Abstract
Transcription factors of the homeodomain-leucine zipper IV (HD-ZIP IV) family play crucial roles in epidermis-related processes. To gain further insight into the molecular function of OUTER CELL LAYER1 (OCL1), 14 target genes up- or down-regulated in transgenic maize (Zea mays) plants overexpressing OCL1 were identified. The 14 genes all showed partial coexpression with OCL1 in maize organs, and several of them shared preferential expression in the epidermis with OCL1. They encoded proteins involved in lipid metabolism, defense, envelope-related functions, or cuticle biosynthesis and include ZmWBC11a (for white brown complex 11a), an ortholog of AtWBC11 involved in the transport of wax and cutin molecules. In support of the annotations, OCL1-overexpressing plants showed quantitative and qualitative changes of cuticular wax compounds in comparison with wild-type plants. An increase in C24 to C28 alcohols was correlated with the transcriptional up-regulation of ZmFAR1, coding for a fatty acyl-coenzyme A reductase. Transcriptional activation of ZmWBC11a by OCL1 was likely direct, since transactivation in transiently transformed maize kernels was abolished by a deletion of the activation domain in OCL1 or mutations in the L1 box, a cis-element bound by HD-ZIP IV transcription factors. Our data demonstrate that, in addition to AP2/EREBP and MYB-type transcription factors, members of the HD-ZIP IV family contribute to the transcriptional regulation of genes involved in cuticle biosynthesis.
The outer-most cell layer or epidermis represents the interface of sessile land plants with their environment and has the somewhat incompatible roles to provide a protective barrier against hostile biotic or abiotic agents and at the same time to allow the exchange of gas, water, and nutrients with the outside world. The bulk of plant organs are covered by ground epidermal cells such as pavement cells on leaves or rhizodermic cells in the root. They show a certain asymmetry in that the cell wall facing the environment is frequently modified or reinforced (Glover, 2000). In addition, some epidermal cells undergo particular developments to form specialized structures such as trichomes or stomatal guard cells on the aerial parts, root hairs in the root, or the aleurone layer in the seed, which are essential for defense, respiration, nutrition, and starch degradation, respectively (Guimil and Dunand, 2007). Over the past few years, a wealth of knowledge has become available on the differentiation of these specialized epidermal cells, highlighting the importance of cell-cell communication, cell lineage, and the formation of particular transcriptional complexes in triggering specialization (Ishida et al., 2008; Nadeau, 2009). In contrast, little is known about the differentiation of ground epidermal cells. An important step forward was the analysis of the shoot epidermal transcriptome in maize (Zea mays) and Arabidopsis (Arabidopsis thaliana) that highlighted the preponderant role of lipid-related functions in the epidermis. Indeed, genes involved in lipid metabolism, cuticle biosynthesis, or biotic/abiotic stress resistance were more abundantly expressed in shoot epidermal cells than in underlying tissues (Nakazono et al., 2003; Suh et al., 2005).
The control of the differentiation and maintenance of epidermal cell fate involves members of the homeodomain-Leu zipper IV (HD-ZIP IV) family of plant-specific transcription factors (Ariel et al., 2007). These proteins are defined by the presence of four highly conserved domains: a homeodomain (HD) associated with a Leu zipper domain (ZIP), a steroidogenic acute regulatory-related lipid transfer domain (START), and a HD-START-associated domain (Mukherjee and Burglin, 2006). The vast majority of characterized HD-ZIP IV genes have an epidermis-specific expression pattern in a variety of species, including Arabidopsis (Lu et al., 1996; Nakamura et al., 2006), cotton (Gossypium hirsutum; Guan et al., 2008), maize (Ingram et al., 2000), rice (Oryza sativa; Ito et al., 2002), and pine tree (Pinus spp.; Ingouff et al., 2001). Functional data concern almost exclusively the 16 HD-ZIP IV genes identified in the Arabidopsis genome (Nakamura et al., 2006), even though a systematic survey of single mutants revealed detectable phenotypes for only three of them. The glabra2 (gl2) mutant is affected in trichome and root hair development, mucilage deposition, and seed oil content (Rerie et al., 1994; Di Cristina et al., 1996; Shen et al., 2006), the homeodomain glabrous11 (hdg11) mutant in trichome branching (Nakamura et al., 2006), and the anthocyaninless2 (anl2) mutant in anthocyanin distribution and root development (Kubo et al., 1999). A more spectacular phenotype was observed in the Arabidopsis thaliana meristem layer1/protodermal factor2 (atml1/pdf2) double mutant, which fails to differentiate a protoderm during embryogenesis and is embryo lethal (Abe et al., 2003). Little is known about HD-ZIP IV target genes, and only four direct target genes have been identified. PDF1, a gene coding for a Pro-rich protein, is directly regulated by ATML1/PDF2 (Abe et al., 2003), and genes coding for the phospholipase D AtPLDζ1, the cellulose synthase CESA5, and the xyloglucan endotransglucosylase XTH17 are directly regulated by GL2 (Ohashi et al., 2003; Tominaga-Wada et al., 2009). The binding of the HD-ZIP IV proteins to these target gene promoters occurs at an 8-bp cis-element called the L1 box, which is thought to be critical for driving epidermis-specific expression (Abe et al., 2001).
