Abstract
Genetic and biochemical studies have shown that selective interactions between the Jun, Fos, and activating transcription factor (ATF) components of transcription factor activating protein 1 (AP-1) exhibit specific and critical functions in the regulation of cell proliferation, differentiation, and survival. For instance, the ratio between c-Jun/c-Fos and c-Jun/ATF2 dimers in the cell can be a determining factor in the cellular response to oncogenic or apoptotic stimuli. Until recently, no methods were available to detect endogenous AP-1 complexes in cells and tissues in situ. Here, we validated the proximity ligation assay (PLA) for its ability to specifically visualize and quantify changes in endogenous c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 complexes by using, among others, partner-selective c-Jun mutants. Furthermore, we examined the levels of c-Jun/AP-1 dimers in cell lines representing different types of human breast cancer and found that aggressive basal-like breast cancer cells can be discriminated from much less invasive luminal-like cells by PLA detection of c-Jun/Fra1 rather than of c-Jun/ATF2 and c-Jun/c-Fos. Also in tumor tissue derived from highly metastatic basal-like MDA-MB231 cells, high levels of c-Jun/Fra1 complexes were detected. Together, these results demonstrate that in situ PLA is a powerful diagnostic tool to analyze and quantify the amounts of biologically critical AP-1 dimers in fixed cells and tissue material.
Protein-protein interactions play crucial roles in development and homeostasis of organisms, tissues, and cells by enabling dynamic interplay between regulatory molecules. The dimeric transcription factor AP-11 offers an example of how selective protein-protein interactions can determine the fate of cells and organisms. The target genes, co-activators, and regulatory factors that AP-1 transcription factors bind to are determined by the composition of the AP-1 dimers, which is dictated by specific interactions between the AP-1 components, the members of the Jun, Fos, and ATF protein families (1–11). The variety of Jun/Fos, Jun/ATF, and ATF/ATF complexes that can be formed in a cell is the result of selective homo- and heterodimerization between these AP-1 components that is predominantly determined by specific amino acids in their leucine zipper domains (4, 12). Importantly, cytokines, growth factors, hormones, and stress stimuli can induce agent-, dose-, and cell context-specific expression patterns of the various Jun, Fos, and ATF family members. This allows these agents to activate distinct and specific programs of AP-1 target genes involved in the stimulus-specific responses (3, 4, 7–9, 11).
Both genetic and biochemical studies have shown that the distinct c-Jun/AP-1 dimers can play specific roles in fibroblast transformation, osteoclast differentiation, and neuronal cell survival in vitro (4, 13–15). In vivo, aberrant levels and/or functioning of, for instance, c-Jun, c-Fos, Fra1, and ATF2 can have major consequences for liver, bone, and skin homeostasis and positively or negative affect carcinogenesis (7, 8, 11, 16–19). c-Jun, Fra1, and ATF2 have also been shown to be involved in breast cancer development and progression (11, 19–25), and Fra1 can enhance human tumor cell motility and invasion (22, 26, 27). However, both in vitro and in vivo, the specific roles of the c-Jun-, c-Fos-, Fra1-, and ATF2-containing AP-1 dimers and the consequences of changes in their relative amounts are hardly known.
The analysis of the role of AP-1 dimer composition in AP-1-regulated processes has been hampered by the absence of an assay that can monitor individual interactions between endogenous AP-1 components in cells or tissues. With the development of the in situ proximity ligation assay (PLA) (28, 29), such a technique has become available. This detection method combines antibody-mediated recognition of two protein targets with proximity-dependent ligation and synthesis of an amplifiable DNA reporter molecule. Amplification of the reporter molecule is established via rolling circle amplification (30), and the resulting rolling circle products (RCPs) are visualized by hybridization with a complementary fluorescence-labeled oligonucleotide probe. Because the DNA reporter remains attached to one of the antibodies, it is possible to detect the location of the interacting proteins in subcellular compartments and tissues (28).
Here, we demonstrate that PLA enables specific in situ detection and quantification of c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 dimers in single cells. We validated the recognition of c-Jun/AP-1 dimers with the PLA procedure by ectopically expressing c-Jun proteins with selective dimerization properties. Furthermore, we visualized and quantified endogenous c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 complexes in human breast cancer cell lines as well as a melanoma line and show that aggressive breast cancer cells can be distinguished from much less invasive cells by PLA detection of c-Jun/Fra1 rather than of c-Jun/ATF2 and c-Jun/c-Fos dimers. We could also detect high levels of c-Jun/Fra1 complexes in mouse tumors derived from the highly metastatic cell line MDA-MB231. This shows that PLA detection of AP-1 dimers might become a powerful novel diagnostic and prognostic tool.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Stimulation
Adenovirus-transformed human embryonic retinoblasts (Ad-HERs) (31) and the cancer cell lines MCF7, T47D, ZR75, MDA-MB231, MDA-MB435, and MDA-MB436 (26) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and antibiotics. For AP-1 activation, cells were treated with 100 nm 12-O-tetradecanoylphorbol-13-acetate (TPA) for 2 h or 1 mm methyl methanesulfonate for 4 h.
Transient Transfection and DNA Constructs
For ectopic expression of c-Jun proteins, subconfluent Ad-HER cells were transfected in 6-well plates with a total amount of 1.5 μg of DNA/well using FuGENE 6 transfection reagent (Roche Applied Science) according to the supplier's protocol. For identification of transfected cells, 35 ng of histone-GFP expression vector was co-transfected. 24 h after transfection, cells were split for Western analysis, immunofluorescence, and/or PLA and grown overnight before treatment. pRSV-Δ6–194 c-JunHA (ΔJHA) and pRSV-c-Jun-m1HA were constructed by replacing the HpaI-KpnI fragments of pRSV-c-JunΔ6–194 (32) and pRSV-c-Jun-m1 (33) with that of pCMV-c-JunHA (34). pCMV-c-Jun-m1HA was created by replacing the PstI-KpnI fragment of pCMV-c-JunHA with the corresponding fragment of pRSV-c-Jun-m1HA.
