Abstract
Plants perceive microorganisms by recognizing microbial molecules known as pathogen-associated molecular patterns (PAMPs) inducing PAMP-triggered immunity (PTI) or by recognizing pathogen effectors inducing effector-triggered immunity (ETI). The hypersensitive response (HR), a programmed cell death response associated with ETI, is known to be inhibited by PTI. Here, we show that PTI-induced HR inhibition is due to direct or indirect restriction of the type III protein secretion system's (T3SS) ability to inject type III effectors (T3Es). We found that the Pseudomonas syringae T3SS was restricted in its ability to inject a T3E-adenylate cyclase (CyaA) injection reporter into PTI-induced tobacco (Nicotiana tabacum) cells. We confirmed this restriction with a direct injection assay that monitored the in planta processing of the AvrRpt2 T3E. Virulent P. syringae strains were able to overcome a PAMP pretreatment in tobacco or Arabidopsis (Arabidopsis thaliana) and continue to inject a T3E-CyaA reporter into host cells. In contrast, ETI-inducing P. syringae strains were unable to overcome PTI-induced injection restriction. A P. syringae pv tomato DC3000 mutant lacking about one-third of its T3E inventory was less capable of injecting into PTI-induced Arabidopsis plant cells, grew poorly in planta, and did not cause disease symptoms. PTI-induced transgenic Arabidopsis expressing the T3E HopAO1 or HopF2 allowed higher amounts of the T3E-CyaA reporter to be injected into plant cells compared to wild-type plants. Our results show that PTI-induced HR inhibition is due to direct or indirect restriction of T3E injection and that T3Es can relieve this restriction by suppressing PTI.
Plants come into contact with a myriad of microorganisms and rely on their innate immune systems to perceive potential microbial infections and induce immune responses. Plant innate immunity can be broadly portrayed as consisting of two major branches, distinguished primarily by their mode of microbe detection. The first branch is activated by extracellular pattern recognition receptors (Boller and Felix, 2009; Nicaise et al., 2009) that perceive broadly conserved molecules called pathogen (microbe)-associated molecular patterns (PAMPs; Medzhitov and Janeway, 1997; Ausubel, 2005). The response induced by this recognition is termed PAMP-triggered immunity (PTI; Jones and Dangl, 2006). A well-characterized example of PTI in plants is the recognition of and subsequent immune response to a small N-terminal region of bacterial flagellin by the FLS2 receptor kinase of Arabidopsis (Arabidopsis thaliana; Felix et al., 1999; Zipfel et al., 2004). Plant resistance (R) proteins activate the second branch of the plant innate immune system by recognizing specific pathogen effector proteins. The response induced by this recognition is termed effector-triggered immunity (ETI; Jones and Dangl, 2006). ETI and PTI induce similar innate immune responses, including ion fluxes, reactive oxygen species (ROS), and callose (β-1,3-glucan) deposition in the cell wall (Tsuda et al., 2008; Boller and Felix, 2009); however, ETI generally also includes the induction of a programmed cell death called the hypersensitive response (HR; Heath, 2000).
The induction of ETI in response to a bacterial plant pathogen is generally due to the recognition of bacterial type III effector (T3E) proteins injected into the plant cell by the pathogen's type III protein secretion system (T3SS; Alfano and Collmer, 1997; Buttner and He, 2009). These recognized T3Es were classically known as avirulence (Avr) proteins because they induced ETI responses sufficient to prevent a normally virulent pathogen from causing disease, thereby rendering it avirulent (Leach and White, 1996). However, it has become increasingly apparent that many T3Es benefit their bacteria by suppressing PTI and ETI (Block et al., 2008; Cui et al., 2009; Guo et al., 2009). Under the current model, plants first developed PTI to reduce microbial colonization of the apoplast. Successful bacterial pathogens countered this by acquiring a T3SS and PTI-suppressing T3Es (Espinosa and Alfano, 2004; Chisholm et al., 2006; Jones and Dangl, 2006).
The bacterial pathogen Pseudomonas syringae infects the aerial parts of many plant species. It displays host specificity, and its strains have been separated into more than 50 pathovars based on the host plants that they infect. For example, P. syringae pv tabaci is virulent in tobacco (Nicotiana tabacum), but it triggers nonhost resistance in Arabidopsis, a plant-microbe interaction referred to as a nonhost interaction. Nonhost resistance describes the resistance observed when all members of a plant species are resistant to a specific pathogen (Thordal-Christensen, 2003; Mysore and Ryu, 2004). While not well understood, both PTI (Li et al., 2005) and ETI (Nissan et al., 2006; Wei et al., 2007) have been shown to play a role in nonhost resistance to bacterial pathogens. In some cases, P. syringae strains display race cultivar resistance. This is generally due to the resistant cultivar possessing an R protein that can recognize a T3E from the pathogen inducing ETI (Bent and Mackey, 2007). One well-studied P. syringae strain is P. syringae pv tomato DC3000, which causes bacterial speck disease on specific tomato (Solanum lycopersicum) cultivars and disease on all ecotypes of Arabidopsis tested. These interactions have been classically referred to as compatible interactions. However, DC3000 triggers nonhost resistance in tobacco and many other plants.
DC3000 contains more than 30 T3Es (Lindeberg et al., 2006; Cui et al., 2009; Cunnac et al., 2009). These are encoded by genes contained within the Hrp pathogenicity island, which also encodes the T3SS apparatus (Alfano et al., 2000), other pathogenicity islands, or as individual genes throughout the genome of DC3000 (Buell et al., 2003; Wei et al., 2007). One molecular tool that has been useful in studying the effect individual T3Es have on plants is the cosmid pHIR11 (Huang et al., 1988). This cosmid encodes a functional T3SS from P. syringae pv syringae 61 and the T3E HopA1. It confers upon nonpathogenic bacteria, such as Pseudomonas fluorescens, the ability to inject HopA1 into plant cells. In tobacco and other plants, injected HopA1 induces ETI, including an HR (Huang et al., 1988; Alfano et al., 1997). The expression of other T3Es in P. fluorescens(pHIR11) enabled them to be screened for the ability to suppress HopA1-induced ETI (Jamir et al., 2004; Guo et al., 2009). Bacterial strains carrying the pHIR11 derivatives pLN18 or pLN1965, both of which lack hopA1 and so no longer induce ETI, were used to determine which T3Es could suppress PTI (Oh and Collmer, 2005; Guo et al., 2009). Collectively, these experiments demonstrated that many P. syringae T3Es possessed the ability to suppress both ETI and PTI.
One PTI suppression assay using P. fluorescens(pLN18) employed by Oh and Collmer (2005) took advantage of earlier observations indicating that PTI could inhibit the ability of the plant to mount an HR in response to an ETI-inducing bacterial strain (Newman et al., 2000; Klement et al., 2003). In this assay, the PTI inducers P. fluorescens(pLN18) or a 22-amino-acid peptide from flagellin (flg22) are infiltrated into Nicotiana benthamiana. Six hours later, the ETI inducer DC3000 is infiltrated in a region of the leaf that overlaps with the earlier infiltration. The HR is typically inhibited in the overlapping region that was pretreated with a PTI inducer. Several T3Es suppressed this inhibition when they were separately delivered at time of pretreatment (Oh and Collmer, 2005). It has been speculated that the probable mechanisms for inhibition of the HR caused by PTI include impairment of delivery of T3Es that induce the HR, modification of the events downstream of T3E recognition, or a shutdown of programmed cell death (Newman et al., 2000).
