Abstract
Two-dimensional LC combined with whole protein and peptide mass spectrometry is used to characterize proteins secreted by methicillin-resistant Staphylococcus aureus COL. Protein identifications were accomplished via off-line protein fractionation followed by digestion and subsequent peptide analysis by reverse phase LC-ESI-LTQ-FT-MS/MS. Peptide MS/MS analysis identified 127 proteins comprising 59 secreted proteins, seven cell wall-anchored proteins, four lipoproteins, four membrane proteins, and 53 cytoplasmic proteins. The identified secreted proteins included various virulence factors of known functions (cytotoxins, enterotoxins, proteases, lipolytic enzymes, peptidoglycan hydrolases, etc.). Accurate whole protein mass measurement (±1.5 Da) of the secreted proteins combined with peptide analysis enabled identification of signal peptide cleavage sites and various post-translational modifications. In addition, new observations were possible using the present approach. Although signal peptide cleavage is highly specific, signal peptide processing can occur at more than one site. Surprisingly, cleaved signal peptides and their fragments can be observed in the extracellular medium. The prediction accuracies of several signal peptide prediction programs were also evaluated.
Staphylococcus aureus, a Gram-positive human pathogen, is the leading cause of nosocomial infections, imposing tremendous economic burden on patients and hospitals throughout the world (1–3). The spectrum of staphylococcal infections is very wide, ranging from minor skin lesions to life-threatening conditions such as bacteremia, pneumonia, endocarditis, osteomyelitis, toxic shock syndrome, and septicemia (2–4). The treatment of staphylococcal infections has become extremely challenging because of its propensity to rapidly evolve antibiotic-resistant strains. The methicillin-resistant staphylococci (MRSA)1 is the most notorious in that it causes an estimated 94,000 life-threatening infections and 19,000 deaths a year in the United States (4, 5). Hospitalization costs associated with MRSA infections are also significant with a mean attributable cost of $35,000 per infection (6). Furthermore, the recent emergence of strains resistant to vancomycin, a glycopeptide antibiotic that is often considered as the last resort drug in treating MRSA infections, has compounded the problem (7–9). Needless to say, it is of paramount importance to discover effective vaccines and to develop new strategies to treat S. aureus infections. This urgency motivated the scientific community to direct significant research effort toward whole genome sequencing of S. aureus strains in the past few years. The wealth of information available from the nine fully annotated and sequenced genomes of S. aureus has provided us with an excellent opportunity to apply powerful technologies including proteomics to gain a comprehensive understanding of the biology of this organism (10–13).
Pathogenesis of S. aureus is complex and involves the synthesis of an array of virulence factors followed by their transport across the cytoplasmic membrane to destinations outside the cell. A majority of the exported proteins in S. aureus are predicted to be secreted via secretory (Sec) pathway, which requires an N-terminal signal peptide (14) at the N terminus of the protein and a cleavage site that is recognized by type I signal peptidases (SPases I) (15). During translocation, or shortly thereafter, the signal peptide of the preprotein is removed by SPase I resulting in the release of the mature protein from the membrane (16, 17). The mature protein may be further modified and is either retained on the cell surface or secreted into the extracellular host milieu. The secreted proteins of S. aureus are postulated to play a prominent role in host infection and are believed to be engaged in tissue damage, invasion and evasion of host immune responses. Therefore, a comprehensive description of secretory proteins (the secretome) of different S. aureus strains is vital to gain insights into its pathogenesis. This information will be valuable in identifying novel virulence factors and should ultimately help in the development of new diagnostic tools and vaccines.
To this end, S. aureus secretory proteins have been identified using a variety of classical techniques including Western blot, ELISA, and one- and two-dimensional gel electrophoresis (1DE/2DE) with N-terminal sequencing (18–20). Although gel-based techniques are well established for separating proteins mixtures, they have several drawbacks including poor reproducibility and sensitivity and limited dynamic range, and they are tedious, labor-intensive, and technically challenging (21–23). Recent proteomics strategies have coupled gel electrophoresis with mass spectrometry (24–27). In these approaches, the peptides resulting from in-gel digestion of excised spots were analyzed by MALDI-TOF-MS or by LC-ESI-MS/MS. A critical drawback of peptide analysis by mass spectrometry is that it provides very limited molecular information about intact secreted proteins. Valuable information regarding loss of signal peptides, signal peptide cleavage sites, post-translational modifications, and protein degradation is usually completely lost. This is particularly true when peptide mass fingerprinting is used. To overcome the shortcomings of the current techniques, a sophisticated gel-free approach combining whole protein and peptide mass spectrometric approaches was used in the present work. To the best of our knowledge, there is only one report in the literature pertaining to whole protein mass analysis of S. aureus secretory proteins. Kawano et al. (18) attempted to identify S. aureus secreted proteins via one-dimensional reverse phase (RP) LC-ESI-MS and N-terminal Edman degradation. Using this approach, they were able to tentatively identify three and four secreted proteins in NCTC 8325 and MRSA 3543 strains, respectively.
In the present study, the secretome of methicillin-resistant Staphylococcus aureus subsp. aureus COL (S. aureus COL) was more comprehensively characterized. Secreted proteins were separated with an in-house constructed automated two-dimensional LC system (28) with on-line fractionation followed by whole protein mass measurement by ESI-Q-TOF-MS. Definitive protein identifications were accomplished via off-line collection of protein fractions followed by protein digestion and subsequent peptide analysis by RPLC-ESI-LTQ-FT-MS/MS. Genome-based signal peptide algorithms predict 71 secretory proteins for S. aureus COL (29). Our peptide analysis successfully identified 59 of these secreted proteins from the culture supernatants of S. aureus COL with average sequence coverage of 79%. In addition, combined information from the two mass spectrometric approaches allowed detailed characterization of 53 of these secreted proteins. The accurate whole protein mass measurement of the secreted proteins allowed verification of signal peptide cleavage sites. Also, the current study provided us with an opportunity to compare the accuracy of several computational tools available for predicting signal peptide cleavage sites. Additionally, we report surprising findings on the presence of cleaved signal peptides and signal peptide fragments in the extracellular medium.
EXPERIMENTAL PROCEDURES
Materials
HPLC grade ACN and urea were purchased from EMD Chemicals (Gibbstown, NJ). Water was purified using a Barnstead/Thermolyne E-Pure system (Barnstead/Thermolyne, Dubuque, IA). 40% aqueous methylamine was obtained from Aldrich. Sodium chloride, formic acid, TFA, and TCA were procured from Mallinckrodt Baker. Ammonium bicarbonate was obtained from Mallinckrodt Baker. Proteomics grade trypsin (T-6567) was purchased from Sigma, and endoproteinase Glu-C was obtained from New England Biolabs (Beverly, MA).
Bacterial Strain and Growth Conditions
The methicillin-resistant S. aureus COL was obtained through the Network on Antimicrobial Resistance in S. aureus (NARSA) program supported under NIAID, National Institutes of Health Contract N01-AI-95359. To prepare stock cultures, a single colony of S. aureus COL was inoculated into 50 ml of sterile brain-heart infusion (BHI) broth (Difco/BD Biosciences) and incubated overnight at 37 °C with rotary aeration (200 rpm). 1 ml of the overnight culture was used to inoculate 200 ml of fresh sterile BHI broth and incubated for 10 h. The resulting culture was aliquoted into 2-ml stocks with 20% by volume glycerol as the cryoprotective agent and stored at −80 °C until required. In a typical experiment, the stock culture was inoculated into 100 ml of sterile BHI broth and grown overnight at 37 °C with rotary aeration (200 rpm). A 2-ml portion of this overnight preculture was then diluted 1:100 in fresh sterile BHI broth and incubated at 37 °C with shaking at 200 rpm. The growth was monitored spectrophotometrically by measuring A600. Bacteria were cultured to stationary growth phase (A600 = 4), which was generally attained in about 8 h.
Precipitation and Preparation of Extracellular Protein Fraction
Bacterial cells were separated from the stationary phase culture by centrifugation at 8500 × g (Sorvall RC-5B centrifuge, DuPont) for 30 min at 4 °C. To remove residual bacteria, the supernatant was filtered using a Stericup®/SteritopTM filtration device with 0.22-μm pore size polyethersulfone membrane (Millipore Corp., Billerica, MA). Soluble proteins in the filtered supernatant were precipitated overnight with 10% (w/v) TCA at 4 °C. The resulting precipitate was pelleted by centrifugation at 8500 × g for 70 min at 4 °C, washed several times with ice-cold acetone, and dried in a SpeedVac vacuum centrifuge (Jouan/Thermo Electron). The protein extract was dissolved in an appropriate amount of 8 m urea, 2 m thiourea solution and centrifuged at 14,100 × g (MiniSpin plus, Eppendorf, Westbury, NY) for 2 min to remove insoluble materials and was immediately used for proteomics analysis. Protein concentration was determined by Bradford assay (30) using bovine serum albumin as a standard.
Live/Dead Staining
500 μl of S. aureus COL stationary phase culture was centrifuged at 16,000 × g for 2 min, and the resulting cell pellet was stained with the LIVE/DEAD® BacLightTM bacterial viability kit (Molecular Probes, Carlsbad, CA) following the protocol provided by the manufacturer and visualized by fluorescence microscopy to detect cell lysis.
Isolation and Preparation of Extracellular Peptide Fraction
To isolate cleaved signal peptides present in the extracellular medium, S. aureus COL was cultured to stationary phase as described above. After removal of bacterial cells by centrifugation, the supernatant was filtered through Microcon Ultracel YM-10 to remove all the high molecular weight proteins (Millipore Corp.). The resulting filtrate was lyophilized and reconstituted in water, and the peptides were isolated, concentrated, and desalted using PepClean C18 spin columns (Pierce). The eluted peptides were dried in a SpeedVac vacuum centrifuge, resuspended in 40 μl of 0.1% TFA in H2O, and analyzed directly without any enzyme pretreatment by nano-RPLC-nano-ESI-LTQ-FT-MS/MS as described below.
Protein Sequence Data
S. aureus COL proteome was obtained from The J. Craig Venter Institute (GenBankTM accession number CP000046.1) (31). Theoretical protein masses were calculated from protein sequences using the Protein Analysis Worksheet Software (PAWS) program (freeware edition for Windows 95/98/NT/2000, ProteoMetrics, LLC, New York, NY). Additional sequence information was obtained from the Swiss-Prot database at ExPASy (32).
Whole Protein Two-dimensional Liquid Chromatography and Mass Spectrometry
S. aureus COL secreted proteins were separated using an automated two-dimensional LC system that has been described previously (28). In a typical analysis, S. aureus COL extracellular protein extract was first separated by strong cation exchange chromatography and fractionated on line using 20 trapping columns. The contents of the trap were then separated by reverse phase chromatography followed by measurement of protein masses by a quadrupole time-of-flight mass spectrometer. A more detailed description of the method is provided in the supplemental data. Mobile phases along with the gradients used are shown in supplemental Tables 1 and 2.
Peptide Nanoliquid Chromatography-Tandem Mass Spectrometry and Data Analysis
A detailed description of peptide analysis and information on MS/MS database search parameters are provided in the supplemental data. Briefly, proteins trapped on each trapping column were eluted with organic mobile phase and collected off line for trypsin or endoprotease Glu-C digestion. Each trap digest was subsequently analyzed by nano-RPLC-nano-ESI-LTQ-FT-MS/MS.
Signal Peptide and Protein Localization Predictions
To predict the presence of signal peptides and signal peptidase I cleavage sites for the proteins identified in the present work, the following prediction tools were used: SignalP-NN (neural network model) and SignalP-HMM (hidden Markov model) version 2.0 (33) and version 3.0 (34), PrediSi (35) (position weight matrix method), and SigCleave (36, 37) (weight matrix method). When required, LipoP version 1.0 (38) and TMHMM program version 2.0 (39) were used to predict lipoproteins and membrane proteins, respectively. PSORTb version 2.0 (40) was used to predict protein subcellular localization.
RESULTS
Protein Identification Strategy
Sibbald et al. (29) used a rigorous approach utilizing a combination of computational tools and an optimized type I signal peptidase (SpsB) recognition search pattern to estimate that 71 extracellular proteins are produced by S. aureus COL.
To identify proteins present in S. aureus COL extracellular medium, proteins were extracted by TCA precipitation from cultures at stationary growth phase, a phase during which extracellular proteins are preferentially expressed (27, 41). The first step in the analysis was to identify the S. aureus COL secretome through peptide analysis. The proteins from each of the C4 trapping columns were digested and analyzed as discussed above. From these peptide data, a total of 127 proteins (supplemental Table 3) were identified in the extracellular medium, and using bioinformatics tools, we classified these proteins into five categories based on their predicted subcellular localization. We classified 59 of the identified proteins as secreted proteins (Table I), and all of these proteins except two contained potential Sec-type signal peptides with SPase I cleavage sites and lacked any cell wall or membrane retention signals. The remaining 68 proteins included seven cell wall-anchored proteins, four lipoproteins, four membrane proteins (supplemental Table 4), and 53 cytoplasmic proteins (supplemental Table 5); these proteins were not predicted to be secreted into the growth medium.
Table I. S. aureus COL secreted proteins identified in present study.
Numbers in the superscript indicate the different forms of the protein identified. Theor., theoretical; Sig P, signal peptide, seq., sequence.
