Abstract
The analysis of cerebrospinal fluid (CSF) is used in biomarker discovery studies for various neurodegenerative central nervous system (CNS) disorders. However, little is known about variation of CSF proteins and metabolites between patients without neurological disorders. A baseline for a large number of CSF compounds appears to be lacking. To analyze the variation in CSF protein and metabolite abundances in a number of well-defined individual samples of patients undergoing routine, non-neurological surgical procedures, we determined the variation of various proteins and metabolites by multiple analytical platforms. A total of 126 common proteins were assessed for biological variations between individuals by ESI-Orbitrap. A large spread in inter-individual variation was observed (relative standard deviations [RSDs] ranged from 18 to 148%) for proteins with both high abundance and low abundance. Technical variation was between 15 and 30% for all 126 proteins. Metabolomics analysis was performed by means of GC-MS and nuclear magnetic resonance (NMR) imaging and amino acids were specifically analyzed by LC-MS/MS, resulting in the detection of more than 100 metabolites. The variation in the metabolome appears to be much more limited compared with the proteome: the observed RSDs ranged from 12 to 70%. Technical variation was less than 20% for almost all metabolites. Consequently, an understanding of the biological variation of proteins and metabolites in CSF of neurologically normal individuals appears to be essential for reliable interpretation of biomarker discovery studies for CNS disorders because such results may be influenced by natural inter-individual variations. Therefore, proteins and metabolites with high variation between individuals ought to be assessed with caution as candidate biomarkers because at least part of the difference observed between the diseased individuals and the controls will not be caused by the disease, but rather by the natural biological variation between individuals.
The analysis of CSF1 is indispensable in the diagnosis and understanding of various neurodegenerative CNS disorders (1–3). CSF is a fluid that has different functions, such as the protection of the brain from outside forces, transport of biological substances, and excretion of toxic and waste substances. It is in close contact with the extracellular fluid of the brain. Therefore, the composition of CSF can reflect biological processes of the brain (4). By discovering the characterization of the proteome and metabolome of CSF we may gain better insight on the pathogenesis of CNS disorders. This would be significant because, for many of these disorders, the etiology is still unclear.
CSF is produced in the ventricles of the brain and in the subarachnoidal spaces. Humans normally produce around 500 mL of CSF each day, and the total volume of CSF at a given time is approximately 150 mL. CSF reflects the composition of blood plasma, although the concentrations of most proteins and metabolites in CSF are lower. However, individual proteins and metabolites can act differently. Active transport from blood and secretion from the brain contribute to the specific composition of CSF. This composition can be disturbed in neurological disorders (5–6). Since CNS-specific proteins and metabolites are typically low in abundance compared with their levels in blood, this change in composition is more likely to be found in CSF because in blood the more abundant plasma proteins can completely mask the signal of the less abundant proteins. Also, if the disease markers do not cross the blood-brain-barrier, then the CSF is the only viable biofluid source. Therefore, CSF might be an excellent source for biomarker discovery for CNS disorders if we follow the hypothesis that neurological diseases induce alterations in CSF protein and metabolite levels.
Analysis of metabolites in CSF has been common practice in clinical chemistry for decades to analyze biomarkers for inborn errors of metabolism. The approaches used are either metabolite profiling of CSF using NMR (7), or targeted analysis of one or a few metabolites using specific analytical methods (8). Metabolomics includes the analysis of metabolites in biofluids by NMR or MS-based approaches, i.e. LC-MS or GC-MS. Several metabolite profiling studies were performed on CSF using NMR, some of which were published only recently (9,10). Surprisingly, very few metabolomics studies using MS-based methods have been performed on CSF to date (11,12). One of the reasons is the fact that the human CSF metabolome has not yet been characterized very well. Many CSF metabolites remain unidentified, and for those that have been identified there is not much known about normal concentration ranges. A systematic categorization of the CSF metabolome is necessary and expected to be beneficial for future biomarker discoveries. Recently, Wishart et al. made a good start in exploring the human CSF metabolome with their computer-aided literature survey that resulted in 308 detectable metabolites in human CSF (13).
The CSF proteome has been characterized to a much larger extent than the CSF metabolome and is currently the topic of investigations in several research groups worldwide. Recently, studies have been published with numerous identities and quantities of CSF proteins. Pan and co-workers were able to identify 2,594 proteins in well-characterized pooled human CSF samples using strict proteomics criteria with a combination of linear trap quadrupole LTQ-FT (Thermo Fisher Scientific, Bremen, Germany) and MALDI TOF/TOF equipment (14). They were also able to quantify several proteins using a targeted LC MALDI TOF/TOF approach (15). Hu et al. have studied the intra- and inter-individual variation in human CSF and found large variations in protein concentrations in six patients by means of two dimensional–gel electrophoresis (16), focusing mainly on the variations within individuals at two different time-points. Although only a limited number of proteins was analyzed, the variation between the time-points was profound, exceeding 200% for seven proteins.
Unique CSF biomarkers may contribute to a deeper understanding of the mechanisms of CNS disorders. However, for this assumption to come true, there are still challenges ahead. Although CSF is not as complex as blood (almost missing the cellular part and the clotting system present in blood), it is expected to consist of thousands of organic- and non-organic salts, sugars, lipids, and proteins. A large part of the CSF consists of a few highly abundant metabolites and proteins, which hamper, if no precautions are undertaken, the identification and quantification of metabolites and proteins that occur in lower amounts. The analysis of the CSF metabolome is complicated because of the diverse chemical nature of metabolites and the lower concentration of metabolites compared with blood. Analytical method development is still required because it is not possible to identify the entire range of CSF metabolites with one single analytical method. Although in proteome research efforts have been made to quantify proteins, metabolomics studies up to now either do not provide quantitative information or they only give information for the most abundant metabolites.
Another challenge is the sample amount obtained by lumbar puncture to collect CSF. Lumbar puncture is an invasive method that is not performed as frequently as blood sampling. However, often after the analysis of various clinical parameters, only a limited amount of CSF sample is available for biomarker discovery. Metabolomics studies are hampered by limited CSF sample amount. Therefore, analytical methods are required that are suitable to handle relatively small sample volumes.
The main objectives of this study were (1) to analyze the variation in CSF protein and metabolite abundances in a number of well-defined individual samples by multiple analytical platforms; and (2) to integrate metabolomics and proteomics to present biological variations in metabolite and protein abundances and compare these with technical variations with the currently used analytical methods. The results will facilitate and increase the application of CSF for future biomarker discovery studies in the field of neurodegenerative diseases and neuro-oncology.
EXPERIMENTAL PROCEDURES
CSF Sampling
CSF samples were obtained by lumbar puncture in the Erasmus University Medical Centre (Rotterdam, the Netherlands). An experienced medical doctor selected 10 samples, which were taken from patients receiving spinal anesthesia before non-neurological surgery. These subjects had no neurological diseases, were not using any medication, and were considered to have neurologically normal CSF. Immediately after sampling, the CSF samples were centrifuged (10 min at 3.000 rpm) to discard cellular elements. The samples were subsequently used for routine CSF diagnostics. This included quantification of total protein concentration by routine clinical chemistry measurements and quantification of the cell count (< 5 white blood cells per mL). The remaining volume of the samples was aliquoted and stored at −80 °C immediately after centrifugation. As a standard procedure, the samples were checked for blood contamination, and any sample in which a hemoglobin or apolipoprotein B100 peptide was identified with a significant score by nanoLC-Orbitrap MS was excluded from the study.
For pooling of the samples (n = 10), the originally obtained samples were thawed on ice and 0.75 mL from each of the samples was joined, resulting in a 7.5 mL pooled CSF sample. This pooled CSF sample was vortexed for 30 seconds and then subdivided into 75 portions of 100 μL in sterile cryogenic vials (Nalgene Nunc Int., Rochester, NY). The portions were immediately frozen at -80 °C. The characteristics of the pooled sample are described in Table 1. This pooled sample was used to assess the technical variation in the proteomics experiments by measuring it five times. For the measurements of the individual patients only nine CSF samples were used because there was insufficient volume of one sample.
