Abstract
The Drosophila melanogaster RNA-induced silencing complex (RISC) forms a large ribonucleoprotein particle on small interfering RNAs (siRNAs) and catalyzes target mRNA cleavage during RNA interference (RNAi). Dicer-2, R2D2, Loquacious, and Argonaute-2 are examples of RISC-associated factors that are involved in RNAi. Holo-RISC is an ∼80 S small interfering ribonucleoprotein, which suggests that there are many additional proteins that participate in the RNAi pathway. In this study, we used siRNA affinity capture combined with mass spectrometry to identify novel components of the Drosophila RNAi machinery. Our study identified both established RISC components and novel siRNA-associated factors, many of which contain domains that are consistent with potential roles in RNAi. Functional analysis of these novel siRNA-associated proteins suggests that these factors may play an important role in RNAi.
Small RNAs can regulate gene expression through a collection of mechanisms broadly termed RNA silencing. Small RNA-mediated silencing mechanisms occur in most species (1–5). The ability to silence the expression of specific genes using small RNAs via RNA interference (RNAi)1 has greatly facilitated our understanding of gene function in eukaryotes. In addition, small RNA-mediated gene silencing has therapeutic potential and holds promise for the treatment of specific diseases (6). Understanding the mechanism of RNAi and identifying the components of the RNAi machinery are essential for harnessing its full potential in both genome-wide screens and therapeutic applications.
Recently, high throughput sequencing technology has revealed the presence of endogenous siRNAs in plant, fly, worm, and mammalian cells (7–16). These endogenous siRNAs target transposable element RNAs, pseudogene RNAs, and protein-coding mRNAs (17). Therefore, the endogenous siRNA pathway seems to have evolved as a mechanism of cellular defense against selfish genetic elements. The roles of these siRNAs in development and cell physiology are poorly understood.
Drosophila melanogaster is a well characterized model system for studying RNAi. In Drosophila, long double-stranded RNAs (dsRNAs) are processed by the endonuclease Dicer-2 into 21-nucleotide siRNAs (18). After processing, these siRNAs form an initiator complex with Dicer-2 and the dsRNA-binding domain (dsRBD)-containing protein R2D2 (19–23). This R2D2-Dicer-2 Initiator (RDI) complex transitions to a larger siRNP called the RISC loading complex (21, 22, 24, 25) and then to pre-RISC (26). Subsequently, pre-RISC matures into holo-RISC, which includes the catalytic activity necessary for target mRNA cleavage (21, 25, 27). The endonuclease subunit responsible for target cleavage in holo-RISC is Argonaute-2 (Ago2) (28, 29), which uses the guide strand of the siRNA duplex to target complementary mRNA sequences for cleavage and degradation.
Studies of the RDI complex strongly suggest that it includes no other proteins besides Dicer-2 and R2D2 (22). Additional proteins such as Ago2 are present in pre-RISC and holo-RISC, but nonetheless the complete compositions of the RISC loading complex, pre-RISC, and holo-RISC are unknown. Furthermore, holo-RISC sediments at ∼80 S during sucrose gradient centrifugation (30). These observations indicate that additional protein factors associate with siRNAs. In this study, we identified siRNA-binding proteins from Drosophila embryo extracts. Target cleavage assays and immunoblotting of our siRNA affinity-selected proteins suggest that we purified active holo-RISC components. Proteomics analysis of the affinity matrix revealed both established and novel siRNA-associated proteins. Functional analyses of a subset of these factors suggest that they play important roles in RNAi.
