Abstract
Proteasome-mediated proteolysis plays a crucial role in many basic cellular processes. In addition to constitutive proteasomes (CPs), which are found in all eukaryotes, jawed vertebrates also express immunoproteasomes (IPs). Evidence suggests that the key role of IPs may hinge on their impact on the repertoire of peptides associated to major histocompatibility complex (MHC) I molecules. Using a label-free quantitative proteomics approach, we identified 417 peptides presented by MHC I molecules on primary mouse dendritic cells (DCs). By comparing MHC I-associated peptides (MIPs) eluted from primary DCs and thymocytes, we found that the MIP repertoire concealed a cell type-specific signature correlating with cell function. Notably, mass spectrometry analyses of DCs expressing or not IP subunits MECL1 and LMP7 showed that IPs substantially increase the abundance and diversity of MIPs. Bioinformatic analyses provided evidence that proteasomes harboring LMP7 and MECL1 have specific cleavage preferences and recognize unstructured protein regions. Moreover, while differences in MIP repertoire cannot be attributed to potential effects of IPs on gene transcription, IP subunits deficiency altered mRNA levels of a set of genes controlling DC function. Regulated genes segregated in clusters that were enriched in chromosomes 4 and 8. Our peptidomic studies performed on untransfected primary cells provide a detailed account of the MHC I-associated immune self. This work uncovers the dramatic impact of IP subunits MECL1 and LMP7 on the MIP repertoire and their non-redundant influence on expression of immune-related genes.
Proteasomes are the main proteases responsible for protein degradation and the production of major histocompatibility complex I (MHC I)1 ligands (1–4). Proteasomes are much more ancient than MHC molecules. Whereas proteasomes are found in all eukaryotes, the MHC appeared only in jawed vertebrates. Proteasomal degradation regulates many basic cellular processes such as cell cycle and division, differentiation and development, response to stress and extracellular effectors, modulation of cell surface receptors, DNA repair, transcriptional regulation and biogenesis of organelles (5, 6). The 20S proteolytic core of the proteasome is hollow and provides an enclosed cavity open at both ends in which proteins are degraded (7). The eukaryotic 20S particle is composed of 14 different subunits organized in a barrel-shaped complex with the stoichiometry α7β7β7α7. Three subunits of the two inner β-rings (β1, β2, and β5) participate directly in peptide bond cleavage.
While all eukaryotes express the above-described constitutive proteasome (CP), gnathostomes (jawed vertebrates) also express another form of proteasome, the immunoproteasome (IP). In IPs, the three catalytic β-subunits expressed in CPs are replaced by three interferon-γ–inducible homologues (immunosubunits): low molecular weight protein (LMP)-2 (or β1i) for β1, multicatalytic endopeptidase complex-like (MECL)-1 (or β2i) for β2, and LMP7 (or β5i) for β5. In gnathostomes, most cells express only CPs under steady state conditions and harbor IPs when exposed to interferon-γ (8). In contrast, dendritic cells (DCs) constitutively express both CPs and IPs. IPs represents half of the proteasome population in immature DCs, and lipopolysaccharide (LPS)–triggered DC maturation slightly increases the IP:CP ratio (9). Hence, in all circumstances, that is, in the absence or presence of infection, DCs express both CPs and IPs (9). IPs are closely linked to the adaptive immune system, being present in all gnathostomes but absent in invertebrates. Phylogenetic analyses revealed that proteasome immunosubunits evolved faster than their constitutive counterparts (10, 11). This finding indicates a functional differentiation between IPs and CPs. However, the ultimate role of IPs, that is, their ecologically relevant and evolutionarily selected function, remains elusive.
It has been assumed that the key role of IPs may hinge on their impact on the repertoire of peptides associated to MHC I molecules. Indeed, cell surface levels of MHC I molecules are reduced in spleen cells from Lmp7−/− and Lmp7−/− Mecl1−/− mice (12). Furthermore, studies of selected epitopes revealed that some MHC I-associated peptides can be generated only by CPs, some only by IPs, and others by both types of proteasomes (7, 13–18). In vitro proteasome digestion experiments suggest that, compared with CPs, IPs have greater efflux and cleavage rates, and generate more N-extended versions of MHC I epitopes (19, 20). In addition, immunosubunits alter proteasome structure and cleavage site preferences (7, 21). Nevertheless, the aforementioned studies cannot predict the overall impact of IPs on the MHC I peptide (MIP) repertoire in vivo, mainly for three reasons. First, in vitro proteasomal digestion may not reproduce in vivo conditions, where most (MIPs) derive from rapidly degraded proteins that translocate into the endoplasmic reticulum a few seconds after cleavage by proteasomes (22, 23). Second, MIP presentation is orchestrated by several steps downstream of proteasomal digestion so that only a small fraction of peptides generated by proteasomes are presented by MHC I molecules (24–26). Finally, previous studies did not take into account potential differences in transcription regulation by CPs and IPs. Whereas CPs clearly regulate transcriptional activation (27–30), the potential impact of IPs on transcription remains unexplored. Conceivably, IPs and CPs might differentially regulate MHC I presentation of a given peptide not only by affecting degradation of the peptide's source protein but also by modulating transcription of the gene encoding that peptide. In this perspective, the goal of our work was to obtain a direct and global evaluation of the impact of IPs on the repertoire of MIPs. To this end, we used a recently described label-free quantitative approach to analyze the MIP repertoire of DCs expressing or not expressing IP subunits MECL1 and LMP7 (31). Also, we analyzed the gene expression profile of those two DC populations using microarrays.
EXPERIMENTAL PROCEDURES
Mice
Mouse cells were prepared on a C57BL/6 background and maintained in a specific pathogen free environment. Wild-type (WT) and β2-microglobulin (β2m)−/− 1 mice were obtained from The Jackson Laboratory. Lmp7−/−Mecl1−/− (dKO) mice were generously provided by Dr T.A. Griffin from the Medicine College of the University of Cincinnati.
Preparation of Bone Marrow-Derived DCs
DCs were generated from WT, double knockout (dKO) and β2m−/− mice bone marrow as previously described (32, 33). Bone marrow cells were extracted from tibia and femur bones of 7- to 9- week-old male mice and plated in 10-cm non tissue culture-treated Petri dishes (BD Bioscience, Mississauga, ON, Canada) at 3 × 106 cells per plate, in 10 mL of complete RPMI 1640 medium (0.048 mmol/L β-mercaptoethanol, 2 mmol/L l-glutamine, 10% fetal bovine serum, 2 mmol/L penicillin-streptavidin) supplemented with 10 ng/mL granulocyte macrophage–colony stimulating factor (GM-CSF, Invitrogen, Burlington, ON, Canada). At days 3 and 6 of culture, 10 mL of cRPMI 1640 medium with 10 ng/mL GM-CSF and 5 mL of cRPMI 1640 medium with 20 ng/mL GM-CSF, respectively, were added. To further induce DC maturation, 1 μg/mL lipopolysaccharide (LPS, Sigma-Aldrich) was added 24 hours before harvesting the non-adherent cells at day 8.
Flow Cytometry
Antibodies used were purchased from BD Bioscience unless stated otherwise. For DC phenotyping, cells were harvested and stained with PECy7-conjugated or FITC-conjugated anti-CD11c (HL3), APC-conjugated anti-CD86 (PO3; BioLegend, San Diego, CA), PE-conjugated anti-IAb (AF6–120.1), APCCy7-conjugated anti-CD11b (AF6–120.1) and PECy5-conjugated anti-CD8α (53–6.7). For MHC I labeling, cells were stained with pure anti-H2Db (B22–249.R1; Cedarlane, Hornby, ON, Canada), pure anti-H2Kb (Y3; ATCC, Manassas, VA), and biotin-conjugated anti-Qa2 (1–1-2) and anti-Qa1b (6A8.6F10.1A6) followed with APC-conjugated streptavidin. H2Kb and H2Db antibodies were coupled with Alexa 647 fluorochrome using the Alexa Fluor 647 Monoclonal Antibody Labeling Kit (Molecular Probes from Invitrogen, Eugene, OR). Cells were incubated with mouse Fc Block CD16/CD32 (BD Bioscience, 2.4G2) before staining. Dead cells were excluded based on propidium iodide staining. Analyses were performed on a BD LSR II flow cytometer using FACSDiva (BD Bioscience) and FCS v.3.0 (De Novo Software, Los Angeles, CA) software.
