Abstract
Ovarian cancers are frequently not diagnosed until advanced stages, resulting in a high case fatality rate. Because of this, more tumor markers, in addition to CA125, for detecting and monitoring ovarian cancer are needed. During a systematic search for potential biomarkers of ovarian cancer, we compared the protein profiles between tumor interstitial fluid and normal interstitial fluid of ovaries, rationalizing that abnormal levels of proteins in tumor interstitial fluid may be detected in peripheral blood and thus serve as easily accessible tumor markers. Here, we show that stress-induced phosphoprotein 1 (STIP1) was secreted by ovarian cancer tissues into the peripheral blood of patients, resulting in a significant increase of serum levels of STIP1 in cancer patients compared with those in age-matched normal controls. Our results further indicated that combined use of CA125 and STIP1 may increase early detection of ovarian cancer. Functionally, recombinant STIP1 significantly induced ERK phosphorylation, promoted DNA synthesis, and increased Ki-67 immunoreactivity in ovarian cancer cells, suggesting that STIP1 in vitro promotes cell proliferation. Colocalization of STIP1 and phospho-ERK in human ovarian cancer tissues also supports an in vivo activation of ERK by STIP1. Further understanding of molecular roles of STIP1 in human ovarian cancer may shed light on its pathophysiology and development of novel therapeutic strategies.
Ovarian cancer is the leading cause of gynecological cancer death. The annual incidence of ovarian cancer is greater than 25,000 in the United Sates with an annual mortality of about 14,000 (1). The high case fatality rate is mainly attributed to the fact that most ovarian cancers are undiagnosed until advanced stages (III or IV) where the cancer has spread beyond the pelvis. Since the first characterization of a monoclonal antibody against a glycoprotein, CA125, in 1981 (2), measurement of serum levels of CA125 has become standard practice for the preoperative evaluation of ovarian masses (3). However, the sensitivity of CA125 is only about 80% for detecting overall ovarian cancer (4) and only about 50% for early stage cancer (5). More tumor markers that may complement CA125 for early detection of ovarian cancer are thus warranted.
Disease proteomics (6, 7), also known as clinical proteomics (8), is the new discipline in which proteomics is applied to better understand disease processes, develop new biomarkers for diagnosis and early detection of disease, and accelerate drug development. A systematic search for potential biomarkers of solid cancer emphasized the comparison of the protein profiles of tumor interstitial fluids (TIFs)1 and nonmalignant interstitial fluid (NIF) (9). The rationale of this approach is that proteins in TIF and NIF readily enter circulation through the lymphatic system; hence, unusual levels of proteins in TIF also may be detected in peripheral blood and may serve as easily accessible biomarkers (10). The search for biomarkers within TIF may circumvent the obstacle against mining the plasma proteome for cancer biomarkers that the levels of plasma abundant proteins always highly exceed those of biomarkers (11). Furthermore, the TIF proteome of ovarian cancers can reflect the interactions between cancer cells and the surrounding stroma cells that are missed when using cancer cell lines in culture systems to obtain the secretome of ovarian cancer (12).
Stress-induced phosphoprotein 1 (STIP1; GeneID 10963; Human Protein Reference Database identifier 05454) is a 62.6-kDa protein also known as heat shock protein (HSP)-organizing protein (13). STIP1 protein contains nine tetratricopeptide repeat (TPR) motifs, which are clustered into domains each consisting of three TPRs, and a nuclear localization signal from amino acid residues 223 to 238 (Human Protein Reference Database). STIP1 acts primarily as an adapter that directs HSP90 to HSP70-client protein complexes in the cytoplasm, but recent evidence suggests that STIP1 can also modulate the chaperone activities of these HSPs (13). The TPR motif is a degenerate 34-amino acid sequence, which forms scaffolds that mediate formation of different protein complexes, participating in RNA splicing, transcription, protein folding, signal transduction, and cell cycle regulation (13, 14). The identification of STIP1 as a potential serum biomarker for ovarian cancer has only been reported by us in the 2008 American Society of Clinical Oncology Annual Meeting (15).
In this biomarker discovery and preclinical validation study, we identified STIP1 among several differentially expressed proteins between TIF and NIF of ovarian tissues. The differential protein levels of STIP1 between ovarian cancer and normal ovaries were confirmed by Western blot analysis and immunohistochemistry. Receiver operating characteristic (ROC) curve analysis of serum levels of STIP1 in ovarian cancer patients and controls suggested a good discriminative capacity of STIP1 for detection of ovarian cancer. Our studies suggest that STIP1 is secreted by ovarian cancer cells into their environment and is functional in promoting cell proliferation.
EXPERIMENTAL PROCEDURES
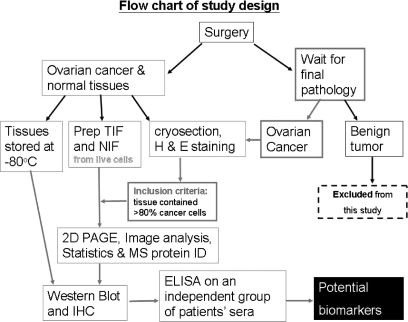
Study Design for Analysis of TIF and NIF
Ovarian tumors and normal ovarian tissues were obtained during surgery for suspected ovarian malignancies and for benign gynecological conditions such as adenomyosis or myoma uteri. Written informed consents were obtained from the participant before surgery. Immediately after the ovarian tumor or control tissues were removed, a gynecological oncologist took a piece of tissue at the size of 1 × 1 × 1 cm3 to the laboratory for the procedures outlined in Fig. 1. A piece of live tissue at about 0.5 × 0.5 × 0.5 cm3 was used to prepare TIF or NIF. On another piece of the same tissue, cryosectioning and subsequent hematoxylin and eosin staining were done to verify that the tissue was at least 80% cancer. The remaining tissues were snap frozen in liquid nitrogen and stored at −80 °C. The TIF, NIF, cryosection, and frozen tissues were prospectively collected. However, each TIF was enrolled into this study for two-dimensional gel analysis only after the pathology report confirmed the diagnosis to be serous epithelial ovarian cancer. Experimental design and the involvement of human subjects in this study were approved by the Institute Review Board of Chang Gung Memorial Hospital (CGMH-IRB numbers 95-0459B, 97-1443C, and 92-142).
Fig. 1.
Study design for identifying potential serum biomarkers for human ovarian cancer. The TIF and NIF were prepared from enrolled cases. However, only those that passed the following criteria were further analyzed with two-dimensional PAGE, Western blot, and immunohistochemistry (the flows are marked with red arrows). The two criteria were: (i) pathological confirmation of ovarian cancer and (ii) that the majority (>80%) of the collected tissue were cancer cells. Prep, prepare; H & E, hematoxylin and eosin; MS protein ID, MS for protein identification; 2D, two-dimensional.
Reagents and Cell Lines
The majority of the chemicals used were purchased from Sigma-Aldrich. Anti-STIP1 antibodies were from Abnova Corp. (Taipei, Taiwan): H00010963-M33 (clone 2E1) was used as the capture antibody, and H00010963-M35 (clone 2E11) was used as the detecting antibody for ELISA. For immunohistochemistry, anti-STIP1 antibody was H00010963-M01 (clone 4B6) (Abnova Corp.), and anti-phospho-ERK was SC-7383 (Santa Cruz Biotechnology). Other reagents included ACN (Merck), methanol (Merck), formalin (Riedel-deHaen, Seelze, Germany), AgNO3 (AppliChem GmbH, Gatersleben, Germany), dithiothreitol (AppliChem, Darmstadt, Germany), iodoacetamide (AppliChem), trypsin (Promega, Madison, WI), and the peptide calibration standard for mass spectrometry (Bruker Daltonics, Bremen, Germany). Human ovarian cancer cell lines TOV112D, TOV21G, OVCAR3, OVCAR8, SKOV3, MDAH2774, PA-1, and ES2 were obtained from ATCC; BR and BG1 were reported previously (16, 17). Cancer cells were cultured in Dulbecco's modified Eagle's medium/F-12 supplemented with 10% fetal bovine serum and antibiotics at 37 °C with 5% CO2.
Preparation of TIF and NIF
Immediately after surgery, a piece of live tissue (0.5 × 0.5 × 0.5 cm3) was minced with sharp scissors, weighed, and rinsed with iced PBS. The tissues were incubated with 800 μl of PBS at 37 °C for 1 h; TIF or NIF was collected in the supernatant after spinning down the tissues at 8000 × g at 4 °C for 20 min. Half the supernatant was desalted with a Bio-Rad cleanup kit and rehydrated with rehydration buffer (8 m urea, 4% CHAPS, 0.02% bromphenol blue), and the other half was aliquoted and stored at −80 °C. After protein concentrations were quantified with the Bio-Rad Bradford protein assay, each TIF or NIF was aliquoted (100 μl/tube) and stored at −80 °C until assayed. The number of freeze-thaw cycles of each tube of biofluid was limited to 2.