EXPERIMENTAL PROCEDURES
Extracts
Drosophila embryo extracts were prepared from wild-type Canton S flies as described (31). Extracts were prepared by lysis of embryos in a Dounce homogenizer in embryo lysis buffer (ELB) (30 mm HEPES, pH 7.5, 100 mm potassium acetate, 2 mm magnesium acetate, 5 mm dithiothreitol). Protein concentrations were determined with a Bradford assay (Bio-Rad).
siRNA Affinity Chromatography
All small RNAs were purchased from Dharmacon. The siRNA sequences were as follows: Pp-luc guide, 5′-UCGAAGUAUUCCGCGUACGUG-3′; Pp-luc passenger, 5′-CGUACGCGGAAUACUUCGAUU-3′; let-7 guide, 5′-UGAGGUAGUAGGUUGUAUAGA-3′; let-7 passenger, 5′-UAUACAACCUACUACCUCAUU-3′. Pp-luc and let-7 guide strand siRNAs (biotinylated at their 3′-ends via an LC-LC linkage) were annealed to their complementary passenger strands in vitro in annealing buffer (30 mm HEPES, pH 7.5, 100 mm potassium acetate, 2 mm magnesium acetate) with each strand present at 40.9 μm. Strands were heated to 95 °C for 2 min and then annealed at 37 °C for 60 min. After the biotinylated siRNAs were annealed, 20.45 nmol of the siRNAs were complexed with 250 μl of streptavidin beads (Pierce) for 30 min at 4 °C with gentle agitation. For control experiments analyzing the association of let-7 or Pp-luc single-stranded guide RNA with proteins, 20.45 nmol of biotinylated guide alone were complexed with 250 μl of avidin beads as described above. Once the siRNA-streptavidin matrix was formed, the beads were washed in ELB four times with 5 bead volumes to remove free siRNAs. After generation of the affinity matrix, the beads were mixed with embryo lysates (prepared as described above) for 30 min at room temperature. After incubation with extract, the affinity matrix was washed by centrifugation in ELB eight times with 10 bead volumes to purify stably associated factors. Immunoblotting was performed with rabbit anti-Dicer-2 antibodies (19) and anti-R2D2 antibodies (Abcam).
Target Cleavage Assays
Drosophila embryo extracts were complexed with siRNA affinity columns and washed as described above. Radiolabeled Pp-luciferase RNA target was then incubated with the affinity-purified beads or negative controls (streptavidin beads alone) for 1 h at 25 °C. Samples were treated with stop buffer (100 mm Tris, pH 7.4, 25 mm EDTA, 50 mm NaCl, 2.1 μg of proteinase K), and total RNA from the cleavage reactions was isolated by TRIzol extraction. The radiolabeled substrate and product RNAs from the cleavage reaction were subjected to electrophoresis in 15% polyacrylamide, 8 m urea denaturing gels and visualized with a PhosphorImager.
Chromatography and Mass Spectrometry
The proteins retained on the siRNA affinity or control chromatography preparations were reduced, alkylated, and trypsinized in 100 mm ammonium bicarbonate as described previously (32). After desalting the samples with a C18 reverse-phase salt trap (Michrom Bioresources Inc.), the peptides were subjected to reverse-phase microcapillary LC-ESI-MS/MS. A 100-μm-inner diameter microcapillary column (New Objective) was packed with 10 cm of 5-μm reverse-phase material (Synergi 4μ Hydro RP80a, Phenomenex). The trypsinized peptides were loaded onto the reverse-phase column equilibrated in buffer A (0.1% formic Acid, 5% acetonitrile) using a packing bomb at 400 p.s.i. The loaded column was placed in line with a Thermo Finnigan LTQ linear ion trap mass spectrometer (Thermo Electron Inc.). The peptides were eluted using a 90-min linear gradient from 0 to 80% buffer B (0.1% formic acid, 80% acetonitrile) at a flow rate of 0.2 μl/min. During the chromatography gradient, the eluted ions were analyzed by one full precursor ion scan (400–2000 m/z) followed by six MS/MS scans of the six most abundant ions. Dynamic exclusion was used to reduce the detection of abundant contaminating peptides. The raw mass spectra were acquired using Xcalibur version 2.2. The raw files were converted to MZXML files using readW version 4.0.2 under default settings. The resulting MZXML files were searched with Sequest version 27 on a SAGE-N Research, Inc. Sorcerer. The converted spectra files were searched against the European Bioinformatics Institute Drosophila proteome database containing 16,496 entries as of August 12, 2007. Sequest was searched with a fragment ion mass tolerance of 1.0 Da, a parent ion tolerance of 1.2 Da, and a single trypsin miscleavage allowance. The iodoacetamide derivative of cysteine was included in the Sequest search parameters as a static modification. Proteins identified from the Sequest Sorcerer search were loaded into Scaffold version 2_04_00. Peptide identifications in Scaffold were accepted if they could be established at greater than 95% probability as specified by the Peptide Prophet algorithm (33) and contained at least three identified peptides in a single sample. Peptides that could not be ascribed to a single protein were grouped into a single protein identification to simplify our analysis. The Scaffold unique protein counts for all samples were exported as a text file, subjected to hierarchical clustering using Cluster version 3.0 (34), and viewed with Treeview (version 1.1.1). All proteins identified are included in supplemental Fig. 1 with the number of unique peptides indicated. Supplemental Fig. 1 includes all of the protein identifications and sequence coverage for each protein in each sample. For proteins with single peptide identifications, the sequence, charge state, parent ion mass, correlation value, and tandem mass spectra are included in supplemental Fig. 2.