Table I. Details on the pooled sample, including gender, average age and average protein concentration.
| Characteristic | Detail |
|---|---|
| Gender | Male 8; Female 2 |
| Mean age (years) | 51 (S.D. = 14) |
| Total protein concentration (g/L) | 0.4 (S.D. = 0.1) |
| Glucose concentration (mmol/L) | 3.3 (S.D. = 0.3) |
The 28 CSF samples from the validation sample set were also taken by an experienced anesthesiologist from patients receiving spinal anesthesia before non-neurological surgery, but these samples were taken at another hospital (Sint Franciscus Gasthuis, Rotterdam, the Netherlands). These subjects had no neurological diseases, were not using any medication, and were considered to have neurologically normal CSF.
The CSF samples used in the experimental sample set were selected by an experienced neurologist and taken from patients undergoing tests for clinical diagnosis. These samples, taken from multiple sclerosis and headache patients were subjected to the same, strict post-sampling procedure as the samples mentioned previously. In these samples no significant difference in protein concentration between the two groups was observed; therefore, there was no leakage in the blood-CSF barrier. All CSF samples used in this study were observed in the morning at approximately10 a.m. The Medical Ethical Committees of the Erasmus University Medical Centre in Rotterdam, The Netherlands, and the Sint Franciscus Gasthuis in Rotterdam, The Netherlands, approved the study protocol and all study participants gave written consent. The average age and protein concentration of the samples in all three sample sets is listed in Table 2, and age, gender and protein concentration of the individual samples is listed in the Supplementary Material.
Table II. Details on the three sample sets: the original sample set (n = 9), the validation sample set (n = 28), and the experimental sample sets (n = 36/42). Age and protein concentration values are averages (standard deviation in brackets). The gender, age and protein concentration of all individual patients is listed in the Supplementary Material.
| Original sample set | Validation sample set | Experimental sample set proteomics | Experimental sample set metabolomics | |
|---|---|---|---|---|
| Gender | 7M/2F | 13M/15F | 13M/23F | 23M/19F |
| Age (years) | 51.0 (14.8) | 44.5 (14.5) | 41.7 (10.8) | 43.5 (12.8) |
| Protein concentration (g/L) | 0.39 (0.12) | 0.37 (0.11) | 0.38 (0.11) | 0.38 (0.13) |
Proteomics
Sample Preparation for NanoLC-Orbitrap MS and MALDI-FT-ICR MS
For measurement of proteins in CSF, samples were enzymatically digested with trypsin to obtain peptides. An amount of 50 μL Rapigest (Waters, Milford, MA) in 50 mmol/L ammonium bicarbonate and 1 μL 100 mmol/L DTT was added to 50 μL CSF. The mixture was heated at 60 °C for 30 minutes, upon which it was cooled down to room temperature in approximately 20 minutes. Iodoacetamide (5 μL of 0.3 mmol/L solution) was added and this mixture was left for 30 minutes in dark at room temperature. Trypsin was added (10 μL, 0.1 mg/mL) and all samples, processed in one batch, were incubated overnight at 37 °C. To stop digestion, 2 μL of a 50% TFA/50% water solution was added. The sample was then incubated for 45 minutes at 37 °C.
NanoLC-Orbitrap MS Analysis
These measurements were performed on an Ultimate 3000 nanoLC system (Dionex, Germering, Germany) online coupled to a hybrid linear ion trap/Orbitrap MS (LTQ Orbitrap XL; Thermo Fisher Scientific, Bremen, Germany). Five microliters of digest were loaded onto a C18 trap column (C18 PepMap, 300 μm ID x 5 mm, 5 μm particle size, 100 Å pore size; Dionex, Amsterdam, The Netherlands) and desalted for 10 minutes using a flow rate of 20 μL/min 0.1% TFA. Then the trap column was switched online with the analytical column (PepMap C18, 75 μm ID x 150 mm, 3 μm particle and 100 Å pore size; Dionex, Amsterdam, The Netherlands) and peptides were eluted with the following binary gradients of solvent A and B: 0–25% solvent B in 120 minutes and 25–50% solvent B in further 60 minutes, where solvent A consisted of 2% acetonitrile and 0.1% formic in water and solvent B consisted of 80% acetonitrile and 0.08% formic acid in water. Column flow rate was set to 300 nL/min. For MS detection, a data-dependent acquisition method was used: high resolution survey scan from 400 - 1800 Th. was performed in the Orbitrap (value of target of automatic gain control (AGC) 106, resolution 30,000 at 400 m/z; lock mass was set to 445.120025 u (protonated (Si(CH3)2O)6) (17)). Based on this survey scan, the five most intensive ions were consecutively isolated (automatic gain control target set to 104 ions) and fragmented by collision-activated dissociation applying 35% normalized collision energy in the linear ion trap. After precursors were selected for MS/MS, they were excluded for further MS/MS spectra for three minutes. Proteins were identified using the Bioworks 3.2 (peak picking by Extract_msn, default settings) software package (Thermo Fisher Scientific, Bremen, Germany), and SEQUEST (Thermo Fisher Scientific, Bremen, Germany), taking the HUPO criteria into account, with XC scores of 1.8, 2.2 and 3.75 for single-, double-, and triple-charged ions, respectively. The database used was the SwissProt-database (version 56.0, human taxonomy (20069 entries)). Carboxymethylation of cysteine (+57.021 u) as fixed and oxidation of methionine (+15.996 u) as variable modifications and tryptic cleavage were considered. The number of allowed missed cleavages was two, the mass tolerance for precursor ions was 10 ppm, and the mass tolerance for fragment ions was 0.5 Da. The cut-off for mass differences with the theoretical mass of the identified peptides was set at 2 ppm.
The Orbitrap data was subsequently analyzed using the Progenesis LC-MS software package (version 2.5, Nonlinear Dynamics, Newcastle-upon-Tyne, United Kingdom), in which the LC runs were aligned and the biological variation between the samples was calculated to assess variation between individuals in this data set. A S/N > 4 and the presence of at least three isotope peaks per peptide were used as a minimum threshold for quantitation. Variation was assessed by comparing the area-under-the-curve of all peptides of a protein. The mean area-under-the-curve, corrected for the total ion current, of all peptides of a protein was compared between the individuals, and the RSD of this value was considered to be the inter-individual variation (listed as RSD [in percentages] in the supplementary material). Technical variation was assessed by performing the same comparison on the five replicas of the pooled sample.
MALDI-FT-ICR MS Analysis
The CSF samples were handled according to the same protocol we reported previously (18), in which the samples were tryptically digested and desalted using C18 material. Using a 2,5-dihydroxybenzoic acid matrix the samples were all measured manually on an APEX IV Qe 9.6 Tesla MALDI-FT-ICR mass spectrometer (Bruker Daltonics, Billarica, MA), using a multishot accumulation as recommended by Mize and Amster, Moyer et al., and O'Connor and Costello (19–21). External mass calibration was applied using a quadratic equation. Quantitative MALDI-FT-ICR has previously been applied to quantify human immunodeficiency virus type 1 protease inhibitors in cell lysates (22) as well as peptides in CSF (18), indicating that quantitative MALDI-FT-ICR methods are readily applicable, which is due to the fact that variation in peak height in MALDI-FT-ICR MS is much more reproducible than in, e.g. MALDI-TOF MS. The sum of the height of 14 omnipresent albumin peaks of each sample was then compared with albumin concentration levels obtained by routine clinical chemistry measurements. Standard deviations of the peak height of the albumin peaks were between 9 and 16% for all 14 albumin peaks.
Biological Variation in an Experimental Setting
To compare the results on the variation of protein abundances found in the neurologically normal individual CSF samples to an experimental setting, an identical experiment was performed on a larger set of samples. A total of 36 CSF samples, obtained from patients with either multiple sclerosis or headaches, was used. These samples, especially those of the multiple sclerosis patients, originated from people who were suffering from neurological problems. Hence, the variation in protein abundance, e.g. immunoglobulin levels, which are known to be elevated in neuro-inflammatory diseases such as multiple sclerosis (23–25), is potentially far more extensive than in the nine well-defined individuals measured previously.