Immunoblot Analyses
Mature DCs were harvested and lysed in RIPA buffer (50 mmol/L Tris-HCl pH 7.4, 1% Nonidet P-40, 0.25% Na-deoxycholate, 150 mmol/L NaCl, 1 mmol/L EDTA) containing a protease inhibitor mixture (Complete; Roche), 1 mmol/L Na3VO4 pH 9 and 5 mmol/L NaF. Samples were resolved by SDS-PAGE and immunoblotted with the following antibodies: anti-β1, anti-β5, anti-LMP2, anti-LMP7 and anti-α5 from Abcam, anti-β2 and anti-MECL1 from Biomol International (Enzo Life Sciences, Plymouth Meeting, PA), anti-calnexin and anti-β-actin (AC-15) from Sigma-Aldrich and anti-H2Kb/H2Db (2G5) from Thermo Scientific (Rockford, IL). After incubation with anti-mouse (BD Bioscience) or anti-rabbit (Cell Signaling, Danvers, MA) horseradish peroxidase–conjugated secondary antibodies, chemiluminescent signal was detected using the GE Healthcare Detection Kit and a LAS-3000 imaging system (Fujifilm, GE Healthcare, Baie d'Urfe, QC, Canada). Band intensities were quantified using the Multi Gauge V3–0 (Fujifilm) software.
Peptide Extraction and MS Analyses
Three biological replicates (5 × 108 DCs per replicate) were prepared from a total of 28 WT mice, 28 dKO mice and 47 β2m−/− mice. MIPs were analyzed as previously reported (31) with minor modifications. MIPs obtained after acid elution (34) were separated using an off-line 1100 series binary LC system (Agilent Technologies, Mississauga, ON, Canada) to remove contaminating species. Peptides were loaded on a homemade SCX column (0.3 mm internal diameter x 45 mm length) packed with strong cation exchange (SCX) bulk material (Polysulfoethyl A™, PolyLC). Peptides were fractionated with a gradient of 0–25% B after 33 minutes, 25–60% B after 35 minutes (Solvent A = 5 mmol/L ammonium formate, 15% acetonitrile, pH3; Solvent B = 2 mol/L ammonium formate, 15% acetonitrile, pH3). MIPs were collected in five consecutive fractions and brought to dryness using a speedvac. MIP fractions were resuspended in 2% aqueous acetonitrile (0.2% formic acid) and analyzed by nanoLC-MS/MS on a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) (31). Full mass spectra were acquired with the Orbitrap analyzer operated at a resolving power of 60,000 (at m/z 400) and collision-activated dissociation tandem mass spectra were acquired in data-dependent mode with the quadrupole linear ion trap analyzer. Mass calibration used either an internal lock mass [protonated (Si(CH3)2O))6; m/z 445.12057] or external calibration using Calmix (caffeine, MRFA and ultramark) and typically provided mass accuracy within 5 ppm for all nanoLC-MS/MS experiments.
MS/MS Sequencing and Peptide Clustering
Data were analyzed using Xcalibur software and peak lists were generated using Mascot distiller (version 2.1.1, www.matrixscience.com). Database searches were performed against an International Protein Index mouse database (version 3.23 containing 51,536 sequences and 24,497,860 residues) using Mascot (version 2.2, www.matrixscience.com) with a mass precursor tolerance of ± 0.05 Da and a fragment tolerance of ± 0.5 Da. Searches were performed without enzyme specificity and a variable modification of oxidized Met. All search results were filtered using an MHC motif filter based on the predicted mouse MHC I allele motifs. Raw data files were converted to peptide maps comprising m/z values, charge state, retention time and intensity for all detected ions above a threshold of 15,000 counts using in-house software (Mass Sense) (31). Peptide maps were aligned and clustered together to profile the abundance of Mascot identified peptides using hierarchical clustering with criteria based on m/z and time tolerance (±0.01 m/z and ±1.5 min). This resulted in a list of non-redundant peptide clusters for all replicates of all samples to be compared. MIPs were further inspected for mass accuracy and MS/MS spectra were validated manually. The Sidekick resource (http://www.bioinfo.iric.ca/sidekick/Main) was used to identify MIP source proteins. The InnateDB resource (35) was used to identify significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated to peptide source genes from DCs and thymocytes. The list of MIPs reported in the present work has been provided to The Immune Epitope Database and Analysis Resource (http://beta.immuneepitope.org/) (36).
Cytotoxicity Assays
In vitro carboxyfluorescein succinimidyl ester (CFSE)-based cytotoxicity assays were performed as previously described with minor modifications (31). CFSE-based assays are more sensitive than classic 51Cr-release cytotoxicity assays (37–39). Briefly, 106 WT and dKO DCs were injected intravenously into mice from both genotypes on days 0 and 7. On day 14, splenocytes from the four immunized mice (WT mice injected with WT or dKO DCs, dKO mice injected with WT or dKO DCs) were used as effector cells in cytotoxicity assays (39). Target cells were concanavalin A treated WT and dKO splenocytes. The percentage of specific lysis was measured as follows: [(number of remaining CFSE+ cells after incubation of target cells alone − number of remaining CFSE+ cells after incubation with effector cells)/number of CFSE+ cells after incubation of target cells alone] × 100.
Bioinformatic Analysis of Cleavage Motifs
All 417 peptides extracted from DCs were used in studies of amino acid usage in MIPs. In analyses of flanking regions, we eliminated peptides that can originate from multiple source proteins with different N- or C-terminal flanking sequences. Peptides used for further analyses of flanking regions (376 for upstream regions, 369 for downstream regions) were ranked according to their WT/dKO fold difference in abundance as determined by MS analyses. We next generated a Euclidean distance matrix to compare amino acid usage at each position. We thereby compared amino acid usage by MIPs located at the left versus the right of each ranked peptide. A bootstrap procedure (100,000 iterations) was performed to evaluate whether the distance measured was significant (p < 0.05 was considered significant). The analysis was performed for each position of the MHC peptides as well as for 10 residues upstream of the peptide N terminus and downstream of the C terminus. For positions that gave a p value < 0.001, we used the Kolmogorov-Smirnov statistical test to determine which specific amino acids were over- or under-represented in WT as opposed to dKO DCs. The R software was used to visualize amino acid distributions (http://www.R-project.org), and the program SEG (http://mendel.imp.ac.at/METHODS/seg.server.html) to determine unstructured regions in source proteins (window size = 12, low complexity = 2.5, high complexity = 2.8) (40). The number of MIPs from unstructured regions in presence or in absence of IP was measured using a Chi-squared test (with p value < 0.05).
Microarrays and Genomic Analyses
Total RNA was extracted from WT and dKO DCs with TRIzol RNA reagent (Invitrogen) as instructed by the manufacturer. Samples were purified using DNase (Qiagen, Mississauga, ON, Canada) and the RNeasy Mini kit (Qiagen), and the overall quality was analyzed with the 2100 Bioanalyzer (Agilent Technologies). Purified RNA (10 μg/sample) was hybridized on MM8 385K NimbleGen chips at the Genomics core facility of the Institute for Research for Immunology and Cancer according to the manufacturer's instruction. Arrays were scanned using a GenePix4000B scanner (Axon Instruments, Molecular Devices, Sunnyvale, CA) at 5 μm resolution. Data were extracted and normalized using the NimbleScan 2.4 extraction software (NimbleGen Systems, Madison, WI). Further microarray analyses were performed using GeneSpring GX 7.3.1. The complete microarray datasets have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession number E-TABM-750. Two-sided Student t test was used to compare transcript abundance in WT versus dKO DCs. Spearman's rank correlation was used to evaluate the relation between MIP abundance and source mRNA expression. The Gene set organization/visualization module of the Web-Based Gene Set Analysis Toolkit (WebGestalt) (41) was used to represent the chromosomal localization of the differentially expressed genes, and the gene enrichment on specific chromosomes was measured using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resource (42).