Here, we show that PTI inhibits the HR on tobacco because it directly or indirectly restricts the ability of P. fluorescens(pLN1965) or DC3000 to inject T3Es based on injection (translocation) assays using T3E-adenylate cyclase (CyaA) fusions. This was confirmed using an independent injection assay that monitored the amount of the cleaved in planta form of the T3E AvrRpt2. Interestingly, this injection restriction was greatly reduced in the compatible interactions between DC3000 and Arabidopsis or between P. syringae pv tabaci 11258 and tobacco. A DC3000 mutant lacking four clusters of T3E genes, which corresponds to 11 T3Es, was less able to inject a T3E-CyaA fusion into PTI-induced Arabidopsis, suggesting that the PTI suppressing activities of the T3E inventory of DC3000 allow it to overcome the injection restriction. Transgenic Arabidopsis plants separately expressing specific T3Es known to be capable of PTI suppression increased the ability of P. fluorescens(pLN1965) to inject a T3E-CyaA fusion into PTI-induced plant cells. Collectively, these data suggest that PTI can directly or indirectly restrict type III injection and PTI suppression by T3Es can relieve this restriction in susceptible plant cells but not plant cells undergoing ETI.
RESULTS
PTI Inhibits the P. fluorescens(pHIR11)-Dependent HR on Tobacco
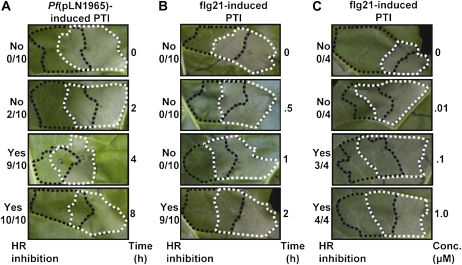
To explore the mechanism underlying PTI inhibition of the HR (Newman et al., 2000; Klement et al., 2003; Oh and Collmer, 2005), we first needed to define the conditions necessary for it to occur in our system. To cause PTI, we used two well-established PTI inducers: one was the nonpathogenic bacterial strain P. fluorescens(pLN1965) (Hauck et al., 2003; Guo et al., 2009), and the other was a 21-amino-acid peptide (flg21) from the N terminus of flagellin (Felix et al., 1999). To cause an HR, we used P. fluorescens(pHIR11), which encodes a functional T3SS, and the T3E HopA1 that elicits an HR on tobacco (Huang et al., 1988). To test HR inhibition, the PTI inducer (pretreatment) was infiltrated into a N. tabacum cv Xanthi (tobacco) leaf. After the specified time interval, the HR inducer was infiltrated into a partially overlapping region of the same leaf. Presence or absence of HR was scored in the overlapping region 48 h after the second infiltration. PTI induction by infiltration of P. fluorescens(pLN1965) at a cell density of 3 × 108 cells/mL at 4 h but not at 2 h before infiltration of P. fluorescens(pHIR11) was sufficient to inhibit the HR (Fig. 1A). Similarly, infiltration with 1 μm flg21 at 2 h but not at 1 h to induce PTI was sufficient to inhibit the pHIR11-dependent HR (Fig. 1B). In addition, a 2-h pretreatment with 0.1 μm flg21 but not with 0.01 μm flg21 could inhibit the pHIR11-dependent HR (Fig. 1C). We used the pretreatment times, bacterial cell densities, and flg21 concentrations established in these experiments for the experiments described below.
Figure 1.
PTI induction times and flagellin concentrations that inhibit the P. fluorescens(pHIR11)-dependent HR in tobacco. PTI was induced by infiltration with 3 × 108 cells/mL of P. fluorescens(pLN1965) or various flg21 concentrations (indicated by black dashed lines). After the specified period, an overlapping infiltration of 3 × 108 cells/mL of the HR-inducing strain P. fluorescens(pHIR11) (indicated by white dashed lines) was performed. The presence or absence of HR in the overlapping region (bordered by white and black dashed lines) was evaluated 48 h after P. fluorescens(pHIR11) infiltration. A, Tobacco leaves were infiltrated with P. fluorescens(pHIR11) 0, 2, 4, or 8 h after having been infiltrated with P. fluorescens(pLN1965). B, Tobacco leaves were infiltrated with P. fluorescens(pHIR11) 0, 0.5, 1, or 2 h after having been infiltrated with 1 μm flg21. C, Tobacco leaves were infiltrated with 0, 0.01, 0.1, or 1 μm of flg21 2 h prior to infiltration with P. fluorescens(pHIR11). The fraction to the left of each image indicates the number of times that the HR was inhibited over the total number of times the assay was performed.
PTI Restricts T3E Injection
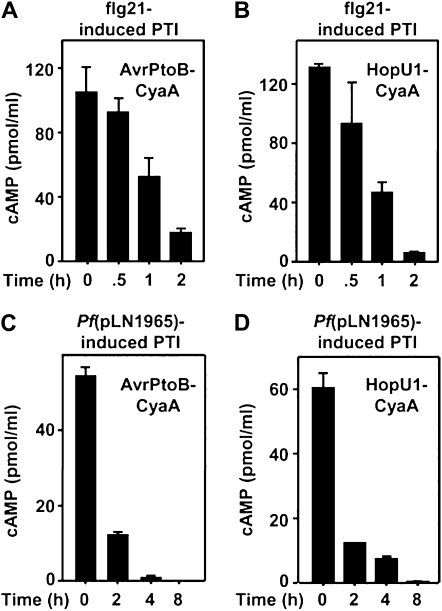
One possible reason why the HR is inhibited by PTI is that the T3Es causing ETI are not entering the plant cells. This could be due to a direct block of the process or an indirect effect on bacterial physiology such that they can no longer inject T3Es. To test whether T3Es were injected into PTI-induced plant cells, we used a CyaA injection (translocation) assay (Sory et al., 1995). This assay determines whether a CyaA fusion protein is injected into eukaryotic cells because CyaA’s ability to produce cAMP is dependent on calmodulin, a protein only present in significant amounts in eukaryotic cells. If a substantial amount of cAMP is detected, it indicates that the CyaA fusion protein was injected. At defined time points after infiltration of the PTI inducer [flg21 or P. fluorescens(pLN1965)] into tobacco, we infiltrated P. fluorescens(pLN1965) carrying an additional construct that encoded the AvrPtoB or HopU1 T3E fused to CyaA. Construct pLN1965 encodes a functional P. syringae T3SS and enables P. fluorescens to inject T3Es. As shown in Figure 2, pretreatment with either PTI inducer greatly restricted the injection of AvrPtoB-CyaA or HopU1-CyaA into tobacco cells as demonstrated by the low levels of cAMP. Importantly, we were unable to detect changes in cAMP levels in plants transiently expressing cyaA after PAMP treatment, indicating that PTI does not directly affect CyaA activity (Supplemental Fig. S1). Moreover, the decrease in cAMP levels in PTI-induced tobacco occurs in the same time frame as HR inhibition, consistent with HR inhibition being due to direct or indirect restriction of T3E injection.
Figure 2.
PTI restricts injection of T3E-CyaA fusion proteins into tobacco cells. PTI was induced by infiltration with 3 × 108 cells/mL P. fluorescens(pLN1965) or 1 μm flg21 at the indicated times prior to an overlapping infiltration of 3 × 108 cells/mL P. fluorescens(pLN1965) expressing AvrPtoB-CyaA or HopU1-CyaA. The level of T3E injection was determined by quantifying cAMP in the overlapping infiltration area 16 h later. cAMP levels in tobacco leaves infiltrated with P. fluorescens(pLN1965 + pavrPtoB-cyaA) pretreated with flg21 (A), P. fluorescens(pLN1965 + phopU1-cyaA) pretreated with flg21 (B), P. fluorescens(pLN1965 + pavrPtoB-cyaA) pretreated with P. fluorescens(pLN1965) (C), or P. fluorescens(pLN1965 + phopU1-cyaA) pretreated with P. fluorescens(pLN1965) (D). Reduced cAMP levels in PTI-induced plant cells show that PTI restricts T3E injection. Standard error bars are shown, and each experiment was repeated three times with similar results.