Cell Culture, Constructs, and dsRNAs
Drosophila S2 cells were cultured in Schneider's Drosophila medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 100 units/ml penicillin (Invitrogen), and 100 units/ml streptomycin (Invitrogen). Firefly and Renilla luciferase constructs, pMT-GL3 and pMT-Ren, respectively, were a gift from R. Andino (35). Firefly and Renilla luciferase expression was analyzed using the Dual-Luciferase reporter assay system (Promega) and analyzed on the Lmax microplate luminometer (Molecular Devices).
dsRNAs were transcribed from PCR products containing the first 500 bp of each gene. Oligonucleotides used for PCR amplification contained the T7 promoter at the 5′-end. dsRNA was made through transcription using the Megascript kit (Ambion) with a 9-h incubation at 37 °C. Resulting dsRNAs were purified by phenol-chloroform extraction, denatured at 75 °C for 15 min, and reannealed by slow cooling to room temperature.
S2 Cell Transfections and Luciferase Assays
S2 cells were plated at a density of ≈3 × 106/well (6-well plates) ≈24 h before use. For bathing, the medium was replaced with 0.5 ml of fresh Drosophila Schneider medium (no fetal bovine serum), and dsRNA was added to a concentration of 5 ng/well. One hour later, an additional 1.5 ml of complete medium was added. After 48 h, the firefly and Renilla luciferase plasmids as well as dsRNA against firefly luciferase were added at a final concentration of 60, 10, and 1 ng/well, respectively, using the Effectine protocol (Qiagen).
For each well, the reporter activity was calculated as a ratio of firefly luciferase (Pp-luc) to Renilla luciferase. Each data point was normalized to the negative control knockdown of white with dsRNA. A statistical analysis of the difference in firefly and Renilla luciferase signal ratios between experimental knockdowns and white knockdowns was performed using a Student's paired t test with a two-tailed distribution.
RESULTS
siRNA Affinity Capture of Drosophila RISC
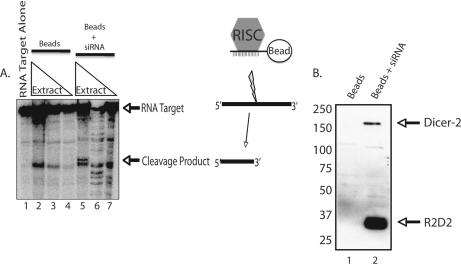
Previous studies have used a model siRNA directed against firefly luciferase (Pp-luc) to direct efficient cleavage of a cognate mRNA target in vitro (21, 36). To capture siRNA-interacting proteins by affinity chromatography, we used a Pp-luc siRNA that was biotinylated on the 3′-end of the guide strand and annealed to a non-biotinylated passenger strand. The resulting biotinylated siRNAs were bound to streptavidin beads and then incubated with three different concentrations of Drosophila embryo extract. After washing, the presence of correctly assembled RNAi effector complexes was assayed through cleavage of a cognate target mRNA (Fig. 1A). Specific target cleavage activity on the affinity matrix required a protein concentration of 30 mg/ml (Fig. 1A, lane 5). Lower concentrations of extract were insufficient for the assembly of functional RISC on the beads (Fig. 1A, lanes 6 and 7). Furthermore, samples that contained avidin beads and embryo extract alone did not generate the specific cleavage product (Fig. 1A, lanes 2–4). These data confirmed that our siRNA affinity approach specifically captured functional RISC.