Two-dimensional PAGE and Image Analysis
The procedures were reported previously (18) but were used with slight modification. TIF or NIF protein (400 μg) was added with 2.5 mg of DTT, 3.3 μl of Biolyte (pH 3–10), and rehydration buffer to a final volume of 330 μl, and the solution was applied to a 17-cm IPG strip (linear pH 5–8; Bio-Rad). The first dimensional electrophoresis using a Bio-Rad Protean IEF cell was carried out with the following program: 20 °C, 50 mA/strip for 12 h, 100 V rapid for 30 min, 500 V slow for 30 min, 1000 V slow for 30 min, 5000 V slow for 30 min, 8000 V slow for 1 h, 58,000 V-h rapid, and 250 V rapid for 24 h. Each IPG strip was alkylated with 3 ml of equilibrium buffer (6 m urea, 30% glycerol, 30 mm Tris, pH 8.8, 0.005% bromphenol blue, 70 mm SDS) in the dark for 20 min before it was applied to the second dimensional electrophoresis with a 12.5% SDS-polyacrylamide gel. The second dimensional electrophoretic program was set to 1000 V, 15 mA/gel, and 250 watts.
The two-dimensional gel was immersed in fix/destain solution (7% acetic acid in 10% methanol) for 2 h, and then the solution was replaced with SYPRO Ruby solution (Molecular Probes, Eugene, OR) for 16 h at room temperature. The destaining process included shaking the gel in fix/destain solution for 2 h followed by rinsing with distilled deionized H2O for 10 min twice, and then the gel was put in distilled deionized H2O until scanned with a ProXPRESS 2D scanner (PerkinElmer Life Sciences). Images were analyzed with the Progenesis work station (Progenesis Discovery, Nonlinear Dynamics, Durham, NC) using the gel that contained the most abundant protein spots as the reference image.
In-gel Enzymatic Digestion and Mass Spectrometry
Procedures were reported previously (18) but were used with the following modifications. The protein spots of interest were manually excised from the SYPRO Ruby-stained two-dimensional gels, destained using 50 mm NH4HCO3 (ammonium bicarbonate) in 50% ACN, and dried in a SpeedVac concentrator. The protein was then digested by incubating overnight at 37 °C with trypsin (Promega; at 5 ng/ml) in 50 mm NH4HCO3, pH 7.8.
Tryptic peptides were extracted from the gel in 1 volume of 0.1% TFA while vortexing for 5 min followed by sonication for 5 min. Crude digest mixtures were concentrated and desalted using mC18 ZipTips (Millipore) followed by eluting in 1.5 ml of matrix (5 mg of α-cyano-4-hydroxycinnamic acid/ml in 50% ACN and 0.1% TFA) for peptide mass fingerprinting by MALDI-TOF mass spectrometry analysis. The mass spectrometer was externally calibrated with peptide calibration standard I (Bruker Daltonics) for each batch of samples to achieve a typical mass measurement accuracy of 50 ppm in the range of 600–4000 m/z. The mass spectra were acquired by FlexControl software (v3.0, Bruker Daltonics) and processed by FlexAnalysis software (v3.0, Bruker Daltonics) to generate a peak list. The sophisticated numerical annotation procedure (SNAP) was used as the peak detection algorithm with a signal to noise ratio threshold >4, and the Savitzky-Golay smoothing algorithm with 0.2 m/z peak width was used. Furthermore, the 2211.10 m/z (trypsin fragments) and 2465.19 m/z (ACTH 18–39) were used for internal calibration and then removed from the peak list before searching against the Mascot search engine (v2.1, Matrix Science Inc., Boston, MA) with the NCBI (NCBInr 20061110, selected for Homo sapiens, 153,289 sequences) database. The search parameters allowed for one missed cleavage, oxidation of methionine, carbamidomethylation of cysteine, and 50-ppm or better mass accuracy for each matching peptide. For the protein identification criteria, probability-based Mowse scores with the p value less than 0.05 were accepted.
Western Blot Analyses and Data Normalization for TIF and NIF
Procedures for Western blot analyses and for the preparation of cell lysates from cancer cell lines were reported previously (19). After protein quantification with the Bradford assay, 70 μg of protein of each TIF and NIF was separated using 10% SDS-polyacrylamide gels and transferred to nitrocellulose membrane (Amersham Biosciences). The membranes were stained with Ponceau S solution (0.2% Ponceau S in 10% acetic acid) for 10 min, washed with water at room temperature for 5 min, and scanned using a high resolution flatbed color scanner (Hewlett-Packard). In this study, we used anti-STIP1, anti-triose-phosphate isomerase 1 (TPI1), anti-ubiquitin carboxyl-terminal esterase L1 (UCHL1), anti-leucine aminopeptidase 3 (LAP3), anti-macrophin 1 isoform 2 (MACF1) (all purchased from Abnova Corp.), and anti-cytokeratin 16 (Acris, Herford, Germany) as first antibodies; corresponding horseradish peroxidase-conjugated second antibodies (Santa Cruz Biotechnology); and enhanced chemiluminescence (Pierce ECL system). For the comparison of STIP1 levels in various human ovarian cancer lines, densitometric data of STIP1 were normalized with β-actin levels in the corresponding specimens. For Western blot analysis of ERK activation, densitometric results of phospho-ERK (Santa Cruz Biotechnology) were normalized with total ERK levels (Santa Cruz Biotechnology).
Because TIF and NIF were secreted from fresh tissues and did not necessarily contain intracellular β-actin, the levels of β-actin in each sample could not be used for normalizing the protein input of each lane. The amount of input protein in each lane was quantified by the whole intensity of each lane using UN-SCAN-IT software (Silk Scientific, Inc., Orem, UT). After quantitation of bands of correct molecular weight on a Western blot with UN-SCAN-IT software, the relative intensity of each sample was normalized by the total protein input of each lane as the Ponceau S staining intensity.
ELISA
ELISA procedures were reported previously (20). In this study, all ELISAs used tetramethylbenzidine as the substrate of peroxidase, and color intensities were detected at A450 nm with a Wallac 1420 VICTOR2 spectrophotometer (PerkinElmer Life Sciences).
Human transferrin levels in TIF and NIF were assayed using a commercial ELISA kit (Alpha Diagnostic, San Antonio, TX). Before measurement, TIF and NIF samples were diluted to 1:500 in PBS. The minimum detection limit of the assay was 50 ng/ml. The intra-assay coefficients of variation (CVs) were 2.2% at 115 ng/ml and 8% at 518 ng/ml (n = 10). The interassay CVs were 8.3% at 108 ng/ml and 6.2% at 499 ng/ml (n = 5).
Human brain-derived neurotrophic factor (BDNF) levels of TIF and NIF were assayed with a commercial kit (R&D Systems, Minneapolis, MN) with a minimal detection limit of 20 pg/ml. The intra-assay CVs of the BDNF ELISA were 5% at 476 pg/ml and 6.2% at 2175 pg/ml (n = 6), and its interassay CVs were 11.3% at 528 pg/ml and 8.1% at 2287 pg/ml (n = 6).
Human serum levels of CA125 were assayed using a commercial Abbott Axsym system (Abbott Park, IL) with a detection sensitivity at 2 units/ml. The intra-assay CVs and interassay CVs at 8.81 units/ml were 16 (n = 6) and 8.1% (n = 20), respectively, and at 44.37 units/ml were 5.87 (n = 6) and 2.13% (n = 20), respectively.
We developed two versions of STIP1 ELISA to determine human serum levels of STIP1 (detailed in the supplemental material). For the newer version of STIP1 ELISA, we used a mouse monoclonal antibody (Abnova Corp.) as the capture antibody that was coated on 96-well plates (F8 MaxiSorp, Nunc A/S, Roskilde, Denmark). Another biotinylated mouse monoclonal antibody (Abnova Corp.) was used as the detection antibody, which could be detected by Amdex streptavidin-peroxidase conjugate, and tetramethylbenzidine substrate was used for color formation. The reaction was stopped by the addition of 100 μl/well 1 n H2SO4. Absorbance was determined at 450 nm in a microplate spectrophotometer (Molecular Devices SpectraMax model 190).