In maize, five of the 17 OUTER CELL LAYER (OCL) genes encoding HD-ZIP IV proteins have been characterized and show an expression pattern restricted to the epidermal or subepidermal layer of various organs (Ingram et al., 2000). Functional data exist for OCL4 involved in anther and trichome development (Vernoud et al., 2009) and OCL1. Dominant negative transgenic lines expressing an OCL1-ENGRAILED fusion show a transient reduction in kernel size, which is possibly caused by a decrease of gibberellin levels (Khaled et al., 2005).
In addition to Arabidopsis and maize, functional data are available in tomato (Solanum lycopersicum), where CUTIN DEFICIENT2 (CD2) is necessary for the biosynthesis of an intact cuticle of the fruit (Isaacson et al., 2009). The cuticle is a protective hydrophobic layer deposited on the external cell wall of epidermal cells in the aerial parts of the plant (Jeffree, 2006). The two major constituents are cutin and waxes. The cutin polymer, a polyester of C16 to C18 fatty acids, represents the structural matrix, which is interspersed and covered by waxes, a mixture of C24 to C34 alcohols, aldehydes, fatty acids, alkanes, ketones, and wax esters (Jenks et al., 2002; Nawrath, 2002; Kunst and Samuels, 2003). Beyond its role in defense (Eigenbrode and Espelie, 1995), and more generally as a mechanical and chemical barrier against biotic and abiotic stress, the plant cuticle is also an efficient means against water loss and sun radiation and allows the control of gas exchanges (Gray et al., 2000; Riederer, 2006). Over the past few years, genetic studies in Arabidopsis have improved our understanding of the enzymatic steps involved in fatty acid elongation and wax biosynthesis (Samuels et al., 2008). In contrast, the mechanisms behind the transport and asymmetric deposition of cuticle components remain poorly understood. For over a decade, many authors hypothesized on the implication of lipid transfer proteins (LTPs) in the transport of cuticular lipids through the cell wall (Kader, 1996). A role of LTPs in cuticle formation has recently been demonstrated by the characterization of mutant Arabidopsis lines lacking LTPG1, which revealed a significant reduction of C29 alkanes in the cuticle (Debono et al., 2009; Lee et al., 2009). Beyond LTPs, there is experimental evidence that the ATP-binding cassette (ABC) transporters ABCG12/CER5 (for ECERIFERUM5) and ABCG11/WBC11 (for WHITE BROWN COMPLEX11) are involved in the transport of wax (CER5) or wax and cutin molecules (WBC11) from their site of synthesis to the cuticle layer (Pighin et al., 2004; Bird et al., 2007).
Here, we provide evidence for a link between the HD-ZIP IV transcription factor OCL1 from maize and certain elements of lipid transport/metabolism, in particular elements needed for cuticle deposition/biosynthesis necessary to make a protective epidermis. We identified 14 direct or indirect target genes of OCL1 and show that the transcriptional activation by OCL1 of a gene coding for an ABC transporter is likely direct and involves an L1 box.