Animals
4–5-week-old female nude mice (BALB/c nu/nu) were purchased from Charles River Laboratories. Animals were housed in individual ventilated cages under sterile condition, and sterile food and water were provided ad libitum. All animal experiments were approved and carried out according to the guidelines provided by the animal welfare committee at the Leiden University Medical Center.
Experimental Tumor Model
1 × 106 freshly harvested MDA-MB231-luc cells (35) were resuspended in 100 μl of sterile PBS and injected subcutaneously into the flank of the mice. 4 weeks later, mice were sacrificed by cervical dislocation, and primary tumors were removed and snap frozen in Optimal Cutting Temperature compound in liquid nitrogen. Cryosections (4 μm) from MDA-MB231-derived tumors were stored at −20 °C until use. Cryosections were stained with Meyer's hematoxylin and eosin to reveal tumor structure.
Western Blot Analysis
Preparation of cell lysates, protein gel electrophoresis, and Western blotting were performed as described previously (36, 37). Antibodies used were anti-c-Jun sc-1694, anti-ATF2 sc-187, and anti-Fra1 sc-605 from Santa Cruz Biotechnology; anti-c-Fos 06-341 from Upstate; and anti-HA11 from BabCo.
PLA
For analysis of cultured cells, cells were grown on collagen-coated Lab-Tek II chamber slides (Nunc, catalog number 154534) for at least 16 h, washed twice with PBS, and fixed in 3.7% formaldehyde in PBS for 15 min at room temperature. Subsequently, the slides were washed with TBS (25 mm Tris, 100 mm NaCl, pH 7.4); incubated for 10 min in 50 mm NH4Cl, TBS; washed with TBS; permeabilized for 15 min in 0.1% Triton X-100 in TBS; and washed with TBST (0.05% Tween 20 in TBS). The slides were then blocked for 2 h with 0.5% Roche Applied Science milk powder in TBST in a humidified chamber at 37 °C and incubated overnight at 4 °C with appropriate combinations of antibodies. After washing with TBST, proximity ligation was performed using the Rabbit PLUS and Mouse MINUS Duolink in situ PLA kits (OLINK Bioscience) according to the manufacturer's protocol. Subsequently, slides were dehydrated, air-dried, and embedded in DAPI-containing Citifluor mounting medium (Citifluor Ltd.). Fluorescence was detected using a Leica DM-RXA microscope. Images were acquired as sets of color images and prepared using PHOTO-PAINT software. For PLA analysis of fresh-frozen MDA-MB231-derived primary tumor tissue, cryosections were fixed with 3.7% formaldehyde in PBS for 15 min, and in situ PLA reactions were performed as described above for cultured cells.
Antibodies used for PLA were mouse anti-c-Jun 2315 (Cell Signaling Technology) or anti-HA11 (BabCo) combined with rabbit anti-ATF2 sc-187 (Santa Cruz Biotechnology), anti-c-Fos 06-341 (Upstate), and anti-Fra1 sc-22794 (Santa Cruz Biotechnology). Similar PLA results were obtained when mouse anti-c-Jun 2315 was combined with rabbit anti-Fra1 sc-605 or anti-c-Fos sc-52 (Santa Cruz Biotechnology), which recognize different Fra1 and c-Fos epitopes than sc-22794 and 06-341, respectively. In tissue sections, combination of mouse anti-Fra1 sc-28310 (Santa Cruz Biotechnology) with rabbit anti-c-Jun sc-1694 (Santa Cruz Biotechnology) or rabbit anti-c-Jun 9165 (Cell Signaling Technology) yielded results similar to those using anti-c-Jun 2315 with anti-Fra1 sc-22794. Single antibody incubations were performed as negative controls. On frozen tissue sections, mouse anti-c-Jun 2315 was combined with rabbit anti-insulin receptor β (Santa Cruz Biotechnology sc-711) as an additional negative control. These negative controls did not give significant PLA signals.
Image Analysis
The RCPs per cell were counted by semiautomated image analysis using the single cell analysis function of BlobFinder software (38). Data are expressed as mean ± S.E. Statistical significance was determined by unpaired Student's two-tailed t test using Excel software. p < 0.05 was considered as statistically significant.
RESULTS
In Situ PLA Detection of Endogenous c-Jun-c-Fos and c-Jun-ATF2 Interactions before and after TPA-induced AP-1 Activation
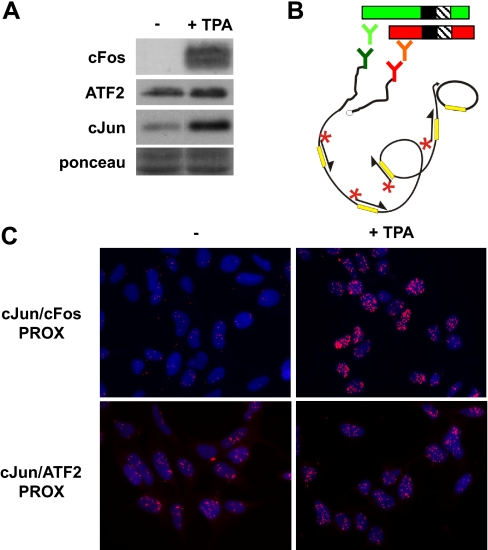
Based on their DNA binding specificities, AP-1 transcription factors can be divided into two subgroups: Jun/Fos dimers, which preferentially bind to the 7-bp “classical” AP-1 sites, and Jun/ATF dimers, which efficiently interact with 8-bp ATF/cAMP-response element-related motifs. Of the two originally identified AP-1 components, c-Jun and c-Fos, c-Fos proteins are only detectable after addition of AP-1-inducing agents in most cell types, whereas c-Jun (and ATF2) proteins are also expressed at detectable levels under non-induced conditions. To ensure visualization of both c-Jun/c-Fos and c-Jun/ATF2 complexes with PLA, we used Ad-HERs that contain relatively high levels of c-Jun/ATF2 activity (31, 36) and treated these cells with the phorbol ester TPA, a potent inducer of AP-1. Western analysis showed that both c-Fos and c-Jun protein levels were strongly induced by TPA in these cells, whereas ATF2 was not significantly affected (Fig. 1A).