Immunohistochemistry (IHC)
Using procedures that were reported previously (21), we did IHC studies on cryosections of human ovarian cancer tissues and normal ovaries from which TIF and NIF were derived, respectively. IHC studies were also done in tissue arrays (SuperBioChips Laboratories, Seoul, Korea) and archival paraffin-embedded ovarian cancer slides of Chang Gung Memorial Hospital. The slides with formalin-fixed, paraffin-embedded tissues sectioned at 4 μm thick were deparaffinized in xylene and rehydrated with a series of graded ethanol. Sections were then stained with a mouse anti-human STIP1 monoclonal antibody (Abnova Corp.) or an anti-phospho-ERK monoclonal antibody (Santa Cruz Biotechnology) using an automated IHC stainer with the Ventana Basic DAB (3,3′-diaminobenzidine) Detection kit (Tucson, AZ) according to the manufacturer's protocol. Counterstaining was performed with hematoxylin.
STIP1 Detection in Cell Culture Supernatant
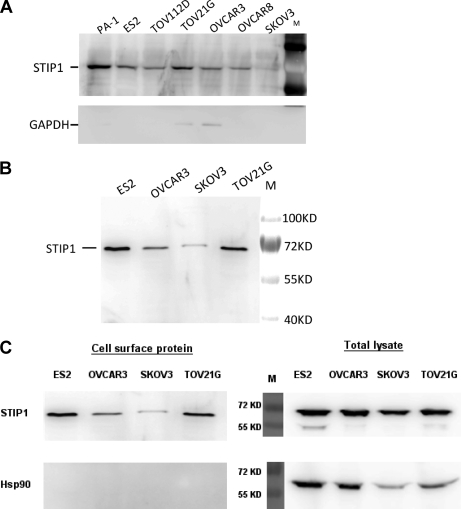
For protein identification in culture supernatant, ovarian cancer cells were cultured overnight in serum-free Opti-MEM (Invitrogen). After centrifugation at 1600 rpm for 10 min to remove cell debris, 45 μl of culture supernatant was analyzed by Western blot with a chicken anti-STIP1 polyclonal antibody (1:1000; Abcam) followed with goat anti-chicken IgY horseradish peroxidase-conjugated antibody (1:3000; Abcam) using the same procedure described above. Mouse anti-GAPDH monoclonal antibody (1:2000; Novus) was used as control to detect whether the intracellular protein had leaked into the culture supernatant.
Biotinylation of Cell Surface Protein to Detect STIP1 as Cell Surface Protein
Cell surface proteins of ovarian cancer cells were isolated using the Pierce Cell Surface Protein Isolation kit (Thermo Scientific) as described by the manufacturer. Briefly, sulfo-NHS-biotin solution was prepared by dissolving one vial of sulfo-NHS-biotin (12 mg/vial) into 48 ml of cold 1× PBS (0.1 m sodium phosphate, 0.15 m NaCl, pH 7.2). Then, cells at 90–95% confluence in a 10-cm dish were incubated with 10 ml of sulfo-NHS-biotin solution for 30 min at 4 °C. After the reaction was quenched by 500 μl of quenching solution/dish, cells were scraped and washed with TBS buffer to remove extra biotin solution. After centrifugation to remove supernatant, cell pellets were lysed in 125 μl of lysis buffer by sonication and clarified by centrifugation, and protein concentration was determined by bicinchoninic acid (BCA) assay. All cell lysates were applied to a NeutrAvidin-agarose column, and biotinyl proteins were then eluted by 100 μl of SDS-PAGE sample buffer with 50 mm DTT. The biotinyl proteins isolated from each 40 μg of total proteins were analyzed by Western blots to determine the amount of cell surface STIP1 protein as described above. Detection of cytoplasmic HSP90 with an anti-HSP90 monoclonal antibody (1:1000; Abcam) was used as a threshold above which was considered to have a possible contamination of intracellular proteins.
Plasmid Construction
The cDNA construct of human STIP1 (GenBankTM accession NM_006819) was purchased from Open Biosystems to serve as DNA template to amplify STIP1 by specific primers with EcoRI and EcoRV linkers in the 5′- and 3′-ends, respectively, for further subcloning into pMT/Bip/V5-His expression vector (Invitrogen). The PCR primers were STIP1-forward primer (5′-TGAGAATTCATGGAGCAGGTCAATGAGCTGAAGGAGAAAGGC-3′) and STIP1-reverse primer (5′-TGAGATATCTCACCGAATTGCAATCAGACCCACATCC-3′), and PCR was performed as follows: 95 °C for 5 min followed by 40 cycles of 95 °C for 30 s, 63 °C for 30 s, and 72 °C for 90 s and finalized at 72 °C for 10 min. The PCR product was purified and digested by EcoRI and EcoRV, which was ligated into the EcoRI/EcoRV-treated pMT/Bip/V5-His vector. Correct construction of pMT/Bip/STIP1 was confirmed by autosequencing analysis (ABI autosequencer).
Insect Cell Culture and Stable Clone Selection
Drosophila S2 cells were cultured in Schneider's medium (Invitrogen) supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Following the manufacturer's protocol, a stable clone of copper-inducible, STIP1-secreting S2 cells was generated by cotransfection of 19 μg of pMT/Bip/STIP1 expression vector and 1 μg of pCoHygro selecting vector (Invitrogen) by using a calcium phosphate transfection kit (Invitrogen) and selected in culture medium containing 500 μg/ml hygromycin (Invitrogen). Secretion of recombinant human STIP1 (rhSTIP1) from the stably transfected clone, S2-STIP1, was induced by addition of 500 μm copper sulfate to the culture medium.
Purification of rhSTIP1
The S2-STIP1 cells were grown at a density of 5 million cells/ml in culture medium without hygromycin, and copper sulfate was added to a final concentration of 500 μm in roller bottles for induction. After induction for 48 h, the cell culture supernatant containing secreted rhSTIP1 was purified using a nickel-nitrilotriacetic acid-agarose column (Invitrogen) according to the manufacturer's protocol. Briefly, nickel-nitrilotriacetic acid-agarose beads were washed twice with distilled water and once with native binding buffer before the cell culture supernatant was applied. Then, the mixture was rocked at 4 °C for 1 h for protein binding followed by washing six times with native wash buffer, and the rhSTIP1 protein was eluted into 8 ml of elution buffer. The eluted fraction was concentrated by using an Amicon centrifugal filter device (Millipore), and the protein concentration was determined by Bradford assay. Concentrated rhSTIP1 solution was separated into small portions and stored at −80 °C.
In Vitro Treatment of Ovarian Cancer Cells with rhSTIP1 for ERK Phosphorylation Assay, BrdUrd Assay, and Ki-67 Immunocytochemistry Study
For the ERK phosphorylation assay, 106 ovarian cancer cells were seeded into a 10-cm-diameter dish overnight. Then, the cells were starved in serum-free medium for 24 h, treated with 0.4 μm rhSTIP1 for 5 min, and lysed with radioimmune precipitation assay buffer. Antibodies for phosphorylated-ERK and total ERK were purchased from Cell Signaling Technology, Inc. (Beverly, MA); the antibody specific for actin was from Sigma.
For BrdUrd assays, ovarian cancer cells were seeded at a density of 104 cells/well in a 96-well plate overnight. After being kept in serum-free medium for 24 h, cells were treated with 0.4 μm rhSTIP1 in the presence of BrdUrd for 24 h. DNA synthesis activity was assayed using a BrdUrd ELISA kit (Roche Applied Science).
For immunocytochemistry studies of Ki-67, ovarian cancer cell lines were cultured on Lab-Tek II chamber slides (Nalge Nunc International, Rochester, NY). After the cells were kept in serum-free Dulbecco's modified Eagle's medium for 24 h, 0.4 μm rhSTIP1 was added to culture medium for another 24 h. At the end of treatment, cells were fixed with 99.9% ethanol, rehydrated with PBS, treated with 3% hydrogen peroxide for 20 min to exhaust intrinsic peroxidase, and permeabilized with 0.1% Triton X-100 (Sigma) for 15 min. Immunostaining for Ki-67 (Thermo Fisher Scientific, Waltham, MA) was performed used a standard protocol similar to the aforementioned IHC protocol.
Statistical Analyses
Using Statistica software (Statsoft, Tulsa, OK), the Kruskal-Wallis test (nonparametric equivalent of analysis of variance) and Mann-Whitney U test (nonparametric equivalent of the t test) were used to analyze continuous variables, whereas a paired t test was used for ERK activity analysis. ROC curve analysis was used to provide sensitivities for given specificity levels of STIP1 (22). For all analyses, p values less than 0.05 were considered statistically significant.
RESULTS
Clinical Information of Included Patients
From January to November 2006, 23 TIF and 19 NIF, including four pairs from the same patients, were prospectively collected. Nine ovarian TIF and 16 non-cancer ovarian NIF specimens were included in this study according to the study design summarized in Fig. 1. The mean age of all nine TIF cases, which were derived from serous epithelial carcinoma, was 60.5 ± 18.7 years old (mean ± S.D.), ranging from 31 to 84 years old (supplemental Table 1). Only the NIF from the ovary that was confirmed to be free of pathology was further analyzed in this study. The mean age of the 16 NIF cases was 47.6 ± 6.4 years old (mean ± S.D.), ranging from 37 to 60 years old (supplemental Table 2). A significant difference (p = 0.0005) was noted between the ages of TIF and NIF. At surgery, none of the patients had received any chemo- or radiotherapy.