Metabolomics
GC-MS Analysis
Human CSF samples from the original sample set (60 μL) were deproteinized by adding 250 μL methanol and subsequently centrifuged for 10 minutes at 10,000 rpm. Human CSF samples from the validation sample set (100 μL) were deproteinized by adding 400 μL methanol. The supernatant was dried under N2 followed by derivatization with methyl-N-(trimethylsilyl)-trifluoroacetamide in pyridine similar to Koek et al (26). During the different steps in the sample Work-up, i.e. before deproteinization, derivatization and injection, different (deuterated) internal standards were added at a level of approximately 20 ng/μL. The final volume was 45 μL for the original sample set and 135 μL for the validation sample set and 1 μL aliquot of the derivatized samples was injected in splitless mode on a HP5-MS 30 m x 0.25 mm x 0.25 μm capillary column (Agilent Technologies, Palo Alto, CA) using a temperature gradient from 70 °C to 320 °C at a rate of 5 °C/min. GC-MS analysis was performed using an Agilent 6890 gas chromatograph coupled to an Agilent 5973 mass selective detector (Agilent Technologies). MS detection was used in electron impact mode and full scan monitoring mode (m/z 15–800). The electron impact for the generation of ions was 70 eV.
Sample work-up was performed in duplicate for the original sample set. For validation, sample set samples were injected in duplicate. For both sample sets, a pooled human CSF sample was analyzed in sextuplicate to determine the analytical error in the analysis of metabolites by GC-MS. Data pre-processing was performed by composing target lists of peaks detected in the samples based on retention time and mass spectra. These peaks were integrated for all samples. All peak areas were subsequently normalized using internal standards. The resulting target lists were used for further statistical analysis. Identities were assigned based on the presence of identical mass spectra using an in-house database.
LC-MS/MS Analysis
To 10 μL of human CSF sample, 10 μL of an internal standard solution containing 13C15N-amino acids was added, followed by the addition of 100 μL of methyl alcohol. The mixture was vortexed for 10 seconds and centrifuged at 10.000 rpm for 10 minutes at 10 °C. The supernatant was dried under N2. The residues were dissolved in 80 μL borate buffer (pH 8.5) and after 10 seconds, vortexing 20 μL of AQC reagent (Waters, Etten-Leur, The Netherlands) was added and the mixture was vortexed immediately. The samples were heated for 10 minutes at 55 °C. After cooling, a 1-μL sample of the reaction mixture was injected into the ultra performance liquid chromatography (UPLC)-MS/MS system.
An ACQUITY UPLCTM system with autosampler (Waters, Milford, MA) was coupled online with a Quattro Premier XE Tandem quadrupole mass spectrometer (Waters) and was used in positive-ion electrospray mode. The instrument was operated under Masslynx data acquisition software (version 4.1; Waters). The samples were analyzed by UPLC-MS/MS using a AccQ-TagTM Ultra 100 mm x 2.1 mm (1.7 μm particle size) column (Waters). A binary gradient system of water - eluent A (10:1, v/v) (AccQ Tag, Waters) and 100% eluent B (AccQ Tag, Waters), was used. Elution of the analytes was achieved by ramping the percentage of eluent B from 0.1 to 90.0% in approximately 9.5 minutes using a combination of both linear and convex profiles. The flow-rate was 0.7 mL/min. The column temperature was maintained at 60 °C, and the temperature of the autosampler tray was set to 10 °C. After each injection, the injection needle was washed with 200 μL strong wash solvent (95% ACN), and 600 μL weak wash solvent (5% ACN).
The Quattro Premier XE was used in the positive-ion electrospray mode and all analytes were monitored in selective reaction monitoring (SRM) using nominal mass resolution (full-width half-maximum 0.7 amu). Next to the derivatization reagent all amino acids were selectively monitored via the transition from the protonated molecule of the AccQ-Tag derivative to the common fragment at m/z 171. Collision energy and collision gas (Ar) pressure were 22 eV and 2.5 mbar, respectively. The complete chromatogram was divided into six time intervals, restricting the number of SRM transitions to follow and allowing quantitative information to be gathered in each segment. Acquired data was evaluated using Quantlynx (Waters). All samples were analyzed in duplicate.
Data-pre-processing was performed by calculating the concentration of 18 amino acids in all samples by peak integration, followed by normalization using relevant internal standards and quantification using external calibration curves. The analytical variation was determined from the duplicate analysis of the samples using weighted regression (http://cran.r-project.org/web/packages/chemCal/index.html) (27).
NMR Analysis
CSF samples from the original sample set (280 μL) were centrifuged (2000g, 15 min) using a filter with a cut-off of 10 kDa (Centrisart I 13239-E) to remove proteins. Next, 25 μL of 8.8 mmol/L TSP-d4 stock solution in D2O was added to 250 μL filtrated CSF to a final concentration of 0.8 mmol/L TSP. The pH of the filtrated CSF was adjusted to approximately 7 (7.0 - 7.1) by adding phosphate buffer (9.7 μl L mol/L, to a final concentration of 35 mnol/L). The final CSF NMR sample (284.7 μL) was then transferred to a Shigemi microcell NMR tube for NMR measurements (also known as non-diluted CSF samples). For the validation sample set, samples (100 μL) were diluted with 270 μL D2O before protein removal.
As a duplicate, for establishing the analytical variation, 100 μL of CSF of the individuals was diluted into 180 μL D20 and subsequently worked up as described above (also known as the diluted CSF samples).
The one-dimensional proton NMR spectra of diluted and non-diluted CSF samples were acquired on an 800 MHz Inova or 600 MHz (Varian Inc., Palo Alto, CA) system equipped with either a 5-mm triple-resonance, XYZ-gradient HCN room-temperature probe or a 5-mm triple-resonance, Z-gradient HCN cold-probe, respectively. Suppression of water was achieved by using WATERGATE (delay: 85 μs) (28) or presaturation. For each one-dimensional proton NMR spectrum, 512 scans of 18,000 data points were accumulated with a spectral width of 9,000 Hz. The acquisition time for each scan was two seconds. Between scans, an eight-second relaxation delay was used. Before spectral analysis, all acquired free induction decays were zero-filled to 64,000 data points, multiplied with a 0.3-Hz line-broadening function, Fourier transformed, and manually phase- and baseline-corrected by using ACD/SpecManager software. Spectra were subsequently transformed to the Chenomx NMR Suite Professional software package version 5.1 for further analysis (Chenomx Inc., Edmonton, Alberta, Canada) (29). Metabolite identification and quantification were performed using the 800-MHz library of metabolite NMR spectra from the Chenomx NMR Suite 5.1 (pH 6–8) for the original sample set (Chenomx NMR Suite 6.1 was used for the validation sample set). The metabolite spectra in the library are predicted based on a database of pure compound spectra acquired using particular pulse sequence and acquisition parameters, e.g. the tn-noesy-presaturation pulse sequence with four-second acquisition time and one second of recycle delay. The Chenomx NMR Suite software fits the spectral signatures (singlets, doublets, triplets etc), i.e. the peak shapes, of a compound from an internal database of reference spectra to the experimental NMR spectrum. The resonance assignments derived from the Chenomx NMR Suite software were further checked against literature spectra. For quantification, Chenomx NMR Suite 5.1 uses the concentration of the known reference signal as calibration (in this case, TSP-d4).
The analytical variation on the individual metabolite concentrations was determined from the NMR analysis of the dilute and non-dilute CSF samples completely independently, and the quintuplicate measurement of the diluted CSF sample of one individual.
Biological Variation in an Experimental Setting
To assess the results on the variation of metabolite abundances found in original and validation sample set, i.e. neurologically normal CSF, an identical experiment was performed using GC-MS on a set of 42 human CSF samples (100 μL), obtained from patients who had multiple sclerosis and other (inflammatory) neurological diseases.
RESULTS
Proteomics
None of the CSF samples was contaminated with plasma and, according to the criteria set, hemoglobin and apolipoprotein B100 were not identified in any of the samples. All sequenced peptides and identified proteins are listed in the supplemental material (including the number of unique peptides per protein and the sequence coverage for all proteins identified with two or more peptides).