Fig. 1.
Capture of Drosophila RISC on siRNA affinity matrix. A, siRNA affinity chromatography samples were tested for target cleavage activity. A radiolabeled Pp-luciferase RNA target was incubated with either no extract (lane 1), 30 mg/ml embryo extract and streptavidin beads (lane 2), 3 mg/ml embryo extract and streptavidin beads (lane 3), or 0.3 mg/ml embryo extract and streptavidin beads (lane 4). In lanes 5–7, 30, 3, or 0.3 mg/ml embryo extract, respectively, was incubated with a Pp-luc-siRNA affinity matrix (streptavidin beads with biotinylated Pp-luc siRNAs) and radiolabeled RNA target. Arrows indicate the positions of the uncleaved RNA target and specific cleavage product. B, 90 mg of embryo extract were complexed with either streptavidin beads (lane 1) or Pp-luc siRNA affinity beads (lane 2). Proteins bound to the affinity matrix were separated by SDS-PAGE and simultaneously immunoblotted for Dicer-2 and R2D2.
Next, we tested our siRNA affinity preparations for the presence of the established RNAi factors Dicer-2 and R2D2. Immunoblotting the siRNA affinity preparations for Dicer-2 and R2D2 confirmed the presence of these RDI complex components (Fig. 1B, lane 2). Dicer-2 and R2D2 were not detected in a parallel sample containing only avidin beads (Fig. 1B, lane 1). Collectively, these results show that we efficiently captured known RNAi factors and functional RISC using siRNA affinity chromatography.
Proteomics Identification of siRNA-associated Factors
To identify additional factors bound to the siRNA affinity purifications, we used a proteomics approach. Proteins from 90 mg of embryo extract were incubated with either Pp-luc siRNA or let-7 siRNA (an siRNA sequence different from Pp-luc) on streptavidin beads. Bona fide RNAi factors should associate with siRNAs regardless of sequence, so requiring proteins to be detected with both siRNAs allows us to enrich for strong candidates. Furthermore, some proteins could conceivably co-purify based on their association with RISC-bound mRNAs that are present in the extract, but this potential problem is minimized by the use of distinct siRNAs (including one (Pp-luc) that lacks any natural mRNA targets in Drosophila) that would likely associate with different populations of mRNAs. As negative controls for these experiments, we incubated either avidin beads or single-stranded Pp-luc or let-7 guide strand alone with embryo extracts. Therefore, our experimental design allows us to distinguish avidin-binding proteins and nonspecific RNA-binding proteins from siRNA duplex-specific factors.
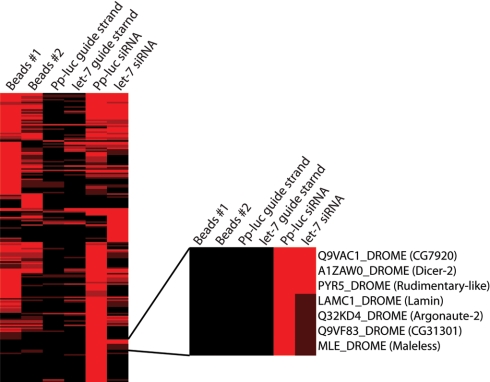
After capture of the proteins on the affinity matrices, the samples and controls were washed, trypsinized, and analyzed by LC-MS/MS mass spectrometry. Importantly, both the Pp-luc and let-7 siRNA purifications showed that Dicer-2 specifically purified with the siRNA affinity matrix but not the avidin beads alone or the guide strand only samples (Fig. 2). Dicer-2 was likely the most abundant protein in the affinity purifications as it was represented by the most total spectra and the most unique peptides and exhibited high sequence coverage among the siRNA-specific binding proteins (supplemental Fig. 1) in agreement with the central role of Dicer-2 in RNAi (21, 30). The dsRBD proteins Loquacious and R2D2 were detected in Pp-luc siRNA affinity purifications but were below the limit of detection in let-7 siRNA affinity purifications (supplemental Fig. 1).