Differentially Expressed Proteins between Human Ovarian TIF and NIF
All TIF and NIF were analyzed after storage in aliquots at −80 °C for 7.0 ± 0.4 months (mean ± S.E.). The mean protein amount of TIF was 3965 ± 1255 μg (mean ± S.E.), ranging from 590 to 13,286 μg, whereas the mean protein amount for NIF was 3166 ± 628 μg (mean ± S.E.), ranging from 525 to 8389 μg. There was no significant difference between protein amounts of TIF and NIF (p = 0.53).
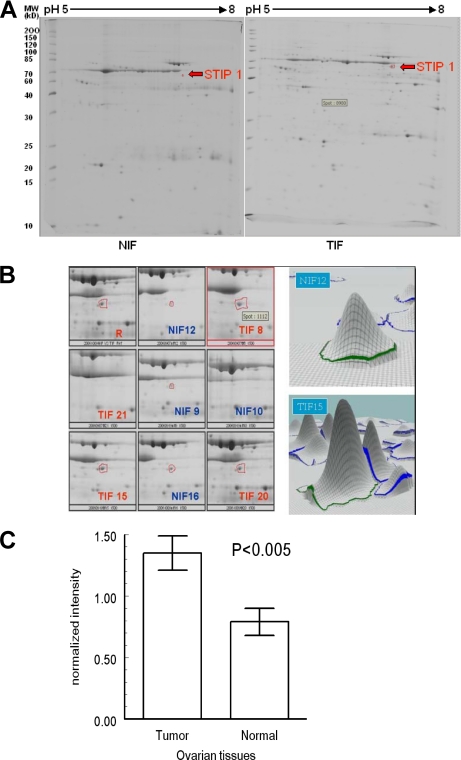
Four TIF and four NIF two-dimensional gels (pH from 5 to 8) stained with SYPRO Ruby were analyzed as one representative pair of two-dimensional gels shown in Fig. 2A. A total of 596 spots were analyzed. Using p < 0.05 as a significant threshold, we identified 10 spots that showed different amounts between TIF and NIF groups in the non-parametric Mann-Whitney U test. Using in-gel digestion and MALDI-TOF mass spectrometry, we identified eight proteins (supplemental Fig. 1 and Table 3). The following proteins were higher in the TIF group: STIP1 (Fig. 2B), LAP3, type I keratin 16, MACF1, BDNF isoform b preproprotein (BDNF1), and TPI1. In the NIF group, however, UCHL1 and transferrin levels were higher (Table I).
Fig. 2.
STIP1 is expressed significantly higher in ovarian TIF than in NIF. A, in this representative pair of TIF and NIF two-dimensional polyacrylamide gels, the protein spots (indicated by arrows) showing differential expression between TIFs and NIFs were identified to be STIP1. B, enlarged images of the STIP1 spots in the four pairs of TIFs and NIFs. The gel that contained the most abundant protein spots was used as the reference image (R). C, the difference in STIP1 between TIF and NIF was significant (Mann-Whitney U test).
Table I. Differentially expressed proteins between TIF and NIF (identified as p value <0.05 with Mann-Whitney U test).
NS, not significant; NA, not applicable.
| Name | Accession number | Theoretical pI/molecular mass (kDa) | Number of unique peptides identified | Sequence coverage | Original results from 4 TIF and 4 NIF | TIF levelsa (n = 9) | NIF levelsa (n = 16) | p values | Confirmed by |
|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||
| STIP1 | gi|5803181 | 6.40/62.60 | 26 | 44 | TIF > NIF | 1.35 ± 0.14 | 0.79 ± 0.11 | p < 0.005 | Western blot |
| LAP3 | gi|4335941 | 7.58/56.01 | 21 | 48 | TIF > NIF | 0.94 ± 0.20 | 1.03 ± 0.15 | NS (p = 0.72) | Western blot |
| Type I keratin 16 | gi|1195531 | 4.99/51.21 | 26 | 53 | TIF > NIF | NA | NA | NA | NA |
| MACF1 | gi|17426164 | 5.04/529.39 | 53 | 14 | TIF > NIF | NA | NA | NA | NA |
| BDNFb | gi|25306235 | 9.21/28.85 | 9 | 35 | TIF > NIF | 21.66 ± 12.03 | 20.36 ± 6.97 | NS (p = 0.92) | ELISAb |
| TPI1 | gi|999892 | 6.51/26.52 | 20 | 85 | TIF > NIF | 1.27 ± 0.11 | 0.82 ± 0.08 | p < 0.005 | Western blot |
| UCHL1 | gi|21361091 | 5.33/24.81 | 12 | 58 | TIF < NIF | 0.84 ± 0.27 | 1.06 ± 0.21 | NS (p = 0.53) | Western blot |
| Transferrinc | gi|4557871 | 6.81/77.00 | 23 | 33 | TIF < NIF | 22.76 ± 3.62 | 34.45 ± 2.72 | p < 0.05 | ELISAc |
a Data are presented as means ± S.E.
b BDNF results of TIF and NIF were normalized by the protein concentration of each specimen, thereby presented as pg/mg.
c Transferrin results of TIF and NIF were normalized by the protein concentration of each specimen, thereby presented as ng/mg.
The differential levels of selected proteins between TIF and NIF were further confirmed in nine TIFs and 16 NIFs using Western blot analysis or ELISA (Table I). An example of quantitative Western blot analysis is shown in supplemental Fig. 2. Twenty-five protein specimens were electrophoresed in three gels. The total protein amount of each sample was measured using Ponceau S staining (upper panel) and was used to normalize levels of the 62-kDa STIP1 band (lower panel). Statistically significant differences between TIF and NIF were identified in STIP1 (Fig. 2C), TPI1, and transferrin (Table I).
STIP1 Expression Is Higher in Ovarian Cancer than in Benign Ovarian Tumors
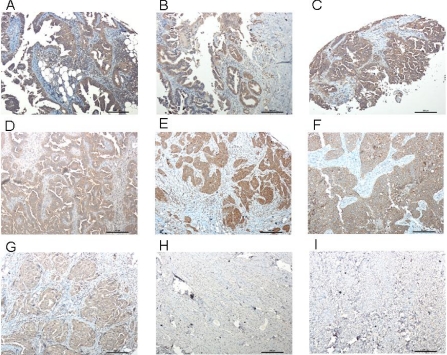
Using immunohistochemical studies, we confirmed that increased intensities of STIP1 immunoreactivities existed in OCT-preserved ovarian cancer tissues and that STIP1 immunoreactivity was not present in normal ovaries (data not shown). We also identified specific expression of STIP1 in formalin-fixed, paraffin-embedded human ovarian cancers, including serous adenocarcinoma and endometrioid carcinoma, but not in benign fibroma and fibrothecoma (Fig. 3).
Fig. 3.
Immunohistochemistry of STIP1 in human ovarian cancer. STIP1 on various types of human ovarian cancers (A–G) and benign tumors (H and I) was specifically detected by an anti-STIP1 monoclonal antibody, shown in brown color. The slides were counterstained by hematoxylin, and a 10× object lens was used for microscopic photography. A, serous papillary carcinoma; B, serous papillary cystadenocarcinoma; C, adenocarcinoma; D, moderately differentiated adenocarcinoma; E, poorly differentiated adenocarcinoma; F, endometrioid carcinoma; G, endometrioid carcinoma; H, fibroma; I, fibrothecoma. Notably, no immunostaining of STIP1 was noted in benign fibroma and fibrothecoma. Scale bars represent 200 μm.
Establishment of ELISA for STIP1 Quantification
Western blot analyses on several human ovarian cancer cell lines indicated that relative levels of STIP1 (normalized by β-actin levels) were highest in BR (1.33) followed by ES2 (1.28), MDAH2774 (1.07), SKOV3 (1.05), TOV112D (0.87), and BG1 (0.76) and lowest in TOV21G (0.69) (not shown). Therefore, we prepared two concentrations of BR ovarian cancer cell lysate to be used as quality controls in the development of ELISA for STIP1. The detection sensitivity of our STIP1 ELISA was 2 ng/ml. Intra-assay CVs of the STIP1 ELISA were 4.6% (n = 6) at 59.5 ng/ml and 5.6% (n = 6) at 16.5 ng/ml. Interassay CVs were 10.6% at 59.5 ng/ml concentration (n = 7) and 9.4% at 16.5 ng/ml (n = 7). In the future, a sufficient pool of sera will be used for determining CV.