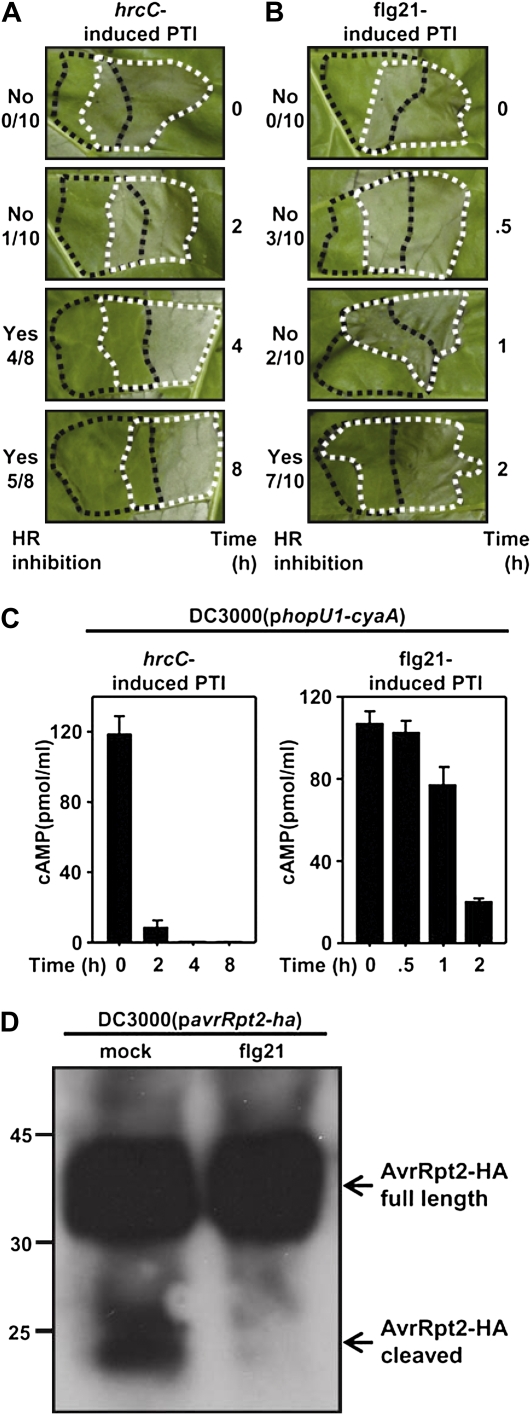
The ability of the nonpathogenic bacterial strain P. fluorescens(pLN1965) to inject T3E-CyaA fusions in tobacco was clearly restricted by the induction of PTI. We next sought to determine the extent that the pathogenic bacterial strain P. syringae pv tomato DC3000 was blocked in its ability to elicit an HR and inject a T3E-CyaA fusion into PTI-induced tobacco plant cells. The PTI inducers used were flg21 and a DC3000 hrcC mutant, which is defective in type III secretion (Hauck et al., 2003; Guo et al., 2009). DC3000 induces nonhost resistance on tobacco and normally causes an HR within 24 h. Using HR inhibition assays, we found that a 2-h pretreatment with flg21 or a 4-h pretreatment with the hrcC mutant were sufficient to prevent the DC3000-induced HR on tobacco (Fig. 3, A and B). In conjunction with this, the ability of DC3000 to inject HopU1-CyaA was also strongly inhibited by a 2-h pretreatment of flg21 or the hrcC strain (Fig. 3C). Thus, PTI also directly or indirectly blocks the ability of DC3000 to inject T3Es into tobacco cells, indicating that this phenotype is not limited to the P. fluorescens(pLN1965) injection system. One explanation for this result is that PTI kills the bacterial cells in the apoplast. To test this, bacterial test strains were infiltrated into tobacco leaves at various time points and cell densities after PTI induction. We were unable to detect any reduction in the number of bacterial cells in PTI-induced plants compared to control plants, suggesting that PTI was not inhibiting the HR or restricting injection simply by causing bacterial cell death (Supplemental Fig. S2). These data suggest that a PTI-induced restriction of injection is responsible for the HR inhibition phenotype. One could envision that reduced T3E injection into tobacco cells would lead to insufficient delivery of recognized T3Es and therefore prevent induction of ETI responses, including the HR.
Figure 3.
The ability of DC3000 to elicit an HR and inject T3E-CyaA fusions in tobacco is inhibited by PTI. A, PTI was induced in tobacco leaves by infiltration with 3 × 108 cells/mL of the DC3000 hrcC mutant 0, 2, 4, or 8 h prior to an overlapping infiltration with 2 × 107 cells/mL of the wild-type DC3000 and scored for HR inhibition. The fraction to the left of each image indicates the number of times that the HR was inhibited over the total number of times the assay was performed. B, PTI was induced in tobacco leaves by infiltration of 1 μm flg21 0, 0.5, 1, or 2 h prior to infiltration with 2 × 107 cells/mL DC3000 and scored for HR inhibition. C, The level of injection was determined by measuring cAMP in tobacco 7 h after infiltration with 3 × 108 cells/mL of DC3000(phopU1-cyaA) in plants pretreated with hrcC or flg21 at the times indicated. D, AvrRpt2-HA is cleaved only when present inside plant cells. PTI was induced in tobacco leaves with a 1 μm flg21 treatment prior to infiltration of DC3000 containing a construct that encodes AvrRpt2-HA. In a water (mock) treatment control cleavage of AvrRpt2-HA can be detected with anti-HA antibodies, but no or reduced amounts of cleaved AvrRpt2-HA can be detected in PTI-induced tobacco tissue. Molecular mass markers in kilodaltons are indicated at the left. PTI induction inhibited the HR and the ability of DC3000 to inject HopU1-CyaA or AvrRpt2-HA into tobacco cells. Each experiment was repeated at least three times with similar results. Standard error bars are shown when appropriate.
PTI Restricts the Injection of AvrRpt2 as PAMP Pretreatment Greatly Reduced the Amount of in Planta AvrRpt2 Cleavage
To confirm that PTI restricted T3E injection using an independent assay, we developed a direct assay to monitor the injection of a T3E after PTI induction. The P. syringae T3E AvrRpt2 has been shown to be processed once it is injected into plant cells by the P. syringae T3SS (Mudgett and Staskawicz, 1999). Wild-type DC3000 was transformed with a construct that encoded AvrRpt2 fused to a hemagglutinin (HA) tag at its C terminus. This strain was infiltrated into tobacco leaf tissue pretreated with flg22 or a mock control. After 6 h, crude plant samples were isolated and subjected to SDS-PAGE and immunoblot analysis. Cleaved AvrRpt2-HA was detected in the mock-treated leaf tissue samples, and greatly reduced amounts of cleaved AvrRpt2-HA were detected in the flg21-pretreated samples infiltrated with DC3000 (Fig. 3D). These results indicate that AvrRpt2 is not injected into PTI-induced plant cells and confirm the findings of the CyaA injection assay. Because the CyaA injection assay data are more easily quantifiable, we chose to use this assay to further define this phenomenon.
