using a capillary isoelectric focusing-based multidimensional separation platform coupled with ESI-tandem MS. Anal. Chem. 77, 6549–6556 [DOI] [PubMed] [Google Scholar]
- 19.Wulfkuhle J. D., Sgroi D. C., Krutzsch H., McLean K., McGarvey K., Knowlton M., Chen S., Shu H., Sahin A., Kurek R., Wallwiener D., Merino M. J., Petricoin E. F., 3rd, Zhao Y., Steeg P. S. (2002) Proteomics of human breast ductal carcinoma in situ. Cancer Res. 62, 6740–6749 [PubMed] [Google Scholar]
- 20.Pasa-Toliæ L., Lipton M. S., Masselon C. D., Anderson G. A., Shen Y., Toliæ N., Smith R. D. (2002) Gene expression profiling using advanced mass spectrometric approaches. J. Mass Spectrom. 37, 1185–1198 [DOI] [PubMed] [Google Scholar]
- 21.Shen Y., Toliæ N., Masselon C., Pasa-Toliæ L., Camp D. G., 2nd, Lipton M. S., Anderson G. A., Smith R. D. (2004) Nanoscale proteomics. Anal. Bioanal. Chem. 378, 1037–1045 [DOI] [PubMed] [Google Scholar]
- 22.Umar A., Luider T. M., Foekens J. A., Pasa-Toliæ L. (2007) NanoLC-FT-ICR MS improves proteome coverage attainable for approximately 3000 laser-microdissected breast carcinoma cells. Proteomics 7, 323–329 [DOI] [PubMed] [Google Scholar]
- 23.Dos Santos A., Thiers V., Sar S., Derian N., Bensalem N., Yilmaz F., Bralet M. P., Ducot B., Brechot C., Demaugre F. (2007) Contribution of laser microdissection-based technology to proteomic analysis in hepatocellular carcinoma developing on cirrhosis. Proteomics Clin. Appl. 1, 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. (1996) Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469 [DOI] [PubMed] [Google Scholar]
- 25.Masselon C. D., Kieffer-Jaquinod S., Brugière S., Dupierris V., Garin J. (2008) Influence of mass resolution on species matching in accurate mass and retention time (AMT) tag proteomics experiments. Rapid Commun. Mass Spectrom. 22, 986–992 [DOI] [PubMed] [Google Scholar]
- 26.Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 27.Dupierris V., Masselon C., Court M., Kieffer-Jaquinod S., Bruley C. (2009) A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 25, 1980–1981 [DOI] [PubMed] [Google Scholar]
- 28.Monroe M. E., Toliæ N., Jaitly N., Shaw J. L., Adkins J. N., Smith R. D. (2007) VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23, 2021–2023 [DOI] [PubMed] [Google Scholar]
- 29.Norbeck A. D., Monroe M. E., Adkins J. N., Anderson K. K., Daly D. S., Smith R. D. (2005) The utility of accurate mass and LC elution time information in the analysis of complex proteomes. J. Am Soc. Mass Spectrom. 16, 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu X., Gharib S. A., Green P. S., Aitken M. L., Frazer D. A., Park D. R., Vaisar T., Heinecke J. W. (2008) Spectral index for assessment of differential protein expression in shotgun proteomics. J. Proteome Res. 7, 845–854 [DOI] [PubMed] [Google Scholar]
- 31.Liu H., Sadygov R. G., Yates J. R., 3rd (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 32.Kearney R. E., Blondeau F., McPherson P. S., Bell A. W., Servant F., Drapeau M., de Grandpre S., Bergeron J. J. M. (2005) Elimination of redundant protein identifications in high throughput proteomics, in 27th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Shanghai, September 1–4, 2005, Vols. 1–7, pp. 4803–4806, Institute of Electrical and Electronic Engineers (IEEE) Engineering in Medicine and Biology Society, Piscataway, NJ: [DOI] [PubMed] [Google Scholar]
- 33.Prakash A., Piening B., Whiteaker J., Zhang H., Shaffer S. A., Martin D., Hohmann L., Cooke K., Olson J. M., Hansen S., Flory M. R., Lee H., Watts J., Goodlett D. R., Aebersold R., Paulovich A., Schwikowski B. (2007) Assessing bias in experiment design for large scale mass spectrometry-based quantitative proteomics. Mol. Cell. Proteomics 6, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 34.Becker R., Lenter M. C., Vollkommer T., Boos A. M., Pfaff D., Augustin H. G., Christian S. (2008) Tumor stroma marker endosialin (Tem1) is a binding partner of metastasis-related protein Mac-2 BP/90K. FASEB J. 22, 3059–3067 [DOI] [PubMed] [Google Scholar]
