Abstract
Systematic study of cell signaling networks increasingly involves high throughput proteomics, transcriptional profiling, and automated literature mining with the aim of assembling large scale interaction networks. In contrast, functional analysis of cell signaling usually focuses on a much smaller sets of proteins and eschews computation but focuses directly on cellular responses to environment and perturbation. We sought to combine these two traditions by collecting cell response measures on a reasonably large scale and then attempting to infer differences in network topology between two cell types. Human hepatocytes and hepatocellular carcinoma cell lines were exposed to inducers of inflammation, innate immunity, and proliferation in the presence and absence of small molecule drugs, and multiplex biochemical measurement was then performed on intra- and extracellular signaling molecules. We uncovered major differences between primary and transformed hepatocytes with respect to the engagement of toll-like receptor and NF-κB-dependent secretion of chemokines and cytokines that prime and attract immune cells. Overall, our results serve as a proof of principle for an approach to network analysis that is systematic, comparative, and biochemically focused. More specifically, our data support the hypothesis that hepatocellular carcinoma cells down-regulate normal inflammatory and immune responses to avoid immune editing.
The development of high throughput methods for detecting genetic and physical interactions among genes and proteins has stimulated interest in methods to infer functional relationships among them and thereby assemble large biological “networks”. These networks are typically represented as node-edge graphs with macromolecules as nodes and interactions as edges (or vertices; Refs. 1 and 2). Protein interaction networks (PINs1 or interactomes; Refs. 1, 3, and 4) are typically deduced directly from systematic two-hybrid (5–7) and affinity purification-mass spectrometry data (8–12) and can encompass the entire proteome. In PINs, edges are undirected (lines rather than arrows) and unsigned (lacking positive or negative labels). Thus, they do not encode substrate-product relationships or specify whether interactions are inhibitory or activating (1). In contrast, edges in protein signaling networks (PSNs) are usually assembled from literature data using manual or automated curation and have directionality and sign. PINs and PSNs often incorporate diverse data from multiple cell types and even different organisms (13, 14). This increases the scope of a network but obscures differences from one cell type to the next despite the obvious value of such comparative insight.
Potentially valuable comparative data are available in the form of transcriptional profiles (15, 16), disease genotypes (17, 18), phosphoproteomic profiles (12, 19), or RNAi screening data (20–22), and many groups are attempting to add this information to PINs to create networks specific to particular cell types or cell states (23–28). In this study, we undertook a complementary approach in which we started with “functional” data characterizing the responses of cells to biological ligands and small molecule drugs and then used inference methods to assemble a network. We exposed primary human hepatocytes and HepG2 liver cancer cells to one of seven growth factors or cytokines in the presence or absence of seven small molecule kinase inhibitors and then measured the levels or states of modification of 17 intracellular proteins or 50 secreted peptides using automated sandwich affinity assays. This yielded a set of ∼26,000 protein state measurements from which interaction graphs could be inferred using multilinear regression (MLR). The resulting graphs directly compare immediate-early signaling downstream of seven transmembrane receptors in normal and transformed liver cells.
Hepatocytes were chosen for comparative pathway analysis of receptor-mediated signaling because liver homeostasis is known to be controlled by endocrine, paracrine, and autocrine ligands that coordinate the fates and functions of multiple cell types (29, 30) including Kupffer cells (specialized liver-resident macrophages), other nonparenchymal cells, and hepatocytes themselves. Data on responses to and secretion of cytokines by hepatocytes are therefore expected to be physiologically informative. The liver is the primary organ in which mammals metabolize nutrients, environmental toxins, and drugs. The liver also plays a critical role in inflammation and innate immunity (29). During the acute phase response, for example, leukocytes recruited to distant sites of inflammation secrete interleukins into the blood, and these interleukins induce the production of acute phase proteins by the liver (31). Many acute phase proteins directly inhibit microbe growth or, like C-reactive protein, promote microbe opsonization and subsequent phagocytosis (31). Liver cells also express receptors that mediate local inflammatory and innate immune responses (32). Toll-like receptors (TLRs), for example, are expressed on hepatocytes, Kupffer cells, and other nonparenchymal cells where they detect antigens commonly associated with infection (e.g. lipopolysaccharide (32)).
Many signal transduction proteins involved in liver biology and HCC have been characterized (e.g. the IL1R, vascular endothelial growth factor A (VEGFA), TLR receptors, and NF-κB and JNK signaling proteins (33–40)), and it has been determined that several broadly expressed cytokines have unusual functions in the liver. Tumor necrosis factor-α (TNFα), for example, is involved in hepatic regeneration, regrowth of an injured liver that takes place following loss of up to 75% of organ mass (41). Chronic inflammatory signaling is observed in many liver diseases and is known to cause progressive cellular transformation and ultimately hepatocellular carcinoma (42). Nonetheless, how diverse signaling proteins are coordinated in normal and diseased liver cells remains poorly understood. Partly as a consequence, HCC has proven difficult to target with existing chemotherapies (43) and remains the third most common cause of cancer death in humans (44). Thus, better understanding of changes in signaling networks that accompany the development of HCC should impact human health.
Whereas transcriptional data can be collected on a genome-wide scale, this is not possible for data on protein levels and modifications: sandwich immune methods such as the xMAP assays used here (45) are limited to <100 analytes (although this can be increased to ∼400 analytes using reverse phase arrays (46–50)). However, immune detection works well with many samples and is sufficiently rapid and sensitive, thus making it practical to perform multiplex assays on primary human tissue at multiple points in time, across multiple ligands, and in the presence or absence of multiple small molecule drugs (this is not yet feasible using phospho-mass spectrometry, which tends to provide data on many analytes under fewer conditions). A key question at the outset of this work was whether biochemical data collectable using existing high throughput biochemical methods would yield useful network insight and uncover interesting new biology. The answer appears to be yes. Comparative regression analysis reveals widespread differences between normal and transformed hepatocytes including up-regulation of prosurvival pathways in tumor cells and unexpected down-regulation of inflammatory, TLR-, and NF-κB-mediated signaling (51, 52). These changes are common to all four liver cancer cell lines examined and appear to reflect general disruption of innate immunity in tumor cells relative to normal hepatocytes.
EXPERIMENTAL PROCEDURES
Cell Culture and Signaling Experiments
HepG2 and Hep3B cells were purchased from ATCC, HuH7 and FOCUS were obtained from J. Wands (Brown University), and fresh primary human hepatocytes were purchased from CellzDirect (Research Triangle Park, NC). Cell lines were passaged up to five times in Eagle's minimum essential medium supplemented with 10% serum; preplated primary hepatocytes were used immediately. Assays were performed on cells in 96-well plates coated with collagen type I (BD Biosciences) with 100 μl of phenol-free Williams' medium E (Sigma-Aldrich) supplemented with 1.0 mm l-glutamine (Invitrogen), 100 nm dexamethasone (Sigma-Aldrich), 5 μg/ml human insulin (Sigma-Aldrich), 5 μg/ml transferrin from human serum (Roche Applied Science), and 5 μg/ml sodium selenite (Sigma-Aldrich). Cells were cultured overnight on collagen, starved for 6 h in 180 μl of Williams' medium E with l-glutamine and dexamethasone, and exposed to kinase inhibitors and ligand cues (prepared as 20× concentration stock solutions). Supernatants were collected and stored in the presence of 0.5% (w/v) BSA, and cells were lysed in 90 μl of manufacturer's buffer (Bio-Rad), and total protein concentrations were quantified using a micro-BCA assay (Pierce). To minimize experimental variability, samples were processed in parallel, and the same batches of cytokines, inhibitors, and assay reagents were used throughout.
xMAP Assays
Multiplexed x-MAP assays were performed on a Luminex 200 system using reagents from Bio-Rad (see Fig. 1). A 17-plex phosphoprotein bead set from Bio-Rad was used to assay phospho-p70S6K (p-p70S6K) (Thr-421/Ser-424), p-CREB (Ser-133), p-p90RSK (Thr-359/Ser-363), p38 (Thr-180/Tyr-182), p-MEK1 (Ser-217/Ser-221), p-JNK (Thr-183/Tyr-185), p-Hsp27 (Ser-78), p-ERK1/2 (Thr-202/Tyr-204 and Thr-185/Tyr-187), p-c-Jun (Ser-63), p-insulin receptor substrate-1 (IRS-1) (Ser-636/Ser-639), p-STAT3 (Tyr-705), p-IκB-α (Ser-32/Ser-36), p-histone H3 (Ser-10), p-p53 (Ser-15), p-GSK-3α/β (Ser-21/Ser-9), and p-Akt (Ser-473). Levels of secreted cytokines were assayed using two sets of Bio-Rad cytokine detection panels: the human group I 27-plex panel and group II 23-plex panel (supplemental Fig. 1). Significant effort was devoted to maximizing the number of measurements that could be obtained from each sample of cells: a 96-well plate assayed for 17 phosphoproteins and 50 cytokines yielded ∼6500 measurements. A significant issue for the phosphoprotein xMAP assay was large differences in the levels of various species combined with the fixed dynamic range of the Luminex 200 photomultiplier tube (low abundance phosphoproteins included p-p38, p-p53, and p-p90RSK, and high abundance signals included p-c-Jun and p-Hsp27). It was necessary to assay different dilutions of cell extract to bring all 17 signals into the linear range of detection.
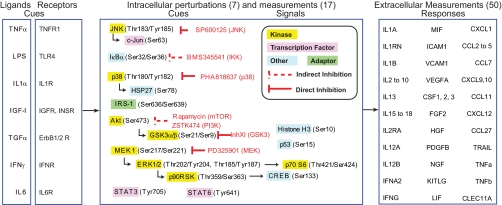
Fig. 1.
Experimental approach. The outline of the experimental approach shows identities of ligand cues and their receptors, intracellular signals assayed using xMAP technology, seven small molecule kinase inhibitors, and 50 secreted cytokines whose levels constituted responses. See Table I and supplemental Tables 2 and 3 for further details.
Data Processing and Multilinear Regression
Primary data were processed and visualized (see also Fig. 2) using the open access MATLAB-based software DataRail (53). Initial pathway maps (see also Fig. 1 and supplemental Fig. 2) were constructed using software and data provided by Ingenuity Systems. For MLR (Fig. 3), data were normalized to the maximum value for each variable across all cell types. To prevent noise from distorting results when measured values were close to the lower detection limit of the Luminex 200 device (1 of 17 intracellular assays and 29 of 50 extracellular assays for the full data set: STAT6; VCAM1; TNFSF10; LTA; CLEC11A; KITLG; NGF; CCL7; CCL11; CCL27; LIF; HGF; interleukins 2–5, 7, 9, 10, 12, 13, and 16–19; IL1A, IL2RA, INFA2, PDGFB, and INFG) were normalized to a value 3 times the noise level of the instrument (∼3 × 166) as determined by taking the standard deviation from repeated measurements of untreated controls.