RESULTS
Identification of 11 OCL1 Target Genes by Microarray Analysis
Eleven target genes of the HD-ZIP IV transcription factor OCL1 were identified by a transcriptome comparison between transgenic maize plants overexpressing OCL1 (OCL1-OE) under the control of the strong cassava vein mosaic virus (CsVMV) promoter and wild-type sister plants. RNA was extracted from the aerial parts of plantlets at 18 d after sowing (DAS) and used to hybridize a genome-wide 59 K microarray. A first gene list of 204 differentially expressed genes was established based on P < 0.01 for the biological triplicate and strong expression differences (logR > 2 or < −2). Using a medium to high spot intensity (logI > 0) as an additional criterion, the list was shortened to 35 candidates. The differential expression was confirmed for 11 of the 35 candidate genes by quantitative reverse transcription (qRT)-PCR experiments based on the same samples that had been used for the initial microarray analysis (Table I; Supplemental Table S1).
Table I. Relative expression levels of confirmed OCL1 target genes in OCL1-OE and OCL1-RNAi plants.
| Oligo IDa and/or Gene Name | Trend in OCL1-OE (Plantlet) | Ratio OCL1-OE/Wild Typeb |
Ratio OCL1-RNAi/Wild Typec | Annotationf | Maize Gene Modelg | Class | ||
| 18-DAS Plantlet | Shoot Apexd | Immature Eard | 18-DAS Plantlete | |||||
| OCL1 | Up | 23.80 | 4.00* | 3.46* | 0.51* | Transcription factor HD-ZIP IV family | GRMZM2G026643 | Transcription factor |
| MZ00005958 | Down | 0.18 | 1.50 | 0.00* | 1.38* | Maize indole-3-glycerol phosphate lyase (Igl) | GRMZM2G015892 | Defense |
| MZ00014373 | Down | 0.10 | 0.39* | 0.04* | 1.05 | MtN3/SALIVA-related transmembrane protein | GRMZM2G179349 | Envelope |
| MZ00018561 | Down | 0.38 | 0.57* | 1.87 | 0.99 | Pro-rich protein; structural constituent of cell wall | GRMZM2G345700 | Envelope |
| MZ00022171 | Up | 20.88 | 0.43 | 0.43 | 1.04 | Male sterility MS5 family protein; contains TPR domain | GRMZM2G075563 | Other |
| MZ00024305 LtpII.12 | Up | 4.34 | 1.94* | 2.40* | 0.58* | Nonspecific lipid transfer protein (nsLTP) type 2 | GRMZM2G387360 | Lipid |
| MZ00024414 | Down | 0.09 | 0.65* | 1.52 | 1.23* | MATE efflux family protein | GRMZM2G339488 | Envelope |
| MZ00028617 | Down | 0.04 | 0.61* | 1.61 | 1.30* | Cytochrome P450; oxygen binding; CYP78A6-like | GRMZM2G034471 | Other |
| MZ00029574 | Up | 7.35 | 1.25* | nd | 0.80* | Carboxylesterase; ATCXE18-like | GRMZM2G104141 | Lipid |
| MZ00030315 | Up | 13.51 | 1.55* | 0.18 | 0.55* | Integral membrane family protein; contains DUF588 | GRMZM2G132128 | Envelope |
| MZ00031783 ZmWBC11a | Up | 6.94 | 1.05 | 1.27* | 0.73* | ABC transporter; ABCG11/COF1/DSO/WBC11-like | GRMZM2G308860 | Lipid |
| MZ00031955 | Up | 7.08 | 2.57* | 2.17* | 1.09 | SEC14/phosphoglyceride transfer family protein | GRMZM2G088501 | Lipid |
| ZmFAR1 | Up | 2.78 | nd | nd | 1.21 | Fatty acyl-CoA reductase (alcohol-forming)/oxidoreductase; FAR1-like | GRMZM2G036217 | Lipid |
| ZmWBC11b | Up | 2.69 | nd | nd | 1.00 | ABC transporter; ABCG11/COF1/DSO/WBC11-like | GRMZM2G096952 | Lipid |
| ZmWBC11c | Up | 2.06 | nd | nd | 0.49* | ABC transporter; ABCG11/COF1/DSO/WBC11-like | GRMZM2G143668 | Lipid |
Identification number of the corresponding oligonucleotide deposited on the microarray.
Mean of a biological triplicate and a technical replicate; the expression values are reported relative to one of the wild-type samples.
Mean of a technical triplicate; the expression values are reported relative to the wild-type samples.
Asterisks indicate a trend of differential expression similar to the one observed in OCL1-OE plantlets. nd, Not determined.
Asterisks indicate a trend of differential expression opposite to the one observed in OCL1-OE plantlets.