| Gene ID | Protein name | Theor. mass | Predicted massa | Observed massb | Mass error | Modifications | Trap location | MS/MS seq. cov.c |
|---|---|---|---|---|---|---|---|---|
| Da | Da | Da | Da | % | ||||
| Cytotoxins | ||||||||
| SACOL1173d | α-Hemolysin precursor (HlY) | 35,973.3 | 33,260.1 | 33,260.6 | 0.5 | −Sig P | Trap 6 | 84 |
| SACOL2003d | β-Hemolysin (Hlb) | 37,237.8 | 33,742.6 | 33,742.8 | 0.1 | −Sig P | Trap 7 | 90 |
| SACOL20221e | δ-Hemolysin (Hld1) | 2,976.6f | 2,976.6f | 2,976.6f | 0.0 | 100 | ||
| SACOL20222e | δ-Hemolysin (Hld2) | 2,976.6f | 3,020.6f | 3,020.6f | 0.0 | +N-terminal formylation, +16 Da | 100 | |
| SACOL2419g | γ-Hemolysin, component A (HlgA) | 34,955.7 | 31,921.9 | Trap 10 | 53 | |||
| SACOL24221d | γ-Hemolysin, component B (HlgB1) | 36,711.0 | 33,392.0 | 33,392.6 | 0.6 | −Sig P, truncated protein | Trap 4 | 69 |
| SACOL24222 | γ-Hemolysin, component B (HlgB2) | 36,711.0 | 33,462.1 | 33,463.6 | 1.5 | −Sig P, truncated protein | Trap 4 | 69 |
| SACOL24223 | γ-Hemolysin, component B (HlgB3) | 36,711.0 | 33,506.2 | 33,507.4 | 1.2 | −Sig P, truncated protein | Trap 4 | 69 |
| SACOL2421d | γ-Hemolysin, component C (HlgC) | 35,625.8 | 32,565.2 | 32,566.2 | 1.0 | −Sig P | Trap 9 | 85 |
| SACOL1880d | Leukotoxin LukD (LukD) | 36,888.9 | 34,158.6 | 34,159.1 | 0.5 | −Sig P | Trap 6 | 53 |
| SACOL1881d | Leukotoxin LukE (LukE) | 34,819.1 | 31,750.4 | 31,751.5 | 1.1 | −Sig P, −C-terminal residue Asn | Trap 8 | 68 |
| SACOL2004d | Leukocidin subunit precursor, putative | 38,686.1 | 35,573.3 | 35,574.4 | 1.1 | −Sig P | Trap 8 | 74 |
| SACOL20061d,h | Aerolysin/leukocidin family protein | 40,434.0 | 37,619.7 | 37,620.8 | 1.1 | −Sig P | Trap 10 | 72 |
| SACOL20062h | Aerolysin/leukocidin family protein | 40,434.0 | 37,418.4 | 37,419.3 | 0.9 | −Sig P | Trap 10 | 73 |
| Superantigenic toxins | ||||||||
| SACOL0442 | Staphylococcal enterotoxin | 23,165.4 | 19,343.9 | 19,345.0 | 1.1 | −Sig P | Trap 8 | 69 |
| SACOL0886d | Staphylococcal enterotoxin (Sek) | 27,727.1 | 24,698.4 | 24,699.6 | 1.2 | −Sig P, −C-terminal residues YKETI | Trap 12 | 82 |
| SACOL0887d | Staphylococcal enterotoxin type I (Sei) | 28,184.6 | 24,846.6 | 24,848.0 | 1.4 | −Sig P, −C-terminal residues TE | Trap 20 | 77 |
| SACOL0907d | Staphylococcal enterotoxin B (Seb) | 31,435.8 | 28,368.0 | 28,368.5 | 0.5 | −Sig P | Trap 8 | 100 |
| Proteases | ||||||||
| SACOL1869d | Serine protease SplA (SplA) | 25,876.2 | 21,853.5 | 21,854.4 | 0.9 | −Sig P | Trap 7 | 70 |
| SACOL1868d | Serine protease SplB (SplB) | 26,096.4 | 22,371.1 | 22,372.0 | 0.9 | −Sig P | Trap 6 | 66 |
| SACOL1867d | Serine protease SplC (SplC) | 26,098.4 | 22,388.0 | 22,388.7 | 0.7 | −Sig P | Trap 4 | 48 |
| SACOL1866d | Serine protease SplD (SplD) | 25,669.1 | 22,010.8 | 22,012.2 | 1.4 | −Sig P, seq. error | Trap 5 | 77 |
| SACOL1865d | Serine protease SplE (SplE) | 25,679.3 | 22,011.9 | 22,013.3 | 1.4 | −Sig P | Trap 7 | 32 |
| SACOL1864d | Serine protease SplF (SplF) | 25,655.1 | 21,941.7 | 21,942.7 | 1.0 | −Sig P | Trap 5 | 93 |
| SACOL1057d,g | V8 protease (SspA) | 36,312.6 | 33,376.1 | Trap 5 | 42 | |||
| SACOL1056d | Cysteine protease precursor SspB (SspB1) | 44,519.0 | 40,649.4 | 40,650.5 | 1.1 | −Sig P | Trap 5 | 78 |
| SACOL1970d,g | Cysteine protease precursor SspB (SspB2) | 44,252.1 | 41,524.8 | Trap 12 | 56 | |||
| SACOL2659d,g | Zinc metalloproteinase aureolysin (Aur) | 56,361.3 | 53,459.8 | Trap 8 | 48 | |||
| Lipolytic enzymes | ||||||||
| SACOL2694d | Lipase1 (Lip1) | 76,675.3 | 73,077.3 | 73,078.0 | 0.8 | −Sig P | Trap 8 | 96 |
| SACOL0317d | Lipase2 (Lip2) | 71,276.8 | 67,152.0 | 67,152.8 | 0.8 | −Sig P | Trap 9 | 71 |
| SACOL0078d | 1-Phosphatidylinositol phosphodiesterase (Plc) | 37,086.7 | 34,127.1 | 34,128.0 | 0.9 | −Sig P | Trap 6 | 90 |
| Peptidoglycan hydrolases | ||||||||
| SACOL0263d | Peptidoglycan hydrolase (LytM) | 35,067.4 | 31,735.5 | 31,736.5 | 1.0 | −Sig P | Trap 4 | 66 |
| SACOL0507d,g | N-Acetylmuramoyl-l-alanine amidase (Sle1) | 35,835.7 | 33,424.9 | Trap 12 | 76 | |||
| SACOL07231d | LysM domain protein | 28,186.8 | 11,848.9 | 11,849.0 | 0.1 | −Sig P, N-terminal fragment | Trap 2 | 100 |
| SACOL07232 | LysM domain protein | 28,186.8 | 13,800.9 | 13,801.8 | 0.9 | −Sig P, C-terminal fragment | Trap 2 | 72 |
| SACOL10621d | Bifunctional autolysin (Atl1) | 137,334.9 | 134,248.2 | 134,249.5 | 1.3 | −Sig P | Trap 9 | 99 |
| SACOL10622 | Bifunctional autolysin (Atl2) | 137,334.9 | 80,786.9 | 80,787.3 | 0.4 | −Sig P, proteolytic processing | Trap 8 | 99 |
| SACOL10623 | Bifunctional autolysin (Atl3) | 137,334.9 | 53,479.5 | 53,479.7 | 0.2 | −Sig P, proteolytic processing | Trap 9 | 99 |
| SACOL2088d | SceD protein (SceD) | 24,096.0 | 21,497.0 | 21,497.7 | 0.7 | −Sig P | Trap 3 | 76 |
| SACOL2584d | Immunodominant antigen A (IsaA) | 24,203.2 | 21,377.9 | 21,377.5 | −0.4 | −Sig P | Trap 3 | 91 |
| SACOL2666d | N-Acetylmuramoyl-l-alanine amidase domain protein | 69,253.2 | 42,250.8 | 42,252.1 | 1.3 | −Sig P, protein degradation | Trap 6 | 89 |
| Da | Da | Da | Da | % | ||||
| Miscellaneous enzymes | ||||||||
| SACOL0303d | Acid phosphatase 5-nucleotidase | 33,351.9 | 30,184.3 | 30,185.0 | 0.7 | −Sig P | Trap 11 | 87 |
| SACOL0860 | Thermonuclease precursor (Nuc) | 25,119.9 | 18,782.3 | 18,782.0 | −0.3 | −Sig P | Trap 7 | 97 |
| SACOL0962d | Glycerophosphoryl diester phosphodiesterase (GlpQ) | 35,310.7 | 32,240.2 | 32,241.0 | 0.8 | −Sig P | Trap 10 | 100 |
| SACOL10711h | Chitinase-related protein | 11,344.8 | 8,720.8 | 8,721.0 | 0.2 | −Sig P | Trap 3 | 91 |
| SACOL10712h | Chitinase-related protein | 11,344.8 | 8,906.9 | 8,907.1 | 0.2 | −Sig P | Trap 3 | 89 |
| SACOL10713h | Chitinase-related protein | 11,344.8 | 9,902.0 | 9,901.6 | −0.4 | −Sig P | Trap 3 | 92 |
| Surface adhesins | ||||||||
| SACOL08581 | Secretory extracellular matrix and plasma-binding protein (Empbp1) | 38,484.9 | 12,890.5 | 12,891.8 | 1.3 | −Sig P, N-terminal fragment | Trap15 | 73 |
| SACOL08582 | Secretory extracellular matrix and plasma-binding protein (Empbp2) | 38,484.9 | 22,709.8 | 22,708.9 | −1.0 | −Sig P, C-terminal fragment | Trap 15 | 53 |
| SACOL0985d | Surface protein, putative | 15,838.2 | 12,859.7 | 12,859.5 | −0.3 | −Sig P | Trap 4 | 100 |
| SACOL1164 | Fibrinogen binding-related protein | 12,596.6 | 9,592.1 | 9,592.5 | 0.4 | −Sig P | Trap 3 | 69 |
| SACOL1168 | Fibrinogen-binding protein (Efb) | 18,764.6 | 15,850.1 | 15,851.0 | 0.9 | −Sig P | Trap 10 | 90 |
| SACOL2002 | Map protein (Map) | 76,945.2 | 73,877.7 | Trap 17 | 31 | |||
| SACOL2019 | SdrH protein, putative (SdrH) | 46,630.0 | 38,083.8 | 38,085.1 | 1.2 | −Sig P, removal of C-terminal residues 377–419 | Trap 3 | 85 |
| SACOL2197d | Surface protein, putative | 15,447.5 | 12,480.0 | 12,480.1 | 0.1 | −Sig P | Trap 3 | 100 |
| SACOL2418d | IgG-binding protein (Sbi) | 50,070.2 | 47,049.7 | 47,049.3 | −0.4 | −Sig P | Trap 9 | 95 |
| SACOL2660 | Immunodominant antigen B (IsaB) | 19,370.2 | 15,785.0 | 15,785.3 | 0.3 | −Sig P | Trap 3 | 100 |
| Unknown functions | ||||||||
| SACOL0270d | Staphyloxanthin biosynthesis protein, putative | 33,032.2 | 30,379.1 | 30,377.9 | −1.2 | −Sig P, −42-Da modification | Trap 12 | 59 |
| SACOL0271 | Virulence factor EsxA (EsxA) | 11,306.2 | 10,905.2 | 10,905.2 | 0.2 | −Met | Trap 3 | 100 |
| SACOL0480 | Hypothetical protein | 11,301.8 | 8,380.3 | 8,380.4 | 0.1 | −Sig P | Trap 9 | 100 |
| SACOL0669d | Conserved hypothetical protein | 18,594.2 | 15,905.0 | 15,905.4 | 0.4 | −Sig P | Trap 6 | 100 |
| SACOL0755d | Conserved hypothetical protein | 16,922.1 | 13,449.9 | 13,449.1 | −0.8 | −Sig P | Trap 7 | 79 |
| SACOL0859 | Hypothetical protein | 17,717.0 | 14,812.5 | 14,813.4 | 0.9 | −Sig P | Trap 7 | 67 |
| SACOL0908d | Hypothetical protein | 20,345.8 | 16,271.1 | 16,271.1 | 0.0 | −Sig P | Trap 5 | 99 |
| SACOL1166d | Hypothetical protein | 15,202.5 | 12,304.0 | 12,304.0 | 0.0 | −Sig P | Trap 5 | 55 |
| SACOL2179 | Conserved hypothetical protein | 32,763.6 | 29,134.0 | 29,135.5 | 1.5 | −Sig P | Trap 8 | 42 |
| SACOL22911d | Staphylococcal secretory antigen SsaA2 | 29,327.1 | 12,211.3 | 12,210.3 | −1.0 | −Sig P, N-terminal fragment | Trap 3 | 98 |
| SACOL22912 | Staphylococcal secretory antigen SsaA2 | 29,327.1 | 14,294.5 | 14,294.9 | 0.4 | −Sig P, C-terminal fragment | Trap 3 | 97 |
| SACOL2295d | Staphyloxanthin biosynthesis protein | 17,424.8 | 14,755.7 | 14,756.7 | 1.0 | −Sig P, +16-Da modification | Trap 3 | 71 |
| SACOL2557 | Conserved domain protein | 16,870.3 | 12,788.5 | 12,789.5 | 1.0 | −Sig P | Trap 6 | 61 |
a Predicted masses were calculated by subtracting the mass of the signal peptide from the theoretical mass. For proteins with additional modifications, recalculated predicted mass is presented in the table.
b Average mass of the protein from replicate analyses.
c Net sequence coverage of the mature protein.
d Protein identified in other S. aureus strains (29).
e Protein identified in the extracellular peptide extract by peptide MS/MS analysis; the reported protein mass was calculated from precursor ion mass.
f Monoisotopic mass.
g Protein could not be identified by whole protein MS analysis.
h Protein with signal peptide cleavages at more than one site.
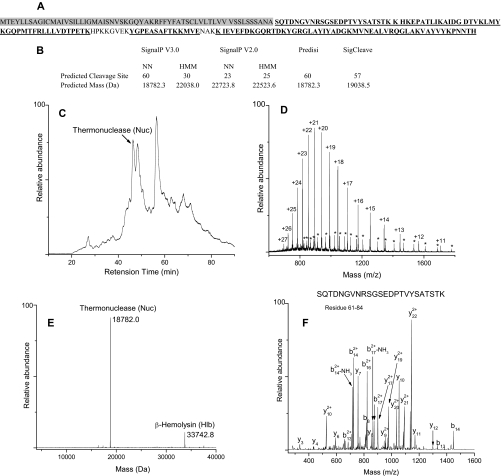
The second step in the analysis of extracellular proteins involved mass measurements of whole proteins captured on each trap by ESI-Q-TOF-MS. Whole protein masses from a particular trap were assigned to molecules that had been identified by peptide analysis of the same trap. The general experimental approach used to identify S. aureus COL extracellular proteins is demonstrated below by using Trap 7 as an example. Peptides corresponding to thermonuclease (Nuc) (Fig. 1A), β-hemolysin (Hlb), SACOL0755, SACOL0859, serine protease SplA (SplA), serine protease SplE (SplE), penicillin-binding protein 2 (MecA), and ribosomal protein L9 (RplI) were heavily populated in Trap 7. Because Nuc, Hlb, SACOL0755, SACOL0859, SplA, and SplE are predicted to undergo post-translational cleavage of the signal peptide, predicted masses for the mature secreted proteins were calculated by obtaining predicted cleavage site information from six signal peptide prediction programs; the resulting list of predicted masses was then matched against the experimental masses derived from whole protein MS analysis of Trap 7 proteins. For example, signal peptide cleavage site prediction of Nuc (theoretical mass of 25119.9 Da) yielded five different predictions (cleavage at residue 23, 25, 30, 57, and 60) resulting in five possible predicted masses for the mature Nuc (Fig. 1B). It is important to point out here that the computational methods used to predict signal peptide cleavage sites frequently provide conflicting predictions as exemplified by this case. Fig. 1C displays the total ion chromatogram of the Trap 7 fraction. Deconvolution of the raw spectrum (Fig. 1D) yields a protein peak at a retention time of 47 min with a mass of 18,782.0 Da (Fig. 1E). This observed mass matches very closely with only one of the possible predicted masses of Nuc (18,782.3 Da) and confirms the signal peptide cleavage site position as Ala60. The cleavage site position of Nuc was further corroborated by identification of the N-terminal peptide of the mature protein by MS/MS analysis as shown in Fig. 1F. Whole protein identification and confirmation of signal peptide cleavage site positions of the other Trap 7 secreted proteins were accomplished similarly. Of the six programs used in the present study, only two (SignalP 3.0-NN and PrediSi) yielded the correct cleavage site for Nuc. Capitalizing on the ability to accurately determine the whole protein mass and hence the cleavage site position of the secreted proteins, we took this opportunity to evaluate the prediction accuracy of different signal peptide prediction programs, and the results are discussed below.
Fig. 1.
Experimental strategy for identification of Nuc in Trap 7. A, combined sequence coverage map of Nuc from trypsin and Glu-C digestion. The underlined amino acids were identified. The shaded region corresponds to the signal sequence. B, signal peptide cleavage site predictions and corresponding predicted masses for Nuc. C, total ion chromatogram of Trap 7 containing a peak corresponding to Nuc. D, raw spectrum of Nuc showing the charge state distribution. Asterisks show charge state distribution of Hlb. E, deconvoluted mass spectrum. F, MS/MS spectrum of N-terminal peptide SQTDNGVNRSGSEDPTVYSATSTK of mature Nuc.