Fig. 2.
Cluster analysis of siRNA-associated proteins from Drosophila embryo extracts. Proteins were detected from two independent replicate samples bound to streptavidin beads, Pp-luc single-stranded guide RNA, let-7 single-stranded guide RNA, or two different siRNA affinity columns (Pp-luc and let-7). The observed proteins were hierarchically clustered and displayed with Treeview. The number of unique peptides corresponding to a given protein is designated by the shade of red. A square in the cluster is black if the protein was not detected in the analysis of the mass spectrometry data. The position of Dicer-2 and other siRNA-binding proteins in the cluster is expanded to the right.
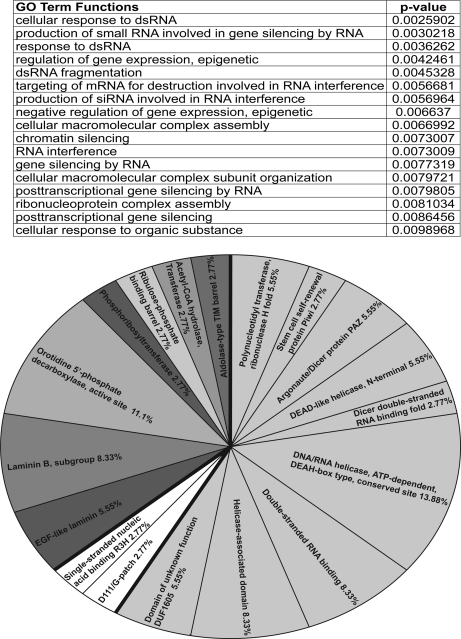
Hierarchical clustering of the samples revealed a group of seven siRNA-specific binding proteins (Fig. 2) that were detected with both siRNAs but not in any of the negative control samples. RNAi factors were strongly enriched in the siRNA-specific group with a statistically significant functional ontology (Fig. 3A). Additionally, a large percentage of the siRNA-associated protein domains were consistent with those of RNAi factors including Piwi, Paz, Helicase, and dsRBD (Fig. 3B). Among these seven proteins were the RNAi factors Dicer-2 and Argonaute-1, reinforcing our isolation of both initiator and effector factors in RNA interference.
Fig. 3.
Gene ontology terms and protein domains among siRNA-binding proteins. A, for the siRNA-specific binding proteins, a list of the FlyMine gene ontology (GO) terms and associated Bonferroni-adjusted p values for statistical significance are shown. B, pie chart of represented protein domains among the seven siRNA-specific binding proteins. EGF, epidermal growth factor.
In addition to these known RNAi factors, we identified Maleless, which contains both Helicase and dsRBD domains. Intriguingly, we identified an RNA-binding protein (encoded by the candidate gene CG31301) that contains both G-patch and R3H domains (Figs. 2 and 3B). G-patch nucleic-acid binding proteins are found within retroviral proteins. Because a subgroup of the predicted domains listed above confers functions consistent with known steps in the RNAi pathway, these proteins represent viable candidates for novel RNAi factors.
Functional Analysis of Novel siRNA-associated Proteins
Our proteomics screen detected seven proteins bound to siRNAs in vitro. To analyze potential functional roles for these proteins in RNAi, we depleted them from Drosophila S2 cells and then tested the ability of those cells to silence a luciferase reporter (35, 37). We restricted our functional analysis to proteins that contain predicted functional domains potentially relevant to RNAi such as nucleases (Argonaute and Dicer proteins), helicases (Maleless), and RNA-binding proteins (CG31301). Proteins such as Lamin, Rudimentary-like (a metabolic enzyme), and other abundant contaminating enzymes were excluded from our analysis. In the Dual-Luciferase assay, cells are first transfected with dsRNAs that target the candidate RNAi factor. After knockdown of the candidate RNAi factor, cells are co-transfected with a Pp-luciferase plasmid, a Renilla luciferase plasmid, and a dsRNA that specifically targets Pp-luciferase mRNA for RNAi. Derepression of the Pp-luciferase signal relative to the Renilla luciferase signal in the knockdown cells (in comparison with nonspecific dsRNA controls) indicates suppression of RNA interference activity, whereas a reduction in the relative Pp-luciferase signal indicates an increase in RNAi activity (35, 37).