Fig. 1.
In situ PLA detection of endogenous c-Jun-c-Fos and c-Jun-ATF2 interactions. A, Ad-HER cells were left untreated (−) or were stimulated (+) with 100 nm TPA. After 2 h, protein extracts were prepared and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Equal loading was confirmed with Ponceau S staining. B, schematic representation of in situ PLA detection of a c-Jun heterodimer using secondary proximity probes. Green and red Ys indicate the primary and oligo-conjugated secondary antibodies specific for c-Jun (red) and its dimer partner (green) depicted at the top of the figure. The long curved black line represents the RCP, including the detection sequence (yellow bars). The full circle at the end of the RCP represents the circular DNA template that resulted from proximity-dependent ligation and was amplified by rolling circle amplification. The detection probes are represented by asterisk-arrows. For a further description, see the text. C, Ad-HER cells were grown on collagen-coated microchamber slides and treated as described above. After fixation, in situ PLA for c-Jun/c-Fos and c-Jun/ATF2 dimers was performed with c-Jun-, c-Fos-, and ATF2-specific antibodies. The detected dimers (cJun/cFos PROX and cJun/ATF2 PROX) are represented by the fluorescent rolling circle products (red dots). PROX, PLA-detected proximity.
For in situ detection of c-Jun/Fos and c-Jun/ATF2 dimers, a modified version of the PLA technique was applied that utilizes unconjugated primary antibodies from different species, one recognizing c-Jun and the other recognizing c-Fos or ATF2, that are detected by secondary antibodies covalently coupled to oligonucleotides, the so-called proximity probes. Only when both primary and secondary antibodies are bound to their respective target molecules can proximity-dependent ligation and rolling circle amplification of a circular DNA reporter molecule occur (Ref. 29 and Fig. 1B). Subsequent hybridization of complementary fluorescence-labeled oligonucleotides to the RCPs visualized the PLA-detected c-Jun-c-Fos or c-Jun-ATF2 interactions as red dots (Fig. 1C). Single antibody control reactions did not result in significant PLA signals (not shown).
In line with the Western results of Fig. 1A, the PLA signals visualizing c-Jun-c-Fos interactions were very low in non-treated cells but strongly induced by TPA (Fig. 1C). In contrast, c-Jun-ATF2 interactions were already easily detected in non-stimulated cells and only weakly enhanced upon TPA treatment (Fig. 1C). This suggests that the induction of c-Jun by TPA causes only a weak increase in the amount of c-Jun/ATF2 dimers.
Validation and Quantification of c-Jun/c-Fos and c-Jun/ATF2 PLA Detection with Dimer-specific c-Jun Mutants
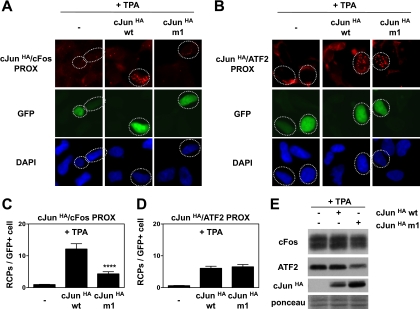
To verify the specificity of the modified version of the PLA protocol for detection of AP-1 dimers, we tested two different types of partner-selective c-Jun proteins: single chain molecules (“tethered” heterodimers) and leucine zipper mutants (13, 33). Indeed, overexpression of c-Jun-ATF2 and c-Jun-c-Fos single chain molecules only gave high PLA signals for the corresponding combinations of c-Jun, ATF2, and c-Fos antibodies (supplemental Fig. S1 and data not shown). Next, we verified this c-Jun-Fos and c-Jun-ATF2 PLA protocol with the c-Jun leucine zipper mutant m1, a mutant that interacts efficiently with ATF2 but has very low affinity for c-Fos (33). For this purpose, cells were transfected with HA-tagged versions of wild type c-Jun or c-Jun-m1 together with a histone-GFP expression vector to identify transfected cells. PLA for the interaction of HA-tagged c-Jun with endogenous c-Fos or ATF2 was subsequently performed in TPA-treated cells using HA and c-Fos or ATF2 antibodies. As expected, detection of RCPs (red dots) with these antibody combinations was dependent on transfection of c-JunHA (Fig. 2, A and B). The number of RCPs in the GFP-positive cell populations was quantified by digital image analysis (Fig. 2, C and D).
Fig. 2.
Validation of PLA detection of c-Jun/c-Fos and c-Jun/ATF2 dimers with a partner-preferring c-Jun mutant. Ad-HER cells were transfected with HA-tagged wild type c-Jun (cJunHA wt), the ATF2-preferring c-Jun mutant m1 (cJunHA m1), or the control vector (−). A histone-GFP vector was cotransfected to identify transfected cells. After 2 days, cells were treated with TPA for 2 h. Subsequently, in situ PLA for c-JunHA-c-Fos interactions (cJunHA/cFos PROX) (A) and c-JunHA-ATF2 interactions (cJunHA/ATF2 PROX) (B) was performed using antibodies against HA and c-Fos (A) or HA and ATF2 (B). Cell nuclei were stained with DAPI (blue); transfected GFP-positive cells (green) are indicated with dashed circles. C and D, in situ PLA signals (red dots) in the GFP-positive cell population (n > 75) were quantified by semiautomated image analysis with BlobFinder software. The average number of RCPs per cell is shown ±S.E. (****, p < 1 × 10−4). E, protein extracts from cells treated as described above were analyzed by SDS-PAGE and immunoblotting with c-Fos-, ATF2-, and HA-specific antibodies. Equal loading of the gel was confirmed with Ponceau S staining. PROX, PLA-detected proximity.