DC3000 But Not P. fluorescens(pLN1965) Can Overcome PTI-Induced Injection Restriction in Arabidopsis
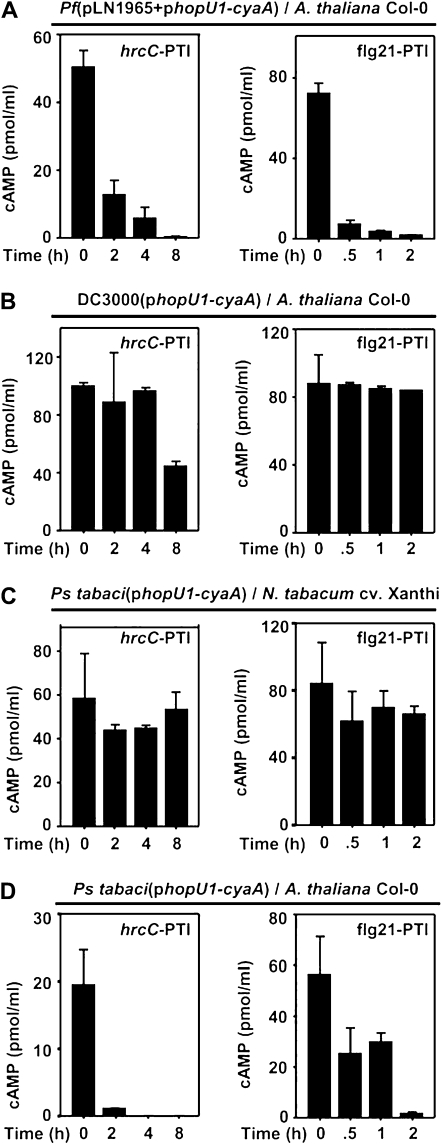
We next sought to determine if this injection restriction also occurred in Arabidopsis. To do this, we tested if the PTI inducers flg21 and hrcC could prevent P. fluorescens(pLN1965) or DC3000 injecting the HopU1-CyaA fusion into Arabidopsis cells. As in tobacco, pretreatment with flg21 or the hrcC mutant reduced P. fluorescens(pLN1965) injection of the HopU1-CyaA fusion into Arabidopsis cells (Fig. 4A). This result was somewhat expected because tobacco and Arabidopsis are known to induce PTI in response to PAMP treatments (Felix et al., 1999), and these plants respond similarly to infiltration of the P. fluorescens(pLN1965) injection system. Interestingly, when we performed similar experiments using DC3000 to inject HopU1-CyaA into Arabidopsis cells, plant leaf tissue contained high amounts of cAMP at all time points, indicating that the HopU1-CyaA fusion continued to be injected by DC3000 after PTI induction (Fig. 4B). These data suggest that in compatible interactions such as that of DC3000 with Arabidopsis, but not in incompatible or nonhost interactions, the bacteria can overcome PTI-induced injection restriction.
Figure 4.
PTI-induced injection restriction occurs in tested P. syringae nonhost interactions but not in compatible interactions. PTI was induced with 3 × 108 cells/mL of the hrcC mutant (left panels) or 1 μm flg21 (right panels). At the times indicated, 3 × 108 cells/mL of bacterial strains expressing HopU1-CyaA were infiltrated into an overlapping area, and cAMP amounts were determined. The following bacteria/plant combinations were used to assess injection capability: A, P. fluorescens(pLN1965 + phopU1-cyaA)/Arabidopsis Col-0; B, DC3000(phopU1-cyaA)/Arabidopsis Col-0, a compatible interaction; C, P. syringae pv tabaci 11258(phopU1-cyaA)/N. tabacum cv Xanthi, a compatible interaction; and D, P. syringae pv tabaci 11258(phopU1-cyaA)/Arabidopsis Col-0, a nonhost interaction. PTI induction severely restricted the ability of bacterial strains in nonhost plants to inject the HopU1-CyaA fusion into plant cells based on the reduced levels of cAMP. However, bacterial strains in host plants were capable of relatively high levels of HopU1-CyaA injection irrespective of PTI induction. Each experiment was repeated at least three times with similar results, and se bars are shown.
PTI Induction Restricts Injection by P. syringae into Nonhost Plant Cells
To determine the extent that host or nonhost interactions influenced the PTI-induced restriction of T3E injection, we electroporated the construct encoding HopU1-CyaA into P. syringae pv tabaci 11258, which has a nonhost interaction with Arabidopsis and a compatible interaction with its host tobacco. In stark contrast to DC3000, this strain was capable of injecting HopU1-CyaA into PTI-induced tobacco cells (Fig. 4C) but not into PTI-induced Arabidopsis cells (Fig. 4D). Normalization of the cAMP levels to the growth of these bacterial strains in Arabidopsis and tobacco within the 16 h time frame of the cAMP measurements did not significantly change these conclusions (Supplemental Fig. S3). Collectively, these data suggest that in compatible interactions, the initial low amount of T3Es injected is capable of suppressing PTI such that normal levels of T3E injection can be achieved. In nonhost interactions, the bacterial T3Es are incapable of suppressing PTI sufficiently to allow normal injection, perhaps because T3Es can be recognized by R proteins and induce ETI, reinforcing the injection restriction or at least preventing T3Es from suppressing PTI. In support of this, several DC3000 T3Es when expressed transiently in tobacco using Agrobacterium tumefaciens induce ETI (Supplemental Fig. S4).
A DC3000 Poly-T3E Mutant Loses the Ability to Inject T3Es into PTI-Induced Arabidopsis and Does Not Grow Well or Cause Disease Symptoms in Planta
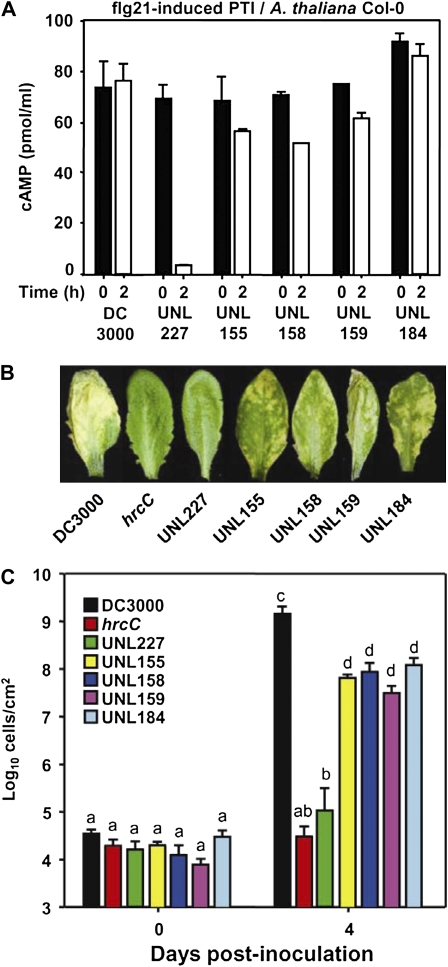
To determine whether T3Es were allowing DC3000 to continue to inject into its susceptible host Arabidopsis despite PTI induction, we made a series of DC3000 mutants lacking different subsets of T3Es. We deleted DNA clusters of T3E genes from DC3000 using a homologous recombination approach that relies on yeast Flp recombinase to act on introduced Flp recombinase target sequences resulting in unmarked mutations (House et al., 2004; Choi and Schweizer, 2005). One DNA cluster we deleted was the exchangeable effector locus (EEL) from the Hrp pathogenicity island (Alfano et al., 2000) resulting in DC3000 mutant UNL155. This mutant lacks the T3E gene hopB1 (Petnicki-Ocwieja et al., 2005). The other T3E gene-containing DNA clusters were pathogenicity islands distributed around the DC3000 genome (Buell et al., 2003), which we gave the following temporary names: Effector pathogenicity island 1 (EPai1) that contains hopD1, hopQ1-1, and hopR1; EPai2 that contains hopAA1-2, hopV1, hopAO1, hopG1, and hopQ1-2; and EPai3 that contains hopF2 and hopU1. DC3000 mutants lacking EPai1, EPai2, or EPai3 were named UNL158, UNL159, and UNL184, respectively. Additionally, we made a DC3000 quadruple mutant lacking the EEL, EPai1, EPai2, and EPai3, which was named UNL227. All of the mutants were confirmed using PCR using primers adjacent to the deleted DNA clusters (Supplemental Fig. S5).