Serum Levels of STIP1 Are Significantly Higher in Patients with Ovarian Cancer than in Age-matched Healthy Controls
To evaluate the usefulness of STIP1 as a serum biomarker for ovarian cancer, we analyzed serum levels of STIP1 between patients with ovarian cancer and healthy controls in 43 age-matched pairs (supplemental Table 4). The serum of each patient with ovarian cancer was collected before surgery when ultrasonographic findings (tumor with septa, cystic tumor with solid parts, size greater than 5 cm in diameter) and/or elevated CA125 levels suggested the possibility of ovarian cancer (CGMH-IRB number 97-1443C). However, the cases were included in this study only after ovarian cancer was confirmed by pathologists. Sera in age-matched healthy controls were selected from the leftovers of another project (CGMH-IRB number 92-142). There was no statistical difference in ages (p = 0.88) between the patients with ovarian cancer (55.0 ± 11.8 years old (mean ± S.D.), n = 43) and the healthy controls (55.4 ± 11.4 years old).
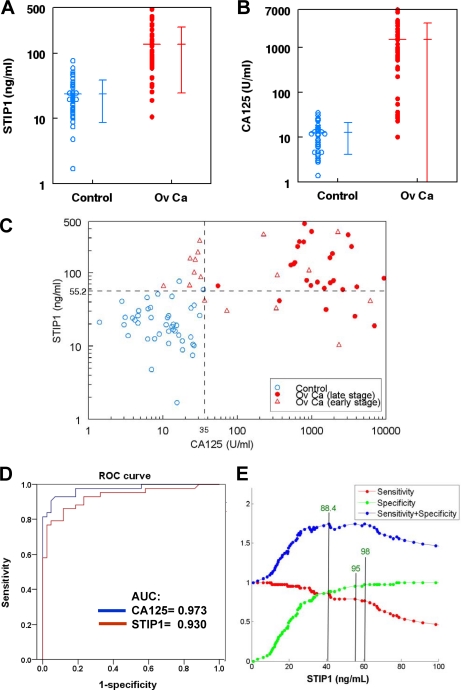
Serum levels of STIP1 in patients with ovarian cancer (137.4 ± 112.7 ng/ml (mean ± S.D.), n = 43) were significantly (p < 0.000001) higher than those in normal controls (23.8 ± 15.1 ng/ml, n = 43) (supplemental Fig. 3). Likewise, serum levels of CA125 (1526.5 ± 2013.6 units/ml, n = 43) in ovarian cancer were significantly (p < 0.00001) higher than those in normal controls (12.6 ± 8.4 units/ml, n = 43) (supplemental Fig. 3). Analyses on logarithmic transformation of either STIP1 (Fig. 4A) or CA125 levels (Fig. 4B) resulted in similar conclusions. Plotting STIP1 against CA125 values on a logarithmic scale in both ovarian cancer patients and controls clearly supported the aforementioned discriminating powers of both biomarkers (Fig. 4C). ROC curve analysis of cancer versus non-cancer groups indicated that the areas under the curve (AUCs) were 0.930 for STIP1 and 0.973 for CA125, which averaged overall levels of specificity (Fig. 4D).
Fig. 4.
Significantly higher concentrations of both STIP1 and CA125 in patients with ovarian cancer than in age-matched healthy controls. A, serum levels of STIP1 in patients with ovarian cancer were significantly (p < 0.000001) higher than those in normal controls (n = 43). Mean ± S.D. is also displayed. B, similarly, serum levels of CA125 in ovarian cancer (Ov Ca) were significantly (p < 0.000001) higher than the serum levels in normal controls (n = 43). C, the STIP1 serum levels of the 43 pairs of cancer patients (red dots, stages III and IV; red triangles, stages I and II) and controls (blue circles) were plotted against CA125 on a logarithmic scale, showing a correlation with r = 0.539 and p < 0.000001. The horizontal dotted line at 55.2 ng/ml represents the STIP1 cutoff value for 95% specificity, whereas the vertical dotted line at 35 units/ml represents the CA125 cutoff value for 95% specificity. D, ROC curve analysis of cancer versus age-matched control groups indicated that the AUCs were 0.930 for STIP1 and 0.973 for CA125. E, three cutoff values of STIP1 (shown by three vertical lines) resulted in the identical maximal sum of specificity and sensitivity: 41.3 ng/ml with 88.4% specificity and 86% sensitivity, 55.2 ng/ml with 95.3% specificity and 79.1% sensitivity, and 60.4 ng/ml with 97.7% specificity and 76.7% sensitivity.
In the 43 pairs of ovarian cancer versus healthy controls, three serum concentrations of STIP1 resulted in the same maximal sum of specificity and sensitivity for detection of ovarian cancer (Fig. 4E). The three STIP1 concentrations were 41.3 ng/ml (86% sensitivity and 88.4% specificity), 55.2 ng/ml (95.3% specificity and 79.1% sensitivity), and 60.4 ng/ml (97.7% specificity and 76.7% sensitivity) (Fig. 4E). In this study, the combined criteria of CA125 >35 units/ml and STIP1 >55 ng/ml could detect all of the human ovarian cancer (100% sensitivity) at a specificity of 95.6% (Fig. 4C). The additional use of STIP1 was especially useful in detecting ovarian cancers of early stages I and II (Fig. 4C, triangle).
In these 43 pairs of ovarian cancer patients and controls, we did not find a significant correlation between ages and serum levels of CA125 (supplemental Fig. 4). On the other hand, there were correlations between ages and serum levels of STIP1 in the healthy control group (p = 0.009) and in the ovarian cancer group (p = 0.044) (supplemental Fig. 5). Analyses on more serum specimens in a larger study are needed to verify the relation between age and STIP1 levels. In these 43 patients with ovarian cancer, there was not a significant difference in the serum STIP1 levels among the four clinical stages of ovarian cancer, but the log(CA125) value in stage I was significantly different from those in stages II and III (supplemental Fig. 6).
STIP1 Is Secreted from Ovarian Cancer Cells
To study whether the increased serum STIP1 levels in cancer patients were actively secreted by ovarian cancer cells or leaked from cancer cells after tissue damage, we determined whether STIP1 was located on the outer cell membrane. After being cultured in serum-free medium for 24 h, the culture media clearly contained STIP1 but no or only small amounts of the intracellular protein GAPDH (Fig. 5A). Although STIP1 is known to be a cytoplasmic protein, results from our isolation of cell membranous proteins using the Pierce Cell Surface Protein Isolation kit (Thermo Scientific) indicated that STIP1 was localized on the outer cell membrane of ovarian cancer cells (Fig. 5B). In the presence of spontaneous cell lysis, as shown in TOV21G and OVCAR3 cells by the presence of GAPDH in culture media (Fig. 5A), our results could not completely rule out the possibility of leakage from cancer tissues as one of the origins of elevated STIP1 serum levels in cancer patients. Therefore, we used the moderate cytosolic levels of HSP90 as the threshold for determining possible contamination with intracellular proteins during the isolation of surface proteins. There was no trace of HSP90 in the isolated cell surface proteins (Fig. 5C, left panel), whereas HSP90 was clearly detected in total cell lysates (Fig. 5C, right panel).
Fig. 5.
STIP1 in ovarian cancer cells is transported to cell membrane for secretion into culture medium. A, after 24-h culture of various ovarian cancer cells, the culture medium supernatant contained STIP1 that was secreted by cultured cells. Absence of GAPDH in the culture media of five cell lines indicates no leakage of intracellular contents into the culture media, suggesting that clearly detected STIP1 in all culture media was from secretion by the cells. In this experiment, however, leakage of intracellular GAPDH was detected in TOV21G and OVCAR3 cells, corresponding to the occurrence of their cell lysis. B, detection of STIP1 on the outer cell membrane of ovarian cancer lines supports its outward transportation. Using the Pierce Cell Surface Protein Isolation kit (Thermo Scientific), we labeled outer membrane proteins on live ovarian cancer cells with biotin, purified them with avidin-agarose beads, and analyzed them with Western blot analysis. C, no trace of HSP90 was identified in the purified cell surface proteins (left panel), whereas HSP90 was clearly detected in the total cell lysates (right panel). M, molecular mass markers.