In agreement with the low affinity of c-Jun-m1 for c-Fos (33), the number of c-JunHA-c-Fos RCPs obtained for this mutant was much lower than the signals obtained for wt c-Jun (Fig. 2, A and C) despite the fact that the expression of c-Jun-m1HA was higher (Fig. 2E). In contrast, c-Jun-wtHA and c-Jun-m1HA gave similar amounts of c-JunHA-ATF2 RCPs (Fig. 2, B and D), although the ATF2 levels were reduced in the cells transfected with c-Jun-m1 (Fig. 2E). These results thus show that the PLA technique displays a high sensitivity in detecting specific c-Jun/AP-1 complexes in situ.
Perturbation of Endogenous c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 Complexes by Interfering c-Jun Mutant
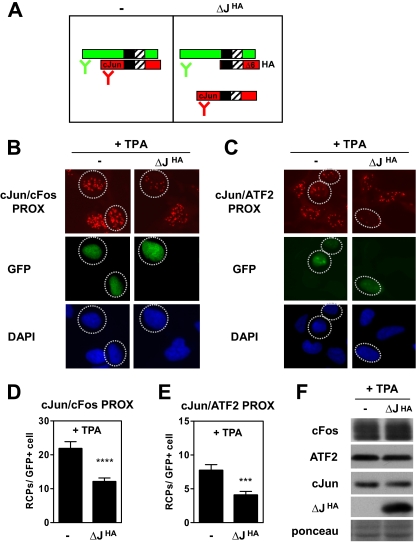
To validate the specificity of the PLA procedure used for detection of endogenous c-Jun dimers, we used the c-Jun transactivation mutant ΔJHA, which lacks amino acids 6–194 (32). This protein can efficiently compete with wild type c-Jun for binding to dimer partners and DNA. Moreover, because of the N-terminal deletion, this truncated c-Jun protein is not recognized by the c-Jun antibody used for PLA detection (Fig. 3A).
Fig. 3.
Reduction of endogenous c-Jun/c-Fos and c-Jun/ATF2 PLA signals by an interfering c-Jun mutant. A, schematic representation of PLA detection of c-Jun (red) interacting with one of its dimer partners (green) in the absence (−) or presence (+) of the competing c-Jun deletion mutant ΔJHA, which lacks the epitope recognized by the c-Jun antibody used in PLA. B–E, Ad-HER cells were transfected with ΔJHA or the control vector (−) and a histone-GFP vector to identify transfected cells. After 2 days, cells were treated with TPA for 2 h. Subsequently, in situ PLA for c-Jun-c-Fos (cJun/cFos PROX) (B) and c-Jun-ATF2 interactions (cJun/ATF2 PROX) (C) was performed. Cell nuclei were stained with DAPI (blue); transfected GFP-positive cells (green) are indicated with dashed circles. D and E, in situ PLA signals in the GFP-positive cell population (n > 75) were quantified by semiautomated image analysis with BlobFinder software. The average number of RCPs per cell is shown ±S.E. (****, p < 1 × 10−4; ***, p < 0.001). F, protein extracts from cells treated as described above were analyzed by SDS-PAGE and immunoblotting with c-Fos-, ATF2-, c-Jun-, and HA-specific antibodies. Equal loading of the gel was confirmed with Ponceau S staining. PROX, PLA-detected proximity.
As shown in Fig. 3, B–E, ΔJHA efficiently reduced the amounts of the endogenous c-Jun-c-Fos and c-Jun-ATF2 interactions detected by PLA upon TPA treatment but did not affect the total levels of c-Jun, c-Fos, and ATF2 as detected by Western analysis (Fig. 3F). This implies that the c-Jun-c-Fos and c-Jun-ATF2 interactions as detected by PLA signals represent bona fide c-Jun/Fos and c-Jun/ATF2 dimers rather than indirect or nonspecific c-Jun-c-Fos or c-Jun-ATF2 proximity.
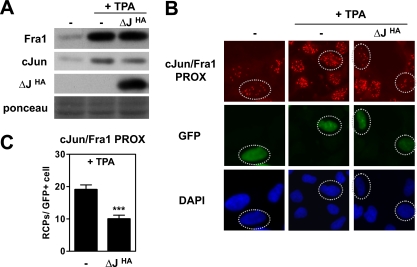
As the Fos family member Fra1 is already expressed under non-stimulated conditions in most cell types, including Ad-HER (Fig. 4A), we next performed proximity ligation analysis for c-Jun-Fra1 interaction. Interaction between endogenous c-Jun and Fra1 could already be detected by PLA in non-stimulated cells but was further increased in TPA-treated cells (Fig. 4B). This was most likely caused by increased c-Jun and Fra1 synthesis (Fig. 4A). Moreover, expression of the dimerization inhibitor ΔJHA efficiently reduced the number of RCPs representing c-Jun-Fra1 interactions (Fig. 4, B and C), indicating that both the c-Jun-c-Fos, c-Jun-ATF2, and c-Jun-Fra1 interactions detected via this PLA method represent genuine dimers.
Fig. 4.
PLA detection and validation of c-Jun/Fra1 dimers. Ad-HER cells were transfected with ΔJHA or the control vector (−) and a histone-GFP vector to identify transfected cells. After 2 days, cells were treated with TPA for 2 h when indicated. A, cell extracts were analyzed by SDS-PAGE and immunoblotting with Fra1-, c-Jun-, and HA-specific antibodies. Equal loading of the gel was confirmed with Ponceau S staining. B, in situ PLA was performed for c-Jun/Fra1 (cJun/Fra1 PROX). Cell nuclei were stained with DAPI (blue); transfected GFP-positive cells (green) are indicated with dashed circles. C, c-Jun/Fra1 PLA signals in the GFP-positive cell population (n > 100) were quantified by semiautomated image analysis with BlobFinder software. The average number of RCPs per cell is shown ±S.E. (****, p < 0.0001). PROX, PLA-detected proximity.