RESULTS
Experimental Design of Peptidomic Studies
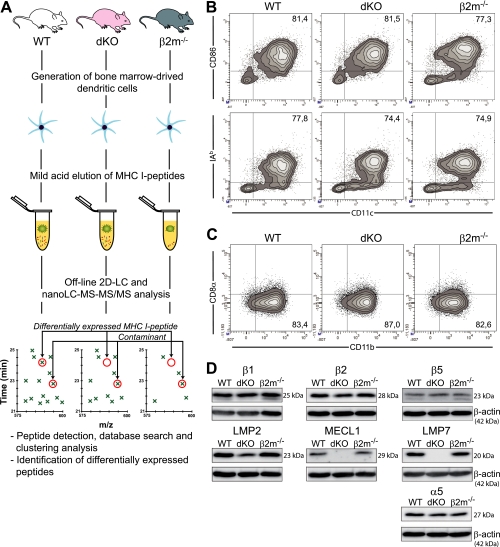
To evaluate the impact of IPs on the MIP repertoire, we elected to study DCs because they are quintessential antigen-presenting cells and constitutively express IPs. Using a recently described label-free quantitative proteomics method (31), we analyzed MIPs eluted from mature WT and double knockout (dKO) DCs (Fig. 1A). WT DCs express both CPs and IPs (alike mature DCs under physiological in vivo conditions (9)) whereas dKO DCs do not express the IP-subunits LMP7 and MECL1 (Fig. 1D). IP subunits are cooperatively incorporated into proteasomes, thereby curtailing the formation of mixed proteasomes containing IP and CP subunits (43). In line with this result, we noted a 50% decrease in the amount of LMP2 protein in dKO cells, suggesting that LMP2 is unstable in the absence of the two other immunosubunits (Fig. 1D). As a negative control we analyzed DCs derived from β2m-deficient mice. Since β2m is essential for formation of stable peptide–MHC I complexes, cells lacking β2m are MHC I–deficient. DCs generated from WT, dKO, and β2m-deficient mice shared a mature (CD11c+CD86+IAb+) myeloid (CD8α−CD11b+) phenotype (Fig. 1B,C).
Fig. 1.
Design of peptidomic studies and characterization of DC populations. (A) Experimental design for isolation and identification of DC MIPs. DCs were generated from the bone marrow of WT, Lmp7−/−Mecl1−/− (dKO) and β2m−/− mice. MIPs were eluted in a mild acid buffer, separated by off-line HPLC, and collected fractions were analyzed by nanoLC-MS-MS/MS. Heat maps displaying m/z, retention time and abundance were generated and analysis of β2m−/− negative controls allowed us to discriminate MIPs from contaminants. Three biological replicates were analyzed for each group. (B) Flow cytometry analysis of bone marrow derived DCs. (C) Expression of CD11b and CD8α on DCs (gated on CD11c+ cells). (D) Immunoblot analysis of proteasomes subunits in total DC lysates. Data are representative of three (C) or four (B, D) independent experiments.
Peptides eluted from DCs were fractionated by off-line LC using a SCX column, then analyzed by nano-LC combined with tandem MS (nanoLC-MS/MS). Peptide maps were generated from each analysis to define the coordinates (m/z, retention time, ion abundance) of identified ions. The corresponding peptide maps were then clustered across conditions and replicate analyses to identify unique peptide ions and profile their changes in abundance. Subtraction of “contaminant peptides” eluted from β2m-deficient cells allowed specific identification of genuine MIPs (Fig. 1A) (31). MS/MS spectra were manually verified for all MIPs. The following softwares were used to associate peptide sequences with specific MHC I allelic products: smm, SYFPEITHI (H2Db and H2Kb), and Rankpep (Qa2) (31). This resulted in the identification of 417 unique MIPs (derived from 389 source proteins) in WT DCs (supplemental Table S1, Fig. S2).
The MIP Repertoire of DCs Conceals a Unique Signature
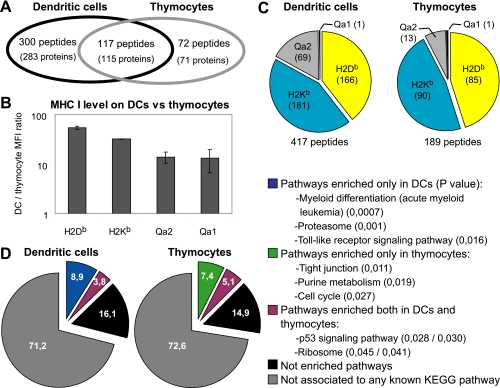
We reported previously that MIPs eluted from thymocytes derived preferentially from transcripts whose abundance was higher in the thymus than in other tissues (31). This suggested that the MIP repertoire might conceal a cell type-specific signature. To directly test this concept, we compared the MIP repertoire of DCs (reported herein) to that of thymocytes (31) derived from the same strain of mice (WT, C57BL/6). We identified more peptides in DCs than in thymocytes (417 versus 189; Fig. 2A). This discrepancy can be attributed, at least in part, to the fact that MHC I molecules are much more abundant on DCs than thymocytes (Fig. 2B). In both cell types we found more peptides associated to MHC Ia (H2Kb, H2Db) than to MHC Ib (Qa1, Qa2) allelic products (Fig. 2C). Of note, the proportion of peptides with a Qa2-binding motif was greater in DCs than in thymocytes (p < 0.01; chi-square test). However, the key finding was that 72 of 189 peptides eluted from thymocytes were not detected in DC eluates, although we recovered less peptides from thymocytes (189) than from DCs (417) (Fig. 2A). This means that about 60% of MIPs present at the surface of thymocytes are also present on DCs whereas 40% are thymocyte-specific. Of 417 peptides recovered from DCs, 117 were shared with thymocytes, whereas 300 were DC-specific.
Fig. 2.
The MIP repertoire conceals a cell–type specific signature. (A) Venn diagram representation of the relation between MIPs (and their source proteins) eluted from C57BL/6 thymocytes and DCs. (B) Cell surface expression of MHC I allelic products was evaluated by flow cytometry. Histogram shows the DC/thymocyte mean fluorescence intensity ratio for H2Db (p = 6 × 10−5; Student t test), H2Kb (p = 6 × 10−7), Qa1 (p = 7 × 10−6) and Qa2 (p = 3 × 10−2) (mean ± S.D. of triplicate experiments). (C) Proportion of peptides associated to different MHC I allelic products in DCs and thymocytes. (D) Pie charts represent the relations between peptide source genes (389 for DCs, 186 for thymocytes) and KEGG pathways. Examples of pathways enriched in the gene datasets are depicted (with p value for enrichment in parentheses).
To further evaluate whether the MIP repertoire might reflect cell type-specific intracellular signaling events, we used the InnateDB resource (35) to analyze pathways catalogued in the KEGG database. We specifically evaluated whether specific KEGG pathways were overrepresented in the group of genes encoding peptides eluted from DCs and/or thymocytes (Fig. 2D). In each dataset (DCs and thymocytes), about 28% of peptide source genes were linked to specific KEGG pathways. Notably, 45% of pathway-connected genes (12.5% of the whole dataset) were associated with pathways significantly overrepresented in the DC and/or thymocyte gene dataset. Many MIP source genes were connected to pathways overrepresented in both DCs and thymocytes (e.g., p53 signaling and ribosome biogenesis). Of special interest, peptide source genes belonging to specific pathways were enriched uniquely in DCs or thymocytes. Many of these pathways reflected the function and differentiation of DCs and thymocytes. For instance, the MIP repertoire of DCs was enriched in peptides whose source genes are involved in myeloid differentiation, proteasome function and Toll-like receptor signaling. Besides, peptide source genes involved in tight junction, purine metabolism and cell cycle were overrepresented in the thymocyte MIP repertoire. For DC and thymocyte gene datasets, a complete list of overrepresented pathways and their constituent genes can be found in supplemental Table S2. Together, these results show that the MIP repertoire conceals a cell type-specific signature that reflects singular functional properties.