Manually improved annotations from SwissProt, GenBank, Trembl, and InterPro databases.
Maize genome release 4a.53 of March 8, 2010 (http://www.maizesequence.org).
A survey of two additional organs suggested that OCL1 was not the only regulatory protein influencing the transcription of its target genes and/or that it might interact with different proteins or via different regulatory cascades in dissected shoot apices and in immature ears. Three target genes (MZ00014373, MZ00024305, and MZ00031955) had the same differential trend in all three organs, while in seven other cases the trend was confirmed only in two of the three organs and in one case only in the original 18-DAS plantlets (Table I). Expression differences between OCL1-OE and the wild type were generally lower in the additional organs, reflecting lower ratios for OCL1 itself, which were probably caused by less efficient transcription off the CsVMV promoter in shoot apices and immature ears. Taken together, we identified 11 genes that were either directly or indirectly up-regulated (six genes) or down-regulated (five genes) by OCL1.
Involvement of OCL1 Target Genes in Lipid Metabolism, Lipid Transfer, and/or Plant Defense
To determine whether the 11 confirmed target genes had similar functions, complete protein sequences were assembled, starting from the 70-nucleotide oligonucleotide deposited on the microarray, exploiting the very rich maize EST data (Messing and Dooner, 2006) as well as the recently established draft of the maize genome sequence (Pennisi, 2008). A group of five genes shared annotations related to lipid metabolism or transport and/or plant defense (Table I). Among them, three up-regulated target genes likely encoded lipid transporters, as they were annotated as nonspecific, type 2 lipid transfer protein (MZ00024305; hereafter named ZmLTPII.12 according to Boutrot et al. [2008]), ABC transporter of the WBC11/ABCG11 clade (MZ00031783), and SEC14/phosphatidylinositol transfer protein (MZ00031955, PITP). Another up-regulated gene (MZ00029574) shared highest identity with AtCXE18 encoding an Arabidopsis carboxylesterase hydrolyzing in vitro short-chain acyl esters (Cummins et al., 2007). Finally, the down-regulated (MZ00005958) Indole-3-glycerol phosphate lyase (Igl) gene had previously been shown to be involved in the tritrophic defense of maize against herbivory (Frey et al., 2000). While a sixth gene (MZ00018561) also carried a plant lipid transfer protein domain, its N-terminal extension made it an atypical LTP and led to a classification as a Pro-rich cell wall-plasma membrane linker protein with lipid-binding capacity. Two other target genes also seemed to have a cell envelope-related function due to their annotations as transmembrane proteins, and more precisely, members of a plant-specific family carrying the DUF588 domain (for domain of unknown function 588; MZ00030315) and of the MtN3/Saliva family (MZ00014373) named after NODULIN3 from Medicago truncatula and SALIVA from Drosophila melanogaster (Gamas et al., 1996). This group was completed by a down-regulated gene (MZ00024414) annotated as a multidrug and toxic compound extrusion (MATE) efflux carrier. The last two genes were predicted to encode a cytochrome P450 of the plant-specific subfamily A most closely related to CYP78A6 from Arabidopsis and a tetratricopeptide repeat (TPR) domain-containing protein similar to the MALE STERILITY5 protein (Glover, 2000). These results suggested that the majority of genes regulated by OCL1 were involved in lipid metabolism or transport and other cell envelope-related functions.
Due to the epidermis-specific expression of OCL1, we scrutinized the 11 annotations for putative epidermis-related functions. In the case of the WBC11-like gene (MZ00031783), which will be called ZmWBC11a hereafter, functional data in a closely related gene clearly suggested an epidermis-related function, since both wax and cutin synthesis are impaired in the Arabidopsis wbc11 mutant (Bird et al., 2007). In order to determine if OCL1 regulated other members of the WBC11 clade in maize, we identified all paralogous genes in the maize genome. Among the four additional WBC11-like genes, ZmWBC11b to ZmWBC11e, the first two showed up-regulation in transgenic 18-DAS plantlets compared with wild-type plantlets (Table I). Interestingly, the three ZmWBC11 genes regulated by OCL1 (ZmWBC11a, ZmWBC11b, and ZmWBC11c) fell into a single clade in the phylogenetic tree of the WBC family, while the remaining two genes, ZmWBC11d and ZmWBC11e, not influenced by OCL1 fell into a sister clade (Supplemental Fig. S1). These results indicated that OCL1 regulated a well-defined subset of genes coding for ABC transporters in maize.