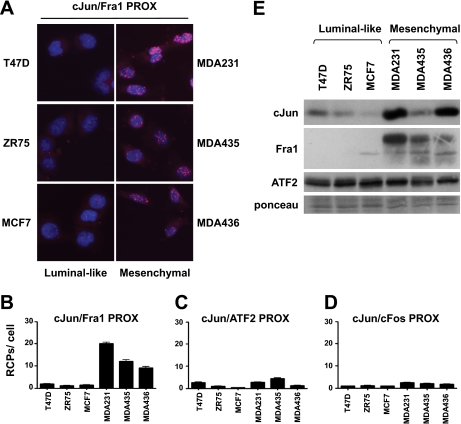
Enhanced Levels of c-Jun/Fra1 Dimers in Basal-like/Mesenchymal Breast Cancer Cells
c-Jun, c-Fos, Fra1, and ATF2 are involved in multiple types of cancer by regulating genes involved in cell proliferation, survival, migration, and invasion (8, 11, 16, 19, 24, 39, 40). Moreover, by controlling DNA repair, DNA damage signaling, and drug resistance, these AP-1 components can influence the response of cancer cells to therapy (41–45). However, it is as yet unknown which AP-1 dimers are functionally involved in these processes and whether quantification of the (relative) levels of these dimers has diagnostic and prognostic value. As a first approach to address this issue, we used the PLA procedure described above to measure the relative amounts of c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 complexes in weakly invasive breast cancer cell lines with luminal-like characteristics (MCF7, T47D, and ZR75) and highly invasive lines with a basal-like/mesenchymal phenotype (MDA-MB231 and MDA-MB436) (26, 46). In addition, MDA-MB435 cells, which were originally described as aggressive breast cancer cells but were later found to be derived from a human melanoma cell line (47), were examined. Strikingly, in all three more invasive and aggressive cancer cell lines, high numbers of c-Jun/Fra1 signals were detected by PLA, whereas much lower numbers of c-Jun/Fra1 RCPs were obtained in the luminal-like cells (Fig. 5, A and B). Similar results were obtained with c-Jun and Fra1 antibodies recognizing different c-Jun and Fra1 epitopes (data not shown). In contrast, the levels of c-Jun/ATF2 and c-Jun/c-Fos signals were low in all cancer lines examined (Fig. 5, C and D).
Fig. 5.
Enhanced levels of c-Jun/Fra1 PLA signals in mesenchymal breast cancer and melanoma cells. A, in situ PLA for c-Jun/Fra1 (cJun/Fra1 PROX) was performed for the luminal-like breast cancer cell lines MCF7, T47D, and ZR75 and the mesenchymal cell lines MDA-MB231, MDA-MB435, and MDA-MB436. Cell nuclei were stained with DAPI (blue). B–D, quantification of the c-Jun/Fra1, c-Jun/ATF2, and c-Jun/c-Fos PLA signals obtained for the cell lines shown in A by semiautomated image analysis with BlobFinder software. The average number of RCPs per cell is shown ±S.E. E, protein extracts from the cell lines described above were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. Equal loading of the gel was confirmed with Ponceau S staining. PROX, PLA-detected proximity.
The high levels of c-Jun/Fra1 PLA signals in the aggressive basal-like cell lines appear to be due to increased expression of either c-Jun, Fra1, or both (Fig. 5E and Refs. 26 and 48). However, there were no significant differences in the ATF2 protein levels in the cell lines examined here, whereas c-Fos was hardly detectable (Fig. 5E and supplemental Fig. S2). The low number of PLA signals detected for c-Jun/c-Fos in these cell lines was not due to a technical problem as treatment of the basal-like MDA-MB231 cells with methyl methanesulfonate strongly induced both c-Fos and c-Jun and also robustly increased the c-Jun/c-Fos-specific PLA signals in these cells (supplemental Fig. S2). In summary, these results indicate that highly invasive basal-like breast cancer cells can be discriminated from less invasive luminal-like cells by PLA detection of c-Jun/Fra1 rather than of c-Jun/ATF2 and c-Jun/c-Fos dimers.
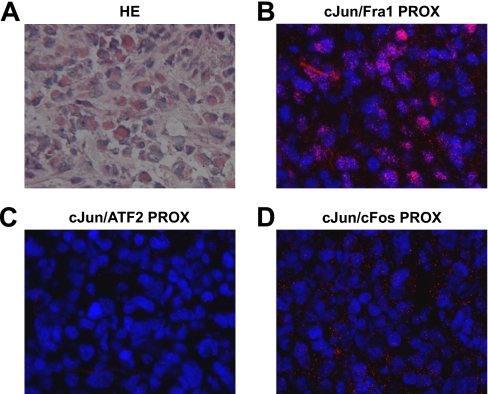
In Situ Detection of Endogenous c-Jun/Fra1 Dimers in MDA-MB231-derived Tumor Tissue
Finally, we tested whether the c-Jun/AP-1 PLA procedure used above could also be utilized to visualize c-Jun/AP-1 dimers in histological tissue sections from breast cancer xenografts. For this purpose, MDA-MB231 cells were injected in the flank of nude mice, and after 4 weeks, the resulting primary tumors were dissected, snap frozen, and processed for cryosectioning. A hematoxylin-eosin-stained frozen section of such an MDA-MB231-derived primary tumor is shown in Fig. 6A. Importantly, c-Jun/Fra1 PLA resulted in strong signals in this tissue (Fig. 6B), but hardly any signal was found for c-Jun/c-Fos and c-Jun/ATF2 (Fig. 6, C and D). Thus, this demonstrates that in situ PLA is a powerful diagnostic tool to analyze and quantify the amounts of biologically critical AP-1 dimers in fixed tissues.
Fig. 6.
In situ detection of c-Jun/Fra1 complexes in MDA-MB231-derived tumor tissue. A, a hematoxylin-eosin (HE)-stained cryosection of a MDA-MB231-derived primary tumor. B–D, in situ PLA for the indicated c-Jun/AP-1 dimers in MDA-MB231-derived tumors. The tissue section was counterstained with DAPI (blue) to visualize the nuclei. PROX, PLA-detected proximity.