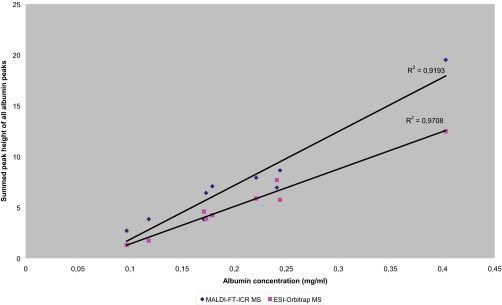
Using MALDI-FT-ICR MS, we analyzed the height of albumin peptide peaks of the nine samples of the original sample set and their correlation to albumin concentration levels in CSF as measured by routine clinical chemistry diagnostics. The sum of the height of 14 omnipresent albumin peaks showed positive correlation to the values measured by clinical chemistry (R2 = 0.919). These values (median: 0.219 g/L, range 0.097–0.403 g/L) clearly show a large variation between individuals, which was also apparent from the differences in height of the peaks measured by MALDI-FT-ICR (Fig. 1). The area under the curve of all peptides identified to be part of albumin in the ESI-Orbitrap experiments was also plotted against the albumin concentration, showing good correlation (R2 = 0.971). RSDs were comparable for all three methods (43.7% for clinical chemistry, 66.7% for MALDI-FT-ICR, and 39.1% for ESI-Orbitrap).
Fig. 1.
Correlation between the measured albumin concentration by clinical chemistry diagnostics and the sum of the height of 14 omnipresent albumin peaks as measured by MALDI-FT-ICR (R2 = 0. 919) and by ESI-Orbitrap (R2 = 0.971).
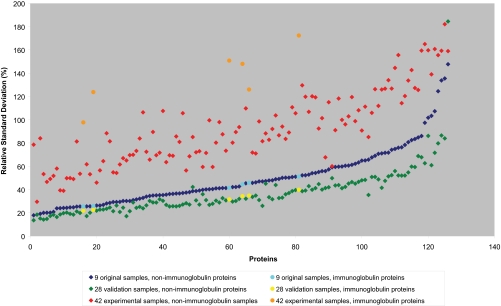
A total of 126 proteins, all identified by multiple peptides and present in all nine normal CSF samples, was analyzed in the nine individual CSF samples by ESI-Orbitrap to assess the variance in protein abundances in CSF, based on the averages of peak heights of all the peptides of a single protein. The RSD ranged from 18% to 148% (median: 43%) in peak height. The far greater part of the examined proteins (119 of 126, i.e. 94.4%) showed less than 100% RSD in average peptide peak height per protein between the nine individual CSF samples. These results were subsequently tested in two larger sample sets, a validation set (28 samples) and an experimental sample set (36 samples), in which similar profiles for the variation in protein abundance, based on the averages of the area-under-the-curve of the peptides in the ESI-Orbitrap, was observed (Fig. 2). In this figure, the variation (in RSD (%)) for all proteins is plotted for all three data sets, and the protein-numbers used here are the same as in the Supplementary Material. The slightly lower RSDs found for the validation sample set are at least partially due to the fact that a longer nanoLC column (50 cm) was used, consequently, more peptides were measured per protein. The correlation of the individual protein variations between the datasets was calculated using these data. This resulted in an R2 of 0.94 for the correlation between the original dataset and the validation dataset, and an R2 of 0.66 for the correlation between the original dataset and the experimental dataset. This is a strong indication that the same proteins have a high inter-individual variation in the healthy CSF patients in both the original and the validation sample set, but that this is quite different in the experimental sample set of patients who have known neurological disorders. In the original sample set, the 126 proteins were observed in all nine normal control samples. In the experimental sample set, 11 proteins were not observed in all 36 samples. This may indicate a greater variance in the experimental samples, whereas in the validation sample set the variation is slightly lower than in the original sample set. In the experimental sample set, the RSD in peptide abundance per protein ranged from 30% to 182% (median: 91%). The greater variation in the experimental sample set is at least partially caused by the sample choice for this set of samples. As referenced earlier, for patients who have multiple sclerosis, it is known that the immunoglobulin levels are elevated. Because this sample set contained both multiple sclerosis CSF samples and samples from patients who had headaches, it is not surprising that many of the proteins with the highest RSD between individuals are all immunoglobulin types and proteins related to inflammatory response, which were indeed elevated in the multiple sclerosis samples. In essence, in the normal controls we observed the biological variation between the individuals, whereas in the experimental sample set both the biological variation as well as the disease-related variation was observed.
Fig. 2.
The proteins sorted by the variation in the original samples paired to the variation in the validation samples and the experimental samples. A trend is clearly visible, but due to the nature of the experimental samples (multiple sclerosis and headaches), the immunoglobulins do not correspond with the overall trend, which is to be expected considering the well-known inflammatory component of multiple sclerosis. Numbers on x axis correlate to protein numbers mentioned in Supplementary Material.
Although all three sample sets are distinct, a number of similarities can be observed. In the sample sets, there is a clear division that can be observed between proteins whose abundances vary highly among individuals and proteins that show a much more limited variation between individuals. Among the proteins that showed limited variation between individual CSF samples were serotransferrin (25% RSD in the original sample set, 18% RSD in the validation sample set and 50% RSD in the experimental sample set), tetranectin (21, 18, and 52%, respectively), and gelsolin (24, 19, and 58%, respectively). Proteins with high variation between individuals in all three sample sets included cadherin-13 (82% RSD in the original sample set, 60% RSD in the validation sample set and 143% RSD in the experimental sample set), contactin-2 (124%, 80 and 156%, respectively), and haptoglobin (135%, 84 and 182%, respectively). The full list of variations between the individuals for the 126 proteins can be found in the Supplementary Material.
Gender and age related inter-individual variation were assessed in the validation sample set. In this sample set the number of males and females was roughly equal (13 males and 15 females) and we defined three different age groups (younger than 35 years of age, between 35 and 50 years of age, and older than 50 years of age). Although both parameters (gender and age) appear to influence the variation, their influence appears limited (complete list of individual protein variations specified for age group and gender is listed in the Supplementary Material). The inter-individual variation in protein abundance appears slightly larger in females than in males, especially for proteins that have a high inter-individual variation, but this is not exclusively the case as there are also proteins of which the variation is higher in males. The same is true for the inter-individual variation of the proteins when comparing age groups. The variation seems slightly larger in the oldest age group, again most clearly for the proteins with the highest inter-individual variation. But, as with age, this is not exclusively so, as there are also proteins that have a higher inter-individual variation in the group of patients below age 35 (full list of age and gender variation in supplemental material (Figs. S1 and S2)). Students t test shows 1.6% and 3.2% of the proteins to be significantly different based on variation of their RSD between males and females and between the age groups, respectively (p < 0.01, Supplementary Material).
Total abundance of most of the measured proteins was slightly higher in males than in females, and, with regard to age, the abundance of most of the measured proteins was highest in the oldest patient group of the validation sample set.
Metabolomics
Three different analytical methods were applied to analyze the individual as well as the pooled CSF samples. The methods included untargeted GC-MS and NMR methods and a targeted LC-MS/MS method specifically for amino acids.
First, a small set of CSF samples, i.e. original sample set consisting of nine samples for GC-MS, eight for LC-MS/MS, and five for NMR, was analyzed by the three methods followed by a larger set of samples, i.e. validation sample set consisting of 28 samples for GC-MS and 27 samples for NMR and LC-MS/MS. The original sample set was used to provide a quick screen of what type of metabolites could be detected and to have a rough idea of biological variation and analytical error. However, for reliable data with respect to biological variation as well as possible gender and age effect and comparison of the three methods, the validation sample set was used.
Analysis of original samples of CSF with GC-MS resulted in a list of 108 metabolites, of which 93 could be identified (see Supplementary Material). The unknown metabolites covered both metabolites that were observed in other biofluids, i.e. plasma and/or urine, as well as metabolites that seemed to be specific for CSF. The metabolites detected by GC-MS cover many different compound classes, i.e. amino acids, organic acids, nucleosides, fatty acids, mono- and disaccharides. Of the 93 identified metabolites, some were only present in trace amounts and were therefore not used for further analysis. Interestingly, all identified metabolites were observed in all samples. The analytical variation for each metabolite was determined from the repeated (n = 6) analysis of the pooled CSF sample. Results show that the analytical variation (< 20%) was less than the biological variation for all metabolites (15 to 85%) (see Supplementary Material). The concentration or relative peak area for each metabolite in the pooled human CSF sample is given in Supplementary Material. As expected, the average concentrations and relative peak areas found for the nine individual CSF samples were very similar to that of the pooled human CSF sample.