- 35.Koopmann J., Thuluvath P. J., Zahurak M. L., Kristiansen T. Z., Pandey A., Schulick R., Argani P., Hidalgo M., Iacobelli S., Goggins M., Maitra A. (2004) Mac-2-binding protein is a diagnostic marker for biliary tract carcinoma. Cancer 101, 1609–1615 [DOI] [PubMed] [Google Scholar]
- 36.Schmidt V. A., Chiariello C. S., Capilla E., Miller F., Bahou W. F. (2008) Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol. Cell. Biol. 28, 1489–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White C. D., Brown M. D., Sacks D. B. (2009) IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 583, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furubo S., Harada K., Shimonishi T., Katayanagi K., Tsui W., Nakanuma Y. (1999) Protein expression and genetic alterations of p53 and ras in intrahepatic cholangiocarcinoma. Histopathology 35, 230–240 [DOI] [PubMed] [Google Scholar]
- 39.Momoi H., Okabe H., Kamikawa T., Satoh S., Ikai I., Yamamoto M., Nakagawara A., Shimahara Y., Yamaoka Y., Fukumoto M. (2001) Comprehensive allelotyping of human intrahepatic cholangiocarcinoma. Clin. Cancer Res. 7, 2648–2655 [PubMed] [Google Scholar]
- 40.Ohashi K., Nakajima Y., Kanehiro H., Tsutsumi M., Taki J., Aomatsu Y., Yoshimura A., Ko S., Kin T., Yagura K.Yoichi Konishi and Hiroshige Nakano (1995) Ki-ras mutations and p53 protein expressions in intrahepatic cholangiocarcinomas: relation to gross tumor morphology. Gastroenterology 109, 1612–1617 [DOI] [PubMed] [Google Scholar]
- 41.Junking M., Wongkham C., Sripa B., Sawanyawisuth K., Araki N., Wongkham S. (2008) Decreased expression of galectin-3 is associated with metastatic potential of liver fluke-associated cholangiocarcinoma. Eur. J. Cancer 44, 619–626 [DOI] [PubMed] [Google Scholar]
- 42.Schmitz K. J., Lang H., Wohlschlaeger J., Sotiropoulos G. C., Reis H., Schmid K. W., Baba H. A. (2007) AKT and ERK1/2 signaling in intrahepatic cholangiocarcinoma. World J. Gastroenterol. 13, 6470–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawase H., Fujii K., Miyamoto M., Kubota K. C., Hirano S., Kondo S., Inagaki F. (2009) Differential LC-MS-based proteomics of surgical human cholangiocarcinoma tissues. J. Proteome Res. 8, 4092–4103 [DOI] [PubMed] [Google Scholar]
- 44.Mikuriya K., Kuramitsu Y., Ryozawa S., Fujimoto M., Mori S., Oka M., Hamano K., Okita K., Sakaida I., Nakamura K. (2007) Expression of glycolytic enzymes is increased in pancreatic cancerous tissues as evidenced by proteomic profiling by two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. Int. J. Oncol. 30, 849–855 [PubMed] [Google Scholar]
- 45.Unwin R. D., Craven R. A., Harnden P., Hanrahan S., Totty N., Knowles M., Eardley I., Selby P. J., Banks R. E. (2003) Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 3, 1620–1632 [DOI] [PubMed] [Google Scholar]
- 46.Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 47.Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Chen Q., Zhang J. T. (2004) Tumor suppressor gene 14–3-3sigma is down-regulated whereas the proto-oncogene translation elongation factor 1delta is up-regulated in non-small cell lung cancers as identified by proteomic profiling. J. Proteome Res. 3, 728–735 [DOI] [PubMed] [Google Scholar]
- 49.Thomas P. A., Kirschmann D. A., Cerhan J. R., Folberg R., Seftor E. A., Sellers T. A., Hendrix M. J. (1999) Association between keratin and vimentin expression, malignant phenotype, and survival in postmenopausal breast cancer patients. Clin. Cancer Res. 5, 2698–2703 [PubMed] [Google Scholar]
- 50.Wei J., Xu G., Wu M., Zhang Y., Li Q., Liu P., Zhu T., Song A., Zhao L., Han Z., Chen G., Wang S., Meng L., Zhou J., Lu Y., Wang S., Ma D. (2008) Overexpression of vimentin contributes to prostate cancer invasion and metastasis via src regulation. Anticancer Res. 28, 327–334 [PubMed] [Google Scholar]