IPs Increase the Abundance and Diversity of MIPs at the Surface of DCs
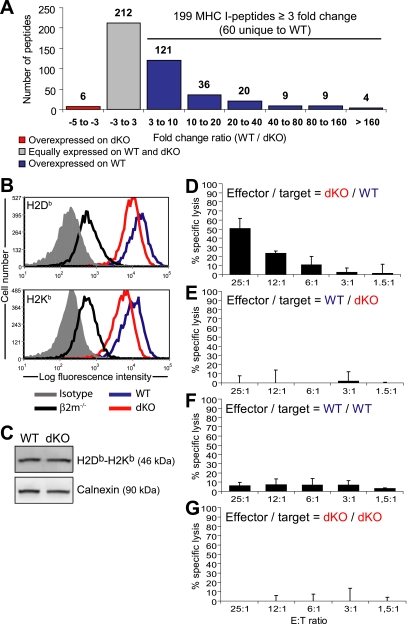
To evaluate the impact of IPs on the MIP repertoire, we compared MIPs eluted from WT and dKO DCs by MS analysis as depicted in Figure 1A. In accordance with previous studies (31), we found that 95% of peptide ions showed a variation of less than ± 3-fold in abundance across biological replicates (n = 3). Therefore, we considered that peptides were differentially presented by MHC I molecules when the fold difference in abundance between WT and dKO DCs was greater than 3. Of the 417 peptides eluted from WT DCs, 212 were expressed at similar levels (within 3-fold) in dKO DCs (Fig. 3A). Remarkably, 199 peptides were overexpressed in WT relative to dKO DCs. Among those 199 peptides, 60 were detected exclusively in WT DCs. Only six peptides were slightly overexpressed (3- to 5-fold) in dKO relative to WT DCs and none were unique to dKO DCs. Peptides with the largest fold differences in abundance are listed in Table I, and the full list of peptides is available in supplemental Table S1. In accordance with our data on MIP abundance, flow cytometry analyses revealed that, as previously shown using WT versus dKO spleen lymphocytes (12), expression of cell surface H2Db and H2Kb was higher by approximately 2.1-fold on WT than dKO DCs (Fig. 3B). However, protein immunoblot analyses on whole cell lysates showed that total cellular amounts of H2Db and H2Kb heavy chains were similar in both types of DCs (Fig. 3C), suggesting that the lower level of surface expressed MHC I on dKO DCs was caused by a limited peptide supply and not an altered level of MHC I molecules available.
Fig. 3.
IPs increase the abundance and diversity of MIPs. (A) Histogram representing the relative abundance in WT and dKO DCs of the 417 peptides identified by MS analyses. (B) WT DCs expressed higher cell surface levels of H2Db (p = 0.04, Student t test) and H2Kb (p = 0.02) than dKO DCs. (C) Immunoblot analysis of H2Db and H2Kb heavy chains in whole DC lysates. Calnexin was used as a loading control. Data are representative of three independent experiments. (D-G) Cytotoxic activity of splenocytes from dKO mice primed against WT DCs (D), from WT mice primed against dKO DCs (E), from WT mice primed against WT DCs (F) and from dKO mice primed against dKO DCs (G) (mean and S.D. for three mice per group).
Table I. Top most overexpressed MIPs in WT relative to dKO DCs.
| No. | m/z | Charge | Mascot score | MHC I allele | Peptide sequence | Gene ID | Gene symbol | Gene Name | WT/dKO ratio |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 520.22 | 2 | 66 | H2Db | GGVVNMYHM | 16913 | Psmb8 | Proteasome subunit, beta type 8 (LMP7) | ≥1181,3 |
| 2 | 460.25 | 2 | 40 | H2Kb | ATRSFPQL | 11750 | Anxa7 | Annexin A7 | ≥380,8 |
| 3 | 624.83 | 2 | 44 | H2Db | HAIRNSFQYL | 170749 | Mtmr4 | Myotubularin related protein 4 | ≥207,3 |
| 4 | 472.28 | 2 | 55 | H2Kb | TIIIFHSL | 14050 | Eya3 | Eyes absent 3 homolog (Dm) | 160,5 |
| 5 | 497.27 | 2 | 43 | H2Kb | VVYIYHSL | 67014 | Mina | Myc induced nuclear antigen | ≥159,3 |
| 6 | 467.28 | 2 | 47 | H2Kb | SALRFLNL | 71728 | Stk11ip | Ser/Thr kinase 11 interacting protein | 120,8 |
| 7 | 479.28 | 2 | 39 | H2Kb | KNVLFSHL | 19084 | Prkar1a | Protein kinase, cAMP dependent regulatory, type I, alpha | ≥109,8 |
| 8 | 524.27 | 2 | 65 | H2Db | RQILNADAM | 66185 | 1110037F02Rik | RIKEN cDNA 1110037F02 gene | ≥96,8 |
| 9 | 496.78 | 2 | 42 | H2Db | SGLLNMTKI | 216445 | Arhgap9 | Rho GTPase activating protein 9 | ≥93,9 |
| 10 | 525.26 | 2 | 29 | H2Kb | HVYYFAHL | 74558 | Gvin1 | GTPase, very large interferon inducible 1 | ≥93,4 |
| 11 | 510.80 | 2 | 56 | H2Kb | VITNFSARI | 68505 | 1110014N23Rik | RIKEN cDNA 1110014N23 gene | ≥83,3 |
| 12 | 507.26 | 2 | 49 | H2Kb | STLIYRNM | 75660 | Lin37 | Lin-37 homolog (C. elegans) | ≥82,1 |
| 13 | 597.28 | 2 | 27 | H2Db | SAIHNFYDNI | 68652 | Map3k7ip2 | MAPKKK 7 interacting protein 2 | ≥80,5 |
| 14 | 523.75 | 2 | 34 | H2Db | SQVYNDAHI | 66923 | Pbrm1 | Polybromo 1 | ≥76,9 |
| 15 | 537.27 | 2 | 40 | H2Kb | LVYQFKEM | 56501 | Elf4 | E74-like factor 4 (ets domain transcription factor) | ≥76,2 |
| 16 | 461.76 | 2 | 62 | H2Db | GAVQNIAHL | 74125 | Armc8 | Armadillo repeat containing 8 | 68,5 |
| 17 | 496.28 | 2 | 32 | H2Kb | ISLEFRNL | 14137 | Fdft1 | Farnesyl diphosphate farnesyl transferase 1 | 68,1 |
| 18 | 506.30 | 2 | 43 | H2Kb | KSYLFQLL | 12566 | Cdk2 | Cyclin-dependent kinase 2 | ≥66,4 |
| 19 | 447.74 | 2 | 53 | H2Db | GGIQNVGHI | 23918 | Impdh2 | Inosine 5′-phosphate dehydrogenase 2 | 54,8 |
| 20 | 493.24 | 2 | 41 | H2Kb | SNYRFEGL | 319955 | Ercc6 | Excision repair cross-complementing rodent repair deficiency, complementation group 6 | ≥46,4 |
| 21 | 479.75 | 2 | 37 | Qa2 | GLMTTVHAI | 640374 | LOC640374 | Similar to GAPDH | 42,0 |
| 22 | 645.34 | 2 | 33 | H2Db | AQYGNILKHVM | 69482 | Nup35 | Nucleoporin 35 | ≥40,3 |
| 23 | 486.78 | 2 | 48 | H2Db | KAPLNIAVM | 667214 | RP23–269N23.3 | Similar to immunity-related GTPase family, cinema 1 | 39,8 |
| 24 | 510.81 | 2 | 54 | H2Db | YGLLNVTKI | 75415 | Arhgap12 | Rho GTPase activating protein 12 | ≥37,6 |
| 25 | 501.27 | 2 | 42 | H2Db | SALANYIHL | 58245 | Gpr180 | G protein-coupled receptor 180 | ≥37,4 |
MIPs that were not detected on dKO cells were attributed the intensity value 15,000, which represents the threshold for limit of detection. Ratios with the symbol ≥ indicate peptides for which the exact fold change could not be measured because they were detected only in WT DCs. See supplemental Table S1 for a complete list of MIPs detected on DCs.