DISCUSSION
The Jun, Fos, and ATF components of transcription factor AP-1 have been implicated in a variety of biological processes, including development, cellular stress responses, and carcinogenesis (1, 2, 8, 16–18). However, much less is known about the specific biological functions of the various homo- and heterodimers formed by these proteins and the consequences of changes in their relative amounts. Analysis of the effect of the AP-1 dimer composition on AP-1 function is complicated by a number of factors. First of all, all AP-1 subfamilies (Jun, Fos, and ATF but also Maf) contain multiple members of which some can also homodimerize, which results in a large number of possible dimer combinations (12). Second, the expression levels of the distinct AP-1 components in a cell at a given time are variable and depend on multiple factors, including, for instance, cell type, cellular context, and the stage of the cell cycle (2, 3, 7, 9). Third, closely related AP-1 components (such as c-Jun and JunB or c-Fos and Fra1) have been shown to be functionally redundant in certain situations (9, 40). Finally, the number of tools to examine (endogenous) AP-1 dimers has been limited.
Some studies have investigated the impact of dimer composition on AP-1 functioning by generating partner-restricted mutants of AP-1 components (4, 5, 12, 15). In addition, covalently linked AP-1 single chain molecules have been generated and analyzed (5, 13, 14). These genetic studies have shown that the functions of AP-1 components in gene activation, cell proliferation, and transformation strongly depend on their dimer partners. However, results obtained with exogenous AP-1 components or dimers should be carefully interpreted because several AP-1 members can regulate their own expression or that of other family members (1–4) and may therefore affect the total pool of AP-1 dimers also indirectly by influencing the availability of “non-targeted” dimer partners.
Methods that have been used to examine (functional) interactions between AP-1 components in cells include co-immune precipitation, mammalian one- or two-hybrid analysis (4), dimer-specific reporters (33), and bimolecular fluorescence complementation (49, 50). These methods have proven to be very powerful but have some disadvantages. Although co-immune precipitation enables detection of interactions between endogenous proteins, the stringency of the buffer is a critical factor: potential artifacts include (partial) loss of weak interactions, “artificial” postlysis dimerization, and/or high background signals. Moreover, due to the necessary cell lysis, only crude indications of the subcellular localization of protein complexes can be obtained. In contrast, the bimolecular fluorescence complementation method provides detailed information on the subcellular localization and interactions of proteins in living cells but requires ectopic expression of proteins fused to GFP domains. Fusion proteins, also required for mammalian hybrid assays, do not necessarily (inter)act and/or localize in the same manner as their native counterparts.
In this study, we show that in situ PLA (28) allows subcellular localization and quantification of the relative amounts of endogenous AP-1 dimers in fixed cells and tissues. Although this method cannot be combined with live cell imaging, it has great potential for the analysis of the roles of the distinct AP-1 dimers in processes such as embryonic development, carcinogenesis, tumor progression, and stress responses. The fact that fixed tissue material can be analyzed makes it possible to study endogenous AP-1 interactions or their disturbances in histopathological tissue sections, something that is not possible with any of the methods mentioned above.
By using single chain dimers, a partner-selective mutant, and a competitive inhibitor of c-Jun dimerization, we have validated the specificity of the modified in situ PLA procedure (29) for detection of c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 dimers. Analysis of an ATF2-preferring c-Jun mutant showed that the PLA technique displays high sensitivity in detecting specific c-Jun/AP-1 complexes in situ. Moreover, the dimerization inhibitor reduced the PLA signals for c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 without affecting the expression of the individual components. In theory, these PLA signals could have been caused by “non-dimer” proximity or dimer-independent binding of two components to (different sites of) a co-activator or scaffold protein. However, such nonspecific proximity is unlikely to be reduced by the competing c-Jun protein. Therefore, we conclude that the detected c-Jun/AP-1 PLA signals represent bona fide dimers.
With the modified in situ PLA procedure, we have also analyzed the changes in the levels of endogenous c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 dimers upon treatment of Ad-HER cells with TPA, a tumor promoter and a well known AP-1-inducing agent. This analysis showed that the levels of c-Jun/ATF2 dimers were only weakly enhanced by TPA, whereas the induction of c-Jun/c-Fos and c-Jun/Fra1 was much more robust. These results nicely reflect the strong TPA induction of c-Jun, c-Fos, and Fra1 protein by TPA.
Aberrant expression and/or functioning of c-Jun, c-Fos, Fra1, and ATF2 has been implicated in breast cancer development and/or progression (20, 23, 26, 27, 51, 52). We therefore used PLA to examine the relative amounts of c-Jun/c-Fos, c-Jun/ATF2, and c-Jun/Fra1 dimers in human breast cancer cell lines and in histological tissue sections from breast cancer xenografts. Importantly, we found that aggressive basal-like breast cancer cells contain much more PLA-detectable c-Jun/Fra1 dimers than less invasive luminal-like breast cancer cells. Moreover, high levels of c-Jun/Fra1, rather than c-Jun/c-Fos or c-Jun/ATF2, were also detected in tissue sections from breast cancer xenografts derived from basal-like MDA-MB231 cells. Notably, similar results were obtained when several other pairs of primary antibodies were used that recognize different c-Jun, c-Fos, and Fra1 epitopes (see “Experimental Procedures” for details). Because of the recent findings on the role of Fra1 in breast cancer invasion and metastasis (Refs. 11, 22, 24, and 26 and references therein), these in situ AP-1 PLA assays will be very useful for the analysis of the role of Fra1 in the tumor microenvironment where it may regulate interactions between tumor cells, cancer-associated fibroblasts, and tumor-associated immune cells. Because related AP-1 components can have similar but also antagonistic functions, it will be interesting to include JunB, JunD, Fra2, and FosB in these PLA assays. In conclusion, the described in situ PLA for AP-1 has the potential to become a powerful diagnostic and prognostic tool and offers a novel approach to study specific AP-1 dimer-dependent mechanisms in vitro and in vivo.
Supplementary Material
Acknowledgments
We are grateful to Bob van de Water, Sylvia Levedec, and all members of the European Network for ENhanced LIGase-based Histochemical Techniques (ENLIGHT), in particular Ola Soederberg, for helpful suggestions and stimulating discussions. We thank Sandra van Heiningen, Frans van de Rijke, and OLINK Bioscience for excellent technical support; Latifa Bakiri and Moshe Yaniv for the single chain c-Jun/AP-1 constructs; and Ton Raap and Ola Soederberg for critical reading of the manuscript.