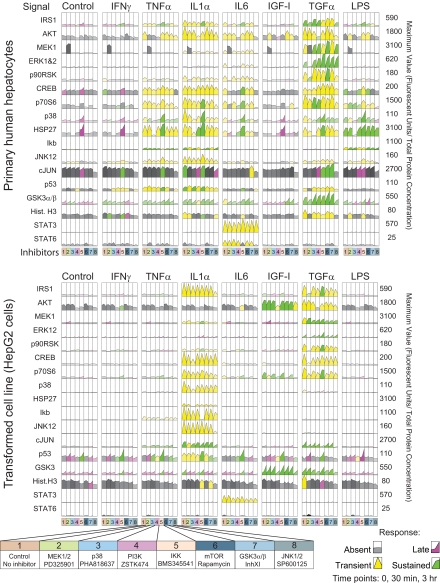
Fig. 2.
Signaling data set from human hepatocytes and HepG2 cells. Rows represent measures of 17 intracellular signals assayed at t = 0, 30 min, and 3 h (relative to ligand addition), and columns represent different ligand cues. For each combination of cue and signal, one of seven kinase inhibitors was applied as indicated in the schematic below the data. Data are coded to highlight no induction (relative to basal activity; gray), transient induction (peaking at 30 min; yellow), sustained induction (equal at 30 min and 3 h; green), or late induction (peaking at 3 h; purple). All data were processed using DataRail software (53). Cytokine data are show in supplemental Fig. 1. Hist., histone; PI3K, phosphatidylinositol 3-kinase.
Fig. 3.
One-step and two-step MLR illuminates widespread differences in common signal transduction pathways between hepatocytes and HepG2 cells. a, schematic of successive stages in MLR as applied to a subset of the data in Fig. 2b (see “Experimental Procedures” and also supplemental Fig. 3 for details). b and c, regression weights (vertical axes) for all cues, kinase inhibitors, and a subset of responses. Green bars depict cue-signal weights (b) and signal-response weights (c) derived from hepatocyte data, and red bars depict those from HepG2 cells. Background colors in each small rectangle code the combination of signal and cue as being more “hepatocyte-like” (green) or more “HepG2-like” (red) based on the relative regression weights; overall, the signals are ordered left to right based on this coding. For example, phosphohistone H3 (HistH3) levels on the extreme left are coded red and are much higher in HepG2 cells relative to hepatocytes, reflecting ongoing cell division in HepG2 cells. Inhibitor data are coded by extent of inhibition; the inhibition of many signals by the JNK inhibitor SP600125 probably reflects off-target effects (116) “X” denotes a measurement that is not meaningful because the cytokine was added exogenously as a cue. Hist., histone; PI3K, phosphatidylinositol 3-kinase; i, inhibitor.
Time-dependent measurements in the original data (see Fig. 3a, first panel) were averaged to create a time-collapsed matrix (Fig. 3a, second panel, Yexp), which is an m × k matrix of m number of dependent variables (total phosphorylation activity) under k conditions. In this study, k = 64 ((7 ligands + 1 control) × (7 kinase inhibitors +1 control)). In the first step, Yexp was constructed to ignore the effects of the inhibitors, thereby making the regression overdetermined and robust; thus, Yexp was deconvolved into a cue-regressed matrix (see Fig. 3a, third panel, WCue·XCue) where XCue corresponds to the presence (=1) or absence (=0) of stimuli and WCue corresponds to an n × m coefficient matrix where n is the number of stimuli (independent variables). The residual between the Yexp and the cue-regressed matrix (Yexp − WCue·XCue) carries information on the effects of inhibitors and was modeled in a second round of MLR using an inhibitor-regressed matrix, WInh·XInh where XInh corresponds to the presence (=1) or absence (=0) of inhibitors and Winh corresponds to a coefficient matrix with the effects of the inhibitors (see Fig. 3a, fourth panel, WInh·XInh). A substantial fraction of the information in the data set was captured through that second step procedure, and the Akaike information criterion (54) dropped from 3.0 (first step) to −46.6 (second step). The residual YRes (see Fig. 3a, fifth panel) represents data that were not captured by either the first or second step of MLR (Yexp − WCue·XCue − WInh·XInh) presumably because they do not conform to our assumption of linearity. The information remaining after two rounds of regression represents the context-specific effects of kinase inhibitors, that is, whether an inhibitor is more or less potent in cells stimulated with one ligand as opposed to another. MLR was also applied to cytokine data by letting phosphoprotein levels constitute the X matrix and cytokine levels constitute the Y matrix; in this case, only a single regression step was required because information on the effects of inhibitors is implicitly “encoded” by the activities of intracellular signals (see supplemental Fig. 3 for cross-validation analysis). The computational demands of the MLR approach were minimal.
Reagents
Ligand Cues
TNFα, insulin-like growth factor-1 (IGF-1), and transforming growth factor (TGF) β1 were obtained from PeproTech; lipopolysaccharide (LPS) and interleukin (IL)-6 were from Sigma-Aldrich; IL1α and TGFα were from R&D Systems; and interferon-γ (IFNγ) was from Roche Diagnostics GmbH. Other than LPS, TLR ligands were obtained from InvivoGen as follows: Pam3CSK4 for TLR1/2, HKLM for TLR2, poly(I-C) for TLR3; Salmonella typhimurium flagellin for TLR5, FSL1-Pam2CGDPKHPKSF for TLR6/2, imiquimod for TLR7, ssRNA40 for TLR8, and ODN2006 for TLR9.
Kinase Inhibitors
Inhibitors for IKK2 (BMS-345541, IMD0354, and TPCA-1), phosphatidylinositol 3-kinase (ZSTK474), GSK3β (inhibitor XI), JNK (SP600125), and mammalian target of rapamycin (rapamycin) were purchased from Calbiochem. Inhibitors for p38 (PHA818637) and MEK (PD325901) were kindly provided by Pfizer Pharmaceuticals. To minimize off-target effects (55), kinase inhibitors were used at concentrations sufficient to inhibit 90% of the phosphorylation of the nominal target as determined by dose-response assays on HepG2 cells (for p38, MEK, IKK, and phosphatidylinositol 3-kinase inhibitors; supplemental Fig. 4) or as obtained from the literature (mammalian target of rapamycin and GSK3 inhibitors (56, 57)). Unless noted otherwise, drug concentrations were as follows: BMS-345541 at 10 μm; ZSTK474 at 2 μm, inhibitor XI at 0.5 μm, SP600125 at 15 μm, rapamycin at 100 nm, PHA818637 at 10 nm, and PD325901 at 5 nm.
Reliability of Signaling Data and Derived Maps
Two issues arise when comparing established cell lines with donor-derived primary cells: (i) experiment-experiment and donor-donor variability and (ii) the purity of the primary cell preparations. To address the first issue, replicated assays of HepG2 cells from different passages and of primary hepatocytes from different donors were compared (supplemental Fig. 5). The coefficient of variation for repeated xMAP measurement of the same set of lysates (technical replicates) was found to be ∼±8%, and data from different HepG2 passages correlated with R2 ∼0.9. Data from different hepatocyte donors correlated with R2 ∼0.8, indicating good donor-to-donor repeatability. As expected, greater variation was observed among primary cells than among different passages of a tumor cell line because of differences in lifestyle and genetic backgrounds of primary human cell donors. However, in those cases in which data overlapped previously published results (e.g. LPS-induced TLR signaling), our data were in agreement with previous findings (58–60). Overall, we conclude that xMAP assays were highly reproducible and that hepatocytes from different donors were quite similar (61) at least with respect to the top ∼50% of the regression weights from MLR that were the focus of the current study. It is likely that weaker interactions are more variable from donor to donor, although we do not have enough data to draw any firm conclusions on this point. Importantly, however, the differences between HCC and primary cells that were the focus of this study were highly significant no matter which hepatocyte sample or HepG2 passage was used for the analysis.
To assess the purity of hepatocyte preparations, we used flow cytometry as described previously (62) with biomarkers specific for hepatocytes, bile epithelial cells, liver endothelial cells, Kupffer cells, and stellate cells (see supplemental Fig. 6a for additional details). Reanalysis of these hepatocyte preparations by flow cytometry showed levels of contaminating cells to be very low: <0.5% for Kupffer cells and even less for stellate and bile epithelial cells. However, Kupffer cells express high levels of TLR and inflammatory receptors and are very active in secretion so that even a small proportion could conceivably make a noticeable contribution to measured cytokines levels. To ensure that the presence of a few Kupffer cells was not a confounding factor in our data, we assayed explicitly for responses characteristic of Kupffer cells: induced secretion of TNFα, IL1β, and IL10 by LPS or IFNγ (63–68). In LPS- or IFNγ-treated hepatocytes from three different donors, levels of TNFα, IL1β, and IL10 were no more than ∼2-fold above background and at least 50-fold below the levels observed in experiments with U937 macrophages (included as a positive control; supplemental Fig. 6b). Thus, we conclude that contamination of hepatocytes by nonparenchymal cells is unlikely to contribute appreciably to the cytokine measurements in the data set.
Availability of Data
The full data set is available at http://www.cdpcenter.org/resources/data/alexopoulos-et-al-2009/. as spreadsheets in MIDAS format (53) that can be directly loaded, processed, and visualized in DataRail or other software tools. Data can be downloaded as supplemental material and are available upon request. Subsets of these data have previously been made available as part of the network inference challenges for the Dialogue on Reverse Engineering Assessments and Methods (DREAM), and these data sets are also available upon request. We describe the precise relationships among these various data sets and the complete set of data provided in this study in supplemental Table 1.