After immunization with WT DCs, dKO mice generated WT-specific cytotoxic T cells (Fig. 3D,G). However, WT mice did not generate cytotoxic effectors against dKO cells (Fig. 3E,F). That unidirectional immunogenicity supports our peptidomic analyses showing that numerous peptides were uniquely detected on WT DCs, whereas no peptides were found only on dKO DCs (Fig. 3A). Furthermore, it is consistent with the previous demonstration that WT splenocytes were immunogenic for Lmp7−/− mice, but not vice versa (44). However, dKO DCs may not be optimal antigen-presenting cells. Indeed, when dKO and WT DCs were coated with exogenous SIINFEKL and injected in mice bearing Kb/SIINFEKL-specific T cells, dKO DCs proved to be immunogenic but less so than WT DCs (supplemental Fig. S1). Collectively, these results show that the presence of IPs has a major impact on the global MIP repertoire, by increasing both the abundance and the diversity of MIPs.
IPs Have Specific Cleavage Preferences
Proteasomal cleavage generates the final C terminus of MIPs whereas their N terminus can be further trimmed by aminopeptidases in the cytosol and the endoplasmic reticulum (22, 25). Proteasomal cleavage can be influenced by approximately five to seven residues flanking the cleavage site on either side (44, 45). To determine whether the MIP repertoire generated in the presence or absence of IPs might reveal discrete cleavage preferences, we analyzed the amino acid composition of MIPs and their flanking residues. The 417 peptides extracted from DCs were used in studies of amino acid frequencies in MIPs. In analyses of flanking regions, we eliminated peptides that can originate from multiple source proteins with different N- or C-terminal flanking sequences. Remaining peptides were ranked according to their WT/dKO fold difference in abundance as determined by MS analyses. Peptides with high WT/dKO ratios are IP-dependent. We next generated a Euclidean distance matrix to compare amino acid usage at each position. Each ranked peptide was used consecutively as a reference. We thereby compared amino acid usage by MIPs having higher versus lower WT/dKO ratios than the reference peptide. A bootstrap procedure (100,000 iterations) was performed to evaluate whether the distance measured was significant. The analysis was performed for each position of the MHC peptides as well as for 10 residues upstream of the peptide N terminus and downstream of the C terminus.
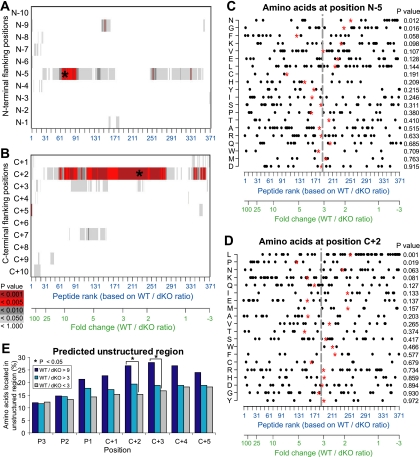
We detected no bias in amino acid frequencies at various positions of the MIPs per se (data not shown). However, we found highly significant deviations of amino acid frequencies at two peptide flanking positions: N-5 upstream of the N terminus and C+2 downstream of the C terminus of MIPs (Fig. 4A,B; p < 0.001). We performed a detailed analysis of amino acid frequencies at positions N-5 and C+2 using the Kolmogorov-Smirnov test. At position N-5, peptides overexpressed in WT cells showed decreased frequencies of glycine and asparagine residues (Fig. 4C). It is not clear how cytosolic and endoplasmic reticulum aminopeptidases processing MIP precursors choose their substrates (46, 47). Therefore, further work is needed to decipher the significance of the amino acid bias at position N-5. For peptides that have high WT/dKO ratios, deviation at position C+2 was particularly dramatic and was characterized by an increased usage of proline and polar residues [lysine and glutamine], with a decreased frequencies of asparagine and hydrophobic residues [leucine and isoleucine] (Fig. 4D). Asparagine and hydrophobic residues are enriched in α-helices and β-sheets, whereas proline and polar residues are enriched in unstructured protein regions, which represent 20–30% of the mammalian proteome (48, 49). The program SEG, which computes sequence complexity, has been used successfully to predict unstructured regions (i.e., lacking secondary and tertiary structures) (40). Using SEG, we found that amino acids C-terminal of IP-dependent peptides (high WT/dKO ratio) derived more frequently from unstructured protein regions than amino acids C-terminal of IP-independent peptides (Fig. 4E). The IP bias toward unstructured protein domains was specific for amino acid residues next to the MIP C terminus (e.g., C+2), and was not detected for the MIP themselves nor for residues upstream of their N terminus (data not shown). We conclude that IPs display specific cleavage preferences and propose that the presence of IPs leads to enhanced MHC I presentation of peptide sequences adjoining the unstructured proteome.
Fig. 4.
IPs have specific cleavage preferences. (A, B) Screen of N terminus (A) and C terminus (B) MIP flanking regions for identification of amino acid preferences in presence or in absence of IPs. Peptides were ranked according to their fold change (WT/dKO intensity ratio). A bootstrap procedure was used to detect significant differences between amino acids flanking peptides overexpressed or not in WT DCs. Color code shows p values computed for each amino acid position, with the strongest p values indicated by a star. (C, D) Distribution of amino acid frequencies at positions N-5 and C+2. Each dot corresponds to one MIP. Red stars represent the mean rank of peptides bearing a specific amino acid residue, and can be compared with the mean rank of the entire peptide dataset (gray line). Relative enrichment of all 20 amino acids as a function of peptides' WT/dKO ratio was estimated with the Kolmogorov-Smirnov statistical test. (E) Amino acid residues located in protein regions predicted to be unstructured by the SEG algorithm. Peptides were classified in three groups (colored bars) as a function of their abundance in WT and dKO DCs (WT/dKO). Proportions were compared using the chi-square test (p = 0.029 and 0.037 for positions C+2 and C+3, respectively).
IPs Have a Non-Redundant Impact on the Transcriptome of DCs
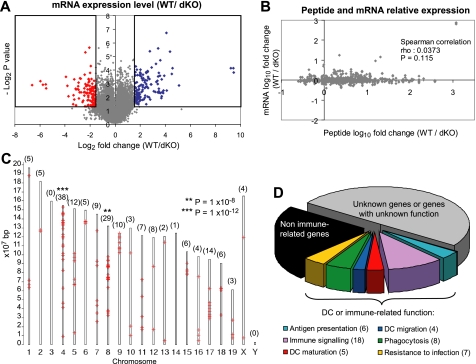
Aside from their role in protein degradation, CPs also regulate transcription (27–30). Whether IPs may regulate transcription differently than CPs is unknown. Thus, we could not assume a priori that peptide overexpression in IP-expressing DCs was due solely to enhanced degradation of peptide source proteins by IPs. Theoretically, IPs might also mold the MIP repertoire by differential regulation of peptide source genes. To test this assumption, we compared the transcriptome of WT and dKO DCs using NimbleGen MM8 385K microarrays. We found that 226 transcripts, representing 171 genes and corresponding to 0.5% of the transcriptome, were differentially expressed between WT and dKO DCs (Fig. 5A; Supplemental Tables S3 and S4). There was no correlation between transcript and MIP abundance (Fig. 5B). We therefore conclude that differential expression of MIPs in WT versus dKO DCs cannot be ascribed to differential transcription of peptide source genes. Nevertheless, a selected set of transcripts was differentially expressed in the presence or absence of IPs. The loci encoding those transcripts were not randomly distributed in the genome. Somewhat unexpectedly, they were clustered in discrete regions located primarily in chromosomes 4, 8, 9, and 17, and practically absent in chromosomes 3, 10, 13, 14, 16, 19, and X. The gene clusters were particularly enriched in chromosomes 4 and 8 (p = 10−12 and 10−8, respectively; Fig. 5C).