- 51.Wu M., Bai X., Xu G., Wei J., Zhu T., Zhang Y., Li Q., Liu P., Song A., Zhao L., Gang C., Han Z., Wang S., Zhou J., Lu Y., Ma D. (2007) Proteome analysis of human androgen-independent prostate cancer cell lines: variable metastatic potentials correlated with vimentin expression. Proteomics 7, 1973–1983 [DOI] [PubMed] [Google Scholar]
- 52.McInroy L., Määttä A. (2007) Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem. Biophys. Res. Commun. 360, 109–114 [DOI] [PubMed] [Google Scholar]
- 53.Jordan M. A., Wilson L. (1998) Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr. Opin. Cell Biol. 10, 123–130 [DOI] [PubMed] [Google Scholar]
- 54.Pawlak G., Helfman D. M. (2001) Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 11, 41–47 [DOI] [PubMed] [Google Scholar]
- 55.Hall A. (2009) The cytoskeleton and cancer. Cancer Metastasis Rev. 28, 5–14 [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi H., Condeelis J. (2007) Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 1773, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan B., Yap C. T., Wang S., Lee C. K., Koh S., Omar M. F., Salto-Tellez M., Kumarasinghe M. P. (2010) Cofilin immunolabelling correlates with depth of invasion in gastrointestinal endocrine cell tumors. Acta Histochem. 112, 101–106 [DOI] [PubMed] [Google Scholar]
- 58.Janke J., Schlüter K., Jandrig B., Theile M., Kölble K., Arnold W., Grinstein E., Schwartz A., Estevéz-Schwarz L., Schlag P. M., Jockusch B. M., Scherneck S. (2000) Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin 1. J. Exp. Med. 191, 1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shields J. M., Rogers-Graham K., Der C. J. (2002) Loss of transgelin in breast and colon tumors and in RIE-1 cells by Ras deregulation of gene expression through Raf-independent pathways. J. Biol. Chem. 277, 9790–9799 [DOI] [PubMed] [Google Scholar]
- 60.Assinder S. J., Stanton J. A., Prasad P. D. (2009) Transgelin: an actin-binding protein and tumour suppressor. Int. J. Biochem. Cell Biol. 41, 482–486 [DOI] [PubMed] [Google Scholar]
- 61.Wittenmayer N., Jandrig B., Rothkegel M., Schlüter K., Arnold W., Haensch W., Scherneck S., Jockusch B. M. (2004) Tumor suppressor activity of profilin requires a functional actin binding site. Mol. Biol. Cell 15, 1600–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heizmann C. W., Fritz G., Schäfer B. W. (2002) S100 proteins: structure, functions and pathology. Front. Biosci. 7, d1356–1368 [DOI] [PubMed] [Google Scholar]
- 63.Schäfer B. W., Heizmann C. W. (1996) The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem. Sci. 21, 134–140 [DOI] [PubMed] [Google Scholar]
- 64.Chen J. H., Ni R. Z., Xiao M. B., Guo J. G., Zhou J. W. (2009) Comparative proteomic analysis of differentially expressed proteins in human pancreatic cancer tissue. Hepatobiliary Pancreat. Dis. Int. 8, 193–200 [PubMed] [Google Scholar]
- 65.Melle C., Ernst G., Schimmel B., Bleul A., Mothes H., Kaufmann R., Settmacher U., Von Eggeling F. (2006) Different expression of calgizzarin (S100A11) in normal colonic epithelium, adenoma and colorectal carcinoma. Int. J. Oncol. 28, 195–200 [PubMed] [Google Scholar]
- 66.Wang G., Wang X., Wang S., Song H., Sun H., Yuan W., Cao B., Bai J., Fu S. (2008) Colorectal cancer progression correlates with upregulation of S100A11 expression in tumor tissues. Int. J. Colorectal Dis. 23, 675–682 [DOI] [PubMed] [Google Scholar]
- 67.Stulík J., Hernychová L., Porkertová S., Knízek J., Macela A., Bures J., Jandik P., Langridge J. I., Jungblut P. R. (2001) Proteome study of colorectal carcinogenesis. Electrophoresis 22, 3019–3025 [DOI] [PubMed] [Google Scholar]
- 68.Ørntoft T. F., Thykjaer T., Waldman F. M., Wolf H., Celis J. E. (2002) Genome-wide study of gene copy numbers, transcripts, and protein levels in pairs of non-invasive and invasive human transitional cell carcinomas. Mol. Cell. Proteomics 1, 37–45 [DOI] [PubMed] [Google Scholar]
- 69.Badea L., Herlea V., Dima S. O., Dumitrascu T., Popescu I. (2008) Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 55, 2016–2027 [PubMed] [Google Scholar]