* This work was supported by the European Union FP6 Program of the European Community (ENLIGHT), Swedish Cancerfonden Grant 090773, and The Netherlands Centre for Biomedical Genetics.
 This article contains supplemental Figs. S1 and S2.
This article contains supplemental Figs. S1 and S2.
1 The abbreviations used are:
- AP-1
- activating protein 1
- ATF
- activating transcription factor
- PLA
- proximity ligation assay
- RCP
- rolling circle product
- TBST
- TBS with Tween
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- Ad-HER
- adenovirus-transformed human embryonic retinoblast
- GFP
- green fluorescent protein
- DAPI
- 4′,6-diamidino-2-phenylindole
- HA
- hemagglutinin
- wt
- wild type.
REFERENCES
- 1.Shaulian E., Karin M. (2002) AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–E136 [DOI] [PubMed] [Google Scholar]
- 2.Shaulian E., Karin M. (2001) AP-1 in cell proliferation and survival. Oncogene 20, 2390–2400 [DOI] [PubMed] [Google Scholar]
- 3.Karin M., Liu Z., Zandi E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 [DOI] [PubMed] [Google Scholar]
- 4.van Dam H., Castellazzi M. (2001) Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene 20, 2453–2464 [DOI] [PubMed] [Google Scholar]
- 5.Diefenbacher M., Sekula S., Heilbock C., Maier J. V., Litfin M., van Dam H., Castellazzi M., Herrlich P., Kassel O. (2008) Restriction to Fos family members of Trip6-dependent coactivation and glucocorticoid receptor-dependent trans-repression of activator protein-1. Mol. Endocrinol. 22, 1767–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciapponi L., Bohmann D. (2002) An essential function of AP-1 heterodimers in Drosophila development. Mech. Dev. 115, 35–40 [DOI] [PubMed] [Google Scholar]
- 7.Hess J., Angel P., Schorpp-Kistner M. (2004) AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 117, 5965–5973 [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Bergami P., Lau E., Ronai Z. (2010) Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat. Rev. Cancer. 10, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mechta-Grigoriou F., Gerald D., Yaniv M. (2001) The mammalian Jun proteins: redundancy and specificity. Oncogene 20, 2378–2389 [DOI] [PubMed] [Google Scholar]
- 10.Suzukawa K., Colburn N. H. (2002) AP-1 transrepressing retinoic acid does not deplete coactivators or AP-1 monomers but may target specific Jun or Fos containing dimers. Oncogene 21, 2181–2190 [DOI] [PubMed] [Google Scholar]
- 11.Verde P., Casalino L., Talotta F., Yaniv M., Weitzman J. B. (2007) Deciphering AP-1 function in tumorigenesis: fra-ternizing on target promoters. Cell Cycle 6, 2633–2639 [DOI] [PubMed] [Google Scholar]
- 12.Vinson C., Myakishev M., Acharya A., Mir A. A., Moll J. R., Bonovich M. (2002) Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol. 22, 6321–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakiri L., Matsuo K., Wisniewska M., Wagner E. F., Yaniv M. (2002) Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 22, 4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakiri L., Takada Y., Radolf M., Eferl R., Yaniv M., Wagner E. F., Matsuo K. (2007) Role of heterodimerization of c-Fos and Fra1 proteins in osteoclast differentiation. Bone 40, 867–875 [DOI] [PubMed] [Google Scholar]
- 15.Yuan Z., Gong S., Luo J., Zheng Z., Song B., Ma S., Guo J., Hu C., Thiel G., Vinson C., Hu C. D., Wang Y., Li M. (2009) Opposing roles for ATF2 and c-Fos in c-Jun-mediated neuronal apoptosis. Mol. Cell. Biol. 29, 2431–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eferl R., Wagner E. F. (2003) AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868 [DOI] [PubMed] [Google Scholar]
- 17.Zenz R., Wagner E. F. (2006) Jun signalling in the epidermis: from developmental defects to psoriasis and skin tumors. Int. J. Biochem. Cell Biol. 38, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 18.Durchdewald M., Angel P., Hess J. (2009) The transcription factor Fos: a Janus-type regulator in health and disease. Histol. Histopathol. 24, 1451–1461 [DOI] [PubMed] [Google Scholar]
- 19.Young M. R., Colburn N. H. (2006) Fra-1 a target for cancer prevention or intervention. Gene 379, 1–11 [DOI] [PubMed] [Google Scholar]
- 20.Bamberger A. M., Methner C., Lisboa B. W., Städtler C., Schulte H. M., Löning T., Milde-Langosch K. (1999) Expression pattern of the AP-1 family in breast cancer: association of fosB expression with a well-differentiated, receptor-positive tumor phenotype. Int. J. Cancer 84, 533–538 [DOI] [PubMed] [Google Scholar]
- 21.Chiappetta G., Ferraro A., Botti G., Monaco M., Pasquinelli R., Vuttariello E., Arnaldi L., Di Bonito M., D'Aiuto G., Pierantoni G. M., Fusco A. (2007) FRA-1 protein overexpression is a feature of hyperplastic and neoplastic breast disorders. BMC Cancer 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y. P., Zhou H., Krueger J., Kaplan C., Liao D., Markowitz D., Liu C., Chen T., Chuang T. H., Xiang R., Reisfeld R. A. (2010) The role of proto-oncogene Fra-1 in remodeling the tumor microenvironment in support of breast tumor cell invasion and progression. Oncogene 29, 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maekawa T., Shinagawa T., Sano Y., Sakuma T., Nomura S., Nagasaki K., Miki Y., Saito-Ohara F., Inazawa J., Kohno T., Yokota J., Ishii S. (2007) Reduced levels of ATF-2 predispose mice to mammary tumors. Mol. Cell. Biol. 27, 1730–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozanne B. W., Spence H. J., McGarry L. C., Hennigan R. F. (2007) Transcription factors control invasion: AP-1 the first among equals. Oncogene 26, 1–10 [DOI] [PubMed] [Google Scholar]
- 25.Yin X., Dewille J. W., Hai T. (2008) A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene 27, 2118–2127 [DOI] [PubMed] [Google Scholar]