Fig. 5.
IPs imprint on the transcriptome. (A) Volcano plot representation of the relative expression of 42,569 transcripts in WT and in dKO DCs. Boxes show transcripts significantly overexpressed in WT (blue) and in dKO (red) DCs across three replicates (p < 0.05; two-sided t test). (B) Spearman correlation between the relative cell surface expression of MIPs (WT/dKO intensity ratio) and the expression of their source transcripts. (C) Chromosomal distribution of differentially expressed genes (red crosses). The number of genes localized on each chromosome is shown in parentheses. Significance of enrichment on chromosomes was measured with a modified Fisher's exact test using DAVID Bioinformatics Resources. (D) Functional classification of differentially expressed genes obtained by a systematic literature review. The number of genes included in each category is shown in parentheses.
To evaluate whether differentially expressed genes might be relevant to DC function, we focused on transcripts for which functional annotation data was available (50% of differentially expressed transcripts; Fig. 5D). Specifically, we excluded transcripts for which no biological data were available, or when the sole available evidence was inferred from electronic annotation that was not assigned by a curator (ND and IEA GO codes) (Fig. 5D). Remarkably, 56% of functionally annotated genes had a demonstrated or putative role in DC function or immune signaling (Fig. 5D). Those 48 genes were aggregated into six functional categories: resistance to infection, antigen presentation, phagocytosis, immune signaling, DC maturation, and DC migration. A complete annotated list of genes belonging to these categories is available in supplemental Table S3. We conclude that IPs have a non-redundant impact on expression of a selected of set of genes that regulate different aspects of DC function.
DISCUSSION
By using a label-free quantitative proteomics approach, we gained valuable insights into the impact of IPs on the molecular composition of the immune self. Our peptidomic studies allowed us to generate a most comprehensive biochemical definition of the MIP repertoire. Since we achieved this using untransfected primary DCs, our data provide a broad and faithful representation of the MHC I–restricted immune self. The present work yielded three major observations. First, the MIP repertoire conceals a cell type-specific signature. Though the MIP repertoire of DCs and thymocytes partially overlap, no less than 40% of their MIPs were cell type–specific. The large proportion of cell type–specific MIPs observed herein argues against the notion that MIPs derive primarily from ubiquitously expressed proteins (50, 51). MIPs unique to DCs or thymocytes reflected cell function and differentiation. The DCs that we studied were antigen-processing cells with a myeloid phenotype whose maturation was induced by LPS (a toll-like receptor–4 ligand). Quite remarkably, the MIP repertoire of those DCs was enriched in peptides encoded by genes regulating proteasome function, myeloid differentiation and toll-like receptor signaling. Thymocytes have a high proliferation index and undergo a myriad of sequential interactions with subpopulations of stromal cells during their three-week journey in the thymus. Their MIP source genes were biased toward cell cycle regulation, purine metabolism and tight junction formation. The notion that a substantial proportion of MIPs are cell type–specific leads us to infer that, at the organismal level, the composition of the MHC I–restricted immune self is highly complex. The cell types studied herein (DCs and thymocytes) are rather closely related in the cell lineage tree because they both derive from hematopoietic stem cells. In the future, it will be interesting to compare the MIP repertoire of DCs to that of non-hematopoietic cells.
Second, our proteomics analyses of MIPs from WT and dKO DCs show that IPs dramatically increase the abundance and diversity of MIPs. In agreement with that, WT DCs were immunogenic for dKO mice, but not vice versa. Our study being based on comparison of WT and dKO DCs, it was aimed specifically at discovering the non-redundant roles of IPs, and not the non-redundant roles of CPs. If anything, our results might slightly underestimate the impact of IPs on the MIP repertoire because, in addition to CPs, DCs from dKO mice may harbor a few mixed proteasomes containing LMP2 admixed with CP β2 and β5 catalytic subunits. It must be realized, however, that eliminating all vestiges of IP activity is not a trivial task. Since the Lmp2 and the Lmp7 genes are closely linked, it is practically impossible to generate triple KO mutants by breeding (LMP7−/−Mecl1−/−) dKO mice with LMP2−/− mice. A plausible alternative would be to treat dKO cells with a pharmacologic LMP2 inhibitor (52). However, this strategy would also be fraught with a caveat: pharmacological inhibitors block the proteolytic activity of IP subunits but not other conformational effects that individual IP subunits have on IP function. Indeed, evidence suggests that incorporation of immunosubunits results in structural changes of the whole 20S IP complexes and thereby influences their biologic properties. For example, a model HBV epitope is not generated in LMP7-deficient cells but can be generated in the presence of a catalytically inactive LMP7 subunit (in which Thr1 is mutated to Ala) (7, 53). Thus, the generation of epitopes like that one would not be blocked by pharmacological inhibitors. Our observation that IPs increase MIP abundance fits well with in vitro proteasome digestion experiments suggesting IPs have greater efflux and cleavage rates than CPs (19, 20) and with the decreased cell surface levels of MHC I molecules on Lmp7−/− and Lmp7−/−Mecl1−/− splenocytes (12). For reasons presented in the Introduction, it was not possible to extrapolate from previous in vitro proteasome digestion experiments the overall impact of IPs on the diversity of the MIP repertoire generated in vivo. Our work now provides a direct and global evaluation of the impact of IPs on MIP diversity. Of 417 peptides eluted from DCs, 199 were overexpressed in WT relative to dKO DCs and 60 were detected exclusively in WT DCs. Thus, about 14% of MIPs (60 of 417) were totally IP-dependent.
Third, our results also suggest that IPs possess distinct cleavage properties that impinge on the MIP of primary DCs. The most salient difference between IP-dependent and -independent peptides was found at position C+2. For IP-dependent peptides, deviation at C+2 was characterized by an increased usage of proline and polar residues with decreased frequencies of asparagine and hydrophobic residues. The most significant difference was the decreased frequency of leucine residues at C+2 in IP-dependent relative to IP-independent peptides. That observation is remarkably coherent with the seminal work conducted by Toes et al. who digested yeast enolase-1 in vitro with CPs or IPs, analyzed fractionated peptide fragments by MS, and found that leucine at C+2 was disfavored by IPs (21). Furthermore, our bioinformatic analyses predict that IPs have a bias toward unstructured protein regions and lead to enhanced MHC I presentation of MIPs adjoining the unstructured proteome. The lack of secondary and tertiary structure confers several properties such as increased interaction surface area, conformational flexibility and accessible posttranslational modification sites (48). Consequently, largely unstructured proteins are especially prone to make promiscuous molecular interactions and their overexpression is particularly dangerous for a cell as it frequently leads to cell death or neoplastic transformation (48, 54). The MIP repertoire allows presentation of only a tiny fraction of the proteome to CD8 T cells (22, 55). Therefore, the bias of IPs toward unstructured protein regions could be of considerable relevance, for example in cancer immunosurveillance.
Integration of our peptidomic data with global profiling of the DC transcriptome revealed that differential expression of MIPs in WT versus dKO DCs cannot be ascribed to differential transcription of peptide source genes. However, we found that IPs have a non-redundant impact on expression of a selected set of transcripts. Of note, IPs may affect expression of numerous other genes redundantly with CPs, but our study design was poised to selectively identify genes differentially regulated by IPs and CPs. Recent evidence suggests that the role of IPs is not limited to processing peptides for MHC presentation (56). For instance, MECL1 is a T-cell–intrinsic factor regulating homeostatic expansion, T cells from dKO mice hyperproliferate in response to polyclonal mitogens, and selective inhibition of LMP7 blocks cytokine production by activated monocytes and T cells (12, 57, 58). The present work suggests that these pleiotropic effects of IPs may be mediated by a non-redundant effect of IPs on gene expression. Differential expression of immune genes could explain why dKO DCs pulsed with optimal levels of exogenous SIINFEKL peptide are less immunogenic than WT DCs (supplemental Fig. S1). Further work is needed to discover how IPs may regulate gene expression. Nevertheless, it is interesting to note that genes on which IPs had a non-redundant effect were clustered in the genome (Fig. 5C). Gene order in eukaryotes is not random. In all well-studied genomes, genes of similar and/or coordinated expression tend to be linked in clusters that can extend up to many megabases (59). Gene clustering often results from the sharing of regulatory elements (60). In line with this, proteasome 20S core particles regulate transcriptional activation by controlling the localization, abundances and activity of transcriptional activators and repressors through proteolytic degradation (27–30). We therefore propose that the non-redundant effect of IPs on gene expression may result from proteolysis of transcriptional modulators or their regulators. The overarching conclusion of our work is that IP subunits MECL1 and LMP7 have more than one non-redundant role. They have a dramatic impact on the MIP repertoire and a heretofore unrecognized impact on expression of immune-related genes. Both of these effects are probably of great importance in adaptive immune responses and may be instrumental in the remarkable conservation of IPs in gnathostomes.