Overlapping Expression of OCL1 with Its Target Genes
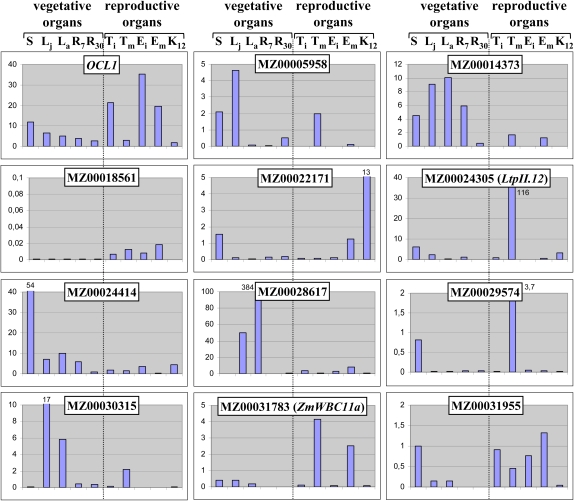
To obtain additional clues with regard to gene function, we established the expression pattern of each confirmed target gene by qRT-PCR in vegetative and reproductive organs as well as during kernel development (Fig. 1; Supplemental Table S2). Three genes of the lipid group (ZmLtpII.12, ZmWBC11a, and MZ0031955) and one of the envelope group (MZ00018561) showed strongest expression in reproductive organs, while the remaining three genes of the envelope group (MZ00014373, MZ00024414, and MZ00030315) and the Cytochrome P450 gene (MZ00028617) were primarily expressed in leaves (Fig. 1). The fourth lipid gene (MZ00029574) and the defense gene (MZ00005958) showed no clear preference for either reproductive organs or leaves. The gene coding for a TPR domain protein (MZ00022171) appeared to be preferentially expressed in the maize kernel. During kernel development, the majority of the 11 target genes showed a peak of expression toward the end of early kernel development, varying between 7 and 12 d after pollination (DAP; Supplemental Table S2). One of the lipid-related genes (MZ00031955) had a second peak during the maturation stage (35–50 DAP). Expression of the Nodulin gene (MZ00014373) was strongest in immature ovules, while the Cytochrome P450 gene (MZ00028617) was up-regulated during dehydration (30–35 DAP).
Figure 1.
Expression profile of OCL1 and its target genes in maize. Real-time RT-PCR experiments were carried out on cDNA prepared from major organs of the maize plant for OCL1 and its 11 target genes identified by microarray experiments. The values are means of a technical replicate. The values of truncated bars are indicated. S, Seedling aerial parts; Lj, leaf juvenile (leaf 4); La, leaf adult (leaf 10); R7, root at 7 DAS; R30, root at 30 DAS; Ti, tassel immature; Tm, tassel mature; Ei, ear immature; Em, ear mature; K12, kernel at 12 DAP. [See online article for color version of this figure.]
In conclusion, most genes clearly showed preferential expression in a limited number of organs. Despite preferences for either leaves or reproductive organs and generally weaker expression in roots, we could not establish an overall pattern common to all genes, although the expression territories of all genes showed at least some overlap with that of OCL1, in particular during kernel development.