Next, the validation sample set was analyzed by GC-MS using a somewhat different sample work-up including different sample volumes leading to a less concentrated extract. As a result, some of the less abundant compounds could not be detected. In total 68 metabolites could be detected and most of them were also detected in the original sample set. The biological variation of these 68 metabolites ranged from 7 to 214%, whereas the analytical error ranged from 1 to 36% and in all cases the analytical error was equal or less than the biological variation (see Table III and Supplementary Material). The biological variation for the metabolites observed with GC-MS in the validation sample set shows a normal distribution, as can be deduced from Figure 3. Multivariate data-analysis using PCA (principal component analysis) showed that overall biological variation was dominant over age and gender effects (see Figs. S3 and S4 in the Supplementary Material).
Table III. Metabolites detected by GC-MS, NMR and LC-MS/MS in the validation sample set and their biological variation.
| Metabolite | GC-MS RSD (%) n = 28 | LC-MS RSD (%) n = 27 | NMR RSD (%) n = 27 |
|---|---|---|---|
| 1,5-anhydro-d-Glucitol | 34 | ||
| 1-methylhistidine | 49 | ||
| 1-monopalmitoylglycerol | 30 | ||
| 1-monostearoylglycerol | 19 | ||
| 2,3-dihydroxybutanoic acid | 28 | ||
| 2,4-dihydroxybutanoic acid | 26 | ||
| 2-aminobutyric acid | 22 | 28 | |
| 2-hydroxybutanoic acid | 35 | 29 | |
| 2-hydroxyisovaleric acid | 24 | 30 | |
| 2-piperidinon | 113 | ||
| 3,4-dihydroxybutanoic acid | 54 | ||
| 3-hydroxybutanoic acid | 106 | 15 | |
| 3-hydroxyisovaleric acid | 31 | 15 | |
| 3-hydroxypropanoic acid | 14 | ||
| 3-methylhistidine | 143 | ||
| Acetic acid | 52 | ||
| Acetoacetic acid | 38 | 26 | |
| Acetone | 20 | ||
| Aconitic acid | 28 | ||
| Alanine | 28 | 32 | 27 |
| Arabinose | 19 | ||
| Arginine | 25 | 19 | |
| Ascorbic acid | 25 | ||
| Asparagine | 22 | ||
| C16:0 Fatty acid | 23 | ||
| C18:0 Fatty acid | 7 | ||
| C18:1 Fatty acid | 58 | ||
| Cholesterol | 30 | ||
| Choline | 24 | ||
| Citric acid | 18 | 15 | |
| Citrulline | 34 | ||
| Creatine | 15 | ||
| Creatinine | 63 | 17 | |
| Dimethylamine | 25 | ||
| Erythronic acid | 28 | ||
| Formic acid | 19 | ||
| Fructose | 38 | 24 | |
| Fucose | 17 | ||
| Galactitol | 23 | ||
| Gluconic acid | 26 | ||
| Glucose | 12 | 12 | |
| Glutamic acid | 30 | ||
| Glutamine | 14 | 18 | |
| Glyceric acid | 14 | ||
| Glycerol | 16 | ||
| Glycerol-galactopyranoside | 22 | ||
| Glycine | 35 | 30 | 24 |
| Glycolic acid | 13 | ||
| Histidine | 16 | 15 | |
| Inositol | 68 | ||
| Inositol related compound | 26 | ||
| Iso-citric acid | 21 | ||
| Iso-leucine | 36 | 30 | 25 |
| Lactic acid | 13 | 14 | |
| Leucine | 22 | 27 | 26 |
| Lysine | 43 | 22 | 16 |
| Mannitol | 26 | ||
| Mannose | 17 | ||
| Meso-erythrytol | 16 | ||
| Methanol | 21 | ||
| Methionine | 57 | 34 | 31 |
| Myo-inositol | 19 | 25 | |
| Ornithine | 46 | ||
| Phenylalanine | 19 | 23 | 30 |
| Phosphoric acid | 214 | ||
| Phosphorylethanolamine | 47 | ||
| Proline | 45 | 49 | |
| Pseudo uridine | 21 | ||
| Pyroglutamic acid | 31 | ||
| Pyruvic acid | 28 | 23 | |
| Quinic acid | 90 | ||
| Ribitol | 30 | ||
| Ribonic acid | 26 | ||
| Ribose | 12 | ||
| Serine | 36 | 17 | |
| sn-Glycerol-3-Phosphate | 48 | ||
| Succinic acid | 23 | ||
| Sucrose | 64 | ||
| Threonic acid | 33 | ||
| Threonine | 38 | 27 | 20 |
| Trimethylamine-N-oxide | 18 | ||
| Tryptophan | 24 | ||
| Tyrosine | 30 | 27 | |
| Urea | 28 | ||
| Uric acid | 69 | ||
| Valine | 33 | 28 | 28 |
| Xanthine | 86 | ||
| Xylonic acid | 45 | ||
| Xylose | 82 |
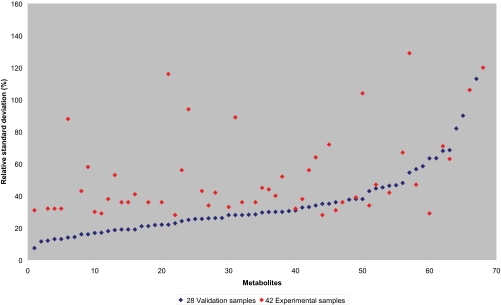
Fig. 3.
Metabolites detected by GC-MS in both the validation (blue) as well as the experimental (red) sample set, sorted by variation in the validation samples paired to the variation in the experimental samples. A trend is visible, but due to the nature of the experimental samples (multiple sclerosis and other neurological disorders), a number of metabolites do not correspond with the overall trend and show significantly higher biological variation. Numbers on x axis correlate to metabolite numbers mentioned in Supplementary Material.
For the experimental sample set of 42 human CSF samples from patients having neurological diseases, a similar profile for the variation in metabolite level could be observed as for the validation sample set from neurologically normal individuals (see Fig. 3 and Supplementary Material). A number of metabolites show a significantly higher RSD for the experimental samples, which is probably caused by the heterogeneity of the experimental CSF samples, as discussed in the proteomics section. Metabolites that show significantly higher RSDs for the experimental samples are 3,4-dihydroxybutanoic acid, fructose, ascorbic acid, glyceric acid, pyruvic acid, and 2-aminobutyric acid. However, there is no clear relation between these metabolites and the neurological disease in the experimental sample set.
For NMR, only five individual CSF samples were analyzed in the original sample set due to limited available sample volumes. Analysis of the CSF samples by NMR resulted in a list of 51 metabolites of which 41 could be quantified (see Supplementary Material). All metabolites observed with NMR were detected in all samples. The biological variation ranged from 8 to 53% whereas the analytical error was between 3 and 9% for all metabolites (Supplementary Material). The concentrations found for the pooled CSF sample were very similar to that of the averages of the individual samples.
The 27 CSF samples in the validation sample set were analyzed by NMR using somewhat different conditions, i.e. more diluted and 600 MHz instead of 800 MHz. However, the same set of metabolites could be quantified in these samples, except for urea. In this sample set the biological variation ranged from 12 to 143% which is somewhat higher compared with the original sample set (see Table III and Supplementary material). The analytical error is in the same range as observed for the original sample set, i.e. 2 to 9%. PCA on the NMR data did not show any significant age or gender effect (see Figs. S5 and S6 in Supplementary Material).