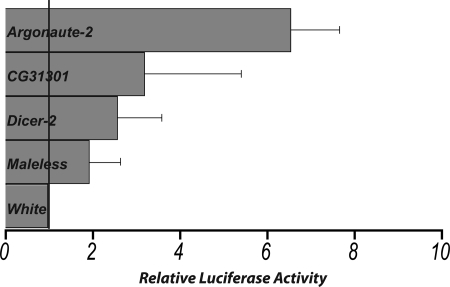
As positive controls, transfection of S2 cells with dsRNAs targeting Dicer-2 or Argonaute-2 resulted in a derepression of luciferase activity relative to the negative control dsRNAs (Fig. 4). Transfection of dsRNAs targeting a subset of the novel siRNA-associated factors identified in our proteomics screen revealed that CG31301, a predicted RNA-binding protein, is likely involved in RNAi (Fig. 3). dsRNAs targeting CG31301 derepressed Pp-luciferase levels ∼3.2-fold relative to a negative control dsRNA directed against the Drosophila gene white (p = 0.048). This derepression is significant in comparison with Dicer-2 (2.6-fold derepression (p = 0.017)) and Argonaute-2 (6.57-fold derepression (p = 0.048)). Additionally, knockdown of Maleless, an RNA helicase critical for sex chromosome dosage compensation, partially compromised RNAi activity (2-fold derepression (p = 0.025)) (Fig. 4). Altogether, these results indicate that our proteomics screen identified both known and novel proteins with biochemical and functional properties consistent with RNAi factors.
Fig. 4.
Functional analysis of siRNA-associated factors. A subset of siRNA-binding proteins was tested for functional roles in RNA interference using the luciferase assay. The luciferase assay tests for derepression of RNAi-inhibited Pp-luciferase after knockdown of a suspected RNAi factor. dsRNAs targeting Argonaute-2 and Dicer-2 were included as positive controls, whereas dsRNA targeting the white gene was included as a negative control. Bars indicate average -fold change in luciferase activities (of independent biological replicates) relative to control cells treated with dsRNA targeting white. Each dsRNA treatment shows the average -fold difference for three different experiments. Candidate gene (CG) numbers were included for genes with no previously ascribed function.
DISCUSSION
Multiple Factors Converge on Distinct siRNAs
Size estimates of RISC suggest that a large number of proteins associate with siRNAs in vitro (30). Consistent with this observation, our results show that Dicer-2, dsRBD proteins (Loqs, R2D2, and Maleless), RNA helicases (Dicer-2 and Maleless), Argonaute proteins, and an additional nucleic acid-binding protein (CG31301) converge on distinct siRNAs to form large siRNP complexes. These results reinforce a model where proteins of multiple apparent functions form an active RISC complex.
Identification of Potential Novel RNAi Factor: CG31301
Our results show that CG31301 associates with two siRNAs of unrelated sequence (Pp-luc and let-7). Additionally, CG31301 is important for efficient RNAi in S2 cells. Taken together, these results strongly suggest that CG31301 functions as a novel RNAi factor. CG31301 contains two predicted protein domains, an R3H domain and a G-patch domain. The R3H domain is found in single-stranded nucleic acid-binding proteins (38), whereas the G-patch domain has been found in some retroviral nucleic acid-binding proteins (39). Interestingly, recent evidence shows that retrotransposons, many of which appear to be derived from retroviruses, are among the primary targets of endogenous siRNAs (7–10, 12, 15, 40, 41). It is tempting to speculate that metazoans could have co-opted retroviral nucleic acid-binding proteins to participate in retrotransposon suppression. Future studies of CG31301 structure and function may illuminate this possibility.