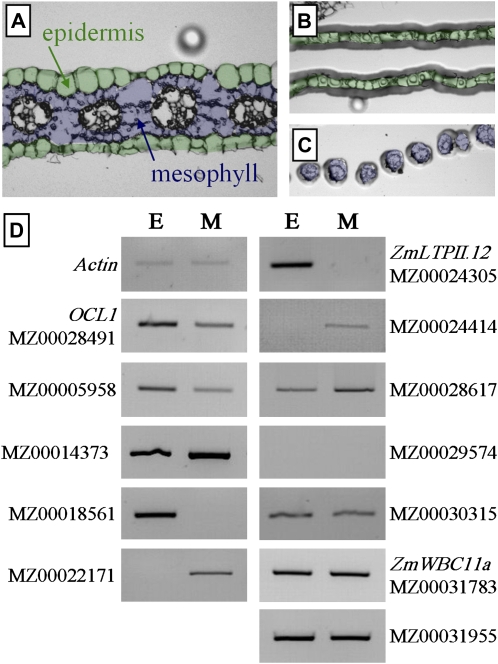
Due to the annotations suggesting epidermis-related functions of the 11 target genes, we performed RT-PCR experiments on epidermal and mesophyll cells captured after laser microdissection of the central part of juvenile leaf 4 (Fig. 2A) in order to reveal any preferential or specific expression in the epidermis. Of the 11 target genes, ZmLTPII.12 (MZ0024305) and the gene encoding a Pro-rich protein (MZ00018561) were specifically expressed in the epidermis, while Igl (MZ00005958) and the gene coding for an integral membrane protein (MZ00030315) showed preferential expression in this layer. Two genes were evenly expressed in both tissues (MZ00031955 and ZmWBC11a), four showed stronger or specific expression in mesophyll cells (MZ00014373, MZ00022171, MZ00024414, and MZ00028617), and one was not detectable in either tissue (MZ00029574). The expression of OCL1 was only preferential but not specific to epidermal cells (Fig. 2), a situation reminiscent of the one observed by in situ hybridization in very young embryos but in contrast with in situ hybridizations on organ primordia or meristematic tissues suggesting epidermis-specific expression (Ingram et al., 1999). In summary, the data further strengthened the hypothesis of a role of OCL1 and the first four genes in epidermis-related functions, but they do not exclude such a role for the remaining genes.
Figure 2.
Expression of OCL1 and its target genes in outer and inner cell layers of leaf 4. A to C, From paraffin-embedded leaf sections (A), epidermal (B) and mesophyll cells (C) were isolated using infrared laser-capture microdissection. D, RT-PCR experiments assessing the expression of OCL1 and its target genes in microdissected epidermal (E) and mesophyll (M) cells. The concentration of the cDNA templates was normalized according to the abundance of the Actin RT-PCR product. [See online article for color version of this figure.]
Changes in Cuticular Wax Composition in OCL1-OE Plants
Since the lipid-related target gene ZmWBC11a belonged to the same orthologous group as AtWBC11, which has a clearly established role in cuticle formation in Arabidopsis, we analyzed the leaves of transgenic lines overexpressing OCL1 for structural modifications of the cuticle. The thickness of the cuticle was measured by transmission electron microscopy and confocal microscopy in transverse sections of juvenile leaves, while the density and shape of wax crystals were assessed by scanning electron microscopy. Neither approach revealed any significant differences between wild-type and OCL1-OE leaves (Supplemental Fig. S2).
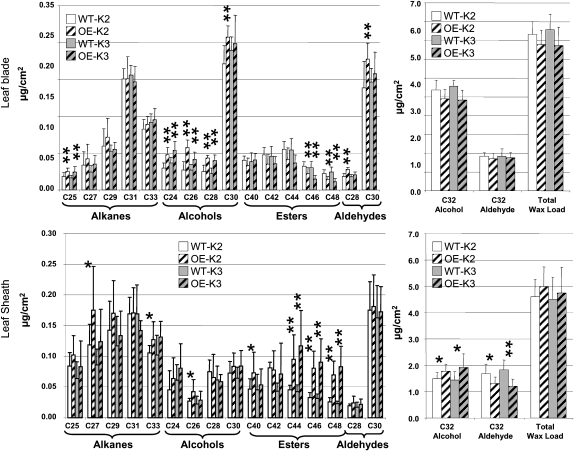
Next, we employed biochemical methods based on chloroform extraction followed by gas chromatography with flame ionization detection or mass spectrometry detection to analyze the quantity and quality of epicuticular waxes from juvenile leaves. Using juvenile leaf 4 from two independent OCL1-OE transformation events (K2 and K3) and their corresponding wild type, we analyzed waxes from both the leaf sheath (the leaf base enveloping the stem) and the leaf blade (the part separated from the stem). No significant difference could be detected in the total wax load for either part of the leaf, but a detailed analysis of the different constituents revealed that the same pools were affected in the two independent OCL1-OE lines (Fig. 3), which showed comparable levels of OCL1 overexpression (19.57-fold for K2 and 23.80-fold for K3). In the leaf sheath, OCL1 overexpression modified the content of the two major components of the waxes with opposite effect: increase in C32 alcohol and decrease in C32 aldehyde. Nevertheless, since these constituents were present at about the same levels and the modifications were similar but in opposite directions, total wax load was not affected. With respect to minor components, strong modifications were consistently detected in the levels of the wax esters, which are made of very long-chain fatty acids and alcohols. In the waxes of both OCL1-OE events, C44 to C48 wax esters were two to three times more abundant. In event K2, this increase in wax ester content was accompanied by higher levels of C26 fatty alcohol and C27 alkane. In the leaf blade, the contents of the two major components (C32 alcohol and C32 aldehyde) were not affected. Among the minor components, C46 and C48 wax esters were reduced (by approximately 30%) rather than increased as in the leaf sheath. Fatty alcohols showed a much broader increase, since the levels of C24 to C28 alcohols were about 30% higher in both OCL1-OE events when compared with the wild type. In addition, the levels of C25 alkane and C28 and C30 aldehyde (event K2 only) were significantly increased in the blade. Altogether, these analyses suggested that overexpression of OCL1 resulted in significant modifications of wax composition, with somewhat different effects in sheath and blade.