STIP1 Promotes Cell Proliferation through Activation of ERK
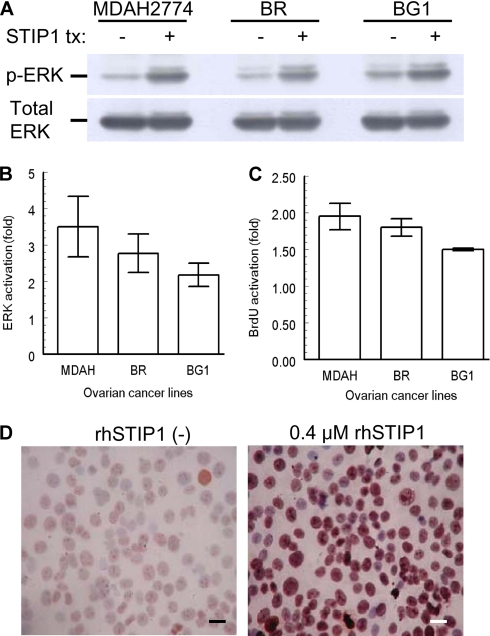
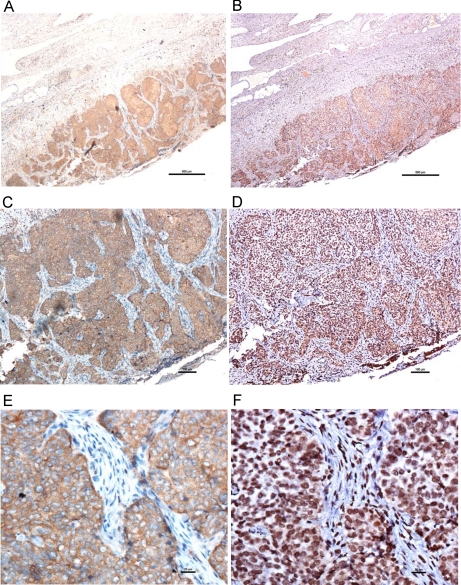
Our stably transfected Drosophila cell clone, S2-STIP1, successfully secreted STIP1 into culture medium. After induction with copper sulfate for 48 h, 400 ml of culture medium yielded about 1000–4000 μg of rhSTIP1, allowing us to do in vitro treatments of ovarian cancer cells with rhSTIP1. To test whether STIP1 in ovarian cancer exerted stimulatory effects similar to those reported in glioblastoma cells (23), we treated ovarian cancer cell lines with 0.4 μm rhSTIP1 for 5 min and analyzed ERK activities. As shown in Fig. 6, A and B, STIP1 significantly induced ERK phosphorylation in three ovarian cancer cell lines (p < 0.05). In multiple ovarian cancer cell lines, treatment with rhSTIP1 for 24 h significantly (p < 0.05) increased BrdUrd incorporation (Fig. 6C) and increased the Ki-67 immunostaining in immunocytochemistry studies (Fig. 6D). Collectively, these results suggest the in vitro function of STIP1 in promoting cell proliferation. Colocalization of STIP1 and phospho-ERK in human ovarian cancer tissues (Fig. 7) also supports an in vivo activation of ERK by STIP1.
Fig. 6.
rhSTIP1 promotes cell proliferation of ovarian cancer cells through ERK activation. A, STIP1 activates ERK in ovarian cancer cells. Ovarian cancer cells were cultured in serum-free medium for 24 h and then treated with 0.4 μm rhSTIP1 for 5 min. Cell lysates were analyzed for ERK phosphorylation (p-ERK) with a phosphorylation-specific anti-ERK antibody. For quantitative comparisons, intensities of phospho-ERK bands were normalized with band intensities of total ERK. tx, treatment. B, significant activation of ERK by rhSTIP1 treatment in three ovarian cancer cell lines. In vitro treatment with rhSTIP1 significantly (p < 0.05) induced ERK phosphorylation in MDAH2774 (3.50 ± 0.83), BR (2.77 ± 0.53), and BG1 (2.18 ± 0.32). Results shown are mean ± S.E. from three independent experiments, each for all three cell lines. C, significant stimulation of BrdUrd (BrdU) incorporation by rhSTIP1 treatment in three ovarian cancer cell lines. In vitro treatment with rhSTIP1 for 24 h significantly (p < 0.05) stimulated BrdUrd incorporation in MDAH2774 (1.95 ± 0.18), BR (1.80 ± 0.12), and BG1 (1.50 ± 0.02). Results shown are mean ± S.E. from three independent experiments, each for all three cell lines. D, rhSTIP1 activates Ki-67 immunostaining in BR ovarian cancer cells. In the representative slides from three independent experiments, Ki-67 immunoreactivity is shown in brown color. The slides were counterstained by hematoxylin, and a 40× object lens was used for microscopic photography. Scale bars represent 20 μm.
Fig. 7.
Immunohistochemical colocalization of STIP1 and phospho-ERK in human ovarian cancer tissues. STIP1 is stained as brown color in A, C, and E, whereas phospho-ERK is stained as dark brown in B, D, and F. Micrographs in A and B, C and D, and E and F were taken with 4×, 10×, and 40× object lenses, respectively. Highly magnified micrographs show that STIP1 is localized in cytoplasm (E) and that phospho-ERK is in the nucleus (F). Scale bars represent 500 (A and B), 100 (C and D), and 20 μm (E and F).
DISCUSSION
In this study, we first performed image analysis and statistics in two groups of two-dimensional gels (four TIF and four NIF) to identify the proteins that showed statistical significance in differential expression. Then, we verified the significant difference of these candidate proteins with more samples (nine TIF and 16 NIF) using additional protein assays such as Western blot analysis and ELISA. Compared with another experimental approach where researchers did many two-dimensional gels immediately followed by protein identification (24), our strategy (Fig. 1) proved to be useful in minimizing the demand of doing two-dimensional gels and protein identification with MALDI-TOF mass spectrometry. However, our approach using only four pairs of gels apparently resulted in a high false discovery rate (three of eight) as shown by LAP3, BDNF, and UCHL1 (Table I).
To our knowledge, this study is the first report that STIP1 is secreted (Fig. 5) by ovarian cancer tissues (Fig. 3) into peripheral blood of cancer patients (Fig. 4). A significant increase of serum levels of STIP1 in ovarian cancer patients compared with age-matched normal controls (Fig. 4) clearly indicates that the up-regulation of STIP1 in ovarian cancer tissues can be assayed in peripheral blood using a rapid, inexpensive ELISA, such as those we developed in this study. Taken together, these results for STIP1 fulfill two criteria of cancer-secreted protein to serve as biomarkers: a high secretion rate by tumor cells and a low base-line concentration in normal blood (12). Analysis of the AUC in the ROC curve is frequently used as an evaluation of the usefulness among different diagnostic methods (22). The AUC of the ROC curve of STIP1 for detecting ovarian cancer was 0.930 (Fig. 4D), supporting the usefulness of STIP1 as a biomarker for ovarian cancer.
The CA125 ovarian cancer antigen is a membrane-associated mucin, the gene product of MUC16 (GeneID 94025) (25, 26). The heavily O-glycosylated transmembrane MUC16 is a multifunctional molecule linked to the actin cytoskeleton (27). Upon identification of mesothelin as a CA125-binding protein, it was shown that CA125 might contribute to metastasis of ovarian cancer to peritoneum by initiating cell attachment to mesothelial epithelium via binding to mesothelin (28–31). On the other hand, STIP1 appeared to be regulated by cell cycle kinases (14) and granzyme B (32).
Given such evidences that STIP1 and CA125 function in distinct biological processes, theoretically, increased levels of STIP1 and CA125 may be used together as complementary biomarkers for ovarian cancer. Indeed, our preliminary results in these 43 pairs of cancer versus healthy controls support the advantage of the combined use of CA125 >35 units/ml and STIP1 >55 ng/ml in detecting ovarian cancer, resulting in 100% sensitivity, 95.3% specificity, 95.6% positive predictive value, and 100% negative predictive value (Fig. 4C). However, we should remain cautious in interpreting these results considering the small sample size of CA125 <35 units/ml (n = 8) and the use of healthy subjects as the controls in our study. In clinical decision-making, it is more challenging to differentiate benign ovarian lesions from ovarian cancer. To further test the efficacy of STIP1 as a complementary test for CA125 as biomarkers for ovarian cancer, analyses on independent groups of serum samples in existing screening studies are needed. International collaborations, such as from the Early Detection Research Network, are invited.
STIP1 has recently been associated with melanoma (33), hepatocellular carcinoma (34), glioma (35), and pancreatic cancer (36). The findings of up-regulation of STIP1, HSP70, and HSP90 members in hepatocellular carcinoma suggest an involvement of heat shock protein families in hepatitis B virus-related carcinogenesis (34). A glioblastoma cell line has been shown to secrete STIP1 into culture medium, and recombinant STIP1 can induce proliferation of glioma cells through activation of ERK and phosphatidylinositol 3-kinase pathways (35). Suppression of STIP1 by short interfering RNA technology has been shown to decrease the invasion activity of pancreatic cancer cell lines (36). The findings that recombinant STIP1 significantly induced ERK phosphorylation, promoted DNA synthesis, and increased Ki-67 immunoreactivity in ovarian cancer cells (Fig. 6) extend the proproliferative role of STIP1 in tumorigenesis of ovarian cancer. Immunohistochemical studies (Fig. 7) also showed that secreted STIP1 stimulates ovarian cancer cells and its microenvironment in autocrine and paracrine fashions, respectively. Our identification of STIP1 in ovarian cancer also supports an evolving concept that biomarkers need not be tissue-specific to be useful because cancer is usually caused by dysregulated pathways; thus, relevant biomarkers may cut across tumor types without showing tissue specificity (11).