- 70.Mohammad H. S., Kurokohchi K., Yoneyama H., Tokuda M., Morishita A., Jian G., Shi L., Murota M., Tani J., Kato K., Miyoshi H., Deguchi A., Himoto T., Usuki H., Wakabayashi H., Izuishi K., Suzuki Y., Iwama H., Deguchi K., Uchida N., Sabet E. A., Arafa U. A., Hassan A. T., El-Sayed A. A., Masaki T. (2008) Annexin A2 expression and phosphorylation are up-regulated in hepatocellular carcinoma. Int. J. Oncol. 33, 1157–1163 [PubMed] [Google Scholar]
- 71.Zimmermann U., Woenckhaus C., Pietschmann S., Junker H., Maile S., Schultz K., Protzel C., Giebel J. (2004) Expression of annexin II in conventional renal cell carcinoma is correlated with Fuhrman grade and clinical outcome. Virchows Arch. 445, 368–374 [DOI] [PubMed] [Google Scholar]
- 72.Pena-Alonso E., Rodrigo J. P., Parra I. C., Pedrero J. M., Meana M. V., Nieto C. S., Fresno M. F., Morgan R. O., Fernandez M. P. (2008) Annexin A2 localizes to the basal epithelial layer and is down-regulated in dysplasia and head and neck squamous cell carcinoma. Cancer Lett. 263, 89–98 [DOI] [PubMed] [Google Scholar]
- 73.Liu J. W., Shen J. J., Tanzillo-Swarts A., Bhatia B., Maldonado C. M., Person M. D., Lau S. S., Tang D. G. (2003) Annexin II expression is reduced or lost in prostate cancer cells and its re-expression inhibits prostate cancer cell migration. Oncogene 22, 1475–1485 [DOI] [PubMed] [Google Scholar]
- 74.Gillette J. M., Chan D. C., Nielsen-Preiss S. M. (2004) Annexin 2 expression is reduced in human osteosarcoma metastases. J. Cell. Biochem. 92, 820–832 [DOI] [PubMed] [Google Scholar]
- 75.Rajasekaran A. K., Rajasekaran S. A. (2003) Role of Na-K-ATPase in the assembly of tight junctions. Am. J. Physiol. Renal Physiol. 285, F388–F396 [DOI] [PubMed] [Google Scholar]
- 76.Barwe S. P., Anilkumar G., Moon S. Y., Zheng Y., Whitelegge J. P., Rajasekaran S. A., Rajasekaran A. K. (2005) Novel role for Na, K-ATPase in phosphatidylinositol 3-kinase signaling and suppression of cell motility. Mol. Biol. Cell 16, 1082–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fleming T. P., Ghassemifar M. R., Sheth B. (2000) Junctional complexes in the early mammalian embryo. Semin. Reprod. Med. 18, 185–193 [DOI] [PubMed] [Google Scholar]
- 78.Klymkowsky M. W., Savagner P. (2009) Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am. J. Pathol. 174, 1588–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zavadil J., Böttinger E. P. (2005) TGF-beta and epithelial-to-mesenchymal transitions. Oncogene 24, 5764–5774 [DOI] [PubMed] [Google Scholar]
- 80.Levy L., Hill C. S. (2006) Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 17, 41–58 [DOI] [PubMed] [Google Scholar]
- 81.Benckert C., Jonas S., Cramer T., Von Marschall Z., Schäfer G., Peters M., Wagner K., Radke C., Wiedenmann B., Neuhaus P., Höcker M., Rosewicz S. (2003) Transforming growth factor beta 1 stimulates vascular endothelial growth factor gene transcription in human cholangiocellular carcinoma cells. Cancer Res. 63, 1083–1092 [PubMed] [Google Scholar]
- 82.Zen Y., Harada K., Sasaki M., Chen T. C., Chen M. F., Yeh T. S., Jan Y. Y., Huang S. F., Nimura Y., Nakanuma Y. (2005) Intrahepatic cholangiocarcinoma escapes from growth inhibitory effect of transforming growth factor-beta1 by overexpression of cyclin D1. Lab. Invest. 85, 572–581 [DOI] [PubMed] [Google Scholar]
- 83.Keshamouni V. G., Jagtap P., Michailidis G., Strahler J. R., Kuick R., Reka A. K., Papoulias P., Krishnapuram R., Srirangam A., Standiford T. J., Andrews P. C., Omenn G. S. (2009) Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-beta-Induced epithelial-mesenchymal transition. J. Proteome Res. 8, 35–47 [DOI] [PubMed] [Google Scholar]
- 84.Guedj N., Zhan Q., Perigny M., Rautou P. E., Degos F., Belghiti J., Farges O., Bedossa P., Paradis V. (2009) Comparative protein expression profiles of hilar and peripheral hepatic cholangiocarcinomas. J. Hepatol. 51, 93–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.