Of the 41 metabolites quantified by NMR in the validation sample set, 21 were also detected by GC-MS. Some of the more volatile metabolites, such as acetone and methanol, can only be analyzed by NMR, showing that despite the overlap, NMR and GC-MS are complementary techniques. Furthermore, NMR can detect a number of metabolites which are difficult to analyze by GC-MS because they cannot be derivatized, e.g. choline, or they can give unstable derivatives, e.g. arginine. On the other hand, a range of metabolites was only observed by GC-MS and not by NMR. In most cases these metabolites either have no proton signal, e.g. uric acid and phosphoric acid, or the concentration is below the detection limit of NMR, e.g. dihydroxybutanoic acids and proline.
The absolute concentrations of amino acids in eight individual CSF samples of the original sample set were determined by a targeted LC-MS/MS. One of the individual CSF samples was omitted due to technical failure. The analytical error is less than the biological variation for all amino acids. The biological variation ranges from 28 to 52%, whereas the analytical error is less than 12% (see Supplementary Material). Despite the differences between samples, all amino acids were present in every individual sample. Again, it can be seen that the concentrations found for the pooled CSF sample were very similar to that of the averages of the individual samples. Glutamic acid and aspartic acid could not be quantified in a reliable way in the CSF samples.
Analysis of the same amino acids in the 27 samples of the validation sample set resulted in similar biological variation, i.e. 14 to 49%, and analytical error, i.e. 1 to 9% (see Table III and Supplementary Material). PCA showed no significant age or gender effects (see Figs. S7 and S8 in Supplementary Material).
Most of the amino acids analyzed by LC-MS/MS were also detected either by GC-MS or NMR. However, one of the advantages of the targeted LC-MS/MS method is the low sample volume required for analysis, i.e. 10 μL versus 60–100 μL for NMR and GC-MS, respectively.
Comparison of the RSD of metabolites that could be analyzed with more than one of the analytical methods, as shown in Table III, shows that on average the biological variation of a metabolite is similar for different methods. Deviations occur mainly for low abundant metabolites, like 3-hydroxybutanoic acid, and metabolites that show relative high analytical errors for certain methods, e.g. creatinine with GC-MS.
DISCUSSION
In this study, we investigated metabolite and protein identities and their abundances and inter-individual variations in abundance in CSF by analyzing a unique and well-defined set of CSF samples and a corresponding pooled CSF sample. Here we have strictly defined criteria to exclude blood contaminated CSF. These criteria warrant that at a certain threshold no contamination is observed; however, contamination not exceeding this threshold can still exist and cannot be ruled out.
Combination of three different analytical techniques for metabolites used in this study resulted in a list of approximately 89 identified metabolites in CSF that can routinely be analyzed, which is approximately one third of the metabolites in CSF present in the human metabolome database (29). It is expected that many of the metabolites which are not detected by NMR and GC-MS are low abundant metabolites, i.e. neurotransmitters, steroids, and eicosanoids, for which more specific, targeted methods are required (30–32). However, these methods often require significant amounts of CSF and should therefore only be used in metabolomics studies when there is evidence that these metabolites are of importance and/or when enough sample volume is available. Furthermore, some metabolites are (almost) absent in normal controls and are only detectable in samples from diseased persons (7).
All endogenous metabolites detected with the three analytical methods in this study were observed in all individual CSF samples. This implies that the qualitative metabolite composition of CSF in normal controls is relatively similar between individuals. Generally, this is also observed in the plasma of healthy persons in contrast to the urine of healthy persons, which is more influenced by dietary intake.
With NMR and LC-MS/MS it was possible to determine the absolute concentration of metabolites. This, in contrast to GC-MS for which metabolites can only be quantified when either internal standards or calibration curves for each metabolite are used, which is practically not feasible; therefore, this method, similar to many other non-targeted methods, is used to measure relative differences in metabolite concentrations between groups or individuals. The absolute concentrations determined by LC-MS/MS and NMR agree well with values reported in literature (13, 33–35). For example, comparison of the concentrations of metabolites detected by NMR with values determined by Wishart et al. (13) and literature values referred to in their paper show that in most cases the values fall within the same concentration range (see Supplementary Material). Furthermore, the concentrations of metabolites determined by more than one method in the validation sample set are also in good agreement.
Although the study of Hu et al. (16) mainly focused on the variation of specific protein abundances within individuals, they concluded that inter-individual variation is far more extensive than intra-individual variation. Yet, in that study, two different stages of Alzheimer's disease were included, which could potentially influence the levels of protein variation between individuals. Here we examined well-defined CSF samples obtained from patients who did not have neurological disorders and we found profound differences in protein abundances between individuals. Characterization of variation of CSF levels of amyloid beta (33) and apolipoprotein E (34) in patients who have Alzheimer's disease have been published, but this is the first attempt to characterize a large number of proteins in CSF of patients who do not have neurological afflictions. Some proteins, such as serotransferrin and fibulin-1, appear to be more constant than others with regards to abundance levels in CSF, as these showed only limited variations in both the sample set of nine non-neurological individuals and the validation set as well as in the experimental sample set. Other proteins, such as contactin-2 and cadherin-13, showed large variations in abundance levels in all three data sets, whereas proteins related to inflammatory response showed the largest variation in the experimental sample set (Fig. 2). This is, in all likelihood, due to the well-known neuroinflammatory component of multiple sclerosis (35–37). The abundance of neuroinflammatory proteins was far higher in the multiple sclerosis samples. This was most obvious in the samples from primary progressive multiple sclerosis patients; therefore, the increased variance of immunoglobulin proteins in this data set is likely due to this group of samples because this type of multiple sclerosis is characterized by continuous inflammation in the CNS, which would result in higher concentrations of inflammation-related proteins.
Additionally, the proteins with high inter-individual variation were not only less abundant proteins, but were also more abundant proteins such as haptoglobin, indicating that these high variations were not caused by measuring at the limit of the detection capabilities of the machines. Proteins specific to the CNS appear to be more variable between individuals than proteins that originate from blood. The majority of the CNS-specific proteins have high inter-individual variations, e.g. neurotrimin and neuroserpin, whereas a large number of the proteins from blood show low inter-individual variation, e.g. serotransferrin and ceruloplasmin. However, there are also blood-specific proteins that display high inter-individual variation, e.g. haptoglobin.
Matching biological samples for age, gender, and protein concentration is an essential step in biomarker research. However, in the validation sample set, we observed that proteins with high inter-individual variance were influenced by gender and age only in a limited way. A good example of this is apolipoprotein E, a protein that is reported to be present at lower concentrations in CSF of patients who have Alzheimer's disease, regardless of age and gender variability (34). Here we found a similar RSD (23–29%) for all variables tested (gender and age-group), suggesting a limited influence of both age and gender on the inter-individual protein abundance of apolipoprotein E.
Although all metabolites detected were present in all individual samples, the concentration of metabolites differed strongly between individuals. For all metabolites, the analytical variation was significantly less than the biological variation. The biological variation was not caused in a significant way by age or gender. The biological variation in this study is about 15% for 70% for the majority of the metabolites which is lower than was observed by Wishart et al. (13), but very similar to the variation reported in the recent study of Crews et al. (38). The main difference between the two studies is the type of CSF sample that was used, i.e. persons without neurological disorders versus patients screened for meningitis. Therefore, it can be concluded that the biological variation for normal controls is, as expected, less than for neurologically diseased individuals.
Lactic acid and glucose, two more abundant metabolites which can be detected by both NMR and GC-MS, show relatively low biological variation, i.e. < 20%, as well as some medium-abundant metabolites such as citric acid and glutamine (Table III). In general, most medium-abundant compounds show an RSD < 30%. Most compounds that show high biological variation are relatively less abundant, e.g. proline, 1- and 3-methylhistidine, and xanthine. Even more interesting is the fact that some less abundant metabolites observed with GC-MS show low biological variation, such as 3-hydroxypropionic acid, arabinose, fucose, glyceric acid, glycerol, glycolic acid, ribose, while others show very high biological variation, e.g. 3-hydroxybutanoic acid, 2-piperidon, inositol, phosphoric acid, sucrose, uric acid, and xylose (see Table III). Both types of metabolites include organic acids and carbohydrates and there is yet no clear biological reason why some metabolites in this study show much higher biological variation than others. The biological variation of the experimental sample set of 42 human CSF samples showed a similar trend as the nine CSF samples, although for a number of metabolites the biological variation was significantly higher (Fig. 3). The latter can of course be attributed to the different diseases of the subjects that might lead to general differences in metabolite levels. Metabolites that showed a significant higher biological variation in the experimental samples could not be directly related to neurological disorders. Further analysis of the data of the experimental samples is necessary to find relations between metabolites and the different types of neurological disorders present in the samples set, including the different stages of multiple sclerosis. More interestingly, for a significant number of metabolites, the biological variation in diseased subjects is similar to that of normal controls, indicating that part of the CSF metabolome is more influenced by person-to-person differences and that the contribution of the diseases is only minor.