Endocytosis of prion protein (GeneID 5621) is reported to be required for ERK signaling induced by STIP1 (37). Although we have not characterized the status of prion protein in ovarian cancer tissues yet, the ovary is reported to be a site of prion protein expression (38) in addition to the central nervous system (39), bone marrow (40), and the immune system (41). Greater understanding of the functional roles of STIP1 and its interaction with prion protein in human ovarian cancer may shed insights into the pathophysiology of ovarian cancer and the development of a novel therapeutic strategy.
Supplementary Material
Acknowledgments
We thank Dr. An-Joy Cheng (Department of Medical Technology, Chang Gung University) for valuable advice; I-Chen Tsai, Jung-Erh Yang, Yen-Tsun Lin, and Ching-Ling Wang (Chang Gung Memorial Hospital) for technical assistance; and Dr. Shihtien T. Wang (Children's Hospital, Medical College of Wisconsin, Milwaukee, WI) for English editing.
* This study was supported by Grants NSC-96-2314-B-182A-124 (to T.-H. W.), NSC-95-2113-M-182A-001 (to J.-K. C. and T.-H. W.), and NSC94-2314-B-182A-137 (to S.-D. C.) from the National Science Council, Taiwan; SMRPG340071 (to T.-H. W.) from the Terry Fox Foundation; and CMRPG360031(to S.-D. C.), CMRPG330313 (to T.-H. W.), CMRPG360952 (to A.-S. C.), and CMRPG340291 (to H.-S. W.) from Chang Gung Memorial Hospital.
 This article contains supplemental Figs. 1–6 and Tables 1–4.
This article contains supplemental Figs. 1–6 and Tables 1–4.
1 The abbreviations used are:
- TIF
- tumor interstitial fluid
- AUC
- area under the curve
- BDNF
- brain-derived neurotrophic factor
- CV
- coefficient of variation
- ERK
- extracellular signal-regulated kinase
- HSP
- heat shock protein
- IHC
- immunohistochemistry
- NIF
- normal interstitial fluid
- ROC
- receiver operating characteristic
- STIP1
- stress-induced phosphoprotein 1
- TPR
- tetratricopeptide repeat
- ACTH
- adrenocorticotropic hormone
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- NHS
- N-hydroxysuccinimide
- rhSTIP1
- recombinant human STIP1
- LAP3
- leucine aminopeptidase 3
- MACF1
- macrophin 1 isoform 2
- TPI1
- triose-phosphate isomerase 1
- UCHL1
- ubiquitin carboxyl-terminal esterase L1.
REFERENCES
- 1.Myers E. R., Havrilesky L. J., Kulasingam S. L., Sanders G. D., Cline K. E., Gray R. N., Berchuck A., McCrory D. C. (2006) Genomic tests for ovarian cancer detection and management. Evid. Rep. Technol. Assess. 145, 1–100 [PMC free article] [PubMed] [Google Scholar]
- 2.Bast R. C., Jr., Feeney M., Lazarus H., Nadler L. M., Colvin R. B., Knapp R. C. (1981) Reactivity of a monoclonal antibody with human ovarian carcinoma. J. Clin. Investig. 68, 1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Calster B., Timmerman D., Bourne T., Testa A. C., Van Holsbeke C., Domali E., Jurkovic D., Neven P., Van Huffel S., Valentin L. (2007) Discrimination between benign and malignant adnexal masses by specialist ultrasound examination versus serum CA-125. J. Natl. Cancer Inst. 99, 1706–1714 [DOI] [PubMed] [Google Scholar]
- 4.Clarke-Pearson D. L. (2009) Clinical practice. Screening for ovarian cancer. N. Engl. J. Med. 361, 170–177 [DOI] [PubMed] [Google Scholar]
- 5.Jacobs I., Bast R. C., Jr. (1989) The CA 125 tumour-associated antigen: a review of the literature. Hum. Reprod. 4, 1–12 [DOI] [PubMed] [Google Scholar]
- 6.Hanash S. (2003) Disease proteomics. Nature 422, 226–232 [DOI] [PubMed] [Google Scholar]
- 7.Hanash S. (2004) HUPO initiatives relevant to clinical proteomics. Mol. Cell. Proteomics 3, 298–301 [DOI] [PubMed] [Google Scholar]
- 8.Celis J. E., Moreira J. M. (2008) Clinical proteomics. Mol. Cell. Proteomics 7, 1779. [DOI] [PubMed] [Google Scholar]
- 9.Celis J. E., Gromov P., Cabezón T., Moreira J. M., Ambartsumian N., Sandelin K., Rank F., Gromova I. (2004) Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol. Cell. Proteomics 3, 327–344 [DOI] [PubMed] [Google Scholar]
- 10.Celis J. E., Moreira J. M., Gromova I., Cabezon T., Ralfkiaer U., Guldberg P., Straten P. T., Mouridsen H., Friis E., Holm D., Rank F., Gromov P. (2005) Towards discovery-driven translational research in breast cancer. FEBS J. 272, 2–15 [DOI] [PubMed] [Google Scholar]
- 11.Hanash S. M., Pitteri S. J., Faca V. M. (2008) Mining the plasma proteome for cancer biomarkers. Nature 452, 571–579 [DOI] [PubMed] [Google Scholar]
- 12.Faça V. M., Hanash S. M. (2009) In-depth proteomics to define the cell surface and secretome of ovarian cancer cells and processes of protein shedding. Cancer Res. 69, 728–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odunuga O. O., Longshaw V. M., Blatch G. L. (2004) Hop: more than an Hsp70/Hsp90 adaptor protein. BioEssays 26, 1058–1068 [DOI] [PubMed] [Google Scholar]
- 14.Longshaw V. M., Chapple J. P., Balda M. S., Cheetham M. E., Blatch G. L. (2004) Nuclear translocation of the Hsp70/Hsp90 organizing protein mSTI1 is regulated by cell cycle kinases. J. Cell Sci. 117, 701–710 [DOI] [PubMed] [Google Scholar]
- 15.Wang T. H., Chao A., Huang H. J., Lai C. H. (2008) Proteomics identification of stress-induced phosphoprotein (STIP1) as a potential biomarker for human ovarian cancer, in 2008 ASCO Annual Meeting Proceedings, Chicago, May 30–June 3, 2008, Abstr. 16533, American Society of Clinical Oncology; Alexandria, VA [Google Scholar]
- 16.Wang T. H., Wang H. S., Ichijo H., Giannakakou P., Foster J. S., Fojo T., Wimalasena J. (1998) Microtubule-interfering agents activate c-Jun N-terminal kinase/stress-activated protein kinase through both Ras and apoptosis signal-regulating kinase pathways. J. Biol. Chem. 273, 4928–4936 [DOI] [PubMed] [Google Scholar]
- 17.Wang T. H., Popp D. M., Wang H. S., Saitoh M., Mural J. G., Henley D. C., Ichijo H., Wimalasena J. (1999) Microtubule dysfunction induced by paclitaxel initiates apoptosis through both c-Jun N-terminal kinase (JNK)-dependent and -independent pathways in ovarian cancer cells. J. Biol. Chem. 274, 8208–8216 [DOI] [PubMed] [Google Scholar]
- 18.Hsieh S. Y., Shih T. C., Yeh C. Y., Lin C. J., Chou Y. Y., Lee Y. S. (2006) Comparative proteomic studies on the pathogenesis of human ulcerative colitis. Proteomics 6, 5322–5331 [DOI] [PubMed] [Google Scholar]
- 19.Wang T. H., Chan Y. H., Chen C. W., Kung W. H., Lee Y. S., Wang S. T., Chang T. C., Wang H. S. (2006) Paclitaxel (Taxol) upregulates expression of functional interleukin-6 in human ovarian cancer cells through multiple signaling pathways. Oncogene 25, 4857–4866 [DOI] [PubMed] [Google Scholar]
- 20.Wang T. H., Chang C. L., Wu H. M., Chiu Y. M., Chen C. K., Wang H. S. (2006) Insulin-like growth factor-II (IGF-II), IGF-binding protein-3 (IGFBP-3), and IGFBP-4 in follicular fluid are associated with oocyte maturation and embryo development. Fertil. Steril. 86, 1392–1401 [DOI] [PubMed] [Google Scholar]