We tested the extent that the DC3000 T3E mutants were altered in their ability to inject a HopU1-CyaA fusion into PTI-induced Arabidopsis plants compared to wild-type DC3000. The DC3000 mutants lacking different T3E gene (UNL155, UNL158, UNL159, and UNL184) were subtly affected in T3E injection but all retained the ability to inject HopU1-CyaA (Fig. 5A). However, the DC3000 quadruple mutant UNL227 was greatly reduced in its ability to inject HopU1-CyaA into PTI-induced Arabidopsis plants based on low cAMP levels in infiltrated Arabidopsis tissue (Fig. 5A). The growth differences between the DC3000 and UNL227 could not account for the different cAMP levels (Supplemental Fig. S3). UNL227 is lacking 11 T3Es normally present in DC3000, which accounts for about one-third of its T3E inventory. This clearly prevented UNL227 from being able to suppress PTI sufficiently to inject T3Es in PTI-induced Arabidopsis. We performed in planta growth assays with the different DC3000 mutants and found that the DC3000 single cluster mutants were only slightly reduced in disease symptoms and in their ability to grow in planta (Fig. 5, B and C). Surprisingly, the DC3000 quadruple mutant UNL227 was dramatically affected in its ability to cause disease symptoms and grow in plants (Fig. 5, B and C). It is remarkable considering the number of extant T3Es that the in planta growth of UNL227 is reduced to a level similar to a DC3000 hrcC mutant, which has a defective T3SS and grows poorly in plants. Additionally, we performed in planta growth assays using wild-type DC3000, a DC3000 hrcC mutant, and the UNL227 mutant on Arabidopsis plants pretreated with flg21 for 2 or 24 h to determine if the flg21 pretreatments had an effect on the ability of these strains to grow in planta. The 2-h flg21 pretreatment did not alter the ability of these strains to grow in Arabidopsis plants (Supplemental Fig. S6). In contrast, a 24-h pretreatment altered the ability of DC3000 to grow in Arabidopsis plants (Supplemental Fig. S6), consistent with earlier reports (Zipfel et al., 2004; Li et al., 2005; Hann and Rathjen, 2007; Guo et al., 2009). These data support the hypothesis that T3Es of virulent bacterial pathogens are responsible for mitigating the PTI-induced injection restriction.
Figure 5.
A DC3000 polyeffector mutant loses its ability to inject T3Es into PTI-induced Arabidopsis. A, Infiltrations of 3 × 108 cells/mL of wild-type DC3000, DC3000 EEL mutant (UNL155), DC3000 effector pathogenicity island (EPai) mutants (UNL158, UNL159, and UNL184), and a DC3000 quadruple mutant (UNL227) each expressing HopU1-CyaA were infiltrated into Arabidopsis after a 0- or 2-h pretreatment with 1 μm flg21, and the production of cAMP was determined. DC3000 single mutants were differentially affected but retained their ability to inject the HopU1-CyaA fusion; however, the DC3000 quadruple mutant was restricted in its T3E injection after PTI induction based on low cAMP levels. B, The virulence of wild-type DC3000, the DC3000 hrcC mutant, the DC3000 single EPai mutants, and the DC3000 quadruple mutant were compared by spray inoculating them at 2 × 107 cells/mL onto untreated Arabidopsis. Virulence was assessed by disease symptoms at 4 d postinoculation (B) and enumeration of bacteria at 0 and 4 d postinfiltration (C). Letters “a” to “d” are statistically different (P < 0.05), and se bars are shown. These data indicate that the quadruple mutant loses the ability to inject T3Es in PTI-induced Arabidopsis, cannot produce disease symptoms, and grows poorly in plant tissue.
PTI Restricts T3E Injection from an ETI-Inducing P. syringae Strain
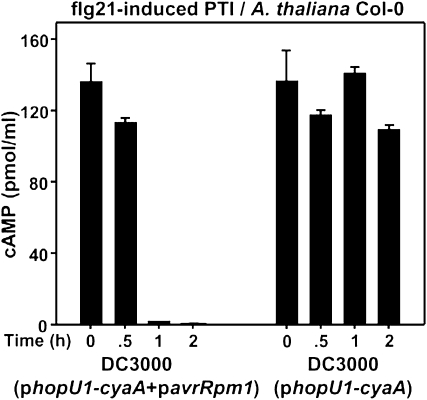
The normally virulent DC3000 strain on Arabidopsis can be converted to an ETI-inducing strain (i.e. avirulent strain) by the introduction of a T3E that is recognized by an Arabidopsis R protein. The T3E AvrRpm1 is recognized by the RPM1 R protein present in Arabidopsis Columbia-0 (Col-0; Grant et al., 1995) resulting in ETI induction. DC3000 carrying avrRpm1 cannot cause disease on Arabidopsis Col-0 due to this race cultivar resistance. We wanted to determine the effect that ETI induction had on the ability of DC3000 to suppress PTI-induced injection restriction. Arabidopsis Col-0 leaves were pretreated with flg21, and at different time points, DC3000(phopU1-cyaA) with or without an avrRpm1 construct was infiltrated into the same leaf regions. DC3000 expressing AvrRpm1 was less able to inject HopU1-CyaA than the DC3000 strain, indicating that AvrRpm1-dependent ETI contributed to the PTI-induced T3E injection restriction (Fig. 6).
Figure 6.
PTI-induced injection restriction occurs in Arabidopsis when DC3000 carries the avirulence gene avrRpm1. PTI was induced with 1 μm flg21. At the times indicated, 3 × 108 cells/mL of DC3000 strains expressing HopU1-CyaA and with or without AvrRpm1 were infiltrated into Arabidopsis Col-0 leaves, and cAMP amounts were determined. T3E injection was severely restricted when DC3000 expressed AvrRpm1, suggesting that ETI contributed to T3E injection restriction. The experiment was repeated three times with similar results, and se bars are shown.
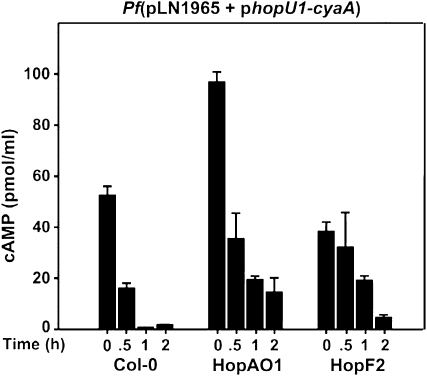
Transgenic Expression of the T3E HopAO1-HA or HopF2-HA in Arabidopsis Relieves PTI-Induced Injection Restriction
We used the P. fluorescens(pLN1965) injection system to determine the amount of the HopU1-CyaA reporter injected into transgenic Arabidopsis plants expressing HopAO1-HA or HopF2-HA, two T3Es that have been shown to suppress PTI (Oh and Collmer, 2005; Underwood et al., 2007; Guo et al., 2009). After 1- or 2-h flg21 pretreatments, both transgenic lines allowed increased amounts of HopU1-CyaA to be injected compared to wild-type controls (Fig. 7). These results show that the PTI suppression activities of at least two T3Es from DC3000 are independently sufficient to partially relieve PTI-induced injection restriction in Arabidopsis. These data in conjunction with the mutant studies above suggest that the combined PTI suppression activity in Arabidopsis of multiple T3Es is required for the complete removal of the injection restriction that is observed in DC3000.
Figure 7.