Figure 3.
Cuticular wax composition of juvenile maize leaves. Total wax load as well as relative amounts of individual compounds from the blade and sheath of juvenile leaves were compared between the OCL1-OE transformation events K2 and K3 (hatched bars) and their respective wild types (WT; white and gray bars). Means and sd indicated by error bars were calculated on seven to 10 biological replicates (Supplemental Table S3). * P < 0.05, ** P < 0.01 as calculated by Student’s t test.
Since in epicuticular waxes of Arabidopsis the production of primary alcohols is catalyzed by the fatty acyl-coenzyme A reductase (FAR) CER4 (Costaglioli et al., 2005; Rowland et al., 2006), we hypothesized that OCL1 could activate the transcription of FAR genes in maize leaf blades. Therefore, we identified the FARs present in the maize genome based on sequence homology to Arabidopsis FARs and examined their expression level in wild-type and OCL1-OE plants. Among the five putative ZmFAR genes detected in the maize genome, only ZmFAR1 showed a differential expression and was up-regulated 2.78-fold in OCL1-OE plants (Table I). While reciprocal blast analyses revealed that the closest relatives of the deduced ZmFAR1 protein sequence in Arabidopsis were CER4 (At4g33790) and At5g22500, with 64% sequence identity each, phylogenetic analyses did not allow establishing orthologous relationships between individual proteins. Four maize sequences including ZmFAR1 were clustered in a clade, and six Arabidopsis sequences including CER4 were clustered in a sister clade (Supplemental Fig. S3). These results indicated that OCL1 may trigger the reduction of fatty acid precursors into primary alcohols through the transcriptional activation of a particular ZmFAR related to AtCER4.
Partial Knockouts of OCL1 Influence Target Gene Expression But Not Wax Composition
To confirm the molecular and phenotypic changes seen in OCL1-OE plants, OCL1-RNAi (for RNA interference) plants under the control of the rice Actin promoter were produced. None of the 14 transformation events showed complete suppression of OCL1 transcript accumulation, and further work focused on line 2, which showed the most efficient OCL1 gene silencing, with a decrease of about 50% of the OCL1 mRNA level (Table I). This knockdown of OCL1 expression was sufficient to affect the expression level of several target genes. In 18-DAS plantlets, eight target genes had a trend opposite to the one observed in OCL1-OE plants (Table I). Again, the lipid group was the main representative, with the ABC transporter genes ZmWBC11a, ZmWBC11c, and ZmLTPII.12 and the Carboxylesterase gene (MZ00029574). Five target genes showed no significant expression difference between OCL1-RNAi plantlets and wild-type siblings, and one gene showed the same trend as in OCL1-OE plants. These data lend further credence to a direct or indirect regulation by OCL1 of the eight genes, with opposite trends in OCL1-OE and OCL1-RNAi plantlets.
Wax composition was analyzed in three independent RNAi events named lines 1, 2, and 3, in which the OCL1 expression level was reduced to 68%, 51%, and 64% of the wild-type level, respectively. A comparative analysis of juvenile leaves from RNAi plants and wild-type siblings revealed significant differences only for wax esters (Supplemental Table S3), which were somewhat difficult to interpret in light of the differences between sheath and blade. No opposite trend to the increase of C24 to C28 alcohols seen in OCL1-OE lines was observed, probably due to the insufficient knockdown of OCL1. In fact, opposite trends are not necessarily expected, since OCL1 expression was only reduced by a factor of 2 in the best OCL1-RNAi line but increased by a factor of 20 in the strongest OCL1-OE line; similarly, the alterations in the expression of the eight target genes with opposite trends were considerably stronger in OCL1-OE lines than in the OCL1-RNAi line.