- 26.Belguise K., Kersual N., Galtier F., Chalbos D. (2005) FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 24, 1434–1444 [DOI] [PubMed] [Google Scholar]
- 27.Milde-Langosch K., Röder H., Andritzky B., Aslan B., Hemminger G., Brinkmann A., Bamberger C. M., Löning T., Bamberger A. M. (2004) The role of the AP-1 transcription factors c-Fos, FosB, Fra-1 and Fra-2 in the invasion process of mammary carcinomas. Breast Cancer Res. Treat. 86, 139–152 [DOI] [PubMed] [Google Scholar]
- 28.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 29.Jarvius M., Paulsson J., Weibrecht I., Leuchowius K. J., Andersson A. C., Wählby C., Gullberg M., Botling J., Sjöblom T., Markova B., Ostman A., Landegren U., Söderberg O. (2007) In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Mol. Cell. Proteomics 6, 1500–1509 [DOI] [PubMed] [Google Scholar]
- 30.Banér J., Nilsson M., Mendel-Hartvig M., Landegren U. (1998) Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 26, 5073–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dam H., Offringa R., Meijer I., Stein B., Smits A. M., Herrlich P., Bos J. L., van der Eb A. J. (1990) Differential effects of the adenovirus E1A oncogene on members of the AP-1 transcription factor family. Mol. Cell. Biol. 10, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angel P., Smeal T., Meek J., Karin M. (1989) Jun and v-jun contain multiple regions that participate in transcriptional activation in an interdependent manner. New Biol. 1, 35–43 [PubMed] [Google Scholar]
- 33.van Dam H., Huguier S., Kooistra K., Baguet J., Vial E., van der Eb A. J., Herrlich P., Angel P., Castellazzi M. (1998) Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 12, 1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treier M., Staszewski L. M., Bohmann D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78, 787–798 [DOI] [PubMed] [Google Scholar]
- 35.Deckers M., van Dinther M., Buijs J., Que I., Löwik C., van derPluijm G., ten Dijke P. (2006) The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 66, 2202–2209 [DOI] [PubMed] [Google Scholar]
- 36.van Dam H., Duyndam M., Rottier R., Bosch A., de Vries-Smits L., Herrlich P., Zantema A., Angel P., van der Eb A. J. (1993) Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 12, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouwens D. M., de Ruiter N. D., van der Zon G. C., Carter A. P., Schouten J., van der Burgt C., Kooistra K., Bos J. L., Maassen J. A., van Dam H. (2002) Growth factors can activate ATF2 via a two-step mechanism: phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 21, 3782–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allalou A., Wählby C. (2009) BlobFinder, a tool for fluorescence microscopy image cytometry. Comput. Methods Programs Biomed. 94, 58–65 [DOI] [PubMed] [Google Scholar]
- 39.Milde-Langosch K. (2005) The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer 41, 2449–2461 [DOI] [PubMed] [Google Scholar]
- 40.Jochum W., Passegué E., Wagner E. F. (2001) AP-1 in mouse development and tumorigenesis. Oncogene 20, 2401–2412 [DOI] [PubMed] [Google Scholar]
- 41.Hamdi M., Popeijus H. E., Carlotti F., Janssen J. M., van der Burgt C., Cornelissen-Steijger P., van de Water B., Hoeben R. C., Matsuo K., van Dam H. (2008) ATF3 and Fra1 have opposite functions in JNK- and ERK-dependent DNA damage responses. DNA Repair 7, 487–496 [DOI] [PubMed] [Google Scholar]
- 42.Bhoumik A., Lopez-Bergami P., Ronai Z. (2007) ATF2 on the double-activating transcription factor and DNA damage response protein. Pigment Cell Res. 20, 498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrlich P., Karin M., Weiss C. (2008) Supreme EnLIGHTenment: damage recognition and signaling in the mammalian UV response. Mol. Cell 29, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potapova O., Basu S., Mercola D., Holbrook N. J. (2001) Protective role for c-Jun in the cellular response to DNA damage. J. Biol. Chem. 276, 28546–28553 [DOI] [PubMed] [Google Scholar]
- 45.MacLaren A., Black E. J., Clark W., Gillespie D. A. (2004) c-Jun-deficient cells undergo premature senescence as a result of spontaneous DNA damage accumulation. Mol. Cell. Biol. 24, 9006–9018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blick T., Widodo E., Hugo H., Waltham M., Lenburg M. E., Neve R. M., Thompson E. W. (2008) Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin. Exp. Metastasis 25, 629–642 [DOI] [PubMed] [Google Scholar]
- 47.Rae J. M., Creighton C. J., Meck J. M., Haddad B. R., Johnson M. D. (2007) MDA-MB-435 cells are derived from M14 melanoma cells—a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res. Treat. 104, 13–19 [DOI] [PubMed] [Google Scholar]
- 48.Zajchowski D. A., Bartholdi M. F., Gong Y., Webster L., Liu H. L., Munishkin A., Beauheim C., Harvey S., Ethier S. P., Johnson P. H. (2001) Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 61, 5168–5178 [PubMed] [Google Scholar]
- 49.Liu H., Deng X., Shyu Y. J., Li J. J., Taparowsky E. J., Hu C. D. (2006) Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. EMBO J. 25, 1058–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu C. D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 51.Maekawa T., Sano Y., Shinagawa T., Rahman Z., Sakuma T., Nomura S., Licht J. D., Ishii S. (2008) ATF-2 controls transcription of Maspin and GADD45 alpha genes independently from p53 to suppress mammary tumors. Oncogene 27, 1045–1054 [DOI] [PubMed] [Google Scholar]
- 52.Vleugel M. M., Greijer A. E., Bos R., van der Wall E., van Diest P. J. (2006) c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum. Pathol. 37, 668–674 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.