Transgenic expression of PTI-suppressing T3Es in Arabidopsis relieves PTI-induced injection restriction. Arabidopsis wild-type plants and Arabidopsis transgenic plants expressing the DC3000 T3Es HopAO1-HA or HopF2-HA were pretreated with 1 μm flg21 for indicated time periods and infiltrated with 3 × 108 cells/mL of P. fluorescens(pLN1965) expressing HopU1-CyaA. The higher amounts of cAMP in PTI-induced transgenic plants compared to the wild-type control indicated that these T3Es can relieve PTI-induced injection restriction. This experiment was repeated at least three times with similar results, and se bars are shown.
flg21-Induced PTI Inhibits the HR and Restricts Type III Injection for a Shorter Time Period Than hrcC-Induced PTI
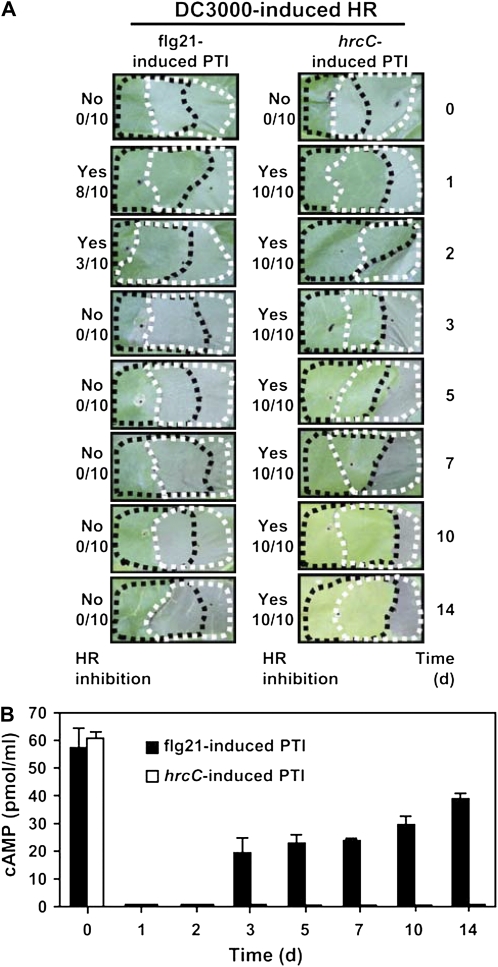
To characterize the persistence of the injection restriction and HR inhibition following PTI induction in tobacco, we performed flg21 and hrcC pretreatments at defined times during a 2-week period before HR induction with DC3000 or injection monitoring with DC3000(phopU1-cyaA). Tobacco pretreated with flg21 inhibited the HR and restricted injection through 2 d. Somewhat surprisingly, the hrcC mutant inhibited the HR and restricted injection through the entire 2-week period (Fig. 8). It is likely that the flg21 PAMP is cleared from the apoplast by endocytosis with FLS2 (Robatzek et al., 2006) and suggests that this process requires 2 d to complete when flg21 is present in the apoplast at 1 μm. The persistent inhibition caused by the hrcC-induced PTI is probably due to the continual lysis of hrcC bacterial cells and release of PAMPs during the 2-week period, preventing the PAMPs from being effectively cleared from the apoplast.
Figure 8.
PTI induced by hrcC inhibits the HR and restricts the injection longer than PTI-induced by flg21. A, PTI was induced in tobacco leaves by infiltration with 1 μm flg21 or 3 × 108 cells/mL of hrcC 0, 1, 2, 3, 5, 7, 10, and 14 d prior to an overlapping infiltration with 2 × 107 cells/mL of DC3000. Pictures were taken 48 h after DC3000 infiltration. The fraction to the left of each image indicates the number of times that the HR was inhibited over the total number of times the assay was performed. B, PTI was induced in tobacco leaves by infiltration with 1 μm flg21 or 3 × 108 cells/mL of hrcC 0, 1, 2, 3, 5, 7, 10, and 14 d prior to an overlapping infiltration with 3 × 108 cells/mL of DC3000(hopU1-cyaA). cAMP levels were measured 7 h after DC3000(hopU1-cyaA) infiltration. PTI induced by flg21 no longer inhibited the HR or restricted T3E injection after 2 d, whereas PTI induced by the hrcC mutant inhibited the HR and restricted injection throughout the 14-d experiment. These experiments were repeated twice with similar results, and se bars are shown.
DISCUSSION
We show here that the PTI-induced inhibition of the HR occurs within 2 h of PAMP pretreatment (Fig. 1). This a reasonably quick response relative to the 12 to 24 h generally required for production of an HR. However, PTI-induced inhibition of the HR appears to require de novo transcription and translation as it was previously reported that the protein synthesis inhibitor cycloheximide restores the HR (Klement et al., 2003). Interestingly, HR inhibition induced by flg21 lasted through 2 d, while HR inhibition induced by intact bacteria persisted over a 2-week period. This difference probably reflects the clearing of flg21 peptide from the apoplast by endocytosis of its receptor FLS2 (Robatzek et al., 2006). In contrast, when bacteria are used to induce PTI, PAMPs continue to be released over an extended time period. Plants often cannot perceive PAMPs while they are part of the bacterial cell. For example, flagellin cannot be detected by the FLS2 receptor kinase while the flagellum filament is intact because the recognized part of the protein is on the inner side of the filament tube and not surface exposed (Zipfel and Felix, 2005). Researchers sometimes use PTI inducers interchangeably. Our results indicate that the choice between PTI inducers flg21 (and likely other purified PAMPs) and intact bacteria may be an important consideration depending on the experimental design.
We show that the PTI-induced inhibition of the HR is due to direct or indirect restriction of T3E injection. The reduced amount of T3Es, a subset of which trigger ETI, entering the plant cell is then insufficient to cause the macroscopic HR. It remains unclear if PTI is directly blocking the T3SS apparatus from injecting T3Es or whether it has a more general effect on the bacteria in the apoplast that prevents them from injecting T3Es. For example, it is intriguing to speculate that the cell wall defenses induced by PTI may strengthen the plant cell wall such that the T3SS pilus can no longer traverse it to inject T3Es. However, the time limit on the effectiveness of at least flg21-induced injection restriction indicates that it is of a transitory rather than permanent nature. The early and spatially limited character of the PTI-induced injection restriction suggests that it may involve ROS, perhaps directly killing bacteria in the apoplast. Indeed, a related study showed that PTI-induced tissue infiltrated with GFP-labeled bacteria had dramatically less fluorescence than untreated plant tissue, which the authors concluded was due to the inhibition of bacterial growth caused by PTI (Oh et al., 2010). However, we were unable to detect any significant reduction of bacterial populations in leaf tissue by direct bacterial counting after PTI induction (Supplemental Fig. S2). We think this discrepancy may be due to reduced GFP production in PTI-induced tissue instead of PTI causing bacterial growth inhibition. It remains possible that a PTI response such as ROS production may affect bacterial transcription or translation preventing the T3SS apparatus from being assembled. In support of this, there has been a report that the expression of Erwinia amylovora T3SS-related genes is altered after treatment with bacterial lipopolysaccharide (Minardi, 1995). We are currently testing whether PTI effects the expression of the T3SS apparatus genes in the P. syringae-Arabidopsis pathosystem.
The PTI-induced inhibition of the HR has previously only been reported in the solanaceous plants tobacco, N. benthamiana, and pepper (Capsicum annuum; Newman et al., 2000; Klement et al., 2003; Oh and Collmer, 2005). PTI-induced inhibition of the HR can also be recapitulated in Arabidopsis leaves (Supplemental Fig. S7), although the small leaf size of Arabidopsis makes overlapping infiltrations difficult. We show here that Arabidopsis PTI restricts T3E injection. Both assays should be added to the limited collection of assays that are currently being used to assess PTI.
PTI in tobacco and Arabidopsis was more effective at restricting T3E injection by P. syringae strains in nonhost than in compatible interactions (Fig. 4). This difference was attributed to the PTI-suppressing activity of T3Es since a DC3000 polyeffector mutant lacking about one-third of its T3E inventory was less able to overcome PTI-induced injection restriction on Arabidopsis compared to wild-type DC3000 (Fig. 5A). PTI-induced transgenic Arabidopsis plants separately expressing two DC3000 T3Es known to suppress PTI relieved injection restriction. Collectively, these results suggest that in compatible interactions, T3Es are apparently injected at a low level during PTI-induced injection restriction. This allows the injected T3Es to suppress PTI in susceptible hosts relieving restriction. Importantly, longer PAMP pretreatments beginning at about 16 h begin to severely restrict T3E injection even in the compatible interaction between DC3000 and Arabidopsis (Supplemental Fig. S8). In addition, a 24-h but not a 2-h PAMP pretreatment allows Arabidopsis to restrict the growth of normally virulent DC3000 (Supplemental Fig. S2). These data indicate that whatever mechanism is causing this phenotype is so substantial by 24 h that it cannot be overcome by virulent P. syringae strains.