Whole protein identification of membrane proteins and cytoplasmic proteins was achieved by directly matching the observed masses with theoretical masses calculated from the protein sequences. In the case of cell wall-anchored proteins and lipoproteins that contain cleavable signal peptides, the strategy used for secreted proteins was applied. For any protein that could not be identified by matching theoretical or predicted masses (after loss of signal peptide) to the observed masses, additional modifications including methylation (+14.03 Da), acetylation (+42.04 Da), oxidation (+15.99 Da), formylation (+27.99 Da), and protein truncation were considered. The PAWS program was used to assign unmatched protein masses derived from whole protein MS analysis to the corresponding truncated secreted proteins. In this approach, an unassigned protein mass from a particular trap was searched against the entire sequence of the suspected protein to determine whether any part of the sequence has a mass that matches the input mass within the experimental error (±1.5 Da).
Post-translational Modifications of Secreted Proteins
Whole protein mass measurements of 20 trap fractions successfully confirmed 53 of the 59 secreted proteins that had been identified by peptide analysis (Table I). Of these, 39 proteins were identified directly by matching the observed masses to the predicted masses calculated by removing a signal peptide. Remaining proteins were identified by considering additional post-translational modifications. Six proteins could not be identified by whole protein analysis probably because of their low abundance or extensive degradation during culture or sample preparation by secreted proteases (20, 26, 42). Only notable secreted proteins exhibiting modifications other than routine signal peptide loss will be discussed in detail below.
Cytotoxins
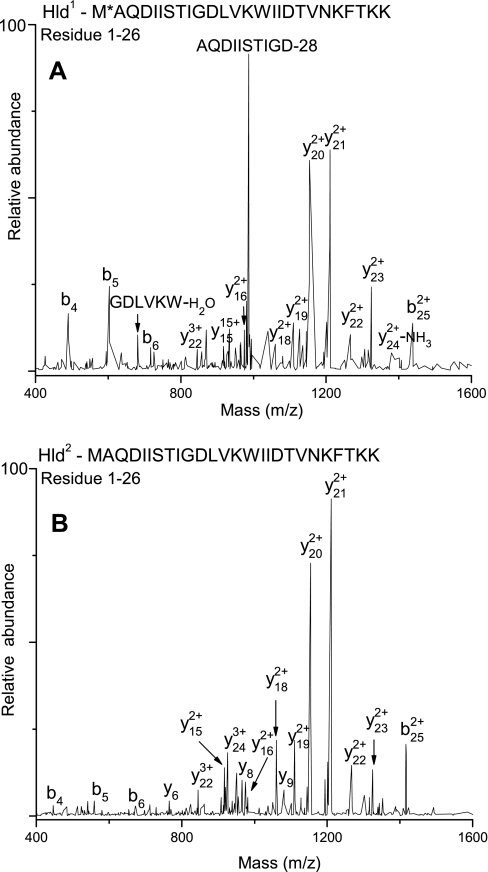
δ-Toxin is a 45 residue (Hld-45; 5009.1 Da) protein that does not contain a classical N-terminal signal peptide. However, the mature form is 26 residues (2978.5 Da) in length, indicating that the first 19 residues constitute the signal peptide (43). Because Hld was identified predominantly as an N-terminal methionine-formylated species, some studies have suggested that the translational start codon has been misassigned such that the native form of Hld is only 26 (Hld-26) residues long and is secreted without a signal peptide (44, 45). In the present study, two forms of Hld (Fig. 2) were identified by peptide MS/MS analysis of extracellular peptide fraction (see “Experimental Procedures”); Hld1 with a monoisotopic mass of 3020.6 Da corresponds to Hld-26 with oxidized N-terminal formylated methionine, and Hld2 (monoisotopic mass of 2976.6 Da) corresponds to Hld-26 with unformylated N-terminal methionine. It is not totally clear whether Hld2 is formed as a result of post-translational deformylation of N-terminal formylated Hld-26 or whether it results from Hld-45 by signal peptide cleavage. The ambiguity arises from the fact that deformylation by peptide deformylase is usually followed by removal of N-terminal methionine by methionine aminopeptidase if the succeeding residue has a small side chain; although Ala is the second residue in Hld-26, we did not find any evidence of methionine removal.
Fig. 2.
Top, LTQ-FT-MS/MS of Hld1. The residue (Met1) marked with an asterisk is the site of modification (formylation and oxidation). Bottom, LTQ-FT-MS/MS of Hld2.
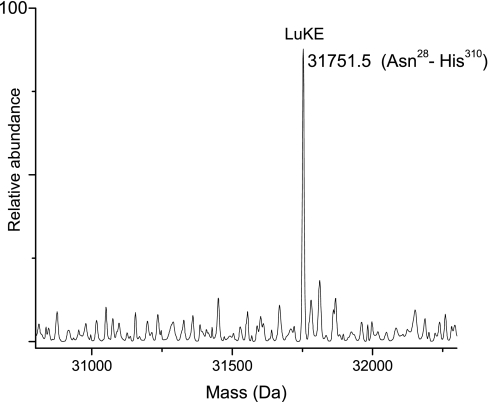
The theoretical mass of leukotoxin LukE (LukE) is 34,819.1 Da. Signal peptide cleavage at Ala27 was predicted by all of the programs, leading to an expected mass of 31,864.5 Da. Several peptides corresponding to LukE (sequence coverage, 68%) including the N-terminal peptide of the mature protein (NTNIENIGDGAEVIKR, residues 28–43) were identified in the Trap 8 tryptic digest. Whole protein mass spectra from Trap 8 did not contain any peak that matched the predicted mass. However, as seen in Fig. 3, a mass of 31,751.5 Da that is 114 Da (±1.5 Da) lower than the predicted mass of LukE was observed in Trap 8. This was identified as LukE with a single C-terminal residue (Asn) truncation. It is most likely that the truncated form resulted from C-terminal degradation by extracellular proteases.
Fig. 3.
Deconvoluted spectrum showing C-terminal truncated form of LukE. The corresponding sequence is shown in parentheses.
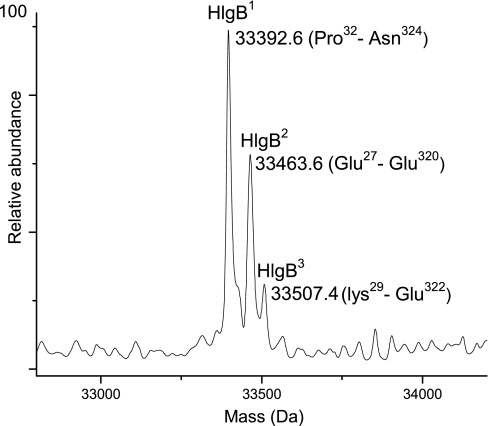
γ-Hemolysin, component B (HlgB) is a 325-residue protein with a theoretical mass of 36,711.0 Da. The predicted signal peptide cleavage site is Ala26, yielding an expected mass of 34,048.7 Da. Peptides corresponding to HlgB were observed in Trap 4 (sequence coverage, 69%); however, whole protein MS analysis of the same trap did not show a mass that matched with the expected mass of HlgB. Instead, three co-eluting protein masses were observed that differed in mass by less than 150 Da, suggesting modified forms of the same protein. The PAWS program revealed that these three protein masses matched well with the three forms of mature HlgB (HlgB1, HlgB2, and HlgB3). As shown in Fig. 4, the observed mass of 33,392.6 Da corresponds to Pro32–Asn324 (HlgB1), the observed mass of 33,463.6 Da corresponds to Glu27–Glu320 (HlgB2), and the observed mass of 33,507.4 Da corresponds to Lys29–Glu322 (HlgB3). The most plausible explanation for the presence of three HlgB forms is truncation of both N-terminal and C-terminal residues due to proteolytic degradation as suggested above for LukE.
Fig. 4.
Deconvoluted spectrum of HlgB showing three truncated forms, HlgB1, HlgB2, and HlgB3. Corresponding sequences are shown in parentheses.
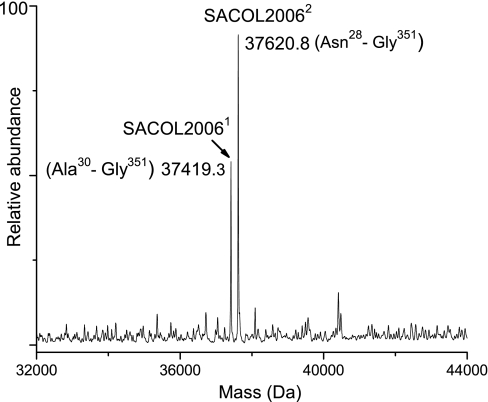
Aerolysin/leukocidin family protein (SACOL2006) has a theoretical mass of 40,434.0 Da, and signal peptide cleavages at Ser29 and Ala27 were predicted for the protein. Peptide analysis of the Trap 10 digest identified SACOL2006 with a sequence coverage of 73%. Interestingly, Trap 10 whole protein MS analysis identified two co-eluting forms of the protein, SACOL20061 (37,419.3 Da) and SACOL20062 (37,620.8 Da), that appear to have formed as a result of signal peptide processing at both Ser29 and Ala27, respectively (Fig. 5). This observation of signal peptide processing at two different sites is quite unusual because signal peptidases generally cleave signal peptides with high fidelity (46). This observation along with another example will be discussed below.
Fig. 5.
Deconvoluted mass spectrum of SACOL2006 suggesting signal peptide processing at two sites. SACOL20061 resulted from signal peptide cleavage at position 29, and SACOL20062 resulted from signal peptide cleavage at position 27.
Superantigenic Toxins
Staphylococcal enterotoxin (Sek) has a theoretical mass of 27,721.1 Da. Signal peptide cleavage at Ala23 was predicted by all the programs, leading to an expected mass of 25,333.1 Da. Peptide analysis of the Trap 12 digest identified Sek with a sequence coverage of 82%. Trap 12 whole protein MS analysis identified Sek as a C-terminal truncated protein because the observed mass of 24,699.6 Da coincided with the mass of the mature Sek protein formed by signal peptide cleavage at position Ala23 and removal of residues YKTI from the C terminus (24,698.4 Da). The signal peptide cleavage at Ala23 was further confirmed by observation of the N-terminal peptide QGDIGIDNLR (residues 24–33). The presence of truncated Sek is most probably due to the degradation of the protein.
The theoretical mass of staphylococcal enterotoxin type I (Sei) is 28,184.6 Da. Signal peptide cleavage at Ala26 was predicted by all the programs, leading to an expected mass of 25,076.9 Da. Sei was identified from the Trap 20 digest with a sequence coverage of 77%. Trap 20 whole protein MS analysis identified Sei as a C-terminal truncated protein with an observed mass of 24,848.0 Da corresponding to signal peptide cleavage at position Ala26 and removal of C-terminal residues TE. The signal peptide cleavage at Ala26 was further confirmed by observation of N-terminal peptide DVGVINLRNFYANYEPE (residues 27–43). C-terminal truncated Sei may be formed by proteolytic degradation.
Proteases
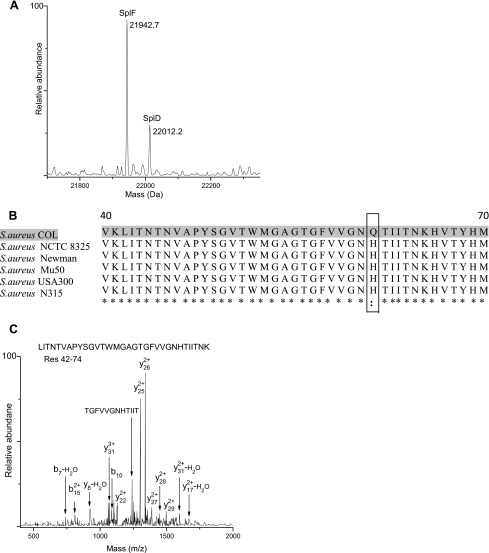
Serine proteases SplA, SplB, SplC, SplE, and SplF and cysteine protease precursor SspB were identified in S. aureus COL extracellular medium by peptide MS/MS as well as whole protein MS analysis, and signal peptide cleavage was the only modification observed. Serine protease SplD was somewhat more interesting. Its theoretical mass is 25,669.1 Da. Signal peptide cleavage at Ala26 was predicted by the prediction programs, yielding an expected mass of 22,001.8 Da. Peptide analysis of the Trap 5 protein digest identified SplD with a high sequence coverage of 77%. Nevertheless, Trap 5 did not yield a whole protein mass that matched the predicted mass. Instead, a recurring mass peak at 22,012.2 Da that is 10.4 Da higher than the predicted mass was observed co-eluting with SplF (Fig. 6A). Because SplD shares 96% sequence identity with SplF (47), we expected it to co-elute with SplF. The observed 10.4 Da mass difference was suspected to be a sequencing error. Examination of single nucleotide substitutions that could account for the observed mass discrepancy yielded two potential candidates for a sequencing error, Ser → Pro with a mass difference of 10 Da and Gln → His with a mass difference of 9 Da. Phylogenetic analysis combined with mass spectrometry has been used previously in our laboratory to identify a large number of sequencing errors in Bacillus subtilis strain 168 (48). A similar approach was utilized in the present study. Multiple amino acid sequence alignment of SplD from different S. aureus strains using the ClustalW program indicated high sequence homology between S. aureus COL and other strains (99.6% sequence identity) and revealed Gln68 → His68 as the plausible sequencing error (Fig. 6B). This was in fact confirmed by MS/MS analysis of the peptide LITNTNVAPYSGVTWMGAGTGFVVGNHTIITNK (residues 42–74) (Fig. 6C).
Fig. 6.
Identification of sequencing error in SplD. A, whole protein mass spectrum of co-eluting SplF and SplD. B, multiple sequence alignment (residues 40–70) of SplD from S. aureus COL and other strains of S. aureus. The suspected sequencing error is shown in the box. An asterisk represents identity among the aligned residues, and two dots represent strong similarity. C, LTQ-FT-MS/MS spectrum of SplD peptide consisting of residues 42–74 that confirm the Gln68 → His68 sequencing error.
A comparison of SplD nucleotide sequences from various S. aureus strains revealed that the CAT codon at position 68 is highly conserved in all strains, suggesting that the observed discrepancy is not a single nucleotide polymorphism. Only SplD of S. aureus COL has a CAA codon at position 68 in the reference genome, and we believe that an A→T nucleotide sequencing error at this position resulted in the observed Gln68 → His68 sequencing error.