Identification of Helicase Associated with siRNAs
We observed that the predicted RNA helicase and dsRBD protein Maleless specifically purified with siRNAs. In our functional assays, knockdown of Maleless modestly compromised RNAi activity. During holo-RISC formation, the guide strand of the siRNA is liberated from the passenger strand to allow hybridization with an RNA target. One mechanism of guide strand liberation is the cleavage of the passenger strand by Ago2 (42, 43). Nevertheless, it is possible that one or more RNA helicases help dissociate the passenger and guide strands (with or without prior passenger strand cleavage), leading to holo-RISC formation. The mammalian protein RNA Helicase A, an apparent paralog of Maleless, has been shown to complex with siRNAs and function in RNAi (44). Therefore, a role for this RNA helicase in RNAi might be evolutionarily conserved.
In summary, we conducted a proteomics screen for siRNA-associated proteins in Drosophila. Our screen identified known RNAi factors (Dicer-2, Loquacious, and R2D2) as well as several novel siRNP candidates. These latter proteins include domains with predicted activities that could play a role in RNAi. Some of these factors appear to be important for efficient RNAi in cultured cells. Future studies will elucidate the precise molecular functions of these novel siRNA-associated factors.
Supplementary Material
Acknowledgments
We thank Qinghua Liu and Raul Andino for sharing reagents and Rich Carthew's laboratory for help with S2 cell culture. We also thank the Chicago Biomedical Consortium for support of the mass spectrometry facilities used in this work. Mass spectrometry experiments were performed at the University of Illinois at Chicago Research Resources Center facility.
* This work was supported, in whole or in part, by National Institutes of Health Grant GM072830 (to E. J. S.).
 This article contains supplemental Figs. 1 and 2.
This article contains supplemental Figs. 1 and 2.
1 The abbreviations used are:
- RNAi
- RNA interference
- RISC
- RNA-induced silencing complex
- siRNA
- small interfering RNA
- siRNP
- small interfering ribonucleoprotein
- dsRNA
- double-stranded RNA
- dsRBD
- dsRNA-binding domain
- RDI
- R2D2-Dicer-2 Initiator
- Ago2
- Argonaute-2
- ELB
- embryo lysis buffer.
REFERENCES
- 1.Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carthew R. W., Sontheimer E. J. (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136, 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malone C. D., Hannon G. J. (2009) Small RNAs as guardians of the genome. Cell 136, 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorek R., Kunin V., Hugenholtz P. (2008) CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6, 181–186 [DOI] [PubMed] [Google Scholar]
- 5.Waters L. S., Storz G. (2009) Regulatory RNAs in bacteria. Cell 136, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castanotto D., Rossi J. J. (2009) The promises and pitfalls of RNA-interference-based therapeutics. Nature 457, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravin A. A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. (2003) The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350 [DOI] [PubMed] [Google Scholar]
- 8.Babiarz J. E., Ruby J. G., Wang Y., Bartel D. P., Blelloch R. (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czech B., Malone C. D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J. A., Sachidanandam R., Hannon G. J., Brennecke J. (2008) An endogenous small interfering RNA pathway in Drosophila. Nature 453, 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghildiyal M., Seitz H., Horwich M. D., Li C., Du T., Lee S., Xu J., Kittler E. L., Zapp M. L., Weng Z., Zamore P. D. (2008) Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320, 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton A., Voinnet O., Chappell L., Baulcombe D. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamura Y., Saito K., Kin T., Ono Y., Asai K., Sunohara T., Okada T. N., Siomi M. C., Siomi H. (2008) Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453, 793–797 [DOI] [PubMed] [Google Scholar]
- 13.Okamura K., Chung W. J., Ruby J. G., Guo H., Bartel D. P., Lai E. C. (2008) The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453, 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruby J. G., Jan C., Player C., Axtell M. J., Lee W., Nusbaum C., Ge H., Bartel D. P. (2006) Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207 [DOI] [PubMed] [Google Scholar]