RESULTS
Constructing Networks of Receptor-mediated Signaling in Primary and Transformed Hepatocytes
To obtain functional data on multiple signaling pathways in liver, we exposed cells to seven extracellular ligand “cues”: two mediators of the acute phase response, TNFα and IL6 (31); the TLR4 agonist LPS (30); two general inflammatory factors active in liver, IL1α and IFNγ (29); and two mitogenic factors, IGF-1 (69) and the ErbB1/2 ligand TGFα (30) (Fig. 1, supplemental Fig. 1, and Table I). Data were also collected from pathways under states of perturbation by pretreating cells with one of seven small molecule kinase inhibitors (at concentrations sufficient to achieve ∼90% target inhibition in HepG2 cells; see “Experimental Procedures” and supplemental Fig. 4). Whole-cell lysates were collected at 30 min and 3 h after ligand exposure to assay the phosphorylation states of intracellular proteins (“signals”; supplemental Table 2), and cell supernatants were assayed at 3 and 24 h poststimulus to measure secreted cytokines (“responses” in the MLR analysis; supplemental Table 3) using xMAP technology (Luminex Corp., Austin, TX). These time points were chosen based on preliminary experiments (performed at six time-points: 0, 15 min, 30 min, 1 h, 3 h, 8 h, and 24 h) in which we looked for the largest changes in protein modification states.
Table I. Identities and biological roles of ligand cues used to create data compendium.
LSECs, Liver sinusoid endothelial cells.
| Name | Concentration | Secreted by | Biological effects |
|---|---|---|---|
| TNFα | 10 ng/ml | Macrophages, T and B cells, monocytes, NK cells | Proinflammatory, immune response, acute phase response |
| IL6 | 100 ng/ml | Antigen-presenting cells, Th2 cells, macrophages, Kupffer cells, LSECs | Proinflammatory, B cell maturation, T lymphocyte activation, acute phase response, insulin resistance |
| IL1α | 100 ng/ml | Macrophages, antigen-presenting cells, T and B cells, monocytes | Proinflammatory, immune response, acute phase response, T cell and macrophage activation |
| IGF-1 | 10 ng/ml | Produced in liver | Insulin pathway |
| TGFα | 2000 units/ml | Many cell types | Major autocrine and paracrine regulator in liver regeneration |
| IFNγ | 100 ng/ml | NK cells | Proinflammatory, stimulates Th1 cells, activates macrophages, inhibits Th2 cell differentiation |
| LPS | 100 ng/ml | Prototypical endotoxin, major component of Gram-negative bacteria | Innate immunity via TLR activation |
The choice of which intracellular proteins and cytokines to assay was governed by prior knowledge of signaling pathways in the liver and by the availability of reagents compatible with multiplex methods. Measurements included levels of activating phosphorylation on cytosolic kinases such as Akt, MEK, and p38; signaling kinase substrates such as Hsp27 (a Prak2-MK2 target) and CREB (a pP90RSK substrate); and adaptor proteins such as IRS-1 (Fig. 1, supplemental Fig. 2, and supplemental Table 2). In total, the data compendium contained 25,856 data points, representing treatment of hepatocytes and HepG2 with seven ligands (plus one negative control) with or without seven small molecule drugs (plus a negative control) followed by single or duplicate assays of 17 intracellular signals at t = 0, 30 min, and 3 h and of 50 secrete cytokines at t = 0, 3, and 24 h (Fig. 2 and supplemental Fig. 1). The absence or poor specificity of some xMAP reagents prevented study of the transforming growth factor-β and Wnt signaling pathways, which are known to be important for liver biology (70, 71), but we intend to add new measurements to the current data set as reagents and assays improve.
Cell Type-specific Maps Reveal Significant Alteration of NF-κB and Prosurvival Pathways
Primary data were consolidated and scaled using DataRail software (53) and analyzed by MLR, a method for determining correlations among multiple variables (72, 73). Our goal was to uncover the most significant differences between primary hepatocytes and HepG2 cells using the simplest methods available. To uncover the effects of cues and inhibitors on signals, we assumed that n independent inputs (e.g. cues and inhibitors) correlated linearly with m dependent output variables (e.g. protein phosphostates). Two types of MLR were performed: (i) one-step MLR involving regression of signals against cytokine levels and (ii) two-step MLR involving regression of cues and inhibitors against signals (see “Experimental Procedures,” Fig. 3a, and supplemental Fig. 3). Two-step regression analysis was performed to ensure that the analysis was overdetermined. More sophisticated analytical approaches are possible and would probably uncover additional differences between cell types, but our cross-validation studies (supplemental Fig. 3) and a recently published analysis of a subset of the data presented here (and made publically available prepublication as part of the DREAM3 competition (74); see supplemental Table 1) show that MLR performs quite well with data such as ours (75).
Following MLR, many differences in regression weights were observed between hepatocytes and HepG2 cells (Fig. 3, b and c). The weights were used to construct node-edge graphs using Cytoscape with nodes corresponding to receptors, intracellular signals, or cytokines and line thickness denoting regression weights (Fig. 4, a and b). As an aid to interpretation, we overlaid the top 25% of correlation weights on a more conventional map of signal transduction prepared using bibliographic data in Ingenuity Systems IPA (Fig. 5; note that this representation is simply a different layout of the data in Fig. 4 but with the regression data added). By depicting the order of action of ligands, receptors, and signaling proteins, this view revealed that whereas hepatocytes were highly responsive to LPS and TNFα HepG2 cells were not. Conversely, HepG2 cells were responsive to IGF-1, but hepatocytes were not (for the intracellular signals examined). Some ligands were active on both cell types (such as IL1α and TGFα), but these ligands nonetheless gave rise to different patterns of signaling in the two cell types. Thus, hepatocytes and HCC cells appear to have extensive differences in the activities and dynamics of canonical signaling pathways.
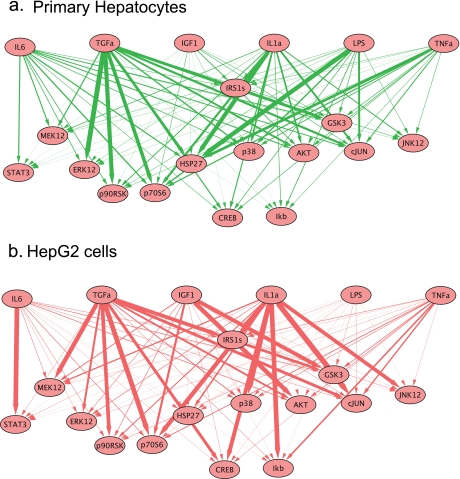
Fig. 4.
Node-edge graphs of signaling pathways reconstructed from data using multilinear regression. Directed node-edge graph of all cue-to-signal regression weights as visualized using the program Cytoscape (117) with receptors (for ligand cues) and signals as nodes. The thickness of each line is directly proportional to the corresponding regression weight. Green denotes primary hepatocytes, and red denotes HepG2 cells.
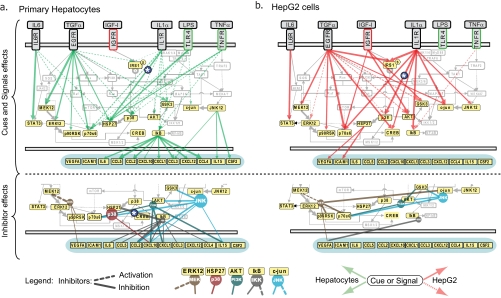
Fig. 5.
Reconstructed node-edge graphs drawn as pathways. Networks were reconstructed using the top 25% of regression weights from node-edge graphs (Fig. 4) and overlaid on a hand-drawn map of relevant pathways. Cue-to-signal regression weights derived from two-step MLR and signal-to-cytokine regression weights derived from one-step MLR are shown in upper panels (cue and signal effects). In lower panels (inhibitor effects), nodes representing inhibitors are positioned above their most proximal target that is also a measured variable (e.g. MEK inhibitor (MEKi) is positioned above ERK, p38 inhibitor is above phospho-Hsp27, etc.) with diameter proportional to the regression weight as obtained from the second step of two-step MLR. Dashed lines in lower panels denote activation, and solid lines denote inhibition. Note that the identities of targets for each drug are predefined (based on the literature), but the inhibition weight is obtained from the data.
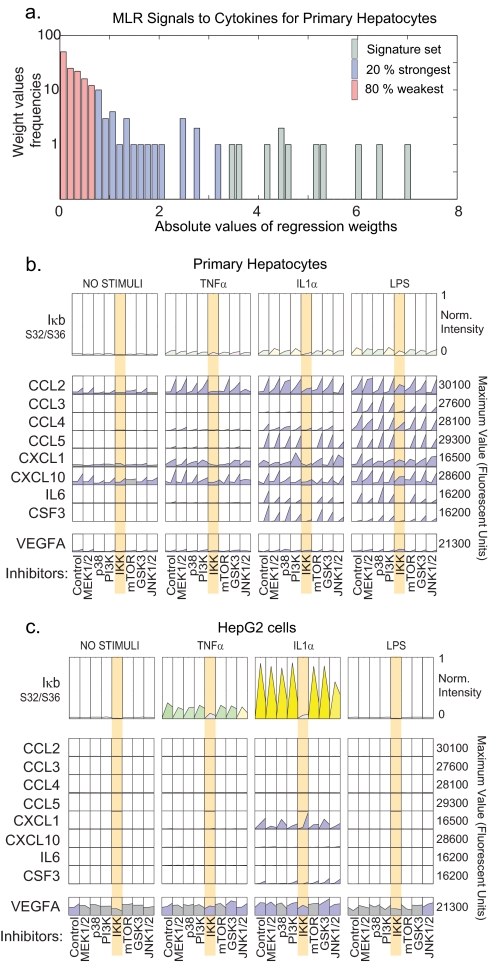
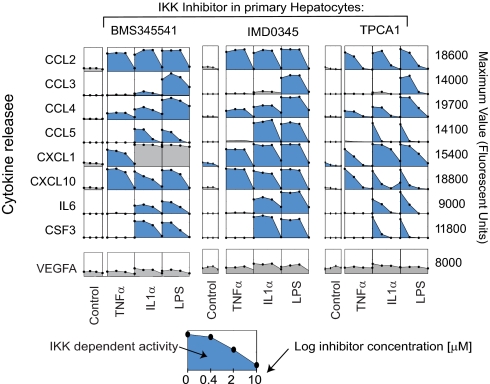
We selected a set of edges in the MLR graph with the greatest differential regression weights between hepatocytes and HepG2 cells for independent experimental confirmation. This set corresponded to coordinated IL1α, TNFα, or LPS-induced secretion of eight cytokines in primary hepatocytes only; these CCR/CCX chemokines included CCL2 through CCL5, CXCL1, CXCL10, IL6, and colony-stimulating factor 3 (CSF3, Fig. 6, b and c). These selectively secreted “hepatocyte signature set” (HSS) proteins are afferent signals that stimulate cellular immune responses by Kupffer cells, circulating macrophages, natural killer (NK) cells, and T cells (Table II). In contrast, ligands associated with antigen-specific immunity such as IL2, IL4, IL5, IL12p40, and IL13 were present at negligible levels (60, 76–78). Release of HSS cytokines from hepatocytes was blocked by three chemically distinct IKK inhibitors, BMS-345541, IMD0345 (79), and TPCA1 (79) (Fig. 7), but secretion of VEGFA, included as a specificity control, was largely unaffected. Thus, secretion of hepatocyte-specific HSS cytokines is dependent on induction of the canonical NF-κB pathway. Although NF-κB was also activated in HepG2 cells, this did not result in HSS cytokine release (see below).