Lipolytic Enzymes
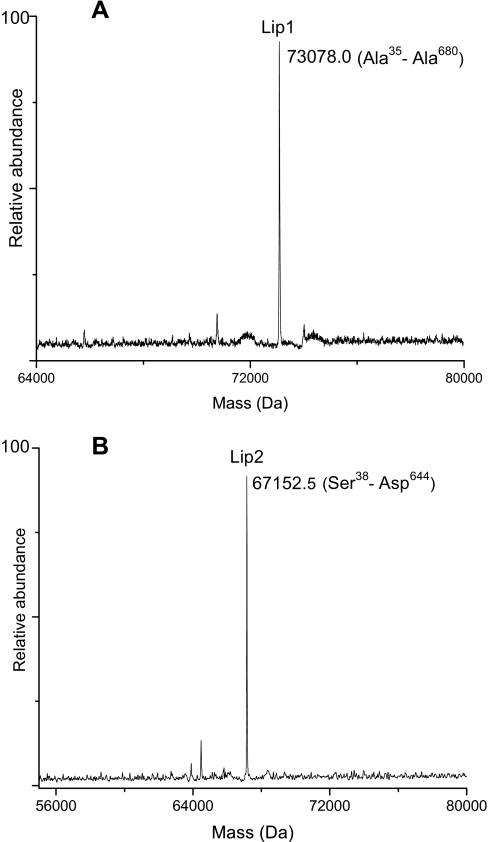
Rollof and Normark (49) have reported that lipase (76 kDa) in S. aureus strain TEN 5 is secreted into the culture medium as a prolipase (82 kDa) after cleavage of the signal peptide; subsequent processing (removal of propeptide) of the prolipase results in a mature lipase (44–45 kDa). It is important to point out that the molecular mass of the observed prolipase is significantly higher than the predicted mass (73 kDa); the reason for the observed mass difference, however, was not explained. In the present study, Lipase1 (Lip1) was identified by peptide analysis of the Trap 8 digest (96% sequence coverage), and Lipase2 (Lip2) was identified from the Trap 9 digest with a sequence coverage of 71%. Lip1 is a 680-residue protein (76,675.3 Da) and is predicted to contain a signal peptide domain (residues 1–34) followed by propeptide domain (residues 35–290) and a mature lipase domain consisting of 390 residues (44,345.3 Da). Similarly, Lip2 (71,276.8 Da) is predicted to contain a signal peptide (37 amino acids), a propeptide (258 amino acids), and a mature lipase with a predicted mass of 44,071.6 Da. The observed mass for Lip1 (73,078.0 Da) obtained from Trap 8 and the observed mass of Lip2 (67,152.8 Da) obtained from Trap 9 matched with the predicted masses corresponding to the signal peptide cleavage at positions 34 (73,077.1 Da) and 37 (67,152.0 Da), respectively, and revealed that after 8 h of growth both Lip1 and Lip2 were present in the extracellular medium as unprocessed proenzymes (Fig. 7). We did not find any evidence of mature lipase forms in the extracellular medium. In contrast to previous studies based on SDS-PAGE and Western blot analysis where the observed masses of staphylococcal prolipases were inexplicably higher than predicted masses (49–51), our study provided accurate mass determination of prolipases, allowing confident identification of the proteins.
Fig. 7.
Deconvoluted mass spectra showing proenzyme forms of Lip1 (A) and Lip2 (B).
Peptidoglycan Hydrolases
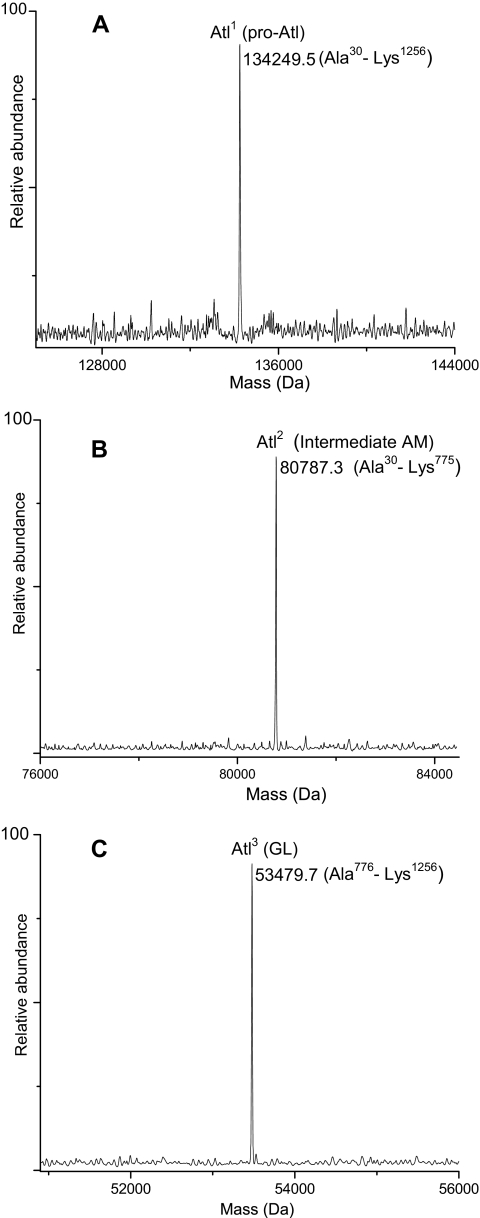
Bifunctional autolysin (Atl; 138 kDa) is a bacteriolytic enzyme capable of causing cell lysis. It consists of two functionally distinct domains. Several studies have reported that Atl undergoes proteolytic processing to generate 62-kDa (amidase) and 51-kDa (glucosaminidase) extracellular lytic enzymes (52, 53). In S. aureus COL, Atl is a 1256-amino acid protein (137,334.9 Da) and is predicted to contain a signal peptide domain (residues 1–29), a propeptide domain (residues 30–198), an N-acetylmuramoyl-l-alanine amidase domain (AM; residues 199–775), and an endo-β-N-acetylglucosaminidase domain (GL; residues 776–1256). The predicted masses of mature AM and GL are 63,008.7 and 53,479.5 Da, respectively. Atl was identified in Traps 8 and 9, and peptide analysis yielded a high sequence coverage of 99%. Whole protein MS analysis of the same traps resulted in the identification of three gene products of Atl (Fig. 8). The observed mass of 134,249.5 Da corresponding to pro-Atl form (Ala30–Lys1256, Atl1) matched closely with the predicted mass (134,248.2 Da), confirming the signal peptide cleavage at Ala29. The other two gene products, Atl2 and Atl3, appear to have formed as a result of proteolytic processing of Atl1. An intermediate form of AM with a mass of 80,787.3 Da (Atl2) corresponding to sequence Ala30–Lys775 was identified. The observed mass of 53,479.7 Da corresponding to the mature GL (Ala776–Lys1256, Atl3) confirmed proteolytic processing at Lys775-Ala776. The mature AM form, however, was not detected in the extracellular medium of S. aureus COL. This suggests that either the intermediate AM form requires more time (>8 h) to undergo proteolytic processing or the enzyme responsible for its processing is not present.
Fig. 8.
Deconvoluted mass spectra showing three Atl gene products identified in the present study. A, Atl1, pro-Atl form. B, Atl2, intermediate AM form. C, Atl3, mature GL form.
LysM domain protein (SACOL0723) has a theoretical mass of 28,186.8 Da (residues 1–265). Signal peptide cleavage at Ala23 and Ala25 was predicted by the programs, yielding expected masses of 25,831.0 and 25,631.8 Da, respectively. Peptide analysis of the Trap 2 digest identified SACOL0723 with a high sequence coverage of 80%. Whole protein MS analysis of Trap 2, however, did not uncover any mass that matched either of the expected masses. Instead, we observed two masses that suggested degradation of SACOL0723 (cleavage between Gly138 and Gly139) into two fragments, SACOL07231 and SACOL07232. The observed mass of SACOL07231 (11,849.0 Da) corresponds to an N-terminal protein fragment (Ser26–Gly138), and the observed mass of SACOL07232 (13,801.0 Da) corresponds to a C-terminal protein fragment (Gly139–His265). The cleavage between Gly138 and Gly139 was further corroborated by identification of C-terminal non-tryptic peptide GYLIMPNQTLQIPNGGSG (residues 121–138) by peptide MS/MS analysis.
The theoretical mass of N-acetylmuramoyl-l-alanine amidase domain protein (SACOL2666) is 69,253.2 Da (609 amino acids). Signal peptide cleavage at Ala27 was predicted by all the programs, leading to the expected mass of 66,309.7 Da. Peptide analysis of the Trap 6 digest identified SACOL2666 with a high sequence coverage of 89%. Whole protein MS analysis of Trap 6 did not provide any mass that was close to the expected mass of SACOL2666. Instead, masses of 15,296.4, 42,252.1, and 7810.9 Da corresponding to fragments Thr29–Thr163, Asp172–Asp546, and Tyr547–Lys619, respectively, were observed, indicating degradation of the protein.
Miscellaneous Enzymes
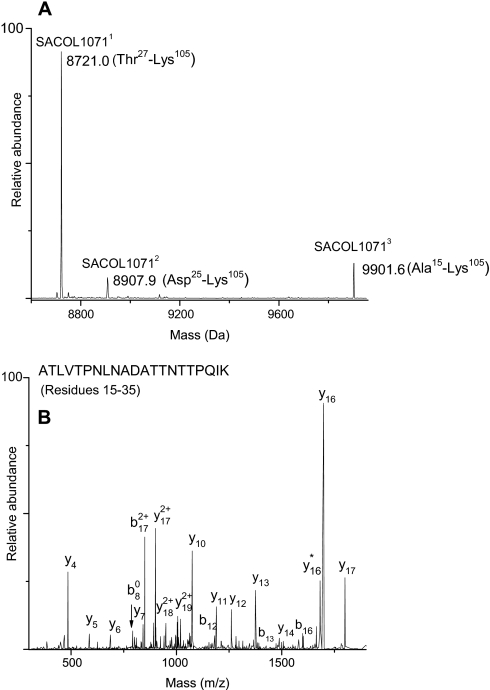
For SACOL1071, signal peptide processing was observed at multiple sites (Fig. 9A). The theoretical mass of SACOL1071 is 11,344.8 Da, and signal peptide cleavage at Ala24 and Ala26 was predicted, leading to expected masses of 8907.1 and 8721.0 Da, respectively. Peptide analysis of the Trap 3 digest identified SACOL1071 with a high sequence coverage of 91%. Trap 3 whole protein MS analysis identified two co-eluting forms of the protein that represented signal peptide processing at Ala26 as well as Ala24, SACOL10711 with an observed mass of 8720.1 Da (Thr27–Lys105) and SACOL10712 with an observed mass of 8907.1 Da (Asp25–Lys105). A comparison of the protein peak intensities suggested that SACOL10711 is the major form present in the extracellular medium. Furthermore, a whole protein mass of 9901.6 Da (SACOL10713) was also observed co-eluting with SACOL10711. This matched the predicted mass of SACOL1071 (9902.0 Da) after removal of N-terminal residues 1–14. This observation implicated another signal peptide processing site for this protein. Identification of the N-terminal non-tryptic peptide ATLVTPNLNADATTNTTPQIK (residues 15–35), which represents the N-terminal peptide of the mature protein via peptide MS/MS analysis, confirmed signal peptide processing at position 14 (Fig. 9B). The general attributes of the 14-residue signal sequence of SACOL1071 do not conform to those typical for a Sec-type signal peptide. The implications of observing multiple cleavage sites in SACOL1071 will be discussed later.
Fig. 9.
A, deconvoluted mass spectrum showing the three forms of SACOL1071 that formed as a result of signal peptide processing at different sites: SACOL10711, signal peptide cleavage at position 26; SACOL10712, signal peptide cleavage at position 26; and SACOL10713, signal peptide cleavage at position 14. B, LTQ-FT-MS/MS spectrum of N-terminal peptide of SACOL10713.
Surface Adhesins
The theoretical mass of secretory extracellular matrix and plasma-binding protein (Empbp) is 38,484.9 Da (residues 1–340). Signal peptide cleavage is predicted at Ala24 and Ala26, yielding expected masses of 35,781.6 and 35,582.4 Da, respectively. Peptide analysis of the Trap 15 digest identified Empbp with a sequence coverage of 65%. Whole protein MS analysis of Trap 15 did not show any protein mass close to the predicted values of mature Empbp. Instead, we observed two masses that suggested degradation of Empbp into two fragments. An observed mass of 12,891.8 Da matched that of an N-terminal protein fragment encompassing residues Ser27–Thr143 (Empbp1), and a peak at 22,708.9 Da matched the mass of a C-terminal protein fragment encompassing residues Gln144-Val340 (Empbp2). The sum of the two fragments (35,600.7 Da) matched closely with the expected mass (35,582.4 + 18 Da) corresponding to signal peptide cleavage at Ala26.
SdrH protein (SdrH) is a 419-residue protein (46,630.4 Da) with a predicted mass of 43,094.9 Da after signal peptide cleavage at position Ala32. SdrH was identified with high sequence coverage (81%) in the digest of Trap 3, but whole protein MS analysis of the same trap did not show any mass that was close to the predicted mass. Instead, we detected a C-terminally truncated protein with an observed mass of 38,085.1 Da (Lys33–Lys376) formed by the removal of residues 377–419. The signal peptide cleavage at position Ala32 was further confirmed via peptide MS/MS analysis by identification of N-terminal peptide KDNLNGEKPTTNLNHNITSPSVNSEMNNNETGTPHESNQTGNEGTGSNSR (residues 33–82). Peptide analysis also indicated that the C-terminal peptides corresponding to the last 44 residues were missing.
Proteins with Unknown Functions
Staphylococcal secretory antigen SsaA2 (SsaA2) has a theoretical mass of 29,327.1 Da (267 residues) and a predicted mass of 26,715.1 Da after signal peptide cleavage at Ala27. Peptides corresponding to SsaA2 were found in Trap 3. However, whole protein MS analysis of the same trap did not provide a mass that corresponded to full-length mature protein. Instead, SsaA2 was identified as two protein fragments, an N-terminal protein fragment with an observed mass of 12,210.3 Da (Ser28–Gly127, SsaA21) and a C-terminal fragment with an observed mass of 14,294.9 Da (Ala131–His267, SsaA22). Furthermore, observation of the peptide ASYSTSSNNVQVTTTMAPSSNGR (residues 131–153) that resulted from non-tryptic cleavage at the N terminus of the peptide confirmed degradation of the protein into two fragments.
Staphyloxanthin biosynthesis protein (SACOL2295) has a theoretical mass of 17,424.8 Da. Signal peptide cleavage is predicted at Ala22 and Ala27, yielding expected masses of 15,297.2 and 14,739.7 Da, respectively. Peptide analysis of the Trap 3 digest identified this protein with a high sequence coverage of 71%. The observed mass (14,756.7 Da) of SACOL2295 obtained from Trap 3 whole protein MS analysis was 17 Da higher than the predicted mass (14,739.7 Da). This led us to propose two modifications for SACOL2295: loss of signal peptide and oxidation of the mature protein. Thiol groups of cysteine residues are known to be sensitive toward oxidation; Wolf et al. (54) have already reported the oxidation of the Cys69 residue in SACOL2295. In this study, it is highly possible that Cys69 is the site of the proposed modification; however, we could not confirm the modification site by peptide MS/MS experiments as the residue was not mapped.
Virulence factor EsxA (EsxA) has a theoretical mass of 11,036.2 Da, and no Sec-type signal sequence is predicted at its N terminus. In the present study, peptide analysis identified EsxA in the Trap 3 digest with high sequence coverage (99%), and there was no evidence of loss of signal peptide from the protein. Burts et al. (55) identified EsxA along with EsxB in S. aureus strain Newman and have shown that they are exported via an ESAT-6 secretion pathway (type VII pathway). Because EsxA of S. aureus COL shares 100% sequence identity with S. aureus Newman, we expect that it is similarly exported. Whole protein MS analysis of Trap 3 did not uncover any mass that was close to the theoretical mass (11,036.2 Da). Instead, we observed a very intense peak at 10,905.2 Da that was 131 Da lower than the theoretical mass, suggesting the removal of N-terminal methionine by methionine aminopeptidase. Identification of the N-terminal methionine truncated peptide AMIKMSPEEIRAKSQSYGQGSDQIRQILSDLTRAQGE (residues 2–38) corroborated this hypothesis. The whole protein MS analysis in combination with peptide analysis definitively confirmed the absence of signal peptide processing in EsxA. In mycobacterium tuberculosis, proteins EsxA and EsxB form a tight 1:1 dimer (56) that is required for stability of the proteins, and this interaction is thought to take place in the cytosol prior to protein export. Burts et al. (55) reported that in S. aureus Newman EsxB is required for the synthesis and secretion of EsxA and vice versa. This led them to believe that EsxA and EsxB also form a heterodimer in S. aureus. However, in the present study, EsxB was not identified despite the fact that EsxA yielded a rather intense signal, suggesting that EsxB may not be required for secretion of EsxA in S. aureus COL. Sundaramoorthy et al. (57) also did not observe heterodimer formation following incubation of S. aureus EsxA and EsxB proteins; instead, EsxA crystallized as a homodimer.