We found that PTI-induced injection restriction was severe in nonhost plants tobacco and Arabidopsis (Fig. 4). It is possible that in nonhost plants the T3Es have an insufficient ability to suppress PTI; however, the severity of the restriction may be due to the induction of ETI. We did find that DC3000 expressing AvrRpm1 was clearly less able to inject the T3E-CyaA reporter than DC3000 not expressing it (Fig. 6). This suggests that ETI induction prevents DC3000 from delaying T3E injection restriction. The induction of ETI may prevent T3Es from acting to suppress PTI and relieving injection restriction. Alternatively, it is possible that ETI may also evoke or enhance injection restriction. We favor the latter explanation because of the extensive overlap between ETI and PTI responses (Navarro et al., 2004; Tsuda et al., 2008). Additionally, when we compared PTI-induced injection restriction initiated by P. fluorescens(pLN1965) (which induces PTI) and P. fluorescens(pLN1965 + pavrRpm1) (which induces PTI and ETI), we found that injection restriction induced by P. fluorescens(pLN1965 + pavrRpm1) occurred more quickly than when it was induced by P. fluorescens(pLN1965) (Supplemental Fig. S9). One important caveat is that we only investigated specific P. syringae interactions and other P. syringae strains may fail to grow on nonhost plants for reasons other than T3E recognition. For example, flagellin from certain P. syringae pathovars induce an HR on nonhost tobacco (Taguchi et al., 2003). We are currently taking several approaches to determine the extent that ETI, in the absence of PTI, can induce injection restriction.
The PTI-induced inhibition of the HR and injection restriction assays should provide a unique perspective on how PTI affects bacteria in the apoplastic environment. This should facilitate the dissection of the plant immune response in such a manner as to potentially identify the mechanism behind these phenomena. For example, do certain plant mutants defective in specific immune responses relieve PTI-induced injection restriction? If this approach is successful, it may help us better understand one of the long-standing questions in molecular phytobacteriology: What plant immune responses are important for successfully defending against bacterial pathogens.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains, plasmids, and primers are listed in Supplemental Tables S1 and S2. Pseudomonas syringae and Pseudomonas fluorescens strains were grown in King's B (KB) medium at 30°C with appropriate antibiotics. The antibiotics were used at the following concentrations (μg/mL): rifampicin (Rf), 100; gentamicin (Gm), 10; tetracycline (Tc), 10; kanamycin (Km), 50; naldixic acid (Nx), 20; and spectinomycin (Sp), 50. Plasmids used in unmarked mutagenesis were as follows: pRK2013 and pRK2073, mobilizing helper plasmids; pBH474, a Flp recombinase encoding plasmid; pMK2016 and pMK2017, both containing FRT cassettes (House et al., 2004). Plasmids used for plant bioassays were pHIR11 (Huang et al., 1988), a cosmid containing the genes for a functional T3SS and the T3E hopA1, and pLN1965 (Guo et al., 2009), a pHIR11 derivative lacking the hopA1 T3E gene. For translocation assays, constructs encoding AvrPtoB or HopU1 C-terminal CyaA fusions were made by LR reactions between pENTR/D-TOPO constructs (Invitrogen) containing each gene and the Gateway vector pLN2193 (Fu et al., 2006), creating the plasmids pLN2250 and pLN2254, respectively.
Plant Bioassays
Wild-type Arabidopsis (Arabidopsis thaliana; Col-0) and transgenic plants were grown in a growth chamber at 24°C on 10-h days. Nicotiana tabacum cv Xanthi (tobacco) plants were grown in greenhouse conditions. HR inhibition assays were done with infiltrations of fresh overnight cultures resuspended in 5 mm MES at pH 5.5 to 3 × 108 cells/mL for P. fluorescens(pHIR11), P. fluorescens(pLN1965), and hrcC and at 2 × 107 cells/mL for DC3000. flg21 was used at 1 μm concentrations in water unless otherwise indicated. Fully expanded leaves of 4-week-old Arabidopsis or 6-week-old tobacco plants were PTI induced by infiltration with flg21, P. fluorescens(pLN1965), or hrcC using a blunt-ended syringe. Then, the HR elicitor, either DC3000 or P. fluorescens(pHIR11), was infiltrated in an overlapping region of the same leaf 0, 0.5, 1, or 2 h after infiltration with flg21 or 0, 2, 4, or 8 h after infiltration with P. fluorescens(pLN1965) or hrcC. Infiltration outlines were marked with a felt-tipped pen, and the overlapping area was assessed 48 h after HR elicitor infiltration for the presence or absence of an HR.
Plant bacterial growth assays were carried out using fresh bacteria grown on KB plates overnight and resuspended in 5 mm MES. PTI induction and bacterial infiltration of tobacco were carried out as in HR inhibition assays. Bacterial growth in Arabidopsis was done by spray inoculation with 2 × 108 cells/mL. Four leaf discs were harvested for each strain at each time point with a 0.8-cm2 cork borer for tobacco and a 0.4-cm2 cork borer for Arabidopsis. The samples were ground in 250 μL of sterile water, serial dilutions were plated on KB with appropriate antibiotics, and colonies were counted to determine bacterial growth. Statistical differences were calculated using single-factor ANOVA.
CyaA Injection Assay
pLN2250 and pLN2254 were transformed via electroporation into P. syringae pv tomato DC3000, P. syringae pv tabaci 11528, P. fluorescens 55, or DC3000 mutant derivatives, and T3E protein expression was confirmed with immunoblots. The CyaA injection assays were performed following a previously published protocol (Schechter et al., 2004). Briefly, tobacco or Arabidopsis were initially challenged to induce PTI in the same manner as in other plant bioassays and were subsequently infiltrated with the strain carrying the T3E-cyaA gene fusion suspended at 3 × 108 cells/mL in 5 mm MES. This strain was infiltrated in an area overlapping the initial infiltration using a blunt-ended syringe. Leaf discs (0.9 cm2) were taken from the area of overlapping infiltration 16 h (7 h for DC3000 in tobacco) after the second infiltration. The samples were ground in liquid nitrogen and resuspended in 0.1 m HCl. Samples were adjusted to 10 ng/μL after quantification of total protein using Bradford assay (Bio-Rad). A direct cAMP (cAMP) immunoassay kit (Assay Design) was used to measure cAMP levels following the manufacturer's instructions.
AvrRpt2 in Planta Processing Injection Assay
This assay was modified from Mudgett and Staskawicz (1999) to fit the experimental conditions. pLN2637 was transformed by electroporation into DC3000. The bacteria were grown overnight on KB media containing appropriate antibiotics, collected, and resuspended in 10 mm MgCl2 at a concentration of 1 × 109 cells/mL. Tobacco leaves were infiltrated with either 1 μm flg21 or water (mock) using a blunt-ended syringe and 2 h later were infiltrated in the same tissue with the bacteria. Six hours after bacterial infiltration, leaf samples were taken with a cork borer, ground in liquid N2 using a pestle and microcentrifuge tube, and then resuspended in 1× phosphate buffered saline containing complete protease inhibitor cocktail (Roche). Soluble protein was collected upon centrifugation at 13,000g for 5 min at 4°C. Protein concentrations were determined using the Bradford assay (Bio-Rad), and all samples were normalized to 800 ng/μL total protein. Protein fractions were mixed 1:1 with 2× sample buffer, boiled for 5 min, and centrifuged at 13,000g for 3 min, and 20 μg of total protein was analyzed by immunoblotting using anti-HA antibodies (Roche).