Fig. 6.
Hepatocytes, but not HepG2 cells, secrete a “signature set” of cytokines in response to inflammatory stimuli. a, distribution of regression weights in “signals to cytokine” MLR from hepatocyte data. The top 25% of weights (as depicted in Fig. 5, a and b) are shown in blue; the most significant weights, corresponding to signature set cytokines, are shown in green. Distributions for all other MLRs are shown in supplemental Fig. 8. b and c, levels of eight signature set cytokines in hepatocytes and HepG2 cells (as indicated) at t = 0, 3, and 24 h following exposure to TNFα, IL1α, or LPS. Phospho-IκB levels, a measure of IKK activity, are shown above the cytokine data with color coding as in Fig. 1. Absence of the signature set cytokine release in HepG2 cells following exposure to TNFα, IL1α, or LPS is shown by orange shading. VEGFA serves as a control. Norm., normalized; PI3K, phosphatidylinositol 3-kinase.
Table II. Identities and probable roles of hepatocyte signature set cytokines secreted in NF-κB-dependent manner by hepatocytes but not HepG2 cells.
DCs, dendritic cells; RANTES, regulated on activation normal T cell expressed and secreted.
| Name | Receptor | Released by | Target cells | Liver function |
|---|---|---|---|---|
| MCP1/ CCL2 | CCR2 | Many cell types | Monocytes, immature DCs | Monocyte and lymphocyte chemotaxis |
| MIP1α/CCL3 | CCR1/CCR5 | Kupffer/dendritic cells, other | Monocytes, immature DCs, Th1 cells, neutrophils | Innate immunity |
| MIP1β/CCL4 | CCR5 | Kupffer/dendritic cells, other | Macrophage, neutrophils | Innate immunity |
| RANTES/CCL5 | CCR1/CCR5 | Kupffer/dendritic cells, other | Monocytes, immature DCs, Th1 cells | Innate immunity |
| GROα/CXCL1 | CXCR2 | Many cell types | Neutrophils | Innate immunity |
| IP10/CXCL10 | CXCR3 | Many cell types | NK cells, Th1 cells | Innate immunity |
| IL6 | IL6R | Many cell types | Hepatocytes | Lymphocyte growth, acute phase response, liver regeneration |
| CSF3 | CSF3R | Macrophages, other | Granulocyte | Recruitment of stem cells in regeneration |
Fig. 7.
Signature cytokine secretion set is NF-κB-dependent. Secretion of the cytokine signature set in hepatocytes (at t = 24 h) is inhibited by three structurally distinct IKK inhibitors. Each subpanel depicts the level of a cytokine (as indicated at left) following exposure to three concentrations (400 nm, 2 μm, and 10 μm) of BMS-345541, IMD0345, or TPCA1 (as indicated above) in hepatocytes stimulated with TNFα, IL1α, or LPS (as indicated below). VEGFA serves as a negative control (gray background).
Differences between hepatocytes and HepG2 cells were also evident in regression weights for factors involved in mitogenesis and cell survival (Fig. 3, b and c). For example, phosphohistone H3, p53, and p-Akt were more heavily weighted in HepG2 cells than hepatocytes (Fig. 3b) as were secreted VEGFA and intercellular adhesion molecule 1 (Fig. 3c). VEGFA is a potent angiogenic molecule known to be up-regulated in HCC and is targeted by a therapeutic antibody (bevacizumab) currently in clinical trials for treatment of liver cancer. Intercellular adhesion molecule 1 has also been implicated in HCC (80). In contrast, p-c-Jun, p-p38, and p-Hsp27 were more heavily weighted in hepatocyte data. Overall then, analysis of signaling data by MLR uncovered many differences in signaling between HepG2 cells and hepatocytes with significant up-regulation of progrowth pathways and dramatic down-regulation of inflammatory and TLR responses in the former.
Confirming Determinants of Connectivity Inferred by MLR
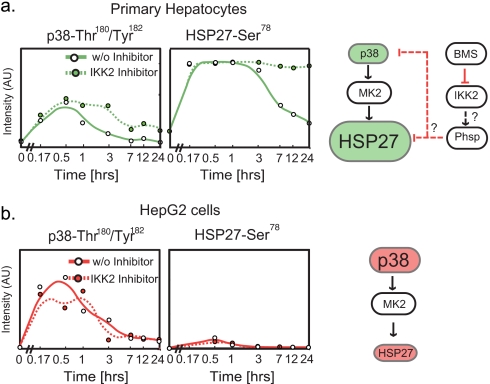
The node-edge graphs generated by MLR have some counterintuitive features. For example, phosphorylation of p38 kinase is a typical response of cells to cytokine treatment, and Hsp27 is an indirect substrate (it is phosphorylated by MK2 and Prak2 kinases, which lie downstream of p38). However, graphs constructed from hepatocyte data link nodes corresponding to five of six receptors (IL1R, TNF receptor, TLR4, IL6R, and epidermal growth factor receptor) and to p-Hsp27 but not to p-p38 (Fig. 5a, white star in blue background). In contrast, in MLR-based graphs for HepG2 cells, IL1R (the only strong p-p38 inducer) was linked to p-p38 but not p-Hsp27 (Fig. 5d). p38 phosphorylation was nonetheless required for Hsp27 phosphorylation in both cell types as evidenced by low p-Hsp27 levels in cells exposed to the selective p38 inhibitor PHA818627 prior to cytokine addition (Fig. 2, a and b, column for p38 inhibitor).
Why is there a difference in connectivity between nodes corresponding to receptors, p38, and substrates in hepatocytes and HepG2 cells? Follow-up experiments revealed that in HepG2 cells p-p38 levels were high following IL1α stimulation, but p-Hsp27 levels were relatively low, whereas in hepatocytes slightly lower p-p38 levels gave rise to dramatically higher p-Hsp27 levels (Fig. 8). These differences are sufficient to explain the observed connectivity and may reflect unequal phosphatase activity in the two cell types or differential regulation of the p38-activated MK2 and Prak kinases. Similarly, a hepatocyte-specific link between p-Hsp27 and IKK arises because BMS-345541 blocks p-Hsp27 dephosphorylation only in hepatocytes, suggesting a cell type-specific role for IKK in phosphatase regulation (Fig. 8, a and b, dashed lines) or a differential expression of IKK isotypes (which are blocked to different degrees by BMS-345541). These data confirm the logic by which MLR assigns differential links between receptors p-p38, p-Hsp27, and IKK in hepatocytes and HepG2 cells. They also suggest a set of hypotheses about differential phosphatase regulation in hepatocytes and HepG2 cells for future study.
Fig. 8.
Validation of pathway comparisons. a and b, confirmation of relationships between phospho-p38 and phospho-Hsp27 levels in IL1α-treated hepatocytes and HepG2 cells. These correspond to the regression links denoted in Fig. 5 as white stars on a blue background. Phosphoprotein levels at seven time points (and t = 0) following treatment with IL1α alone (solid lines) or IL1α in combination with the IKK2 inhibitor BMS-345541 (BMS) (dashed line) were measured in hepatocytes and HepG2 cells. Following IL1α treatment, phospho-Hsp27 was stimulated to a greater extent in hepatocytes than in HepG2 cells even though p38 is more highly phosphorylated in HepG2 cells. Moreover, dephosphorylation of Hsp27 at t >3 h was BMS-345541-sensitive in hepatocytes but not HepG2 cells, confirming the primary MLR data and suggesting that in hepatocytes dephosphorylation of p38 and Hsp27 following IL1α treatment is IKK2-dependent, possibly via an IKK2-activated phosphatase (Phsp); w/o, without; A.U., absorbance units.
Innate Immune Responses and NF-κB Signaling Are Altered in HCC Cells
As noted above, dramatic differences were evident between hepatocytes and HepG2s in responsiveness to LPS, TNFα, and IL1α, particularly with respect to induced secretion of HSS cytokines (Figs. 5, a and b, and 6, b and c, compare the relative line weights for edges connecting IκB to HSS cytokines). Induced secretion of HSS cytokines was strongly IKK-dependent in hepatocytes (Fig. 7), consistent with the central role of NF-κB in immune priming (81). Thus, both hepatocytes and HepG2 cells are active in IKK/NF-κB signaling (as determined by TNFα- and IL1α-induced phosphorylation of IκB Ser-32/Ser-36 sites involved in IκB degradation and NF-κB activation; Fig. 6b), but only hepatocytes secrete detectable HSS cytokines.
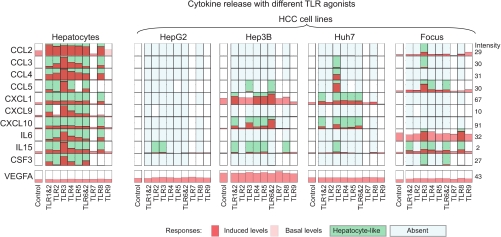
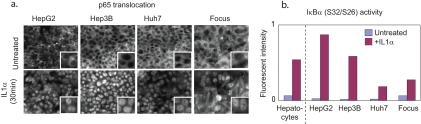
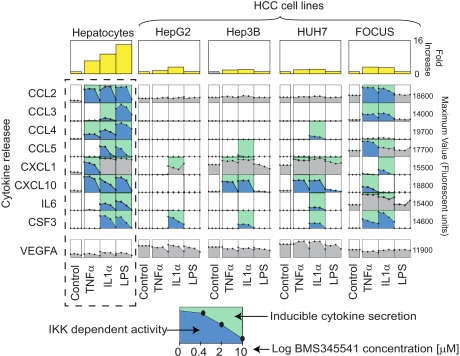
To determine whether the observed defect in LPS-induced HSS cytokine secretion in HepG2 cells is reflective of a general defect in TLR signaling in HCC cell lines, we analyzed three additional liver cancer cell lines: among these, FOCUS cells are considered relatively differentiated as compared with HepG2 cells, whereas Hep3B and HuH7 lines represent intermediate degrees of differentiation (52, 82). When FOCUS, Hep3B, and HuH7 lines were compared with HepG2 cells and primary hepatocytes across a panel of TLR agonists (Fig. 9), we observed that whereas primary hepatocytes were broadly responsive (as assayed by release of HSS cytokines) all four HCC lines were much less so (Fig. 9). Thus, the lack of HepG2 responsiveness to LPS in the MLR-based network was also observed in other HCC cell lines for multiple TLR agonists. Nonetheless, IL1α treatment of Hep3B, HuH7, and FOCUS cells caused dramatic increases in IκB Ser-32/Ser-36 phosphorylation and rapid translocation of NF-κB p65 into the nucleus, a response also present in HepG2 cells (NF-κB p65 localization was determined by immunofluorescence microscopy; Fig. 10, a and b). In addition, Western blotting showed NF-κB family members p50, p65, p105, and c-Rel to be expressed in HCC lines at similar, or slightly higher, levels than in hepatocytes (supplemental Fig. 7). Thus, the absence of HSS secretion in HCC cells (Fig. 11) does not reflect simple absence or inactivity of the IKK/NF-κB pathway but rather a subtle shift in the spectrum of genes induced by NF-κB.