The predicted mass of SACOL0270 is 30,421.1 Da following signal peptide cleavage at Ala24. Peptides corresponding to this protein were found in Trap 12; however, whole protein MS analysis of the same trap did not show any mass that matched the predicted value within the experimental error. Instead, an unmatched mass of 30,377.9 Da was observed that we believe is SACOL0270. The observed mass discrepancy of −42 Da is most probably due to arginine modification. Hydrolysis of arginine to form ornithine is a well known modification that results in a mass shift of −42 Da.
Post-translational Modifications of Non-secretory Proteins
Similar to secreted proteins, post-translational modifications of cell wall-anchored proteins, membrane proteins, lipoproteins, and cytoplasmic proteins were characterized, and a detailed discussion on the observed modifications is provided in the supplemental data.
Stable Cleaved Signal Peptides and Signal Peptide Fragments
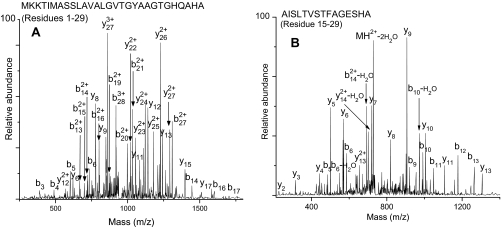
Several reports have suggested that after cleavage of a signal peptide from a preprotein rapid removal and degradation of the signal peptide is important for proper functioning of the export machinery (58, 59). Nevertheless, peptide analysis of some trap fractions indicated the presence of stable cleaved signal peptides and signal peptide fragments derived from a few secreted proteins. Because TCA does not precipitate peptides efficiently (60), we suspected that there may be more peptides in the extracellular medium than those identified in the TCA protein extract. Using the procedure outlined under “Experimental Procedures,” we attempted to isolate the peptides present in the S. aureus COL stationary phase culture. Indeed, RPLC-ESI-LTQ-FT-MS/MS of the peptide extract revealed the presence of several peptides including stable cleaved signal peptides of five proteins, Sle1, SACOL0723, SceD, IsaA, and SACOL2295 (supplemental Table 6), and signal peptide fragments of 18 secreted proteins (Table II). As an example, MS/MS spectra of cleaved signal peptide of IsaA and signal peptide fragment of SACOL1164 identified in the present study are shown in Fig. 10. It is noteworthy that all of the observed signal peptide fragments are from C-terminal portions of respective signal peptides containing SPase I-cleaved sites and appear to have formed by cleavage in the hydrophobic region of the signal peptide. To account for the signal peptide fragments observed in the present study, we have proposed cleavage sites in the SPase I-processed signal peptides as shown in Table II. The implications of these observations will be discussed later.
Table II. Observed signal peptide fragments and proposed cleavage sites to account for fragments detected.
Peptide Mascot scores are provided in supplemental Table 7.
| Gene ID | Observed signal peptide fragments | Proposed cleavage sites for degradation of signal peptide |
|---|---|---|
| SACOL0480 | VGVLATGVVGYGNQADA | MKFKKVLVATAM↓VGVLATGVVGYGNQADA |
| SACOL0723a | FAITATSGAAAFLTHHDAQA | MKKLA↓FAITATSGAAAFLTHHDAQA |
| SACOL0860 | VLTLVVVSSLSSSANA | MTEYLLSAGICMAIVSILLIGMAISNVSKGQYAKRFFYFATSCL↓VLTLVVVSSLSSSANA |
| SACOL0908 | ALVLTTVGSGFHSSSNYNGINNVAKA | MNKKLLTRTLIAS↓ALVLTTVGSGFHSSSNYNGINNVAKA |
| SACOL1062 | LTLVGSAVTAHQVQA | MAKKFNYKLPSMVA↓LTLVGSAVTAHQVQA |
| SACOL1164 | AISLTVSTFAGESHA | MKKNFIGKSILSIA↓AISLTVSTFAGESHA |
| SACOL1864 | TILTSITGVGTTMVEGIQQTAKA | MNKNIIIKSIAAL↓TILTSITGVGTTMVEGIQQTAKA |
| SACOL1868 | TILTSVTGIGTTLVEEVQQTAKA | MNKNVVIKSLAAL↓TILTSVTGIGTTLVEEVQQTAKA |
| SACOL2003 | ANLLLVGALTDNSAKA | MVKKTKSNSLKKVATLAL↓ANLLLVGALTDNSAKA |
| SACOL2088a | SLAVGLGIVAGNAGHEAHA | MKKTLLAS↓SLAVGLGIVAGNAGHEAHA |
| SACOL2197 | LGLLSTVGAALPSHEASA | MKLKSFVTATLA↓LGLLSTVGAALPSHEASA |
| SACOL2291 | AGFATIAIASGNQAHA | MKKIATATIAT↓AGFATIAIASGNQAHA |
| SACOL2295a | ATTLTAGIGTALVGQAYHADA | MKKLVT↓ATTL↓TAGIGTALVGQAYHADA |
| TAGIGTALVGQAYHADA | ||
| SACOL2418 | TITLATMISNGEAKA | MKNKYISKLLVGAA↓TITLATMISNGEAKA |
| SACOL2421 | SVSLLAPLANPLLENAKA | MLKNKILTTTL↓SVSLLAPLANPLLENAKA |
| SACOL2557 | AVLFSADFTYQSVEQTHQSHA | MEYKKILIRLLIAF↓AVLFSADFTYQSVEQTHQSHA |
| SACOL2584a | IMASSLAVALGVTGYAAGTGHQAHA | MKKT↓IMA↓SSL↓AVALGVTGYAAGTGHQAHA |
| SSLAVALGVTGYAAGTGHQAHA | ||
| AVALGVTGYAAGTGHQAHA | ||
| SACOL2660 | GTLIGVTVVENSAPTSKQAQA | MNKTSKVCVAATLAL↓GT↓LIGVTVVENSAPTSKQAQA |
| LIGVTVVENSAPTSKQAQA |
a Stable cleaved signal peptide also identified in the extracellular medium by LTQ-FT-MS/MS analysis.
Fig. 10.
A, LTQ-FT-MS/MS spectrum of stable cleaved signal peptide of Isa. B, LTQ-FT-MS/MS spectrum of signal peptide fragment of SACOL1164.
Signal Peptide Prediction Accuracy
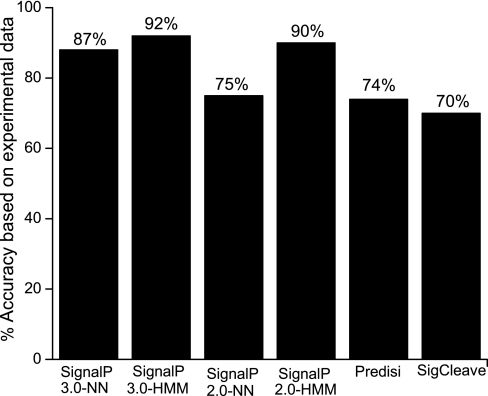
The prediction accuracy of computational programs commonly used to predict signal peptides and cleavage site position has been debated. Few studies have evaluated the performance of signal peptide prediction programs using experimentally verified signal peptide data from different organisms (61–63). To the best of our knowledge, the suitability of the commonly used prediction programs to predict secretory proteins and signal peptide cleavage sites in S. aureus has not been reported. Results from the present study are shown in Fig. 11. It is evident from the figure that SignalP 3.0-HMM (92%), SignalP 2.0-HMM (90%), and SignalP 2.0-NN (87%) are superior in predicting the correct cleavage sites. In contrast, the prediction accuracies of SignalP 2.0-NN (75%), PrediSi (74%), and SigCleave (70%) are substantially lower. Although we observed only a slight improvement in the performance of SignalP 3.0-HMM compared with SignalP 2.0-HMM (older version), there is a major performance improvement in SignalP 3.0-NN in comparison with SignalP 2.0-NN. False negatives were observed for PrediSi (7%), SigCleave (5%), and SignalP-NN (2%), whereas there were none from SignalP-HMM programs. In addition, we tested the prediction programs on a negative set of 58 non-secretory proteins. False positive predictions were observed for all the programs: SignalP 3.0-HMM, 5%; SignalP 2.0-HMM, 7%; SignalP 3.0-NN, 10%; SignalP 2.0-NN, 12%; PrediSi, 4%; and SigCleave, 28%. It is noteworthy that false positive predictions from SigCleave were particularly high. Based on our results, SignalP 3.0-HMM appears to be the best program in predicting the signal peptide cleavage sites accurately. This observation is different from that reported by Zhang and Henzel (63) who found that SignalP 2.0-NN gave the best result when tested on an experimentally (Edman analysis) verified data set consisting of 270 recombinant human proteins. Recently, Gupta et al. (62) identified signal peptide cleavage sites for 94 proteins in a comprehensive proteomics analysis of Gram-negative bacterium Shewanella oneidensis MR-1 via LC-MS/MS. They tested two programs, PrediSi and SignalP. From their results, it appears that PrediSi performed better than SignalP. The varying performances of prediction programs may be associated with the different organisms, eukaryotes, Gram-negative prokaryotes, and Gram-positive prokaryotes, studied. These observations strongly indicate the need for experimental data on signal peptides as it will help fine tune the existing programs.
Fig. 11.
Prediction accuracies of various signal peptide prediction programs.
DISCUSSION
Secreted Proteins of S. aureus COL and Their Post-translational Modifications
Secretory proteins of S. aureus are of particular importance to virulence and pathogenesis. Depending on the S. aureus strain, ∼70–90 proteins can be expected to be secreted into the extracellular milieu (29). Several proteomics studies have investigated the identification of S. aureus extracellular proteins produced by different strains using a variety of gel-based techniques. Bernardo et al. (19) identified 13 extracellular proteins produced by S. aureus ATCC 29213 and ATCC 43300 using 1DE and MALDI-TOF-MS. Using 2DE and N-terminal sequencing, Nakano et al. (42) identified 29 proteins in MRSA strains. Ziebandt et al. (27) identified 18 and 19 proteins from culture supernatants of S. aureus COL and RN6390, respectively, using N-terminal sequencing or MALDI-TOF-MS. Using a combination of techniques (MALDI-TOF-MS/N-terminal sequencing/LC-MS/MS), Ziebandt et al. (26) also studied the influence of accessory gene regulator (agr) and alternative sigma factor σB (sigB) on the expression of extracellular proteins in RN6390 and RN6911 and identified a total of 43 proteins including secreted, cell wall-associated, and cytoplasmic proteins. Using 1DE and 2DE with LC-MS/MS, MALDI-TOF-MS, and SEDI-MS/MS, Pocsfalvi et al. (64) identified 119 proteins in S. aureus ATCC 14458 including 22 secreted proteins containing potential signal peptides. Burlak et al. (24) have reported the identification of 256 proteins from extracellular extracts of S. aureus MW2 and LAC using 2DE and automated direct infusion MS/MS. However, only 38 of these were predicted to be secreted based on the presence of putative signal peptides. Similarly, Jones et al. (25) reported the identification of 541 proteins from culture supernatants of S. aureus UAMS-1 using 1DE and nano-LC-MS/MS of which only 41 proteins have predicted export signals for secretion into the extracellular milieu. Although previous studies confirmed the presence of a given protein in the S. aureus extracellular milieu, they failed to provide a detailed characterization of the proteins. This is a particular weakness of peptide-based analyses because only a fraction of the total theoretical peptide population of a given protein may be identified. The present study provides a comprehensive picture of the secretome of S. aureus COL by identification of the proteins and characterization of their post-translational modifications.
All but two of the 59 secreted proteins identified in the present study were predicted to possess Sec-type signal peptides, and we were able to verify the signal peptide loss in these proteins; this confirmed that they were exported via a Sec-dependent pathway. Also, we confirmed that EsxA, which is known to be exported via the ESAT-6 pathway, does not contain a cleavable signal peptide, and the only modification observed was the removal of N-terminal methionine.
In a majority of proteins, signal peptide loss was the only modification observed. Other observed modifications included proteolytic processing, N-terminal formylation, methionine removal, oxidation, formation of ornithine, and protein truncation. Degradation of a few secreted proteins observed in the present study indicated proteolytic activity in the culture supernatants. This observation has been reported by several investigators, and it has been suggested that the secreted proteins are degraded by the action of their secreted proteases during culture and sample preparation (20, 26, 42, 65–67). Degradation of proteins by extracellular proteases has also been reported in other microorganisms; in B. subtilis, it has been demonstrated that mutants lacking proteases exhibit a substantial increase in the abundance of various extracellular proteins compared with the wild type (65–67). Degradation of extracellular proteins may be due to slow or incorrect post-translational folding of the proteins or to the presence of exposed protease recognition sequences in the folded protein (52, 54). It may also be a means of nutrient recycling for survival (20, 66, 68). However, in the present study, a majority of the proteins were refractory to nonspecific protease activity because they were identified as intact proteins.
Predicted Versus Observed Secreted Proteins of S. aureus COL
52 S. aureus COL secreted proteins of the 71 predicted proteins (29) were unambiguously identified by LTQ-FT-MS/MS analysis from three or more peptides with a Mascot score above the threshold of significance. Seven predicted secretory proteins, putative uncharacterized protein (SACOL0129), exotoxin 3 (SACOL0468), exotoxin (SACOL0470), exotoxin 3 (SACOL0478), surface protein (SACOL0479), cell wall hydrolase (SACOL1264), and hypothetical protein (SACOL1870), were identified from one or two peptide sequences (Mascot score, p < 0.05) and were not included in the list of identified secreted proteins because of the stringent protein identification criteria used in the present study. These proteins are apparently present in the extracellular medium in low abundance. The remaining 12 predicted proteins not detected in the extracellular medium are probably not secreted by S. aureus COL under the conditions studied or are present in trace amounts. There is no evidence from the published literature to indicate that these proteins are indeed secreted by S. aureus COL. Recent proteomics data on membrane, cell wall, and extracellular proteins of B. subtilis revealed that a good number of proteins that are predicted to contain cleavable Sec-type signal peptide and an SPase I recognition site are not secreted into the medium but are in fact retained in the membrane (69); this could be the case for the predicted proteins not identified in the present study. Furthermore, five proteins that were not predicted to be secreted because of the presence of transmembrane domains (Nuc and SACOL0442), the presence of Thr in the −1 position (SACOL2179) or +1 position (SACOL1071), or the presence of Tyr in the +1 position (SACOL0270) relative to the cleavage site (29) were identified in the present study by whole protein and peptide MS analysis as being secreted proteins released into the medium by removal of Sec-type signal peptides. This indicates that Thr in the −1 and +1 positions and Tyr in the +1 position are accepted by S. aureus SPase I; a discussion of the amino acid residues accepted by SPase I at positions −3 to +1 relative to the signal peptide cleavage site and the frequency of their occurrence is presented below.