- 15.Tam O. H., Aravin A. A., Stein P., Girard A., Murchison E. P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R. M., Hannon G. J. (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z., Johansen L. K., Gustafson A. M., Kasschau K. D., Lellis A. D., Zilberman D., Jacobsen S. E., Carrington J. C. (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS. Biol. 2, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golden D. E., Gerbasi V. R., Sontheimer E. J. (2008) An inside job for siRNAs. Mol. Cell 31, 309–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamore P. D., Tuschl T., Sharp P. A., Bartel D. P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33 [DOI] [PubMed] [Google Scholar]
- 19.Liu Q., Rand T. A., Kalidas S., Du F., Kim H. E., Smith D. P., Wang X. (2003) R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301, 1921–1925 [DOI] [PubMed] [Google Scholar]
- 20.Liu X., Jiang F., Kalidas S., Smith D., Liu Q. (2006) Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 12, 1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pham J. W., Pellino J. L., Lee Y. S., Carthew R. W., Sontheimer E. J. (2004) A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117, 83–94 [DOI] [PubMed] [Google Scholar]
- 22.Pham J. W., Sontheimer E. J. (2005) Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J. Biol. Chem. 280, 39278–39283 [DOI] [PubMed] [Google Scholar]
- 23.Tomari Y., Matranga C., Haley B., Martinez N., Zamore P. D. (2004) A protein sensor for siRNA asymmetry. Science 306, 1377–1380 [DOI] [PubMed] [Google Scholar]
- 24.Li H., Trang P., Kim K., Zhou T., Umamoto S., Liu F. (2006) Effective inhibition of human cytomegalovirus gene expression and growth by intracellular expression of external guide sequence RNA. RNA 12, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomari Y., Du T., Haley B., Schwarz D. S., Bennett R., Cook H. A., Koppetsch B. S., Theurkauf W. E., Zamore P. D. (2004) RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116, 831–841 [DOI] [PubMed] [Google Scholar]
- 26.Kim K., Lee Y. S., Carthew R. W. (2007) Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamura K., Ishizuka A., Siomi H., Siomi M. C. (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song J. J., Smith S. K., Hannon G. J., Joshua-Tor L. (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 [DOI] [PubMed] [Google Scholar]
- 29.Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J. J., Hammond S. M., Joshua-Tor L., Hannon G. J. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 [DOI] [PubMed] [Google Scholar]
- 30.Lee Y. S., Nakahara K., Pham J. W., Kim K., He Z., Sontheimer E. J., Carthew R. W. (2004) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 [DOI] [PubMed] [Google Scholar]
- 31.Tuschl T., Zamore P. D., Lehmann R., Bartel D. P., Sharp P. A. (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13, 3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders S. L., Jennings J., Canutescu A., Link A. J., Weil P. A. (2002) Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22, 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 34.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saleh M. C., van Rij R. P., Hekele A., Gillis A., Foley E., O'Farrell P. H., Andino R. (2006) The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8, 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nykänen A., Haley B., Zamore P. D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107, 309–321 [DOI] [PubMed] [Google Scholar]
- 37.Dorner S., Lum L., Kim M., Paro R., Beachy P. A., Green R. (2006) A genomewide screen for components of the RNAi pathway in Drosophila cultured cells. Proc. Natl. Acad. Sci. U.S.A. 103, 11880–11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grishin N. V. (1998) The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem. Sci. 23, 329–330 [DOI] [PubMed] [Google Scholar]
- 39.Aravind L., Koonin E. V. (1999) G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24, 342–344 [DOI] [PubMed] [Google Scholar]
- 40.Drinnenberg I. A., Weinberg D. E., Xie K. T., Mower J. P., Wolfe K. H., Fink G. R., Bartel D. P. (2009) RNAi in budding yeast. Science 326, 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehwinkel J., Natalin P., Stark A., Brennecke J., Cohen S. M., Izaurralde E. (2006) Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol. Cell. Biol. 26, 2965–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matranga C., Tomari Y., Shin C., Bartel D. P., Zamore P. D. (2005) Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 [DOI] [PubMed] [Google Scholar]
- 43.Rand T. A., Petersen S., Du F., Wang X. (2005) Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629 [DOI] [PubMed] [Google Scholar]
- 44.Robb G. B., Rana T. M. (2007) RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell 26, 523–537 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.