- 21.Chao A., Wang T. H., Lee Y. S., Hsueh S., Chao A. S., Chang T. C., Kung W. H., Huang S. L., Chao F. Y., Wei M. L., Lai C. H. (2006) Molecular characterization of adenocarcinoma and squamous carcinoma of the uterine cervix using microarray analysis of gene expression. Int. J. Cancer 119, 91–98 [DOI] [PubMed] [Google Scholar]
- 22.Schulzer M. (1994) Diagnostic tests: a statistical review. Muscle Nerve 17, 815–819 [DOI] [PubMed] [Google Scholar]
- 23.Americo T. A., Chiarini L. B., Linden R. (2007) Signaling induced by hop/STI-1 depends on endocytosis. Biochem. Biophys. Res. Commun. 358, 620–625 [DOI] [PubMed] [Google Scholar]
- 24.Yi X., Luk J. M., Lee N. P., Peng J., Leng X., Guan X. Y., Lau G. K., Beretta L., Fan S. T. (2008) Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol. Cell. Proteomics 7, 315–325 [DOI] [PubMed] [Google Scholar]
- 25.Yin B. W., Lloyd K. O. (2001) Molecular cloning of the CA125 ovarian cancer antigen: identification as a new mucin, MUC16. J. Biol. Chem. 276, 27371–27375 [DOI] [PubMed] [Google Scholar]
- 26.Yin B. W., Dnistrian A., Lloyd K. O. (2002) Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int. J. Cancer 98, 737–740 [DOI] [PubMed] [Google Scholar]
- 27.Blalock T. D., Spurr-Michaud S. J., Tisdale A. S., Heimer S. R., Gilmore M. S., Ramesh V., Gipson I. K. (2007) Functions of MUC16 in corneal epithelial cells. Invest. Ophthalmol. Vis. Sci. 48, 4509–4518 [DOI] [PubMed] [Google Scholar]
- 28.Rump A., Morikawa Y., Tanaka M., Minami S., Umesaki N., Takeuchi M., Miyajima A. (2004) Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J. Biol. Chem. 279, 9190–9198 [DOI] [PubMed] [Google Scholar]
- 29.Gubbels J. A., Belisle J., Onda M., Rancourt C., Migneault M., Ho M., Bera T. K., Connor J., Sathyanarayana B. K., Lee B., Pastan I., Patankar M. S. (2006) Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol. Cancer 5, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scholler N., Garvik B., Hayden-Ledbetter M., Kline T., Urban N. (2007) Development of a CA125-mesothelin cell adhesion assay as a screening tool for biologics discovery. Cancer Lett. 247, 130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergan L., Gross J. A., Nevin B., Urban N., Scholler N. (2007) Development and in vitro validation of anti-mesothelin biobodies that prevent CA125/Mesothelin-dependent cell attachment. Cancer Lett. 255, 263–274 [DOI] [PubMed] [Google Scholar]
- 32.Bredemeyer A. J., Carrigan P. E., Fehniger T. A., Smith D. F., Ley T. J. (2006) Hop cleavage and function in granzyme B-induced apoptosis. J. Biol. Chem. 281, 37130–37141 [DOI] [PubMed] [Google Scholar]
- 33.Carta F., Demuro P. P., Zanini C., Santona A., Castiglia D., D'Atri S., Ascierto P. A., Napolitano M., Cossu A., Tadolini B., Turrini F., Manca A., Sini M. C., Palmieri G., Rozzo A. C. (2005) Analysis of candidate genes through a proteomics-based approach in primary cell lines from malignant melanomas and their metastases. Melanoma Res. 15, 235–244 [DOI] [PubMed] [Google Scholar]
- 34.Sun W., Xing B., Sun Y., Du X., Lu M., Hao C., Lu Z., Mi W., Wu S., Wei H., Gao X., Zhu Y., Jiang Y., Qian X., He F. (2007) Proteome analysis of hepatocellular carcinoma by two-dimensional difference gel electrophoresis: novel protein markers in hepatocellular carcinoma tissues. Mol. Cell. Proteomics 6, 1798–1808 [DOI] [PubMed] [Google Scholar]
- 35.Erlich R. B., Kahn S. A., Lima F. R., Muras A. G., Martins R. A., Linden R., Chiarini L. B., Martins V. R., Moura Neto V. (2007) STI1 promotes glioma proliferation through MAPK and PI3K pathways. Glia 55, 1690–1698 [DOI] [PubMed] [Google Scholar]
- 36.Walsh N., O'Donovan N., Kennedy S., Henry M., Meleady P., Clynes M., Dowling P. (2009) Identification of pancreatic cancer invasion-related proteins by proteomic analysis. Proteome Sci. 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caetano F. A., Lopes M. H., Hajj G. N., Machado C. F., Pinto Arantes C., Magalhães A. C., Vieira Mde P., Américo T. A., Massensini A. R., Priola S. A., Vorberg I., Gomez M. V., Linden R., Prado V. F., Martins V. R., Prado M. A. (2008) Endocytosis of prion protein is required for ERK1/2 signaling induced by stress-inducible protein 1. J. Neurosci. 28, 6691–6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathivanan S., Ahmed M., Ahn N. G., Alexandre H., Amanchy R., Andrews P. C., Bader J. S., Balgley B. M., Bantscheff M., Bennett K. L., Björling E., Blagoev B., Bose R., Brahmachari S. K., Burlingame A. S., Bustelo X. R., Cagney G., Cantin G. T., Cardasis H. L., Celis J. E., Chaerkady R., Chu F., Cole P. A., Costello C. E., Cotter R. J., Crockett D., DeLany J. P., De Marzo A. M., DeSouza L. V., Deutsch E. W., Dransfield E., Drewes G., Droit A., Dunn M. J., Elenitoba-Johnson K., Ewing R. M., Van Eyk J., Faca V., Falkner J., Fang X., Fenselau C., Figeys D., Gagné P., Gelfi C., Gevaert K., Gimble J. M., Gnad F., Goel R., Gromov P., Hanash S. M., Hancock W. S., Harsha H. C., Hart G., Hays F., He F., Hebbar P., Helsens K., Hermeking H., Hide W., Hjernø K., Hochstrasser D. F., Hofmann O., Horn D. M., Hruban R. H., Ibarrola N., James P., Jensen O. N., Jensen P. H., Jung P., Kandasamy K., Kheterpal I., Kikuno R. F., Korf U., Körner R., Kuster B., Kwon M. S., Lee H. J., Lee Y. J., Lefevre M., Lehvaslaiho M., Lescuyer P., Levander F., Lim M. S., Löbke C., Loo J. A., Mann M., Martens L., Martinez-Heredia J., McComb M., McRedmond J., Mehrle A., Menon R., Miller C. A., Mischak H., Mohan S. S., Mohmood R., Molina H., Moran M. F., Morgan J. D., Moritz R., Morzel M., Muddiman D. C., Nalli A., Navarro J. D., Neubert T. A., Ohara O., Oliva R., Omenn G. S., Oyama M., Paik Y. K., Pennington K., Pepperkok R., Periaswamy B., Petricoin E. F., Poirier G. G., Prasad T. S., Purvine S. O., Rahiman B. A., Ramachandran P., Ramachandra Y. L., Rice R. H., Rick J., Ronnholm R. H., Salonen J., Sanchez J. C., Sayd T., Seshi B., Shankari K., Sheng S. J., Shetty V., Shivakumar K., Simpson R. J., Sirdeshmukh R., Siu K. W., Smith J. C., Smith R. D., States D. J., Sugano S., Sullivan M., Superti-Furga G., Takatalo M., Thongboonkerd V., Trinidad J. C., Uhlen M., Vandekerckhove J., Vasilescu J., Veenstra T. D., Vidal-Taboada J. M., Vihinen M., Wait R., Wang X., Wiemann S., Wu B., Xu T., Yates J. R., Zhong J., Zhou M., Zhu Y., Zurbig P., Pandey A. (2008) Human Proteinpedia enables sharing of human protein data. Nat. Biotechnol. 26, 164–167 [DOI] [PubMed] [Google Scholar]
- 39.Collinge J. (2001) Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 24, 519–550 [DOI] [PubMed] [Google Scholar]
- 40.Dodelet V. C., Cashman N. R. (1998) Prion protein expression in human leukocyte differentiation. Blood 91, 1556–1561 [PubMed] [Google Scholar]
- 41.Li R., Liu D., Zanusso G., Liu T., Fayen J. D., Huang J. H., Petersen R. B., Gambetti P., Sy M. S. (2001) The expression and potential function of cellular prion protein in human lymphocytes. Cell. Immunol. 207, 49–58 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.