Fig. 9.
Release of signature set cytokines is profoundly impaired in HCC cells. Levels of signature set cytokines at t = 24 h following exposure of primary hepatocytes and four HCC cell lines to nine TLR agonists (see “Experimental Procedures” for details) are shown. Red bars denote induction of a cytokine significantly above background levels (shown in pink). Light blue background denotes cytokines that are inducibly secreted in hepatocytes but absent in HCC cell lines, whereas green background denotes a hepatocyte-like secretion.
Fig. 10.
NF-κB pathway remains functional in HCC cells. Nuclear translocation of NF-κB in IL1α-treated cells as assayed by immunofluorescence microscopy (right panels) is shown. Induction of phospho-IκB (Ser-32/Ser-36) levels in HCC cells and hepatocytes was determined following exposure to IL1α for 30 min.
Fig. 11.
Release of signature set cytokines remains NF-κB-dependent. Secretion of signature set cytokines at t = 24 h in four HCC cells in the presence and absence of IKK inhibitor BMS-345541 is shown. Data for primary hepatocytes (as indicated by the dashed box) are reproduced from Fig. 6 for sake of comparison. Each subpanel depicts the level of a cytokine (as indicated at left) in cells stimulated with TNFα, IL1α, or LPS (indicated below) and three BMS-345541 concentrations. Green background denotes inducible cytokine secretion, and blue denotes inhibition by BMS-345541. Gray denotes cytokine levels that were neither cue-inducible nor BMS-345541-inhibitable. The yellow bar graphs above show the integrated levels of all signature set cytokines for each stimulus in the absence of BMS-345541.
DISCUSSION
Here we describe an approach to reconstructing signaling networks from biochemical data on cellular responsiveness to ligands and drugs. Three primary findings emerge from comparing immediate-early signaling networks in primary hepatocytes and transformed HepG2 cells. First, the degree of cellular responsiveness to a spectrum of growth factors, inflammatory ligands, and TLR agonists is very different with primary cells responding more to inflammatory cytokines such as TNFα and transformed cells responding more to growth factors such as IGF. Second, even when both cell types are sensitive to the same ligand (e.g. epidermal growth factor or IL1α) the extent to which specific immediate-early signaling pathways are activated is strikingly different. Third, patterns of induced cytokine secretion, a component of autocrine and paracrine signaling in the liver, are also different even when a common transcription factor (NF-κB) is involved. These differences provide new insight into hepatocyte transformation, suggest specific features of TLR-mediated innate immunity that merit further study in real tumors, and demonstrate the utility of reverse engineering biochemical data as a means to compare signal transduction in diseased and healthy cells of diverse origin.
Cell Type-specific Protein Signaling Networks
Considerable effort is currently devoted to reconstructing mammalian metabolic and signaling pathways on a large scale using manual (13, 83, 84) or automated (14) literature mining or high throughput two-hybrid (5–7), affinity purification-mass spectrometry (8–11), genetic interaction (85, 86), or RNAi screening data (20–22). Most of these efforts emphasize integration of data from many cell types and even multiple organisms as a means to generate a comprehensive interaction graph for a “prototypical” cell (indeed, creating a proteome-wide interaction network is a goal of the human interactome project (3, 87)). Such approaches sacrifice information on differences from one cell type to the next to achieve maximum scope, but regulatory networks vary significantly with cell state and type (88). In this study, we focused on creating smaller networks that are cell-specific and focused on the receptor-mediated signaling processes known to play an important role in oncogenic transformation. Other efforts to assemble cell-specific networks have involved gene expression (16, 89), chromatin immunoprecipitin in combination with DNA microarrays (90), addition of expression data to PINs (27, 91, 92), or use of gene ontology annotations in combination with interaction data (93). The current work is distinguished from previous studies in its use of biochemical rather than genetic data, the inclusions of intracellular and extracellular measurements, and its explicitly comparative measurement of responses to ligands and drugs. This yields PSNs with lower connectivity than prototypical PSNs but significantly better predictivity (94). Despite their small scope, cell-specific biochemical networks are likely to have significant advantages for understanding disease processes and for studying the properties of signaling networks from an information-theoretic perspective (95).
Perturbation-rich Rather than Measurement-rich Data Sets
Pathway reconstruction benefits from as much data as possible, and this requirement, in combination with a focus on primary human cells that are available in limited quantities, mandated our use of microscale immunoassays. However, such assays are available for only a limited number of proteins; the Luminex xMAP methods used in the current work permit ∼20 phosphoprotein and ∼50 cytokine measurements per well of a 96-well plate, a much smaller number of measurements than would be possible using expression profiling. At the outset of this work, it was unclear whether such a limited set of measurements would yield useful network inference. Our apparent success likely reflects the value of data collection across a spectrum of cell states (exposure to different cytokines), each perturbed in multiple ways using a panel of kinase inhibitors. Thus, although the number of data points per state is relatively small, the number of states is large (∼50 per cell line). Moreover, measurements concentrate on proteins known to be involved in signal transduction, most of which exhibited significant changes in state or level across the data set. In contrast, the majority of genes in an expression microarray do not change in response to a particular biological cue.
Extending the approaches described here should enable rapid reconstruction of larger and more precise comparative signaling graphs. Examining ligands in combination, studying the effects of different extracellular matrices on signaling, comparing immediate-early and late responses, and complementing small molecule drugs with RNAi all represent direct extensions of our approach. The availability of appropriate data will always be a critical issue, but as sensitivity and throughput improve, it will be possible to supplement immunoassays with phosphoprotein SILAC (stable isotope labeling with amino acids in cell culture) (96) and other mass spectrometry methods (12) that yield information on proteins for which no immune reagents exist. The potential of mass spectrometry to reconstruct cell-specific networks has been demonstrated recently for EphB1- and ephrin-B1-expressing cells grown in coculture (19). However, experience to date strongly suggests that effective network inference will require data sets involving rich combinations of cell states and perturbations rather than simply collecting more measurements made under a limited set of conditions. Thus, it will probably be necessary to adapt existing mass spectrometry methods so that many samples can be compared even if this entails a reduction in the number of analytes tracked.
Defects in NF-κB- and TLR-mediated Signaling in HCC Cell Lines
The largest difference in regression weights between graphs of hepatocyte and HepG2 cells involves responsiveness to inflammatory factors and LPS. A set of eight IKK (and NF-κB)-dependent cytokines was secreted at a high level by hepatocytes exposed to TNFα, IL1α, and TLR agonists but was largely absent in supernatants from similarly treated HepG2 cells and three other HCC cell lines examined. These eight coordinately expressed HSS cytokines and chemokines are principally efferent regulators of immune cells and are strongly induced by IL1α, TNFα, or LPS, all afferent signals for primary hepatocytes (Table II). Connections between tumor and immune cells are myriad and complex (34, 42, 97, 98). Tumor-associated macrophages, for example, secrete factors that act as mitogens for cancer cells (99, 100). Suppression of inflammation (by drugs for example) interferes with tumor growth under these circumstances. In other cases, immune cells act in an antioncogenic fashion, apparently by recognizing and killing transformed cells (101). To survive, tumor cells must adapt to and avoid such immune surveillance (102–104). The changes we observed in HCC cells appear to reflect this latter paradigm: by damping down TLR-induced secretions of factors that mobilize and attract immune cells, HCC cells are presumably able to avoid immune recognition.
Apparent differences in the function of NF-κB in primary hepatocytes and HCC cell lines are consistent with a broad and complex literature implicating IKK/NF-κB in liver cancer. For example, deletion of IKKβ in the mouse increases hepatocellular carcinogenesis (39), and this also appears to be true of IKKγ (34); NF-κB itself appears to function as a tumor promoter (42) and plays an important role in viral induction of liver cancer (105, 106). Unfortunately, genetic studies in the mouse have not, as yet, revealed precisely which NF-κB functions are altered in transformed relative to normal liver cells. Our data suggest that HCC cells have evolved to exploit the transforming potential of NF-κB while down-regulating the NF-κB functions involved in immune activation. Kupffer cells and other liver-resident macrophages are tasked with removing apoptotic hepatocytes that are damaged in the course of responding to toxins and infectious agents (107). To accomplish this, Kupffer cells require priming, and this depends on cytokines secreted by hepatocytes. To become tumorigenic, transformed hepatocytes almost certainly need to avoid such immune surveillance, and we speculate that they therefore evolve to reduce secretion of immunostimulatory cytokines, which are represented in our data by HSS cytokines.
The profound defects in TLR signaling we observed in HCC cells have not, to our knowledge, been described previously but are consistent with the hypothesis that innate immunity plays an important role in suppressing tumorigenesis (102, 108, 109). We do not know precisely why HCC cells are unresponsive to a wide range of TLR agonists, but RNA levels for TLR receptor and a set of downstream signaling molecules appear to be lower in HCC cell lines than in primary hepatocytes (as assayed by RT-PCR; supplemental Table 4). However, low responsiveness to ligands was observed even when the cognate TLR was expressed at apparently normal levels, suggesting that other factors must be involved. In addition, defects in TLR expression cannot of course explain why secretion of HSS cytokine is low in HCC following exposure to cytokines such as IL1α or TNFα. It might be possible to assay for reduced TLR expression in primary liver cancers, although this would require controlling for tumor infiltration by macrophages, which express very high levels of TLRs, potentially masking receptor down-regulation in hepatocytes.