A comparison of secreted proteins identified in the present study with those identified in various S. aureus strains (29) showed an overlap between identified proteins and revealed potential vaccine and drug candidates. Of the 56 secreted proteins that have been identified in other S. aureus strains (29), 48 are encoded in S. aureus COL. 43 of these (Table I) were identified in the present study by three or more peptides, and three proteins (SACOL0478, SACOL0479, and SACOL1870) were identified by one or two peptides. Two proteins (SACOL0209 and SACOL2691), however, were not identified.
Signal Peptides and Cleavage Sites of S. aureus COL Secreted Proteins
Table III lists the signal sequences of 59 S. aureus COL proteins (secretory and cell wall-associated proteins) identified in the present study. In Gram-positive bacteria, SPase I recognizes residues at positions −3 and −1 with respect to the cleavage site, and Ala-X-Ala is the most common sequence preceding the signal peptide cleavage site (70). It is evident from Table III that the signal sequences of S. aureus COL proteins identified in the present study all contain the N-, H-, and C-domains of a typical Sec-type signal peptide. The length of the signal peptides varies from 23 to 60 amino acids with an average of 31 residues. Table IV lists the residues accepted at and around the verified SPase I cleavage sites of S. aureus COL proteins identified in the present study. In a majority of the proteins, Ala is predominantly preferred at −3 (77%) and −1 (97%) positions. Residues Val (10%) and Ser (10%) are also accepted at the −3 position and occur with a higher frequency than Thr, Leu, and Ile (2%). With respect to the −1 position, residues Ser and Thr are also accepted but with a markedly lower frequency (2%) than Ala. The residues found in −3 and −1 positions of S. aureus COL signal sequences are small and uncharged; this is in agreement with the assumption that side chains of residues at the −1 and −3 positions are bound in two shallow hydrophobic substrate-binding pockets (S1 and S3) of the active site of SPase I (71). In contrast, the side chain of the residue at position −2 is thought to be pointing outward from the enzyme. As a consequence, a variety of residues appear to be tolerated at the −2 position including Lys, Asn, Gln, His, Asp, Ser, Glu, Leu, Phe, Tyr, Gly, and Arg with a preference for Lys (23%). There appears to be a preference for Ala (31%) at the +1 position. Other residues including Ser, Glu, Lys, Asp, Gln, Thr, Phe, Asn, Tyr, and Leu were also accepted in the +1 position. Approximately 78% of S. aureus COL secreted proteins identified in the present study possess signal sequences that contain a helix-breaking residue (mostly glycine) in the middle of the H-domain, and about 50% contain a helix-breaking residue (proline or glycine) at position −7 to −4 relative to the predicted processing site for SPase I. Helix-breaking residues found at the end of the H-domain are thought to facilitate cleavage by SPase I (71).
Table III. Signal sequences and observed and predicted signal peptide cleavage sites of S. aureus COL proteins (secretory and cell wall-associated proteins) identified in present study.
| Gene ID | Signal peptide with SPase Ia (−3 to −1) | SPase I +1 | Cleavage site |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Obsd. clvg. siteb | SignalP 3.0 |
SignalP 2.0 |
PrediSi | SigCleave | |||||
| NN | HMM | NN | HMM | ||||||
| SACOL0024 | MKALLLKTSVWLVLLFSVMGLWQVSNA | Ala | 27 | 27 | 27 | 27 | 29 | 27 | 27 |
| SACOL0050 | MNKNSKKKLDFLPNKLNKYSIRRFTVGTASILVGATLIFGVANDQAEA | Ala | 48 | 48 | 48 | 48 | 48 | 48 | 48 |
| SACOL0078 | MKKCIKTLFLSIILVVMSGWYHSAHA | Ser | 26 | 26 | 26 | 26 | 26 | 26 | 26 |
| SACOL0263 | MEDVLYMKKLTAAAIATMGFATFTMAHQADA | Ala | 31 | 31 | 31 | 29 | 31 | 31 | 31 |
| SACOL0095 | MKKKNIYSIRKLGVGIASVTLGTLLISGGVTPAANA | Ala | 36 | 36 | 36 | 36 | 36 | 36 | 36 |
| SACOL0119 | MKKLATVGSLIVTSTLVFSSMPFQNAHA | Asp | 28 | 28 | 28 | 28 | 28 | 28 | 28 |
| SACOL0270 | MKKTILLTMTTLTLFSMSPNSAQA | Tyr | 24 | 24 | 24 | 24 | 24 | 24 | 24 |
| SACOL0303 | MNKISKYIAIASLSVAVTVSAPQTTNSTAFA | Lys | 31 | 31 | 31 | 21 | 31 | 29 | 20 |
| SACOL0317 | MLRGQEERKYSIRKYSIGVVSVLAATMFVVSSHEAQA | Ser | 37 | 37 | 37 | 37 | 37 | 37 | 37 |
| SACOL0442 | MFKKYDSKNSIVLKSILSLGIIYGGTFGIYPKADA | Ser | 35 | 35 | 35 | None | 35 | None | None |
| SACOL0480 | MKFKKVLVATAMVGVLATGVVGYGNQADA | Lys | 29 | 29 | 29 | 29 | 29 | None | 19 |
| SACOL0507 | MQKKVIAAIIGTSAISAVAATQANA | Ala | 25 | 25 | 25 | 25 | 25 | 25 | 25 |
| SACOL0610 | MINRDNKKAITKKGMISNRLNKFSIRKYTVGTASILVGTTLIFGLGNQEAKA | Ala | 52 | None | 52 | 52 | 52 | 52 | 52 |
| SACOL0669 | MKKLLTASIIACSVVMGVGLVNTSAEA | Ala | 27 | 27 | 27 | 27 | 27 | 27 | 27 |
| SACOL0723 | MKKLAFAITATSGAAAFLTHHDAQA | Ser | 25 | 25 | 25 | 23 | 25 | 25 | 25 |
| SACOL0755 | MTVKNLFLGFVAVILTVCLIGLLILATNEDALA | Lys | 33 | 33 | 33 | 33 | 33 | 33 | 33 |
| SACOL0856 | MNMKKKEKHAIRKKSIGVASVLVGTLIGFGLLSSKEADA | Ser | 39 | 39 | 39 | 39 | 39 | 39 | 39 |
| SACOL0858 | MKKKLLVLTMSTLFATQIMNSNHAKA | Ser | 26 | 26 | 26 | 24 | 26 | 24 | 26 |
| SACOL0859 | MKRKVLVLTMGVICATQLWHSNHANA | Leu | 26 | 26 | 26 | 24 | 33 | 24 | 26 |
| SACOL0860 | MTEYLLSAGICMAIVSILLIGMAISNVSKGQYAKRFFYFATSCLVLTLVVVSSL SSSANA | Ser | 60 | 23 | 25 | 60 | 30 | 60 | 57 |
| SACOL0886 | MKKLISILLINIIILGVSNSASA | Gln | 23 | 23 | 23 | 23 | 23 | 23 | 23 |
| SACOL0887 | MNKIFRILTVSLFFFTFLIKNNLAYA | Asp | 26 | 26 | 26 | 26 | 26 | 26 | 26 |
| SACOL0907 | MYKRLFISHVILIFALILVISTPNVLA | Glu | 27 | 27 | 27 | 27 | 27 | 27 | 27 |
| SACOL0908 | MNKKLLTRTLIASALVLTTVGSGFHSSSNYNGINNVAKA | Ser | 39 | 24 | 39 | 24 | 39 | 30 | 28 |
| SACOL0962 | MTNSSKSFTKFMAASAVFTMGFLSVPTAGA | Glu | 30 | 30 | 30 | 30 | 30 | 30 | 30 |
| SACOL0985 | MKLKSFITVTLALGMIATTGATVAGNEVSA | Ala | 30 | 30 | 30 | 30 | 30 | 30 | 21 |
| SACOL1056 | MNSSCKSRVFNIISIIMVSMLILSLGAFANNNKAKA | Asp | 36 | 36 | 36 | 36 | 36 | 36 | 29 |
| SACOL1062 | MAKKFNYKLPSMVALTLVGSAVTAHQVQA | Ala | 29 | 29 | 29 | 29 | 29 | 29 | 29 |
| SACOL1071c | MNKLLQSLSALGVSATLVTPNLNADA | Thr | 26 | 26 | 24 | 26 | 24 | 26 | 24 |
| SACOL1071c | MNKLLQSLSALGVSATLVTPNLNA | Asp | 24 | 26 | 24 | 26 | 24 | 26 | 24 |
| SACOL1140 | MTKHYLNSKYQSEQRSSAMKKITMGTASIILGSLVYIGADSQQVNA | Ala | 46 | 39 | 46 | 46 | 46 | None | 46 |
| SACOL1164 | MKKNFIGKSILSIAAISLTVSTFAGESHA | Gln | 29 | 29 | 29 | 29 | 29 | 29 | 29 |
| SACOL1166 | MKKNITKTIIASTVIAAGLLTQTNDAKA | Phe | 28 | 28 | 28 | 28 | 28 | 28 | 28 |
| SACOL1168 | MKNKLIAKSLLTLAAIGITTTTIASTADA | Ser | 29 | 29 | 29 | 29 | 29 | 29 | 26 |
| SACOL1173 | MKTRIVSSVTTTLLLGSILMNPVANA | Ala | 26 | 26 | 26 | 26 | 26 | 26 | 26 |
| SACOL1864 | MNKNIIIKSIAALTILTSITGVGTTMVEGIQQTAKA | Glu | 36 | 36 | 36 | 36 | 36 | 29 | 24 |
| SACOL1865 | MNKNIIIKSIAALTILTSVTGVGTTVVEGIQQTAKA | Glu | 36 | 36 | 36 | 36 | 36 | 36 | None |
| SACOL1866 | MNKNIIIKSIAALTILTSITGVGTTVVDGIQQTAKA | Glu | 36 | 36 | 36 | 36 | 36 | 36 | None |
| SACOL1867 | MNKNIVIKSMAALAILTSVTGINAAVVEETQQIANA | Glu | 36 | 25 | 24 | 24 | 36 | 25 | 21 |
| SACOL1868 | MNKNVVIKSLAALTILTSVTGIGTTLVEEVQQTAKA | Glu | 36 | 36 | 36 | 36 | 36 | 36 | 36 |
| SACOL1869 | MNKNVMVKGLTALTILTILTSLGFAENISNQPHSIAKA | Glu | 38 | 38 | 38 | 38 | 38 | 30 | 23 |
| SACOL1880 | MKMKKLVKSSVASSIALLLLSNTVDA | Ala | 26 | 26 | 26 | 26 | 26 | 26 | 26 |
| SACOL1881 | MFKKKMLAATLSVGLIAPLASPIQESRA | Asn | 28 | 28 | 28 | 28 | 28 | 28 | 28 |
| SACOL2003 | MVKKTKSNSLKKVATLALANLLLVGALTDNSAKA | Glu | 34 | 34 | 34 | 34 | 34 | 34 | 34 |
| SACOL2004 | MIKQLCKNITICTLALSTTFTVLPATSFA | Lys | 29 | 29 | 29 | 29 | 29 | 29 | 29 |
| SACOL2006c | MKNKKRVLIASSLSCAILLLSAATTQA | Asn | 27 | 27 | 29 | 29 | 29 | 29 | 29 |
| SACOL2006c | MKNKKRVLIASSLSCAILLLSAATTQANS | Ala | 29 | 27 | 29 | 29 | 29 | 29 | 29 |
| SACOL2019 | MSYHWFKKMLLSTSILILSSSSLGLATHTVEA | Lys | 32 | 32 | 32 | 32 | 32 | 32 | 56 |
| SACOL2088 | MKKTLLASSLAVGLGIVAGNAGHEAHA | Ser | 27 | 27 | 27 | 23 | 27 | 27 | 27 |
| SACOL2179 | MKKIFVIITTLLAVAIIIGSIIMVVFSQRQAQT | Phe | 33 | 31 | 31 | 31 | 29 | 31 | 60 |
| SACOL2197 | MKLKSFVTATLALGLLSTVGAALPSHEASA | Asp | 30 | 30 | 30 | 30 | 30 | 30 | 30 |
| SACOL2291 | MKKIATATIATAGFATIAIASGNQAHA | Ser | 27 | 27 | 27 | 27 | 27 | 27 | 27 |
| SACOL2295 | MKKLVTATTLTAGIGTALVGQAYHADA | Ala | 27 | 27 | 27 | 27 | 27 | 22 | 27 |
| SACOL2418 | MKNKYISKLLVGAATITLATMISNGEAKA | Ser | 29 | 29 | 29 | 27 | 29 | 29 | 29 |
| SACOL2419 | MIKNKILTATLAVGLIAPLANPFIEISKA | Glu | 29 | 29 | 29 | 29 | 29 | 29 | 29 |
| SACOL2421 | MLKNKILTTTLSVSLLAPLANPLLENAKA | Ala | 29 | 29 | 29 | 29 | 29 | 29 | 29 |
| SACOL2422 | MKMNKLVKSSVATSMALLLLSGTANA | Glu | 26 | 26 | 26 | 26 | 26 | 26 | 26 |
| SACOL2557 | MEYKKILIRLLIAFAVLFSADFTYQSVEQTHQSHA | Ala | 35 | 35 | 35 | 26 | 35 | None | 19 |
| SACOL2584 | MKKTIMASSLAVALGVTGYAAGTGHQAHA | Ala | 29 | 29 | 29 | 29 | 29 | 29 | 29 |
| SACOL2660 | MNKTSKVCVAATLALGTLIGVTVVENSAPTSKQAQA | Ala | 36 | 36 | 36 | 27 | 36 | 27 | 36 |
| SACOL2694 | MKSQNKYSIRKFSVGASSILIATLLFLSGGQAQA | Ala | 34 | 34 | 34 | 34 | 34 | 34 | 34 |
a Residues at positions −3 to −1 relative to the verified SPase I cleavage sites are shown in bold.
b Signal peptide cleavage site positions were experimentally determined by whole protein MS analysis, peptide MS/MS analysis (identification of the N-terminal peptide of the mature protein), or both.
c Proteins with signal peptide cleavages at more than one site.