In general, the biological variation of metabolites which were detected by more than one analytical method showed good agreement (see Table III). Also in general, amino acids determined by LC-MS/MS and NMR showed less than 10% difference in RSD and the same was true, with some exceptions, for metabolites analyzed by both GC-MS and NMR.
The work discussed above showed that metabolomics (i.e. non-targeted analysis of as many metabolites as possible) of CSF is possible with a combination of analytical techniques currently available. Depending on the amount of CSF available and existing knowledge with respect to the biological question, a combination of non-targeted and targeted analytical methods is preferred to cover different classes of metabolites ranging from more to less abundant metabolites. The metabolites that were detected in CSF seemed to be similar between normal controls, although the concentration of metabolites can differ between individuals up to 60% depending on the specific metabolite.
Conclusion
We conclude that for most proteins the biological variation in two sets of normal control CSF samples, i.e. samples from patients who do not have any significant neurological disorders, appears to be limited, e.g. serotransferrin with RSD 25% (original sample set) and 18% (validation sample set), which includes a technical variation of approximately 20%. The majority of the identified proteins show less than 60% RSD. However, for 28% of the identified proteins, the RSD is greater than 60% and for a limited number of proteins (5% of total) the inter-individual variation is extensive (RSD > 100%, original sample set). The results of extensive inter-individual variation for 5% of the identified proteins is not limited to less abundant proteins. Several more abundant protein shows extensive biological variation, e.g. haptoglobin with RSD of 135% in the original sample set (Table IV).
Table IV. Proteins with high and low biological variation in normal control CSF samples (original sample set).
| Low variation between individuals |
High variation between individuals |
||||
|---|---|---|---|---|---|
| Accession number, protein | Biological variation (%) | Technical variation (%) | Accession number, protein | Biological variation (%) | Technical variation (%) |
| P01011, Alpha-1-antichymotrypsin | 26 | 17 | Q9BYH1, Seizure 6-like protein | 102 | 21 |
| P07339, Cathepsin D | 26 | 17 | Q8TCZ2, Voltage-dependent calcium channel subunit alpha-2/delta-1 | 103 | 19 |
| P23142, Fibulin-1 | 27 | 16 | P54764, Ephrin type-A receptor 4 | 107 | 19 |
| P02774, Vitamin d-binding protein | 29 | 21 | Q02246, Contactin-2 | 124 | 19 |
| P17900, Ganglioside GM2 activator | 29 | 23 | P00738, Haptoglobin | 135 | 28 |
| P02749, Beta-2-glycoprotein | 30 | 18 | Q86YZ3, Hornerin | 148 | 28 |
Metabolomics analysis on the same CSF samples showed that the biological variation for most CSF metabolites is limited, especially compared with the proteomics results. Only a few metabolites were observed with a biological RSD > 70%. However, within the group of metabolites that could be quantified with the different analytical methods, substantial differences in RSDs could be observed between individual metabolites. For example, glucose, a more abundant metabolite, showed an RSD of only 12%, whereas for acetic acid an RSD of 52% was observed.
These results show that for CSF biomarker discovery research it is essential to have an understanding of the biological variation between normal controls because observation of differential abundance between controls and diseased individuals must necessarily be weighed against known inter-individual variations in normal controls. Proteins and metabolites showing high RSD in healthy CSF samples ought to be assessed with caution as a candidate biomarker because a large part of the observed difference will not be caused by the disease under investigation, but to the natural biological variation between individuals.
Supplementary Material
Acknowledgments
This study was performed within the framework of Top Institute Pharma project number D4–102.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CSF
- cerebrospinal fluid
- CNS
- central nervous system
- RSD
- relative standard deviation
- GC-MS
- gas chromatography MS
- NMR
- nuclear magnetic resonance
- SRM
- selective reaction monitoring
- UPLC
- ultra performance liquid chromatography
- PCA
- principal component analysis.
REFERENCES
- 1. Frankfort S. V., Tulner L. R., van Campen J. P., Verbeek M. M., Jansen R. W., Beijnen J. H. (2008) Amyloid beta protein and tau in cerebrospinal fluid and plasma as biomarkers for dementia: a review of recent literature. Curr. Clin. Pharmacol. 3, 123–131 [DOI] [PubMed] [Google Scholar]
- 2. Helbok R., Broessner G., Pfausler B., Schmutzhard E. (2009) Chronic meningitis. J. Neurol. 256, 168–175 [DOI] [PubMed] [Google Scholar]
- 3. Lewczuk P., Hornegger J., Zimmermann R., Otto M., Wiltfang J., Kornhuber J. (2008) Neurochemical dementia diagnostics: assays in CSF and blood. Eur. Arch. Psychiatr. Clin. Neurosci. 258 Suppl 5, 44–49 [DOI] [PubMed] [Google Scholar]
- 4. Romeo M. J., Espina V., Lowenthal M., Espina B. H., Petricoin E. F., 3rd, Liotta L. A. (2005) CSF proteome: a protein repository for potential biomarker identification. Expert Rev. Proteomics. 2, 57–70 [DOI] [PubMed] [Google Scholar]
- 5. Johnston I., Teo C. (2000) Disorders of CSF hydrodynamics. Childs Nerv. Syst. 16, 776–799 [DOI] [PubMed] [Google Scholar]
- 6. Taniguchi M., Okayama Y., Hashimoto Y., Kitaura M., Jimbo D., Wakutani Y., Wada-Isoe K., Nakashima K., Akatsu H., Furukawa K., Arai H., Urakami K. (2008) Sugar chains of cerebrospinal fluid transferrin as a new biological marker of Alzheimer's disease. Dementia Geriatric Cog Dis. 26, 117–122 [DOI] [PubMed] [Google Scholar]
- 7. Moolenaar S., von Engelke U., Hoenderop S., Morava E., van der Graaf M., Wevers R. (2002) Handbook of 1H-NMR spectroscopy in inborn errors of metabolism, Heilbronn, SPS Publications [Google Scholar]
- 8. Baran R., Reindl W., Northen T. R. (2009) Mass spectrometry based metabolomics and enzymatic assays for functional genomics. Curr. Opin. Microbiol 12, 547–552 [DOI] [PubMed] [Google Scholar]
- 9. Lindon J. C., Nicholson J. K., Everett J. R. (1999) NMR spectroscopy of biofluids. In: Webb G. A. ed. Annual Reports on NMR Spectroscopy, London, Academic Press; 1999: pp. 1–88 [Google Scholar]
- 10. Lutz N. W., Viola A., Malikova I., Confort-Gouny S., Audoin B., Ranjeva J. P., Pelletier J., Cozzone P. J. (2007) Inflammatory multiple-sclerosis plaques generate characteristic metabolic profiles in cerebrospinal fluid. PLos One. 2, e595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawashima H., Oguchi M., Ioi H., Amaha M., Yamanaka G., Kashiwagi Y., Takekuma K., Yamazaki Y., Hoshika A., Watanabe Y. (2006) Primary biomarkers in cerebral spinal fluid obtained from patients with influenza-associated encephalopathy analyzed by metabolomics. Int J Neurosci. 116, 927–936 [DOI] [PubMed] [Google Scholar]