Supplementary Material
Acknowledgments
We thank Dr T. A. Griffin for providing the LMP7−/−Mecl1−/− mice, Jean-Philippe Laverdure for help with bioinformatic analyses and Martin Giroux for thoughtful suggestions. We are grateful to the staff of the following core facilities at the Institute for Research in Immunology and Cancer (IRIC) for their outstanding support: Animal facility, Bioinformatics, Flow cytometry, Genomics, and Proteomics.
* This work was supported by Terry Fox New Frontiers Program in Cancer Immunotherapy (grant# 018005) and the Canadian Cancer Society Research Institute (grant # 019475). DdV and DPG are supported by training grants from the CIHR. MHF was supported by a studentship from the FRSQ. CP and PT hold Canada Research Chairs in Immunobiology, and Proteomics and Bioanalytical Spectrometry, respectively. SM holds the CIBC Breast Cancer Research Chair at Université de Montréal. IRIC is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canada Foundation for Innovation, and the FRSQ.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MHC
- major histocompatibility complex
- β2m
- β2-microglobulin
- CFSE
- carboxyfluorescein succinimidyl ester
- CP
- constitutive proteasome
- DC
- dendritic cell
- dKO
- Lmp7−/−Mecl1−/− double-knockout mice
- IP
- immunoproteasome
- LPS
- lipopolysaccharide
- MHC
- major histocompatibility complex
- Lmp
- low molecular weight protein
- nanoLC-MS/MS
- nano-LC combined with tandem MS
- Mecl
- multicatalytic endopeptidase complex–like
- MIP
- MHC I-associated peptide
- SCX
- strong cation exchange
- WT
- wild-type.
REFERENCES
- 1.Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 2.Wherry E. J., Golovina T. N., Morrison S. E., Sinnathamby G., McElhaugh M. J., Shockey D. C., Eisenlohr L. C. (2006) Re-evaluating the generation of a “proteasome-independent” MHC class I-restricted CD8 T cell epitope. J. Immunol. 176, 2249–2261 [DOI] [PubMed] [Google Scholar]
- 3.Loureiro J., Ploegh H. L. (2006) Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv. Immunol. 92, 225–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shastri N., Cardinaud S., Schwab S. R., Serwold T., Kunisawa J. (2005) All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol. Rev. 207, 31–41 [DOI] [PubMed] [Google Scholar]
- 5.Glickman M. H., Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 6.Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 7.Kloetzel P. M. (2001) Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2, 179–187 [DOI] [PubMed] [Google Scholar]
- 8.Heink S., Ludwig D., Kloetzel P. M., Kruger E. (2005) IFN-γ-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc. Natl. Acad. Sci. U.S.A. 102, 9241–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macagno A., Kuehn L., de Giuli R., Groettrup M. (2001) Pronounced up-regulation of the PA28α/β proteasome regulator but little increase in the steady-state content of immunoproteasome during dendritic cell maturation. Eur. J. Immunol. 31, 3271–3280 [DOI] [PubMed] [Google Scholar]
- 10.Hughes A. L. (1997) Evolution of the proteasome components. Immunogenetics 46, 82–92 [DOI] [PubMed] [Google Scholar]
- 11.Kesmir C., van Noort V., de Boer R. J., Hogeweg P. (2003) Bioinformatic analysis of functional differences between the immunoproteasome and the constitutive proteasome. Immunogenetics 55, 437–449 [DOI] [PubMed] [Google Scholar]
- 12.Zaiss D. M., de Graaf N., Sijts A. J. (2007) The proteasome immunosubunit MECL-1 is a T cell intrinsic factor influencing homeostatic expansion. Infect. Immun. 76, 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sewell A. K., Price D. A., Teisserenc H., Booth B. L. J., Gileadi U., Flavin F. M., Trowsdale J., Phillips R. E., Cerundolo V. (1999) IFN-γ exposes a cryptic cytotoxic T lymphocyte epitope in HIV-1 reverse transcriptase. J. Immunol. 162, 7075–7079 [PubMed] [Google Scholar]
- 14.Morel S., Lévy F., Burlet-Schiltz O., Brasseur F., Probst-Kepper M., Peitrequin A. L., Monsarrat B., Van Velthoven R., Cerottini J. C., Boon T., Gairin J. E., Van den Eynde B. J. (2000) Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity 12, 107–117 [DOI] [PubMed] [Google Scholar]
- 15.Chen W., Norbury C. C., Cho Y., Yewdell J. W., Bennink J. R. (2001) Immunoproteasomes shape immunodominance hierarchies of antiviral CD8+ T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 193, 1319–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapiro J., Claverol S., Piette F., Ma W., Stroobant V., Guillaume B., Gairin J. E., Morel S., Burlet-Schiltz O., Monsarrat B., Boon T., Van den Eynde B. J. (2006) Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J. Immunol. 176, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 17.Deol P., Zaiss D. M. W., Monaco J. J., Sijts A. J. A. M. (2007) Rates of processing determine the immunogenicity of immunoproteasome-generated epitopes. J. Immunol. 178, 7557–7562 [DOI] [PubMed] [Google Scholar]
- 18.Dannull J., Lesher D. T., Holzknecht R., Qi W., Hanna G., Seigler H., Tyler D. S., Pruitt S. K. (2007) Immunoproteasome down-modulation enhances the ability of dendritic cells to stimulate anti-tumor immunity. Blood 110, 4341–4350 [DOI] [PubMed] [Google Scholar]
- 19.Cascio P., Hilton C., Kisselev A. F., Rock K. L., Goldberg A. L. (2001) 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 20, 2357–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishto M., Luciani F., Holzhutter H. G., Bellavista E., Santoro A., Textoris-Taube K., Franceschi C., Kloetzel P. M., Zaikin A. (2008) Modeling the in vitro 20S proteasome activity: the effect of PA28-αβ and of the sequence and length of polypeptides on the degradation kinetics. J. Mol. Biol. 377, 1607–1617 [DOI] [PubMed] [Google Scholar]
- 21.Kloetzel P.-M., Ossendorp F. (2004) Proteasome and peptidase function in MHC-class-I mediated antigen presentation. Curr. Opin. Immunol. 16, 76–81 [DOI] [PubMed] [Google Scholar]
- 22.Yewdell J. W., Reits E., Neefjes J. (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nature Rev. Immunol. 3, 952–961 [DOI] [PubMed] [Google Scholar]
- 23.Eisenlohr L. C., Huang L., Golovina T. N. (2007) Rethinking peptide supply to MHC class I molecules. Nat. Rev. Immunol. 7, 403–410 [DOI] [PubMed] [Google Scholar]
- 24.York I. A., Mo A. X. Y., Lemerise K., Zeng W., Shen Y., Abraham C. R., Saric T., Goldberg A. L., Rock K. L. (2003) The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity 18, 429–440 [DOI] [PubMed] [Google Scholar]
- 25.Hammer G. E., Kanaseki T., Shastri N. (2007) The final touches make perfect the peptide-MHC class I repertoire. Immunity 26, 397–406 [DOI] [PubMed] [Google Scholar]
- 26.Wearsch P. A., Cresswell P. (2007) Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat. Immunol. 8, 873–881 [DOI] [PubMed] [Google Scholar]
- 27.Lipford J. R., Deshaies R. J. (2003) Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat. Cell Biol. 5, 845–850 [DOI] [PubMed] [Google Scholar]
- 28.Lipford J. R., Smith G. T., Chi Y., Deshaies R. J. (2005) A putative stimulatory role for activator turnover in gene expression. Nature 438, 113–116 [DOI] [PubMed] [Google Scholar]