Table IV. Amino acid residues around confirmed SPase I cleavage site of S. aureus COL proteins.
| Position −3 |
Position −2 |
Position −1 |
Position +1 |
||||
|---|---|---|---|---|---|---|---|
| Residue | Frequency | Residue | Frequency | Residue | Frequency | Residue | Frequency |
| Ala | 47 (77%) | Lys | 14 (23%) | Ala | 59 (97%) | Ala | 19 (31%) |
| Val | 6 (10%) | Ann | 11 (18%) | Ser | 1 (2%) | Ser | 12 (20%) |
| Ser | 6 (10%) | Gln | 8 (13%) | Thr | 1 (2%) | Glu | 11 (18%) |
| Thr | 1 (2%) | His | 7 (12%) | Lys | 5 (8%) | ||
| Leu | 1 (2%) | Asp | 8 (13%) | Asp | 4 (7%) | ||
| Ser | 3 (5%) | Gln | 2 (3%) | ||||
| Glu | 3 (5%) | Thr | 2 (3%) | ||||
| Leu | 2 (3%) | Phe | 2 (3%) | ||||
| Phe | 2 (3%) | Asn | 2 (3%) | ||||
| Tyr | 1 (2%) | Tyr | 1 (2%) | ||||
| Gly | 1 (2%) | Leu | 1 (2%) | ||||
| Arg | 1 (2%) | ||||||
Signal Peptide Processing at Two Cleavage Sites by SPase I
Whole protein MS analysis of SACOL1071 and SACOL2006 revealed processing of the signal peptide at more than one site. This observation is very interesting as signal peptide cleavage by SPase I is generally considered to be highly specific (72, 73), and reports on the observation of signal peptide cleavage at multiple sites in wild-type proteins are very rare. The reasons for the high fidelity of SPase I are not clearly understood. An important requirement for cleavage by SPase I is the presence of amino acids with small neutral side chains at positions −1 and −3 in the C-region; the −3 position is less restrictive than the −1 position (33). This is also evident from Table IV. SACOL1071 and SACOL2006 each have more than one potential cleavage site that is in compliance with the substrate specificity of SPase I and may compete for recognition. The N-terminal signal sequence MNKLLQSLSALGVSATLVTPNLNA24 ↓DA26↓ of SACOL1071 contains the ubiquitous AXA motif (−3 to −1 position) as well as an LXA motif (−5 to −3 position) with observed cleavages (↓) at the −3 and −1 positions. The weighted average signal intensities in the whole protein mass spectrometry experiments suggest that signal peptide processing at the −1 position (82%) is preferred over the −3 position (18%). Similarly, the N-terminal sequence of SACOL2006, MKNKKRVLIASSLSCAILLLSAATTQA27 ↓NS2↓ also contains two potential sites for signal peptide cleavage, the AXS sequence (−3 to −1 position) and the TXA sequence (−5 to −3 position) with observed cleavages (↓) at the −1 and −3 position, respectively. The weighted average of signal intensities of the two mature proteins derived from whole protein MS experiments suggests that processing at the −3 position (64%) is preferred over the −1 position (36%). From our results, it appears that SPase I is capable of processing the signal peptide at more than one site depending on the availability of an alternate potential site close by (−3 and −1 positions in the two examples). The differences in the nature of residues at −1, −3, and −5 positions; the proximity of the cleavage site to the hydrophobic core; the β-turn structure; and the residues that are capable of breaking α-helix or β-strand structure probably play a role in the efficiency of processing at different sites (74). A close inspection of the signal sequences of the proteins identified in this study (Table III) indicates that there are 11 additional sequences that have SPase I-compatible residues in the −5 to −3 positions. However, signal peptide cleavage at only one site was detected in these cases. The observed cleavages at two sites for SACOL1071 and SACOL2006 may be due to some unique characteristics of their signal sequences. In general, prokaryotic signal peptides possess redundant information in that they contain more than one potential cleavage site, more than one basic residue in the N-region, and a longer H-region than required (75, 76). It is not clear why alternate sites for signal peptide cleavage exist or whether there is some biological significance to this.
Possible Secretion of SACOL1071 via Alternate Secretory Pathway
Chitin, a homopolymer of β-1,4-N-acetyl-d-glucosamine (GlcNAc), is one of the most abundant natural polymers. Several bacterial species secrete chitinolytic enzymes and chitin-binding proteins that are thought to degrade chitin (77, 78). SACOL1071, a putative chitinase B protein, has a predicted Sec-type signal peptide and was identified in this study as a mature protein formed by removal of this peptide. In addition, another mature form of the protein, SACOL10713, also was identified by whole protein and peptide analysis. It appears to have been secreted by cleavage of a 14-residue N-terminal segment (Fig. 9B). This 14-residue signal peptide, MNKLLQSLSALGVS, does not resemble any known signal peptide, suggesting secretion of SACOL10713 by an alternate pathway. In Gram-negative Pseudomonas aeruginosa bacteria, Folders et al. (79) identified a chitinase C (ChiC) protein in the extracellular medium following cleavage of an 11-residue signal peptide (MIRIDFSQLHQ). ChiC also does not contain a typical N-terminal sequence. Because SACOL1071 has a functional Sec-dependent export route, it would be intriguing if it were secreted by an alternate pathway.
Non-secretory Proteins in Extracellular Medium
The presence of non-secretory proteins in the extracellular medium has been reported by several investigators for a number of organisms including S. aureus (24, 65, 80). The possible explanation for the presence of cytoplasmic proteins in the extracellular medium is cell lysis during growth; this has been visually confirmed in the present study by fluorescence microscopy using the live/dead staining method. Although only 3.0 ± 0.5% of the cells in the stationary phase culture were lysed, that is sufficient to detect the very abundant proteins listed in supplemental Table 5. The rest of the non-secretory proteins were most probably released into the extracellular medium by proteolytic processing, shedding, or cell wall turnover as suggested by other investigators (69, 81).
Stable Cleaved Signal Peptides and Signal Peptide Fragments
We have reported our unexpected findings on the presence of cleaved stable signal peptides and signal peptide fragments in the extracellular medium. In Escherichia coli, two signal peptide peptidases, a membrane-bound protease IV and cytoplasmic oligopeptidase A, have been identified (83). It has been proposed that protease IV initially cleaves the SPase I-processed signal peptides by making endoproteolytic cuts, and the products of the initial cleavage may diffuse or be transported back into the cytoplasm and further degraded into amino acids by oligopeptidase A and other cytoplasmic enzymes. Similar proteins have also been identified in B. subtilis (84). However, homologous proteins have not been found in S. aureus (29). Because we have identified fragments of signal peptides, degradation of cleaved signal peptides must also happen in S. aureus by some unknown proteases. All the signal peptide fragments observed in the present study appear to have formed by endoproteolytic cleavage at hydrophobic residues including Leu, Thr, Ala, Val, Phe, and Met (Table II), suggesting that the protease responsible for processing signal peptide in S. aureus may have a preference for hydrophobic residues and that its substrate specificity is similar to protease IV of E. coli (83). As mentioned earlier, only C-terminal signal peptide fragments and not N-terminal signal peptide fragments were identified in the extracellular medium in the present study. This suggests that an endoproteolytic cut in the hydrophobic region of the signal peptides releases the C-terminal signal peptide fragments into the extracellular medium, whereas the N-terminal fragments are retained in the membrane or released into the cytoplasm or completely degraded.
The observation of cleaved stable signal peptides in the extracellular medium raises some interesting questions. Because various reports have suggested that after cleavage from the preprotein rapid degradation of signal peptide is important for proper functioning of the export machinery (58, 59), why are the observed cleaved signal peptides stable? Also, because their long hydrophobic cores should lead to peptide retention in the membrane, how are they released into the medium? What is the biological role of the released signal peptide and signal peptide fragments? The fact that the signal peptides of Sle1, SACOL0723, SceD, IsaA, and SACOL2295 were detected in the culture supernatant indicates that they have high solubility in water. Also the Zyggregator algorithm (85) (which analyzes aggregation propensities of proteins) predicts that the cleaved signal peptides identified in the present study should not have the propensity to aggregate. It is possible that certain unique biophysical properties allow the observed cleaved signal peptides to be readily released from the membrane into the extracellular medium after cleavage from the preprotein.
It is important to note here that the fate of cleaved signal peptides and their fragments is not clearly understood, particularly for prokaryotes. However, recent studies have suggested that cleaved signal sequences and their fragments may have important biological functions including signaling. For instance, bacteriocin release protein, an E. coli lipoprotein, is processed very slowly by signal peptidase II to yield a mature bacteriocin release protein and a stable signal peptide that mediate the translocation of cloacin DF13 (86). In eukaryotes, the N-terminal signal peptide fragments of preprolactin and human immunodeficiency virus type 1 p-gp160 were found to be released into the cytoplasm and bound to calmodulin/Ca2+, suggesting that the liberated signal peptide fragments may influence signal transduction pathways in the cell (87). As stated by Martoglio and Dobberstein (88), signal sequences are more than just simple greasy peptides possessing a wealth of functional information. It can be conjectured that cleaved signal peptides and signal peptide fragments of S. aureus COL released into the extracellular medium also have the potential to perform important roles. Tristan et al. (82) have proposed that the signal peptide of Panton-Valentine leukocidin LUKS (LukS-PV), a component of hetero-oligomeric pore-forming toxin (PVL) in S. aureus PVL-positive strains, mediates increased adhesion to extracellular matrix components. According to their hypothesis, LukS-PV signal peptide is released from the membrane after cleavage by SPase I and attaches to the cell wall through its unique C-terminal (TSFHESKA) sequence, thus exposing the positively charged N terminus to the extracellular matrix molecules. S. aureus COL does not encode PVL gene; therefore, we could not confirm the release of LukS-PV signal peptide. Also, none of the signal peptides identified in the present study share homology with LukS-PV signal peptide.
Conclusions
We have presented a new approach to study S. aureus extracellular proteins utilizing the combination of whole protein MS analysis and peptide MS/MS analysis that enabled us to provide the most comprehensive view of S. aureus extracellular proteins reported to date. We identified 59 secreted proteins in S. aureus COL and characterized their post-translational modifications. Accurate determination of signal peptide cleavage sites of S. aureus COL secreted proteins allowed us evaluate the prediction accuracies of several programs. Because S. aureus COL secreted proteins are potential drug targets or vaccine candidates, the signal peptide cleavage site information provided by this study will be useful in making constructs and recombinant proteins and in protein engineering. This information will also extend our current knowledge base on experimentally verified signal peptide cleavage sites and may assist in improving prediction accuracies of the programs. We also detected signal peptide processing at multiple sites and the release of cleaved signal peptides and signal peptide fragments into the extracellular medium. These observations are unusual, and their biological significance is yet to be understood. The current approach should be useful in secretome analysis of other S. aureus strains and other pathogenic organisms.
Supplementary Material
Acknowledgments
We thank Dr. Malcolm Winkler and Lokto Sham for help with the live/dead staining analysis.
* This work was supported in part by the Indiana Metabolomics and Cytomics Initiative (METACyt) Initiative of Indiana University, funded in part through a major grant from Lilly Endowment, Inc., and by National Science Foundation Grants CHE-0518234 and CHE-0832651.
 This article contains supplemental Tables 1–7.
This article contains supplemental Tables 1–7.
1 The abbreviations used are:
- MRSA
- methicillin-resistant S. aureus
- SPase I
- type I signal peptidase
- 1DE
- one-dimensional gel electrophoresis
- 2DE
- two-dimensional gel electrophoresis
- BHI
- brain-heart infusion
- SCX
- strong cation exchange
- RP
- reverse phase
- PAWS
- Protein Analysis Worksheet Software
- AM
- N-acetylmuramoyl-l-alanine amidase domain
- GL
- endo-β-N-acetylglucosaminidase domain
- NN
- neural network model
- HMM
- hidden Markov model
- LukS-PV
- Panton-Valentine leukocidin LUKS
- PVL
- Panton-Valentine leukocidin
- N
- amino terminus
- C
- Carboxy terminus
- H
- Hydrophobic region
- LTQ
- Linear Trap Quadrupole.
REFERENCES
- 1.Diekema D. J., Pfaller M. A., Schmitz F. J., Smayevsky J., Bell J., Jones R. N., Beach M. (2001) Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY antimicrobial surveillance program, 1997–1999. Clin. Infect. Dis. 32, S114–S132 [DOI] [PubMed] [Google Scholar]
- 2.Lowy F. D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 3.Noskin G. A., Rubin R. J., Schentag J. J., Kluytmans J., Hedblom E. C., Jacobson C., Smulders M., Gemmen E., Bharmal M. (2007) National trends in Staphylococcus aureus infection rates: impact on economic burden and mortality over a 6-year period (1998–2003). Clin. Infect. Dis. 45, 1132–1140 [DOI] [PubMed] [Google Scholar]
- 4.Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., Townes J. M., Craig A. S., Zell E. R., Fosheim G. E., McDougal L. K., Carey R. B., Fridkin S. K. (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 5.Klein E., Smith D. L., Laxminarayan R. (2007) Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 13, 1840–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stetson M. G., Vautour R. J. (2006) Cost effectiveness of screening for methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococcus (VRE) in an intensive care unit at a 200-bed community hospital with low prevalence of these antibiotic-resistant organism. Poster Presented at Association for Professionals in Infection Control and Epidemiology, Tampa; June 11–15; 5–39 [Google Scholar]
- 7.Chang S., Sievert D. M., Hageman J. C., Boulton M. L., Tenover F. C., Downes F. P., Shah S., Rudrik J. T., Pupp G. R., Brown W. J., Cardo D., Fridkin S. K. (2003) Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. New Engl. J. Med. 348, 1342–1347 [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan A., Dick J. D., Perl T. M. (2002) Vancomycin resistance in staphylococci. Clin. Microbiol. Rev. 15, 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitener C. J., Park S. Y., Browne F. A., Parent L. J., Julian K., Bozdogan B., Appelbaum P. C., Chaitram J., Weigel L. M., Jernigan J., McDougal L. K., Tenover F. C., Fridkin S. K. (2004) Vancomycin-resistant Staphylococcus aureus in the absence of vancomycin exposure. Clin. Infect. Dis. 38, 1049–1055 [DOI] [PubMed] [Google Scholar]
- 10.Kuroda M., Ohta T., Uchiyama I., Baba T., Yuzawa H., Kobayashi I., Cui L., Oguchi A., Aoki K., Nagai Y., Lian J., Ito T., Kanamori M., Matsumaru H., Maruyama A., Murakami H., Hosoyama A., Mizutani-Ui Y., Takahashi N. K., Sawano T., Inoue R., Kaito C., Sekimizu K., Hirakawa H., Kuhara S., Goto S., Yabuzaki J., Kanehisa M., Yamashita A., Oshima K., Furuya K., Yoshino C., Shiba T., Hattori M., Ogasawara N., Hayashi H., Hiramatsu K. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240 [DOI] [PubMed] [Google Scholar]
- 11.Baba T., Takeuchi F., Kuroda M., Yuzawa H., Aoki K., Oguchi A., Nagai Y., Iwama N., Asano K., Naimi T., Kuroda H., Cui L., Yamamoto K., Hiramatsu K. (2002) Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359, 1819–1827 [DOI] [PubMed] [Google Scholar]