Data Analysis and Network Inference
By emphasizing functional responses and comparing multiple cell types, this study describes an experimental approach to reverse engineering signaling networks that is a significant extension to existing network inference paradigms. We leverage a relatively limited set of high throughput biochemical assays by maximizing the number of conditions under which assays are performed and using systematic perturbation (with well characterized small molecule drugs) to uncover cause-effect relationships among receptors, intracellular kinases, and cytokine secretion.
Sophisticated computational methods have been developed to construct gene and protein networks using mutual information theory (110), Bayesian analysis (111, 112), and related probabilistic methods. These approaches are necessary with genome-scale expression data in which the number of measurements is very large but the signal-to-noise ratio is relatively low. In contrast, our data comprise many fewer measurements, but the signal-to-noise ratio is good largely because we use prior knowledge to select combinations of ligands, drugs, and biochemical assays likely to be informative. Bayesian network inference has also been used to infer networks from protein data collected by flow cytometry (113, 114) and micro-Western assays (115). Whereas the flow cytometry data in Sachs et al. (114) involve many independent single cell measurements across a restricted set of signaling kinases and the micro-Western data modeled in Ciaccio et al. (115) involve dense temporal sampling of a restricted set of receptor-proximal processes, the data in our study cover a wider range of biological processes from receptor phosphorylation to transcriptional induction. We speculate that with our data Bayesian inference is less successful (albeit only in preliminary studies) than simpler regression methods because we sampled networks less densely than Sachs et al. (114) and Ciaccio et al. (115). Whether this is true should be revealed by future DREAM (74) competitions: we will continue to provide data to DREAM with the goal of determining precisely which inference methods are optimal for reconstructing signaling networks from different types of cell response data.
We have shown recently that immediate-early signal transduction can effectively be analyzed using logical modeling and data similar in structure to the data in this study. We assemble an initial logical model from literature-derived PSNs and then optimize the model against experimental data. The results are models that are significantly more predictive than the starting PSN but 2–3-fold less highly connected (94). Logic-based modeling provides mechanistic insight that is missing from MLR, but our approach requires some prior knowledge about network topology; this is largely absent in the case of regulated cytokine secretion. In the future, we envision hybrid models that combine logical or probabilistic elements for pathways we seek to represent in mechanistic detail (particularly early response pathways that are well annotated in the literature) and less detailed regression-based elements for downstream events such as cytokine secretion (for which regulatory mechanisms are less well understood). However, we emphasize that even the simple regression-based methods applied in this study are sufficient to uncover widespread differences between immediate-early signaling networks in primary and tumor cells. The two most significant differences appear to involve inflammation and innate immunity. (i) In contrast to primary hepatocytes, HCC cells lack a robust response to TLR ligands. (ii) NF-κB can be activated in both hepatocytes and HCC cells by inflammatory cytokines such as IL1α, but the spectrum of genes induced by NF-κB is very different in the two types of cells specifically with regard to cytokines that might induce Kupffer cell priming. In both cases, we speculate that the changes reflect evolution of HCC cells so that their abnormality goes unnoticed by Kupffer and other immune cells.
Supplementary Material
Acknowledgments
We thank R. Ward, D. deGraaf, J. Xu, M. Zhang, H. Nguyen, A. Smith, J. Wands, B. Hendriks, C. Espelin, J. Wagner, M. Hasan, E. Farazi, and J. Chung for help with methods and the manuscript.
* This work was supported, in whole or in part, by National Institutes of Health Grants GM68762 and CA112967. This work was also supported by a grant from Pfizer Inc.
 This article contains supplemental Figs. 1–8 and Tables 1–4.
This article contains supplemental Figs. 1–8 and Tables 1–4.
1 The abbreviations used are:
- PIN
- protein interaction network
- PSN
- protein signaling network
- RNAi
- RNA interference
- MLR
- multilinear regression
- TLR
- toll-like receptor
- HCC
- hepatocellular carcinoma
- VEGFA
- vascular endothelial growth factor A
- JNK
- c-Jun N-terminal kinase
- TNF
- tumor necrosis factor
- CREB
- cAMP-response element-binding protein
- p-
- phospho-
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- ERK
- extracellular signal-regulated kinase
- IGF
- insulin-like growth factor
- TGF
- transforming growth factor
- LPS
- lipopolysaccharide
- IFN
- interferon
- IKK
- IκB kinase
- DREAM
- Dialogue on Reverse Engineering Assessments and Methods
- IRS-1
- insulin receptor substrate-1
- HSS
- hepatocyte signature set
- NK
- natural killer.
REFERENCES
- 1.Pieroni E., de la Fuente van Bentem S., Mancosu G., Capobianco E., Hirt H., de la Fuente A. (2008) Protein networking: insights into global functional organization of proteomes. Proteomics 8, 799–816 [DOI] [PubMed] [Google Scholar]
- 2.Joughin B. A., Cheung E., Karuturi R., Saez-Rodriguez J., Lauffenburger D. A., Liu E. T. (2009) Cellular signaling networks, in Systems Biomedicine: Concepts and Perspective (Liu T., Lauffenburger D. A. eds) Academic Press; New York [Google Scholar]
- 3.Cusick M. E., Klitgord N., Vidal M., Hill D. E. (2005) Interactome: gateway into systems biology. Hum. Mol. Genet. 14, R171–R181 [DOI] [PubMed] [Google Scholar]
- 4.Russell R. B., Aloy P. (2008) Targeting and tinkering with interaction networks. Nat. Chem. Biol. 4, 666–673 [DOI] [PubMed] [Google Scholar]
- 5.Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 6.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 7.Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 8.Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 9.Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 10.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 11.Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gstaiger M., Aebersold R. (2009) Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 10, 617–627 [DOI] [PubMed] [Google Scholar]
- 13.Bauer-Mehren A., Furlong L. I., Sanz F. (2009) Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol. Syst. Biol. 5, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen L. J., Saric J., Bork P. (2006) Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 7, 119–129 [DOI] [PubMed] [Google Scholar]
- 15.Carro M. S., Lim W. K., Alvarez M. J., Bollo R. J., Zhao X., Snyder E. Y., Sulman E. P., Anne S. L., Doetsch F., Colman H., Lasorella A., Aldape K., Califano A., Iavarone A. (2010) The transcriptional network for mesenchymal transformation of brain tumours. Nature 463, 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amit I., Garber M., Chevrier N., Leite A. P., Donner Y., Eisenhaure T., Guttman M., Grenier J. K., Li W., Zuk O., Schubert L. A., Birditt B., Shay T., Goren A., Zhang X., Smith Z., Deering R., McDonald R. C., Cabili M., Bernstein B. E., Rinn J. L., Meissner A., Root D. E., Hacohen N., Regev A. (2009) Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pleasance E. D., Cheetham R. K., Stephens P. J., McBride D. J., Humphray S. J., Greenman C. D., Varela I., Lin M. L., Ordóñez G. R., Bignell G. R., Ye K., Alipaz J., Bauer M. J., Beare D., Butler A., Carter R. J., Chen L., Cox A. J., Edkins S., Kokko-Gonzales P. I., Gormley N. A., Grocock R. J., Haudenschild C. D., Hims M. M., James T., Jia M., Kingsbury Z., Leroy C., Marshall J., Menzies A., Mudie L. J., Ning Z., Royce T., Schulz-Trieglaff O. B., Spiridou A., Stebbings L. A., Szajkowski L., Teague J., Williamson D., Chin L., Ross M. T., Campbell P. J., Bentley D. R., Futreal P. A., Stratton M. R. (2010) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhaak R. G., Hoadley K. A., Purdom E., Wang V., Qi Y., Wilkerson M. D., Miller C. R., Ding L., Golub T., Mesirov J. P., Alexe G., Lawrence M., O'Kelly M., Tamayo P., Weir B. A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H. S., Hodgson J. G., James C. D., Sarkaria J. N., Brennan C., Kahn A., Spellman P. T., Wilson R. K., Speed T. P., Gray J. W., Meyerson M., Getz G., Perou C. M., Hayes D. N.; Cancer Genome Atlas Research Network (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jørgensen C., Sherman A., Chen G. I., Pasculescu A., Poliakov A., Hsiung M., Larsen B., Wilkinson D. G., Linding R., Pawson T. (2009) Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science 326, 1502–1509 [DOI] [PubMed] [Google Scholar]
- 20.Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. (2003) Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 [DOI] [PubMed] [Google Scholar]
- 21.Friedman A., Perrimon N. (2007) Genetic screening for signal transduction in the era of network biology. Cell 128, 225–231 [DOI] [PubMed] [Google Scholar]
- 22.Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A. G. (2006) Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 38, 896–903 [DOI] [PubMed] [Google Scholar]
- 23.Shapira S. D., Gat-Viks I., Shum B. O., Dricot A., de Grace M. M., Wu L., Gupta P. B., Hao T., Silver S. J., Root D. E., Hill D. E., Regev A., Hacohen N. (2009) A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor I. W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D., Bull S., Pawson T., Morris Q., Wrana J. L. (2009) Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 27, 199–204 [DOI] [PubMed] [Google Scholar]