Unmarked Mutagenesis
The construction of the DNA cluster deletion mutants was done by unmarked mutagenesis (House et al., 2004). Plasmids and primers are listed in Supplemental Tables S1 and S2, respectively. DNA clusters containing T3E genes were identified in P. syringae pv tomato DC3000 based on the presence of type III promoters using the Artemis Genome Viewer (Sanger Institute). For each DNA cluster, the 2.5-kb upstream (US) DNA sequence and 2.0-kb downstream (DS) DNA sequence were amplified by PCR using the following Gateway-compatible primer sequences: for the EEL, the primers P2443/P2444 and P2445/P2446 were used; for EPai3, the primers P2404/P2405 and P2406/P2407 were used; for EPai1, the primers P2091/P2092 and P2093/P2094 were used; and primers P2097/2098 and P2099/P2100 were used for EPai2. The purified PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen). The entry vectors were then recombined by LR reactions into pMK2017 for US sequences and pMK2016 for DS sequences.
The plasmids containing the US flanking sequences were integrated into DC3000 chromosome using biparental mating with selection on KB plates containing Rf and Tc. Positive colonies were then confirmed for proper integration of the plasmid with PCR using P1790 and the relevant US reverse primer and checked for UV fluorescence, Sp sensitivity, and lack of growth at 37°C. The confirmed single integrant DC3000 mutants were subsequently triparental mated with DH5α containing the DS sequence plasmid and HB101(pRK2013). Colonies were selected on KB plates containing Rf, Sp, and Tc and tested for UV fluorescence and lack of growth at 37°C. Proper integration of both plasmids flanking the targeted DNA cluster was confirmed with PCR using P1790 with the relevant right border reverse primer and P1789 with the relevant left border reverse primer.
Upon confirmation of double integrants, excision of the integrated plasmids at FRT sites was conducted. Triparental mating of the DC3000 double integrant mutants DB3.1(pBH474) and DB3.1(pRK2073) was performed and colonies selected on KB plates containing Rf and Gm. Excision of the integrated plasmids was confirmed by testing for sensitivity to Sp and Tc. Isolated colonies were streaked onto KB plates containing Rf and 5% Suc to select against retention of pBH474. After deletion by the Flp recombinase enzyme, a 0.2-kb FRT scar remained in place of the targeted DNA cluster. As a final measure of proper excision, the DC3000 mutants were confirmed with PCR using primers designed to anneal 0.5 kb within the border region, resulting in a 1.2-kb PCR product.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. CyaA remains functional in PTI-induced tissue.
Supplemental Figure S2. Growth of DC3000(phopU1-cyaA) was not affected by PAMP pretreatment.
Supplemental Figure S3. cAMP production normalized for bacterial growth.
Supplemental Figure S4. Several DC3000 T3Es elicit an HR-like response when their DNAs are delivered with Agrobacterium.
Supplemental Figure S5. Confirmation of DC3000 DNA cluster mutants.
Supplemental Figure S6. The induction of PTI by flg21 at 24 h but not 2 h before inoculation with DC3000 inhibits bacterial growth in Arabidopsis.
Supplemental Figure S7. PTI induction inhibits AvrRpm1-induced HR in Arabidopsis.
Supplemental Figure S8. Time course of the restriction of HopU1-CyaA injection into PTI-induced Arabidopsis plant cells.
Supplemental Figure S9. PTI- and ETI-induced Arabidopsis plants appear to restrict T3E injection more quickly than when PTI only is induced.
Supplemental Table S1. Strains and plasmids used in this study.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank members of the Alfano laboratory for fruitful discussions that led to many of the experiments described in this manuscript.
References
- Alfano JR, Charkowski AO, Deng W, Badel JL, Petnicki-Ocwieja T, van Dijk K, Collmer A. (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA 97: 4856–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A. (1997) The type III (Hrp) secretion pathway of plant pathogenic bacteria: Trafficking harpins, Avr proteins, and death. J Bacteriol 179: 5655–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Kim HS, Delaney TP, Collmer A. (1997) Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in an hrp-dependent manner within tobacco cells. Mol Plant Microbe Interact 10: 580–588 [DOI] [PubMed] [Google Scholar]
- Ausubel FM. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Block A, Li G, Fu ZQ, Alfano JR. (2008) Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol 11: 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D, He SY. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol 150: 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Choi KH, Schweizer HP. (2005) An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5: 30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Xiang T, Zhou JM. (2009) Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol 11: 1453–1461 [DOI] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A. (2009) Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr Opin Microbiol 12: 53–60 [DOI] [PubMed] [Google Scholar]
- Espinosa A, Alfano JR. (2004) Disabling surveillance: bacterial type III secretion system effectors that suppress innate immunity. Cell Microbiol 6: 1027–1040 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Alfano JR. (2006) Pseudomonas syringae HrpJ is a type III secreted protein that is required for plant pathogenesis, injection of effectors, and secretion of the HrpZ1 harpin. J Bacteriol 188: 6060–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR. (2009) The majority of the type III effectory inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann DR, Rathjen JP. (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J 49: 607–618 [DOI] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, He SY. (2003) A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- House BL, Mortimer MW, Kahn ML. (2004) New recombination methods for Sinorhizobium meliloti genetics. Appl Environ Microbiol 70: 2806–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Schuurink R, Denny TP, Atkinston MM, Baker CJ, Yucel I, Hutcheson SW, Collmer A. (1988) Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J Bacteriol 170: 4748–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamir Y, Guo M, Oh H-S, Petnicki-Ocwieja T, Chen S, Tang X, Dickman MB, Collmer A, Alfano JR. (2004) Identification of Pseudomonas syringae type III effectors that suppress programmed cell death in plants and yeast. Plant J 37: 554–565 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Klement Z, Bozso Z, Kecskes ML, Besenyei E, Arnold C, Ott PG. (2003) Local early induced resistance of plants as the first line of defence against bacteria. Pest Manag Sci 59: 465–474 [DOI] [PubMed] [Google Scholar]
- Leach JE, White FF. (1996) Bacterial avirulence genes. Annu Rev Phytopathol 34: 153–179 [DOI] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM. (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. (2006) Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact 19: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA. (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell 91: 295–298 [DOI] [PubMed] [Google Scholar]
- Minardi P. (1995) Altered expression of Erwinia amylovora hrp genes in tobacco leaves pretreated with bacterial protein-lipopolysaccharide complexes. J Phytopathol 143: 199–205 [Google Scholar]
- Mudgett MB, Staskawicz BJ. (1999) Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol Microbiol 32: 927–941 [DOI] [PubMed] [Google Scholar]
- Mysore KS, Ryu CM. (2004) Nonhost resistance: how much do we know? Trends Plant Sci 9: 97–104 [DOI] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, von Roepenack E, Daniels M, Dow M. (2000) Lipopolysaccharides and the plant responses to phytopathogenic bacteria. Mol Plant Pathol 1: 25–31 [DOI] [PubMed] [Google Scholar]
- Nicaise V, Roux M, Zipfel C. (2009) Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol 150: 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan G, Manulis-Sasson S, Weinthal D, Mor H, Sessa G, Barash I. (2006) The type III effectors HsvG and HsvB of gall-forming Pantoea agglomerans determine host specificity and function as transcriptional activators. Mol Microbiol 61: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Oh HS, Collmer A. (2005) Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J 44: 348–359 [DOI] [PubMed] [Google Scholar]
- Oh HS, Park DH, Collmer A. (2010) Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity. Mol Plant Microbe Interact 23: 727–739 [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, van Dijk K, Alfano JR. (2005) The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J Bacteriol 187: 649–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol 186: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sory MP, Boland A, Lambermont I, Cornelis GR. (1995) Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA 92: 11998–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F, Shimizu R, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. (2003) Post-translational modification of flagellin determines the specificity of HR induction. Plant Cell Physiol 44: 342–349 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H. (2003) Fresh insights into processes of nonhost resistance. Curr Opin Plant Biol 6: 351–357 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. (2008) Interplay between MAMP-triggered and SA-mediated defense responses. Plant J 53: 763–775 [DOI] [PubMed] [Google Scholar]
- Underwood W, Zhang S, He SY. (2007) The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J 52: 658–672 [DOI] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51: 32–46 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Felix G. (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8: 353–360 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.