- 29.Collins G. A., Tansey W. P. (2006) The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16, 197–202 [DOI] [PubMed] [Google Scholar]
- 30.Bhaumik S. R., Malik S. (2008) Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit Rev. Biochem. Mol. Biol. 43, 419–433 [DOI] [PubMed] [Google Scholar]
- 31.Fortier M. H., Caron E., Hardy M. P., Voisin G., Lemieux S., Perreault C., Thibault P. (2008) The MHC class I peptide repertoire is molded by the transcriptome. J. Exp. Med. 205, 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells J. W., Darling D., Farzaneh F., Galea-Lauri J. (2005) Influence of interleukin-4 on the phenotype and function of bone marrow-derived murine dendritic cells generated under serum-free conditions. Scand. J. Immunol. 61, 251–259 [DOI] [PubMed] [Google Scholar]
- 33.Son Y. I., Egawa S., Tatsumi T., Redlinger R. E., Jr., Kalinski P., Kanto T. (2002) A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J. Immunol. Methods 262, 145–157 [DOI] [PubMed] [Google Scholar]
- 34.Brochu S., Baron C., Hetu F., Roy D. C., Perreault C. (1995) Oligoclonal expansion of CTLs directed against a restricted number of dominant minor histocompatibility antigens in hemopoietic chimeras. J. Immunol. 155, 5104–5114 [PubMed] [Google Scholar]
- 35.Lynn D. J., Winsor G. L., Chan C., Richard N., Laird M. R., Barsky A., Gardy J. L., Roche F. M., Chan T. H., Shah N., Lo R., Naseer M., Que J., Yau M., Acab M., Tulpan D., Whiteside M. D., Chikatamarla A., Mah B., Munzner T., Hokamp K., Hancock R. E., Brinkman F. S. (2008) InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 4, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters B., Sette A. (2007) Integrating epitope data into the emerging web of biomedical knowledge resources. Nat. Rev. Immunol. 7, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barchet W., Oehen S., Klenerman P., Wodarz D., Bocharov G., Lloyd A. L., Nowak M. A., Hengartner H., Zinkernagel R. M., Ehl S. (2000) Direct quantitation of rapid elimination of viral antigen-positive lymphocytes by antiviral CD8+ T cells in vivo. Eur. J. Immunol. 30, 1356–1363 [DOI] [PubMed] [Google Scholar]
- 38.Lecoeur H., Fevrier M., Garcia S., Riviere Y., Gougeon M. L. (2001) A novel flow cytometric assay for quantitation and multiparametric characterization of cell-mediated cytotoxicity. J. Immunol. Methods 253, 177–187 [DOI] [PubMed] [Google Scholar]
- 39.Jedema I., van der Werff N. M., Barge R. M., Willemze R., Falkenburg J. H. (2004) New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood 103, 2677–2682 [DOI] [PubMed] [Google Scholar]
- 40.Ferron F., Longhi S., Canard B., Karlin D. (2006) A practical overview of protein disorder prediction methods. Proteins 65, 1–14 [DOI] [PubMed] [Google Scholar]
- 41.Zhang B., Kirov S., Snoddy J. (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33(Web Server Issue), W741–W748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 43.Groettrup M., Standera S., Stohwasser R., Kloetzel P. M. (1997) The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc. Natl. Acad. Sci. U.S.A. 94, 8970–8975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toes R. E., Nussbaum A. K., Degermann S., Schirle M., Emmerich N. P., Kraft M., Laplace C., Zwinderman A., Dick T. P., Muller J., Schonfisch B., Schmid C., Fehling H. J., Stevanovic S., Rammensee H. G., Schild H. (2001) Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 194, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schatz M. M., Peters B., Akkad N., Ullrich N., Martinez A. N., Carroll O., Bulik S., Rammensee H. G., van E. P., Holzhutter H. G., Tenzer S., Schild H. (2008) Characterizing the N-terminal processing motif of MHC class I ligands. J. Immunol. 180, 3210–3217 [DOI] [PubMed] [Google Scholar]
- 46.Reits E., Neijssen J., Herberts C., Benckhuijsen W., Janssen L., Drijfhout J. W., Neefjes J. (2004) A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity 20, 495–506 [DOI] [PubMed] [Google Scholar]
- 47.Hammer G. E., Gonzalez F., Champsaur M., Cado D., Shastri N. (2006) The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat. Immunol. 7, 103–112 [DOI] [PubMed] [Google Scholar]
- 48.Gsponer J., Futschik M. E., Teichmann S. A., Babu M. M. (2008) Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322, 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gsponer J., Madan B. M. (2009) The rules of disorder or why disorder rules. Prog. Biophys. Mol. Biol. 99, 94–103 [DOI] [PubMed] [Google Scholar]
- 50.Hughes A. L., Hughes M. K. (1995) Self peptides bound by HLA class I molecules are derived from highly conserved regions of a set of evolutionarily conserved proteins. Immunogenetics 41, 257–262 [DOI] [PubMed] [Google Scholar]
- 51.Engelhard V., Brickner A., Zarling A. (2002) Insights into antigen processing gained by direct analysis of the naturally processed class I MHC associated peptide repertoire. Mol. Immunol. 39, 127–137 [DOI] [PubMed] [Google Scholar]
- 52.Ho Y. K., Bargagna-Mohan P., Wehenkel M., Mohan R., Kim K. B. (2007) LMP2-specific inhibitors: chemical genetic tools for proteasome biology. Chem. Biol. 14, 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gileadi U., Moins-Teisserenc H. T., Correa I., Booth B. L., Jr., Dunbar P. R., Sewell A. K., Trowsdale J., Phillips R. E., Cerundolo V. (1999) Generation of an immunodominant CTL epitope is affected by proteasome subunit composition and stability of the antigenic protein. J. Immunol. 163, 6045–6052 [PubMed] [Google Scholar]
- 54.Vavouri T., Semple J. I., Garcia-Verdugo R., Lehner B. (2009) Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138, 198–208 [DOI] [PubMed] [Google Scholar]
- 55.Caron É., Charbonneau R., Huppé G., Brochu S., Perreault C. (2005) The structure and location of SIMP/STT3B account for its prominent imprint on the MHC I immunopeptidome. Int. Immunol. 17, 1583–1596 [DOI] [PubMed] [Google Scholar]
- 56.Groettrup M., Kirk C. J., Basler M. (2010) Proteasomes in immune cells: more than peptide producers? Nat. Rev. Immunol. 10, 73–78 [DOI] [PubMed] [Google Scholar]
- 57.Caudill C. M., Jayarapu K., Elenich L., Monaco J. J., Colbert R. A., Griffin T. A. (2006) T cells lacking immunoproteasome subunits MECL-1 and LMP7 hyperproliferate in response to polyclonal mitogens. J. Immunol. 176, 4075–4082 [DOI] [PubMed] [Google Scholar]
- 58.Muchamuel T., Basler M., Aujay M. A., Suzuki E., Kalim K. W., Lauer C., Sylvain C., Ring E. R., Shields J., Jiang J., Shwonek P., Parlati F., Demo S. D., Bennett M. K., Kirk C. J., Groettrup M. (2009) A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 15, 781–787 [DOI] [PubMed] [Google Scholar]
- 59.Hurst L. D., Pal C., Lercher M. J. (2004) The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5, 299–310 [DOI] [PubMed] [Google Scholar]
- 60.Sproul D., Gilbert N., Bickmore W. A. (2005) The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 6, 775–781 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.