- 25.Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mani K. M., Lefebvre C., Wang K., Lim W. K., Basso K., Dalla-Favera R., Califano A. (2008) A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol. Syst. Biol. 4, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gersten M., Alirezaei M., Marcondes M. C., Flynn C., Ravasi T., Ideker T., Fox H. S. (2009) An integrated systems analysis implicates EGR1 downregulation in simian immunodeficiency virus encephalitis-induced neural dysfunction. J. Neurosci. 29, 12467–12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Przytycka T. M., Singh M., Slonim D. K. (2010) Toward the dynamic interactome: it's about time. Brief. Bioinformatics 11, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gershwin M. E., Vierling J. M., Manns M. P. (eds) (2007) Liver Immunology: Principles and Practice, 1st Ed., Humana Press, Totowa, NJ [Google Scholar]
- 30.Gao B., Jeong W. I., Tian Z. (2008) Liver: An organ with predominant innate immunity. Hepatology 47, 729–736 [DOI] [PubMed] [Google Scholar]
- 31.Moshage H. (1997) Cytokines and the hepatic acute phase response. J. Pathol. 181, 257–266 [DOI] [PubMed] [Google Scholar]
- 32.Seki E., Brenner D. A. (2008) Toll-like receptors and adaptor molecules in liver disease: update. Hepatology 48, 322–335 [DOI] [PubMed] [Google Scholar]
- 33.Mee C. J., Farquhar M. J., Harris H. J., Hu K., Ramma W., Ahmed A., Maurel P., Bicknell R., Balfe P., McKeating J. A. (2010) Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology 138, 1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luedde T., Beraza N., Kotsikoris V., van Loo G., Nenci A., De Vos R., Roskams T., Trautwein C., Pasparakis M. (2007) Deletion of NEMO/IKK[gamma] in Liver Parenchymal Cells Causes Steatohepatitis and Hepatocellular Carcinoma. Cancer Cell 11, 119–132 [DOI] [PubMed] [Google Scholar]
- 35.Zhai Y., Shen X. D., O'Connell R., Gao F., Lassman C., Busuttil R. W., Cheng G., Kupiec-Weglinski J. W. (2004) Cutting edge: TLR4 activation mediates liver ischemia/reperfusion inflammatory response via IFN regulatory factor 3-dependent MyD88-independent pathway. J. Immunol. 173, 7115–7119 [DOI] [PubMed] [Google Scholar]
- 36.Li K., Chen Z., Kato N., Gale M., Jr., Lemon S. M. (2005) Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 280, 16739–16747 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Kato N., Hoshida Y., Yoshida H., Taniguchi H., Goto T., Moriyama M., Otsuka M., Shiina S., Shiratori Y., Ito Y., Omata M. (2003) Interieukin-1 beta gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology 37, 65–71 [DOI] [PubMed] [Google Scholar]
- 38.Eferl R., Ricci R., Kenner L., Zenz R., David J. P., Rath M., Wagner E. F. (2003) Liver tumor development: c-Jun antagonizes the proapoptotic activity of p53. Cell 112, 181–192 [DOI] [PubMed] [Google Scholar]
- 39.Maeda S., Kamata H., Luo J. L., Leffert H., Karin M. (2005) IKK beta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 121, 977–990 [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi R., Yano H., Iemura A., Ogasawara S., Haramaki M., Kojiro M. (1998) Expression of vascular endothelial growth factor in human hepatocellular carcinoma. Hepatology 28, 68–77 [DOI] [PubMed] [Google Scholar]
- 41.Fausto N., Campbell J. S., Riehle K. J. (2006) Liver regeneration. Hepatology 43, S45–S53 [DOI] [PubMed] [Google Scholar]
- 42.Pikarsky E., Porat R. M., Stein I., Abramovitch R., Amit S., Kasem S., Gutkovich-Pyest E., Urieli-Shoval S., Galun E., Ben-Neriah Y. (2004) NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 431, 461–466 [DOI] [PubMed] [Google Scholar]
- 43.Simonetti R. G., Liberati A., Angiolini C., Pagliaro L. (1997) Treatment of hepatocellular carcinoma: a systematic review of randomized controlled trials. Ann. Oncol. 8, 117–136 [DOI] [PubMed] [Google Scholar]
- 44.Parkin D. M., Bray F., Ferlay J., Pisani P. (2005) Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 [DOI] [PubMed] [Google Scholar]
- 45.Vignali D. A. (2000) Multiplexed particle-based flow cytometric assays. J. Immunol. Methods 243, 243–255 [DOI] [PubMed] [Google Scholar]
- 46.Sevecka M., MacBeath G. (2006) State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat. Methods 3, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korf U., Lobke C., Sahin O., Haller F., Sultmann H., Arlt D., Poustka A. (2009) Reverse-phase protein arrays for application-orientated cancer research. Proteomics Clin. Appl. 3, 1140–1150 [DOI] [PubMed] [Google Scholar]
- 48.van Oostrum J., Calonder C., Rechsteiner D., Ehrat M., Mestan J., Fabbro D., Voshol H. (2009) Tracing pathway activities with kinase inhibitors and reverse phase protein arrays. Proteomics Clin. Appl. 3, 412–422 [DOI] [PubMed] [Google Scholar]
- 49.Spurrier B., Honkanen P., Holway A., Kumamoto K., Terashima M., Takenoshita S., Wakabayashi G., Austin J., Nishizuka S. (2008) Protein and lysate array technologies in cancer research. Biotechnol. Adv. 26, 361–369 [DOI] [PubMed] [Google Scholar]
- 50.Kornblau S. M., Tibes R., Qiu Y. H., Chen W., Kantarjian H. M., Andreeff M., Coombes K. R., Mills G. B. (2009) Functional proteomic profiling of AML predicts response and survival. Blood 113, 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knowles B. B., Howe C. C., Aden D. P. (1980) Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 209, 497–499 [DOI] [PubMed] [Google Scholar]
- 52.Lee H. C., Tian B., Sedivy J. M., Wands J. R., Kim M. (2006) Loss of Raf kinase inhibitor protein promotes cell proliferation and migration of human hepatoma cells. Gastroenterology 131, 1208–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saez-Rodriguez J., Goldsipe A., Muhlich J., Alexopoulos L. G., Millard B., Lauffenburger D. A., Sorger P. K. (2008) Flexible informatics for linking experimental data to mathematical models via DataRail. Bioinformatics 24, 840–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akaike H. (1974) A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 [Google Scholar]
- 55.Weiss W. A., Taylor S. S., Shokat K. M. (2007) Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat. Chem. Biol. 3, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Neill D. J., Shen L., Prouty C., Conway B. R., Westover L., Xu J. Z., Zhang H. C., Maryanoff B. E., Murray W. V., Demarest K. T., Kuo G. H. (2004) Design, synthesis, and biological evaluation of novel 7-azaindolyl-heteroaryl-maleimides as potent and selective glycogen synthase kinase-3beta (GSK-3beta) inhibitors. Bioorg. Med. Chem. 12, 3167–3185 [DOI] [PubMed] [Google Scholar]
- 57.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mawet E., Shiratori Y., Hikiba Y., Takada H., Yoshida H., Okano K., Komatsu Y., Matsumura M., Niwa Y., Omata M. (1996) Cytokine-induced neutrophil chemoattractant release from hepatocytes is modulated by Kupffer cells. Hepatology 23, 353–358 [DOI] [PubMed] [Google Scholar]
- 59.Migita K., Abiru S., Nakamura M., Komori A., Yoshida Y., Yokoyama T., Daikoku M., Ueki T., Takii Y., Yano K., Yastuhashi H., Eguchi K., Ishibashi H. (2004) Lipopolysaccharide signaling induces serum amyloid A (SAA) synthesis in human hepatocytes in vitro. FEBS Lett. 569, 235–239 [DOI] [PubMed] [Google Scholar]
- 60.Rowell D. L., Eckmann L., Dwinell M. B., Carpenter S. P., Raucy J. L., Yang S. K., Kagnoff M. F. (1997) Human hepatocytes express an array of proinflammatory cytokines after agonist stimulation or bacterial invasion. Am. J. Physiol. Gastrointest. Liver Physiol. 273, G322–G332 [DOI] [PubMed] [Google Scholar]
- 61.Klingmüller U., Bauer A., Bohl S., Nickel P. J., Breitkopf K., Dooley S., Zellmer S., Kern C., Merfort I., Sparna T., Donauer J., Walz G., Geyer M., Kreutz C., Hermes M., Gotschel F., Hecht A., Walter D., Egger L., Neubert K., Borner C., Brulport M., Schormann W., Sauer C., Baumann F., Preiss R., MacNelly S., Godoy P., Wiercinska E., Ciuclan L., Edelmann J., Zeilinger K., Heinrich M., Zanger U. M., Gebhardt R., Maiwald T., Heinrich R., Timmer J., von Weizsäcker F., Hengstler J. G. (2006) Primary mouse hepatocytes for systems biology approaches: a standardized in vitro system for modelling of signal transduction pathways. Syst. Biol. 153, 433–447 [DOI] [PubMed] [Google Scholar]
- 62.Cosgrove B. D., Cheng C., Pritchard J. R., Stolz D. B., Lauffenburger D. A., Griffith L. G. (2008) An inducible autocrine cascade regulates rat hepatocyte proliferation and apoptosis responses to tumor necrosis factor-alpha. Hepatology 48, 276–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwabe R. F., Seki E., Brenner D. A. (2006) Toll-like receptor signaling in the liver. Gastroenterology 130, 1886–1900 [DOI] [PubMed] [Google Scholar]
- 64.Bojes H. K., Germolec D. R., Simeonova P., Bruccoleri A., Schoonhoven R., Luster M. I., Thurman R. G. (1997) Antibodies to tumor necrosis factor alpha prevent increases in cell replication in liver due to the potent peroxisome proliferator, WY-14,643. Carcinogenesis 18, 669–674 [DOI] [PubMed] [Google Scholar]
- 65.Tsutsui H., Matsui K., Okamura H., Nakanishi K. (2000) Pathophysiological roles of interleukin-18 in inflammatory liver diseases. Immunol. Rev. 174, 192–209 [DOI] [PubMed] [Google Scholar]
- 66.Seki E., Tsutsui H., Nakano H., Tsuji N., Hoshino K., Adachi O., Adachi K., Futatsugi S., Kuida K., Takeuchi O., Okamura H., Fujimoto J., Akira S., Nakanishi K. (2001) Lipopolysaccharide-induced IL-18 secretion from murine Kupffer cells independently of myeloid differentiation factor 88 that is critically involved in induction of production of IL-12 and IL-1beta. J. Immunol. 166, 2651–2657 [DOI] [PubMed] [Google Scholar]
- 67.Knolle P., Schlaak J., Uhrig A., Kempf P., Meyer zum Büschenfelde K. H., Gerken G. (1995) Human Kupffer cells secrete IL-10 in response to lipopolysaccharide (LPS) challenge. J. Hepatol. 22, 226–229 [DOI] [PubMed] [Google Scholar]
- 68.Olinga P., Merema M. T., de Jager M. H., Derks F., Melgert B. N., Moshage H., Slooff M. J., Meijer D. K., Poelstra K., Groothuis G. M. (2001) Rat liver slices as a tool to study LPS-induced inflammatory response in the liver. J. Hepatol. 35, 187–194 [DOI] [PubMed] [Google Scholar]