- 12.Holden M. T., Feil E. J., Lindsay J. A., Peacock S. J., Day N. P., Enright M. C., Foster T. J., Moore C. E., Hurst L., Atkin R., Barron A., Bason N., Bentley S. D., Chillingworth C., Chillingworth T., Churcher C., Clark L., Corton C., Cronin A., Doggett J., Dowd L., Feltwell T., Hance Z., Harris B., Hauser H., Holroyd S., Jagels K., James K. D., Lennard N., Line A., Mayes R., Moule S., Mungall K., Ormond D., Quail M. A., Rabbinowitsch E., Rutherford K., Sanders M., Sharp S., Simmonds M., Stevens K., Whitehead S., Barrell B. G., Spratt B. G., Parkhill J. (2004) Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101, 9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsay J. A., Holden M. T. G. (2004) Staphylococcus aureus: Superbug, super genome? Trends Microbiol. 12, 378–385 [DOI] [PubMed] [Google Scholar]
- 14.Blobel G. (1980) Intracellular protein topogenesis. Proc. Natl. Acad. Sci. U. S. A. 77, 1496–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalbey R. E., Lively M. O., Bron S., van Dijl J. M. (1997) The chemistry and enzymology of the type I signal peptidases. Protein Sci. 6, 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormick J. K., Yarwood J. M., Schlievert P. M. (2001) Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55, 77–104 [DOI] [PubMed] [Google Scholar]
- 17.Novak P., Ray P. H., Dev I. K. (1986) Localization and purification of two enzymes from Escherichia coli capable of hydrolyzing a signal peptide. J. Biol. Chem. 261, 420–427 [PubMed] [Google Scholar]
- 18.Kawano Y., Ito Y., Yamakawa Y., Yamashino T., Horii T., Hasegawa T., Ohta M. (2000) Rapid isolation and identification of staphylococcal exoproteins by reverse phase capillary high performance liquid chromatography-electrospray ionization mass spectrometry. FEMS Microbiol. Lett. 189, 103–108 [DOI] [PubMed] [Google Scholar]
- 19.Bernardo K., Fleer S., Pakulat N., Krut O., Hünger F., Krönke M. (2002) Identification of Staphylococcus aureus exotoxins by combined sodium dodecyl sulfate gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2, 740–746 [DOI] [PubMed] [Google Scholar]
- 20.Kawano Y., Kawagishi M., Nakano M., Mase K., Yamashino T., Hasegawa T., Ohta M. (2001) Proteolytic cleavage of staphylococcal exoproteins analyzed by two-dimensional gel electrophoresis. Microbiol. Immunol. 45, 285–290 [DOI] [PubMed] [Google Scholar]
- 21.Corthals G. L., Wasinger V. C., Hochstrasser D. F., Sanchez J. C. (2000) The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis 21, 1104–1115 [DOI] [PubMed] [Google Scholar]
- 22.Gygi S. P., Corthals G. L., Zhang Y., Rochon Y., Aebersold R. (2000) Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. U. S. A. 97, 9390–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Issaq H. J., Conrads T. P., Janini G. M., Veenstra T. D. (2002) Methods for fractionation, separation and profiling of proteins and peptides. Electrophoresis 23, 3048–3061 [DOI] [PubMed] [Google Scholar]
- 24.Burlak C., Hammer C. H., Robinson M. A., Whitney A. R., McGavin M. J., Kreiswirth B. N., Deleo F. R. (2007) Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell. Microbiol. 9, 1172–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones R. C., Deck J., Edmondson R. D., Hart M. E. (2008) Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J. Bacteriol. 190, 5265–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziebandt A. K., Becher D., Ohlsen K., Hacker J., Hecker M., Engelmann S. (2004) The influence of agr and sB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 4, 3034–3047 [DOI] [PubMed] [Google Scholar]
- 27.Ziebandt A. K., Weber H., Rudolph J., Schmid R., Höper D., Engelmann S., Hecker M. (2001) Extracellular proteins of Staphylococcus aureus and the role of SarA and sB. Proteomics 1, 480–493 [DOI] [PubMed] [Google Scholar]
- 28.Karty J. A., Running W. E., Reilly J. P. (2007) Two dimensional liquid phase separations of proteins using online fractionation and concentration between chromatographic dimensions. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 103–113 [DOI] [PubMed] [Google Scholar]
- 29.Sibbald M. J., Ziebandt A. K., Engelmann S., Hecker M., de Jong A., Harmsen H. J., Raangs G. C., Stokroos I., Arends J. P., Dubois J. Y., van Dijl J. M. (2006) Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70, 755–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Read S. M., Northcote D. H. (1981) Minimization of variation in response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal. Biochem. 116, 53–64 [DOI] [PubMed] [Google Scholar]
- 31.Gill S. R., Fouts D. E., Archer G. L., Mongodin E. F., Deboy R. T., Ravel J., Paulsen I. T., Kolonay J. F., Brinkac L., Beanan M., Dodson R. J., Daugherty S. C., Madupu R., Angiuoli S. V., Durkin A. S., Haft D. H., Vamathevan J., Khouri H., Utterback T., Lee C., Dimitrov G., Jiang L., Qin H., Weidman J., Tran K., Kang K., Hance I. R., Nelson K. E., Fraser C. M. (2005) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Streptococcus epidermidis strain. J. Bacteriol. 187, 2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boeckmann B., Bairoch A., Apweiler R., Blatter M. C., Estreicher A., Gasteiger E., Martin M. J., Michoud K., O'Donovan C., Phan I., Pilbout S., Schneider M. (2003) The SWISS-PROT protein knowledge-base and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 34.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 35.Hiller K., Grote A., Scheer M., Münch R., Jahn D. (2004) PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 32, W375–W379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Heijne G. (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14, 4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Von Heijne G. (1987) Sequence Analysis in Molecular Biology: Treasure Trove on Trivial Pursuit, pp. 429–436, Academic Press, San Diego, CA [Google Scholar]
- 38.Juncker A. S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cserzö M., Wallin E., Simon I., von Heijne G., Elofsson A. (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10, 673–676 [DOI] [PubMed] [Google Scholar]
- 40.Gardy J. L., Laird M. R., Chen F., Rey S., Walsh C. J., Ester M., Brinkman F. S. L. (2005) PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21, 617–623 [DOI] [PubMed] [Google Scholar]
- 41.Vandenesch F., Kornblum J., Novick R. P. (1991) A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173, 6313–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakano M., Kawano Y., Kawagish M., Hasegawa T., Iinuma Y., Oht M. (2002) Two-dimensional analysis of exoproteins of methicillin-resistant Staphylococcus aureus (MRSA) for possible epidemiological applications. Microbiol. Immunol. 46, 11–22 [DOI] [PubMed] [Google Scholar]
- 43.Jeljaszewicz J., Mlynarczyk A., Mlynarczyk G. (2000) Exoproteins in Staphylococcus aureus virulence. Bull. Pol. Acad. Sci. Biol. Sci. 48, 77–98 [Google Scholar]
- 44.Fitton J. E., Dell A., Shaw W. V. (1980) The amino acid sequence of the delta hemolysin of Staphylococcus aureus. FEBS Lett. 115, 209–212 [DOI] [PubMed] [Google Scholar]
- 45.Janzon L., Löfdahl S., Arvidson S. (1989) Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 219, 480–485 [DOI] [PubMed] [Google Scholar]
- 46.Carlos J. L., Paetzel M., Brubaker G., Karla A., Ashwell C. M., Lively M. O., Cao G., Bullinger P., Dalbey R. E. (2000) The role of the membrane-spanning domain of type I signal peptidases in substrate cleavage site selection. J. Biol. Chem. 275, 38813–38822 [DOI] [PubMed] [Google Scholar]
- 47.Reed S. B., Wesson C. A., Liou L. E., Trumble W. R., Schlievert P. M., Bohach G. A., Bayles K. W. (2001) Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect. Immun. 69, 1521–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lauber M. A., Running W. E., Reilly J. P. (2009) B. subtilis ribosomal proteins: structural homology and post-translational modifications. J. Proteome Res. 8, 4193–4206 [DOI] [PubMed] [Google Scholar]
- 49.Rollof J., Normark S. (1992) In vivo processing of Staphylococcus aureus lipase. J. Bacteriol. 174, 1844–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Götz F., Verheij H. M., Rosenstein R. (1998) Staphylococcal lipases: molecular characterization, secretion, and processing. Chem. Phys. Lipids 93, 15–25 [DOI] [PubMed] [Google Scholar]
- 51.Nikoleit K., Rosenstein R., Verheij H. M., Götz F. (1995) Comparative biochemical and molecular analysis of the Staphylococcus hyicus, Staphylococcus aureus and a hybrid lipase. Indication for a C-terminal phospholipase domain. Eur. J. Biochem. 228, 732–738 [PubMed] [Google Scholar]
- 52.Foster S. J. (1995) Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177, 5723–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oshida T., Sugai M., Komatsuzawa H., Hong Y. M., Suginaka H., Tomasz A. (1995) A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U.S.A. 92, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf C., Hochgräfe F., Kusch H., Albrecht D., Hecker M., Engelmann S. (2008) Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8, 3139–3153 [DOI] [PubMed] [Google Scholar]
- 55.Burts M. L., Williams W. A., DeBord K., Missiakas D. M. (2005) EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U.S.A. 102, 1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renshaw P. S., Lightbody K. L., Veverka V., Muskett F. W., Kelly G., Frenkiel T. A., Gordon S. V., Hewinson R. G., Burke B., Norman J., Williamson R. A., Carr M. D. (2005) Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 24, 2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundaramoorthy R., Fyfe P. K., Hunter W. N. (2008) Structure of Staphylococcus aureus EsxA suggests a contribution to virulence by action as a transport chaperone and/or adaptor protein. J. Mol. Biol. 383, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koren R., Burstein Y., Soreq H. (1983) Synthetic leader peptide modulates secretion of proteins from microinjected Xenopus oocytes. Proc. Natl. Acad. Sci. U.S.A. 80, 7205–7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L., Tai P. C., Briggs M. S., Gierasch L. M. (1987) Protein translocation into Escherichia coli membrane vesicles is inhibited by functional synthetic signal peptides. J. Biol. Chem. 262, 1427–1429 [PubMed] [Google Scholar]
- 60.Yvon M., Chabanet C., Pélissier J. P. (1989) Solubility of peptides in trichloroacetic acid (TCA) solutions. Hypothesis on the precipitation mechanism. Int. J. Pept. Protein Res. 34, 166–176 [DOI] [PubMed] [Google Scholar]
- 61.Leversen N. A., de Souza G. A., Målen H., Prasad S., Jonassen I., Wiker H. G. (2009) Evaluation of signal peptide prediction algorithms for identification of mycobacterial signal peptides using sequence data from proteomic methods. Microbiology 155, 2375–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta N., Tanner S., Jaitly N., Adkins J. N., Lipton M., Edwards R., Romine M., Osterman A., Bafna V., Smith R. D., Pevzner P. A. (2007) Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 17, 1362–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z., Henzel W. J. (2004) Signal peptide prediction based on analysis of experimentally verified cleavage sites. Protein Sci. 13, 2819–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pocsfalvi G., Cacace G., Cuccurullo M., Serluca G., Sorrentino A., Schlosser G., Blaiotta G., Malorni A. (2008) Proteomic analysis of exoproteins expressed by enterotoxigenic Staphylococcus aureus strains. Proteomics 8, 2462–2476 [DOI] [PubMed] [Google Scholar]
- 65.Antelmann H., Tjalsma H., Voigt B., Ohlmeier S., Bron S., van Dijl J. M., Hecker M. (2001) A proteomic view on genome-based signal peptide predictions. Genome Res. 11, 1484–1502 [DOI] [PubMed] [Google Scholar]
- 66.Westers L., Westers H., Zanen G., Antelmann H., Hecker M., Noone D., Devine K. M., van Dijl J. M., Quax W. J. (2008) Genetic or chemical protease inhibition causes significant changes in the Bacillus subtilis exoproteome. Proteomics 8, 2704–2713 [DOI] [PubMed] [Google Scholar]
- 67.Aziz R. K., Pabst M. J., Jeng A., Kansal R., Low D. E., Nizet V., Kotb M. (2004) Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51, 123–134 [DOI] [PubMed] [Google Scholar]
- 68.Sarvas M., Harwood C. R., Bron S., van Dijl J. M. (2004) Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1694, 311–327 [DOI] [PubMed] [Google Scholar]
- 69.Tjalsma H., van Dijl J. M. (2005) Proteomics-based consensus prediction of protein retention in a bacterial membrane. Proteomics 5, 4472–4482 [DOI] [PubMed] [Google Scholar]
- 70.Schneewind O., Missiakas D. (2007) What genomics has taught us about gram-positive protein secretion and targeting, in Bacterial Pathogenomics (Pallen M. J., Nelson K. E., Preston G. M. eds) pp. 301–326, American Society for Microbiology (ASM) Press, Washington, DC [Google Scholar]
- 71.Paetzel M., Dalbey R. E., Strynadka N. C. (1998) Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature 396, 186–190 [DOI] [PubMed] [Google Scholar]
- 72.Sekizaki T., Takai S., Egawa Y., Ikeda T., Ito H., Tsubaki Shiro. (1995) Sequence of the Rhodococcus equi gene encoding the virulence-associated 15–17-kDa antigens. Gene 155, 135–136 [DOI] [PubMed] [Google Scholar]
- 73.Folz R. J., Nothwehr S. F., Gordon J. I. (1988) Substrate specificity of eukaryotic signal peptidase. Site-saturation mutagenesis at position −1 regulates cleavage between multiple sites in human pre(Delta pro)apolipoprotein A-II. J. Biol. Chem. 263, 2070–2078 [PubMed] [Google Scholar]
- 74.Fikes J. D., Barkocy-Gallagher G. A., Klapper D. G., Bassford P. J., Jr. (1990) Maturation of Escherichia coli maltose-binding protein by signal peptidase I in vivo. Sequence requirements for efficient processing and demonstration of an alternate cleavage site. J. Biol. Chem. 265, 3417–3423 [PubMed] [Google Scholar]
- 75.Puziss J. W., Fikes J. D., Bassford P. J., Jr. (1989) Analysis of mutational alterations in the hydrophilic segment of the maltose-binding protein signal peptide. J. Bacteriol. 171, 2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fikes J. D., Bankaitis V. A., Ryan J. P., Bassford P. J., Jr. (1987) Mutational alterations affecting the export competence of a truncated but fully functional maltose-binding protein signal peptide. J. Bacteriol. 169, 2345–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bassler B. L., Yu C., Lee Y. C., Roseman S. (1991) Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J. Biol. Chem. 266, 24276–24286 [PubMed] [Google Scholar]
- 78.Watanabe T., Kimura K., Sumiya T., Nikaidou N., Suzuki K., Suzuki M., Taiyoji M., Ferrer S., Regue M. (1997) Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179, 7111–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Folders J., Algra J., Roelofs M. S., van Loon L. C., Tommassen J., Bitter W. (2001) Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J. Bacteriol. 183, 7044–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Målen H., Berven F. S., Fladmark K. E., Wiker H. G. (2007) Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7, 1702–1718 [DOI] [PubMed] [Google Scholar]
- 81.Tjalsma H., Antelmann H., Jongbloed J. D., Braun P. G., Darmon E., Dorenbos R., Dubois J. Y., Westers H., Zanen G., Quax W. J., Kuipers O. P., Bron S., Hecker M., van Dijl J. M. (2004) Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68, 207–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tristan A., Benito Y., Montserret R., Boisset S., Dusserre E., Penin F., Ruggiero F., Etienne J., Lortat-Jacob H., Lina G., Bowden M. G., Vandenesch F. (2009) The signal peptide of Staphylococcus aureus Panton Valentine leukocidin LukS component mediates increased adhesion to heparan sulfates. PLoS One 4, e5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Novak P., Dev I. K. (1988) Degradation of a signal peptide by protease IV and oligopeptidase A. J. Bacteriol. 170, 5067–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bolhuis A., Matzen A., Hyyryläinen H. L., Kontinen V. P., Meima R., Chapuis J., Venema G., Bron S., Freudl R., van Dijl J. M. (1999) Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J. Biol. Chem. 274, 24585–24592 [DOI] [PubMed] [Google Scholar]
- 85.Tartaglia G. G., Vendruscolo M. (2008) The Zyggregator method for predicting protein aggregation propensities. Chem. Soc. Rev. 37, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 86.van der Wal F. J., Oudega B., Kater M. M., ten Hagen-Jongman C. M., de Graaf F. K., Luirink J. (1992) The stable BRP signal peptide causes lethality but is unable to provoke the translocation of cloacin DF13 across the cytoplasmic membrane of Escherichia coli. Mol. Microbiol. 6, 2309–2318 [DOI] [PubMed] [Google Scholar]
- 87.Martoglio B., Graf R., Dobberstein B. (1997) Signal peptide fragments of preprolactin and HIV-1 p-gp 160 interact with calmodulin. EMBO J. 16, 6636–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martoglio B., Dobberstein B. (1998) Signal sequences: more than just greasy peptides. Trends Cell Biol. 8, 410–415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.