- 12. Myint K. T., Aoshima K., Tanaka S., Nakamura T., Oda Y. (2009) Quantitative profiling of polar cationic metabolites in human cerebrospinal fluid by reversed-phase nanoliquid chromatography/mass spectrometry. Analytical Chem. 81, 1121–1129 [DOI] [PubMed] [Google Scholar]
- 13. Wishart D. S., Lewis M. J., Morrissey J. A., Flegel M. D., Jeroncic K., Xiong Y., Cheng D., Eisner R., Gautam B., Tzur D., Sawhney S., Bamforth F., Greiner R., Li L. (2008) The human cerebrospinal fluid metabolome. J. Chromatogr. 871, 164–173 [DOI] [PubMed] [Google Scholar]
- 14. Pan S., Zhu D., Quinn J. F., Peskind E. R., Montine T. J., Lin B., Goodlett D. R., Taylor G., Eng J., Zhang J. (2007) A combined dataset of human cerebrospinal fluid proteins identified by multi-dimensional chromatography and tandem mass spectrometry. Proteomics. 7, 469–473 [DOI] [PubMed] [Google Scholar]
- 15. Pan S., Rush J., Peskind E. R., Galasko D., Chung K., Quinn J., Jankovic J., Leverenz J. B., Zabetian C., Pan C., Wang Y., Oh J. H., Gao J., Zhang J., Montine T., Zhang J. (2008) Application of targeted quantitative proteomics analysis in human cerebrospinal fluid using a liquid chromatography matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometer (LC MALDI TOF/TOF) platform. J Proteome Res. 7, 720–730 [DOI] [PubMed] [Google Scholar]
- 16. Hu Y., Malone J. P., Fagan A. M., Townsend R. R., Holtzman D. M. (2005) Comparative proteomic analysis of intra- and interindividual variation in human cerebrospinal fluid. Mol. Cell. Proteomics. 4, 2000–2009 [DOI] [PubMed] [Google Scholar]
- 17. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 18. Stoop M. P., Dekker L. J., Titulaer M. K., Lamers R. J., Burgers P. C., Sillevis Smitt P. A., van Gool A. J., Luider T. M., Hintzen R. Q. (2009) Quantitative matrix-assisted laser desorption ionization-fourier transform ion cyclotron resonance (MALDI-FT-ICR) peptide profiling and identification of multiple-sclerosis-related proteins. J Proteome Res. 8, 1404–1414 [DOI] [PubMed] [Google Scholar]
- 19. Mize T. H., Amster I. J. (2000) Broad-band ion accumulation with an internal source MALDI-FTICR-MS. Analytical Chem. 72, 5886–5891 [DOI] [PubMed] [Google Scholar]
- 20. Moyer S. C., Budnik B. A., Pittman J. L., Costello C. E., O'Connor P. B. (2003) Attomole peptide analysis by high-pressure matrix-assisted laser desorption/ionization Fourier transform mass spectrometry. Analytical chemistry. 75, 6449–6454 [DOI] [PubMed] [Google Scholar]
- 21. O'Connor P. B., Costello C. E. (2000) Application of multishot acquisition in Fourier transform mass spectrometry. Analytical Chem. 72, 5125–5130 [DOI] [PubMed] [Google Scholar]
- 22. van Kampen J. J., Burgers P. C., de Groot R., Osterhaus A. D., Reedijk M. L., Verschuren E. J., Gruters R. A., Luider T. M. (2008) Quantitative analysis of HIV-1 protease inhibitors in cell lysates using MALDI-FTICR mass spectrometry. Analytical Chem. 80, 3751–3756 [DOI] [PubMed] [Google Scholar]
- 23. Frequin S. T., Barkhof F., Lamers K. J., Hommes O. R., Borm G. F. (1992) CSF myelin basic protein, IgG and IgM levels in 101 MS patients before and after treatment with high-dose intravenous methylprednisolone. Acta Neurol Scandinavica. 86, 291–297 [DOI] [PubMed] [Google Scholar]
- 24. Presslauer S., Milosavljevic D., Brucke T., Bayer P., Hubl W. (2008) Elevated levels of kappa free light chains in CSF support the diagnosis of multiple sclerosis. J Neurol. 255, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 25. Williams K. C., Ulvestad E., Hickey W. F. (1994) Immunology of multiple sclerosis. Clinical Neurosci 2, 229–245 [PubMed] [Google Scholar]
- 26. Koek M. M., Muilwijk B., van der Werf M. J., Hankemeier T. (2006) Microbial metabolomics with gas chromatography/mass spectrometry. Analytical Chem. 78, 1272–1281 [DOI] [PubMed] [Google Scholar]
- 27. Massart L. M., Vandenginste B. G. M., Buydens L. M. C., De Jong S., Lewi P. J., Smeyers-Verbeke J. Handbook of Chemometrics and Qualimetrics, Part A, Amsterdam, Elsevier, 1997, pp. 186–188 [Google Scholar]
- 28. Sweatman B. C., Farrant R. D., Holmes E., Ghauri F. Y., Nicholson J. K., Lindon J. C. (1993) 600 MHz 1H-NMR spectroscopy of human cerebrospinal fluid: effects of sample manipulation and assignment of resonances. J Pharmaceut Biomedic Analysis. 11, 651–664 [DOI] [PubMed] [Google Scholar]
- 29. Wishart D. S., Tzur D., Knox C., Eisner R., Guo A. C., Young N., Cheng D., Jewell K., Arndt D., Sawhney S., Fung C., Nikolai L., Lewis M., Coutouly M. A., Forsythe I., Tang P., Shrivastava S., Jeroncic K., Stothard P., Amegbey G., Block D., Hau D. D., Wagner J., Miniaci J., Clements M., Gebremedhin M., Guo N., Zhang Y., Duggan G. E., Macinnis G. D., Weljie A. M., Dowlatabadi R., Bamforth F., Clive D., Greiner R., Li L., Marrie T., Sykes B. D., Vogel H. J., Querengesser L. (2007) HMDB: the Human Metabolome Database. Nucleic Acids Res. 35, D521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim Y. S., Zhang H., Kim H. Y. (2000) Profiling neurosteroids in cerebrospinal fluids and plasma by gas chromatography/electron capture negative chemical ionization mass spectrometry. Analytical Biochem. 277, 187–195 [DOI] [PubMed] [Google Scholar]
- 31. Obata T., Nagakura T., Maeda H., Yamashita K., Maekawa K. (1999) Simultaneous assay of prostaglandins and thromboxane in the cerebrospinal fluid by gas chromatography-mass spectrometry-selected ion monitoring. J. Chromatogr. B. Biomed. Sci. Appl. 731, 73–81 [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez S., Vio K., Wagner C., Barria M., Navarrete E. H., Ramirez V. D., Perez-Figares J. M., Rodriguez E. M. (1999) Changes in the cerebrospinal-fluid monoamines in rats with an immunoneutralization of the subcommissural organ-Reissner's fiber complex by maternal delivery of antibodies. Exp Brain Res. Experimentelle Hirnforschung. 128, 278–290 [DOI] [PubMed] [Google Scholar]
- 33. Bateman R. J., Wen G., Morris J. C., Holtzman D. M. (2007) Fluctuations of CSF amyloid-beta levels: implications for a diagnostic and therapeutic biomarker. Neurology. 68, 666–669 [DOI] [PubMed] [Google Scholar]
- 34. Lefranc D., Vermersch P., Dallongeville J., Daems-Monpeurt C., Petit H., Delacourte A. (1996) Relevance of the quantification of apolipoprotein E in the cerebrospinal fluid in Alzheimer's disease. Neurosci. Lett. 212, 91–94 [DOI] [PubMed] [Google Scholar]
- 35. Compston A., Coles A. (2002) Multiple sclerosis. Lancet. 359, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 36. Frohman E. M., Eagar T., Monson N., Stuve O., Karandikar N. (2008) Immunologic mechanisms of multiple sclerosis. Neuroimaging Clinics North Am. 18, 577–588, ix [DOI] [PubMed] [Google Scholar]
- 37. Uccelli A., Pedemonte E., Narciso E., Mancardi G. (2003) Biological markers of the inflammatory phase of multiple sclerosis. Neurol. Sci. 24, (suppl 5), S271–274 [DOI] [PubMed] [Google Scholar]
- 38. Crews B., Wikoff W. R., Patti G. J., Woo H. K., Kalisiak E., Heideker J., Siuzdak G. (2009) Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Analytical Chem. 81, 8538–8544 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.