- 69.Alexia C., Fallot G., Lasfer M., Schweizer-Groyer G., Groyer A. (2004) An evaluation of the role of insulin-like growth factors (IGF) and of type-I IGF receptor signalling in hepatocarcinogenesis and in the resistance of hepatocarcinoma cells against drug-induced apoptosis. Biochem. Pharmacol. 68, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 70.Weng H. L., Ciuclan L., Liu Y., Hamzavi J., Godoy P., Gaitantzi H., Kanzler S., Heuchel R., Ueberham U., Gebhardt R., Breitkopf K., Dooley S. (2007) Profibrogenic transforming growth factor-beta/activin receptor-like kinase 5 signaling via connective tissue growth factor expression in hepatocytes. Hepatology 46, 1257–1270 [DOI] [PubMed] [Google Scholar]
- 71.Takigawa Y., Brown A. M. (2008) Wnt signaling in liver cancer. Curr. Drug Targets 9, 1013–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He J., Zelikovsky A. (2006) MLR-tagging: informative SNP selection for unphased genotypes based on multiple linear regression. Bioinformatics 22, 2558–2561 [DOI] [PubMed] [Google Scholar]
- 73.Pavesi A. (1999) Relationships between transcriptional and translational control of gene expression in Saccharomyces cerevisiae: a multiple regression analysis. J. Mol. Evol. 48, 133–141 [DOI] [PubMed] [Google Scholar]
- 74.Prill R. J., Marbach D., Saez-Rodriguez J., Sorger P. K., Alexopoulos L. G., Xue X., Clarke N. D., Altan-Bonnet G., Stolovitzky G. (2010) Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS One 5, e9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duvenaud D., Eaton D., Murphy K., Schmidt M. (2010) Causal learning without DAGs. J. Mach. Learn. Res. Workshop Conf. Proc. 6, 177–190 [Google Scholar]
- 76.Harvey C. E., Post J. J., Palladinetti P., Freeman A. J., Ffrench R. A., Kumar R. K., Marinos G., Lloyd A. R. (2003) Expression of the chemokine IP-10 (CXCL10) by hepatocytes in chronic hepatitis C virus infection correlates with histological severity and lobular inflammation. J. Leukoc. Biol. 74, 360–369 [DOI] [PubMed] [Google Scholar]
- 77.Apolinario A., Majano P. L., Alvarez-Pérez E., Saez A., Lozano C., Vargas J., García-Monzón C. (2002) Increased expression of T cell chemokines and their receptors in chronic hepatitis C: relationship with the histological activity of liver disease. Am. J. Gastroenterol. 97, 2861–2870 [DOI] [PubMed] [Google Scholar]
- 78.Makishima H., Ito T., Momose K., Nakazawa H., Shimodaira S., Kamijo Y., Nakazawa Y., Ichikawa N., Ueno M., Kobayashi H., Kitano K., Saito H., Kiyosawa K., Ishida F. (2007) Chemokine system and tissue infiltration in aggressive NK-cell leukemia. Leuk. Res. 31, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 79.Kamon J., Yamauchi T., Muto S., Takekawa S., Ito Y., Hada Y., Ogawa W., Itai A., Kasuga M., Tobe K., Kadowaki T. (2004) A novel IKKbeta inhibitor stimulates adiponectin levels and ameliorates obesity-linked insulin resistance. Biochem. Biophys. Res. Commun. 323, 242–248 [DOI] [PubMed] [Google Scholar]
- 80.Yoong K. F., McNab G., Hübscher S. G., Adams D. H. (1998) Vascular adhesion protein-1 and ICAM-1 support the adhesion of tumor-infiltrating lymphocytes to tumor endothelium in human hepatocellular carcinoma. J. Immunol. 160, 3978–3988 [PubMed] [Google Scholar]
- 81.Akira S., Takeda K. (2004) Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 82.He L., Isselbacher K. J., Wands J. R., Goodman H. M., Shih C., Quaroni A. (1984) Establishment and characterization of a new human hepatocellular carcinoma cell line. In Vitro 20, 493–504 [DOI] [PubMed] [Google Scholar]
- 83.Cusick M. E., Yu H., Smolyar A., Venkatesan K., Carvunis A. R., Simonis N., Rual J. F., Borick H., Braun P., Dreze M., Vandenhaute J., Galli M., Yazaki J., Hill D. E., Ecker J. R., Roth F. P., Vidal M. (2009) Literature-curated protein interaction datasets. Nat. Methods 6, 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G. R., Wu G. R., Matthews L., Lewis S., Birney E., Stein L. (2005) Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 33, D428–D432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roguev A., Bandyopadhyay S., Zofall M., Zhang K., Fischer T., Collins S. R., Qu H., Shales M., Park H. O., Hayles J., Hoe K. L., Kim D. U., Ideker T., Grewal S. I., Weissman J. S., Krogan N. J. (2008) Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322, 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) The genetic landscape of a cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ideker T., Valencia A. (2006) Bioinformatics in the human interactome project. Bioinformatics 22, 2973–2974 [DOI] [PubMed] [Google Scholar]
- 88.Luscombe N. M., Babu M. M., Yu H., Snyder M., Teichmann S. A., Gerstein M. (2004) Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431, 308–312 [DOI] [PubMed] [Google Scholar]
- 89.Basso K., Margolin A. A., Stolovitzky G., Klein U., Dalla-Favera R., Califano A. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390 [DOI] [PubMed] [Google Scholar]
- 90.Odom D. T., Dowell R. D., Jacobsen E. S., Nekludova L., Rolfe P. A., Danford T. W., Gifford D. K., Fraenkel E., Bell G. I., Young R. A. (2006) Core transcriptional regulatory circuitry in human hepatocytes. Mol. Syst. Biol. 2, 2006.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myers C. L., Troyanskaya O. G. (2007) Context-sensitive data integration and prediction of biological networks. Bioinformatics 23, 2322–2330 [DOI] [PubMed] [Google Scholar]
- 92.Bossi A., Lehner B. (2009) Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 5, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rachlin J., Cohen D. D., Cantor C., Kasif S. (2006) Biological context networks: a mosaic view of the interactome. Mol. Syst. Biol. 2, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saez-Rodriguez J., Alexopoulos L. G., Epperlein J., Samaga R., Lauffenburger D. A., Klamt S., Sorger P. K. (2009) Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol. Syst. Biol. 5, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barabási A. L., Oltvai Z. N. (2004) Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113 [DOI] [PubMed] [Google Scholar]
- 96.Ong S. E., Schenone M., Margolin A. A., Li X., Do K., Doud M. K., Mani D. R., Kuai L., Wang X., Wood J. L., Tolliday N. J., Koehler A. N., Marcaurelle L. A., Golub T. R., Gould R. J., Schreiber S. L., Carr S. A. (2009) Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc. Natl. Acad. Sci. U.S.A. 106, 4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoffmann A., Xia Y., Verma I. M. (2007) Inflammatory tales of liver cancer. Cancer Cell 11, 99–101 [DOI] [PubMed] [Google Scholar]
- 98.Dunn G. P., Old L. J., Schreiber R. D. (2004) The immunobiology of cancer immunosurveillance and immunoediting. Immunity 21, 137–148 [DOI] [PubMed] [Google Scholar]
- 99.Hoffmann A., Baltimore D. (2006) Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 210, 171–186 [DOI] [PubMed] [Google Scholar]
- 100.Karin M., Greten F. R. (2005) NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5, 749–759 [DOI] [PubMed] [Google Scholar]
- 101.Koebel C. M., Vermi W., Swann J. B., Zerafa N., Rodig S. J., Old L. J., Smyth M. J., Schreiber R. D. (2007) Adaptive immunity maintains occult cancer in an equilibrium state. Nature 450, 903–907 [DOI] [PubMed] [Google Scholar]
- 102.Wang T., Niu G., Kortylewski M., Burdelya L., Shain K., Zhang S., Bhattacharya R., Gabrilovich D., Heller R., Coppola D., Dalton W., Jove R., Pardoll D., Yu H. (2004) Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 10, 48–54 [DOI] [PubMed] [Google Scholar]
- 103.Kortylewski M., Kujawski M., Wang T., Wei S., Zhang S., Pilon-Thomas S., Niu G., Kay H., Mulé J., Kerr W. G., Jove R., Pardoll D., Yu H. (2005) Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 11, 1314–1321 [DOI] [PubMed] [Google Scholar]
- 104.Teicher B. A. (2007) Transforming growth factor-beta and the immune response to malignant disease. Clin. Cancer Res. 13, 6247–6251 [DOI] [PubMed] [Google Scholar]
- 105.Seeger C., Mason W. S. (2000) Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64, 51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Llovet J. M., Burroughs A., Bruix J. (2003) Hepatocellular carcinoma. Lancet 362, 1907–1917 [DOI] [PubMed] [Google Scholar]
- 107.Murray K., Messner D., Kowdley K. (2006) Mechanisms of hepatocyte detoxification, in Physiology of the Gastrointestinal Tract (Barrett K. E., Ghishan F. K., Merchant J. L., Said H. M., Wood J. D., Johnson L. R. eds) 4th Ed., pp. 1483–1504, Academic Press, New York [Google Scholar]
- 108.Pardoll D. (2003) Does the immune system see tumors as foreign or self? Annu. Rev. Immunol. 21, 807–839 [DOI] [PubMed] [Google Scholar]
- 109.Xue W., Zender L., Miething C., Dickins R. A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S. W. (2007) Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Margolin A. A., Nemenman I., Basso K., Wiggins C., Stolovitzky G., Dalla Favera R., Califano A. (2006) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7, Suppl. 1, S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Friedman N., Linial M., Nachman I., Pe'er D. (2000) Using Bayesian networks to analyze expression data. J. Comput. Biol. 7, 601–620 [DOI] [PubMed] [Google Scholar]
- 112.Hartemink A. J., Gifford D. K., Jaakkola T. S., Young R. A. (2001) Using graphical models and genomic expression data to statistically validate models of genetic regulatory networks. Pac. Symp. Biocomput. 422–433 [DOI] [PubMed] [Google Scholar]
- 113.Sachs K., Gifford D., Jaakkola T., Sorger P., Lauffenburger D. A. (2002) Bayesian Network Approach to Cell Signaling Pathway Modeling. Sci. STKE 2002, pe38. [DOI] [PubMed] [Google Scholar]
- 114.Sachs K., Perez O., Pe'er D., Lauffenburger D. A., Nolan G. P. (2005) Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 [DOI] [PubMed] [Google Scholar]
- 115.Ciaccio M. F., Wagner J. P., Chuu C. P., Lauffenburger D. A., Jones R. B. (2010) Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat. Methods 7, 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bertelsen M., Sanfridson A. (2005) Inflammatory pathway analysis using a high content screening platform. Assay Drug Dev. Technol. 3, 261–271 [DOI] [PubMed] [Google Scholar]
- 117.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.