Transactivation of ZmWBC11a and ZmLtpII.12 by OCL1 after Transient Transformation of Maize Kernels
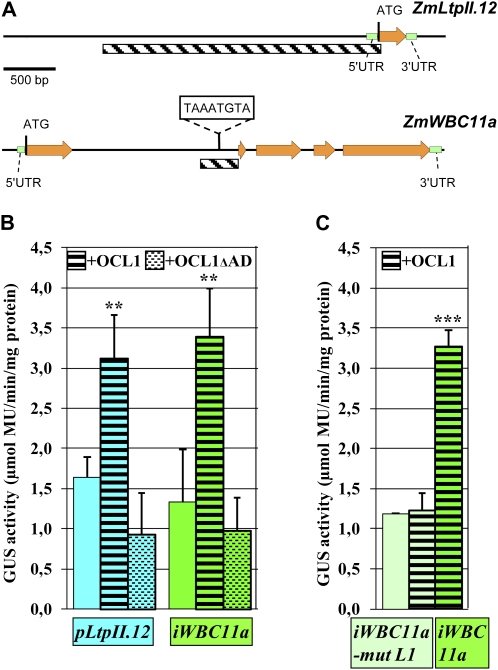
In order to discriminate between direct and indirect OCL1 target genes, we obtained the genomic sequence of each target gene using maize genome resources (www.maizegenome.org) and scanned the upstream and intron sequences for the presence of an L1 or L1-like (L1L) box. The asymmetric L1 box 5′-TAAATG(C/T)A-3′ is a cis-element showing a gel shift in the presence of the HD-ZIP IV transcription factors ATML1 (Abe et al., 2001), PDF2 (Abe et al., 2003), and GL2 (Ohashi et al., 2003), while the slightly longer palindromic L1L box 5′-GCATTAAATGC-3′ has been defined as the consensus-binding site of recombinant HD-ZIP IV proteins HDG7, HDG9, and ATML1 in PCR-assisted DNA selection assays (Nakamura et al., 2006). The only L1 or L1L box detected in the 11 genomic sequences was located near the end of the first intron of ZmWBC11a (Fig. 4), which became a good candidate to be a direct target gene of OCL1. The presence of regulatory elements in introns is not rare, one of the best characterized examples being intron 1 of AGAMOUS in Arabidopsis (Sieburth and Meyerowitz, 1997).
Figure 4.
Transactivation of ZmLTPII.12 and ZmWBC11a by OCL1 in maize kernels. A, Genomic structures of ZmLTPII.12 and ZmWBC11a indicating the region fused to the GUS reporter gene (hatched). UTR, Untranslated region. B and C, Quantification of GUS activity from transiently transformed maize kernels. B, The reporter construct alone (no motif), with OCL1 (black stripes) or with OCL1 lacking its activation domain (AD; stippled stripes), was used for particle bombardment. C, An iWBC11a reporter construct mutated in the L1 box (light green) or with an intact L1 box (dark green) was used. Means and sd indicated by error bars were calculated on three biological replicates. Each replicate represented a pool of 36 bombarded kernels. ** P < 0.01, *** P < 0.001.
To further investigate the possible binding of OCL1 to regulatory regions of target genes, we chose the L1 box-containing intron of ZmWBC11a and the upstream region of the epidermis-specific ZmLtpII.12 (Fig. 4A). Immature maize kernels were cobombarded with fusions of the respective regulatory regions to a GUS reporter gene and a second construct expressing OCL1 under the control of the constitutive CsVMV promoter. Quantification of GUS activity by the 4-methylumbelliferyl β-d-glucuronide test revealed a basal level of fluorescence after transient transformation with the reporter constructs iWBC11a::GUS and pLtpII.12::GUS on their own (Fig. 4B). When cotransformed with OCL1, we observed a marked increase in GUS activity indicating transactivation by OCL1. In contrast, the cotransformation of pLtpII.12::GUS or iWBC11a::GUS with OCL1ΔAD coding for an inactive form of OCL1 lacking its activation domain (N. Depège-Fargeix, personal communication) failed to transactivate the transcription of the ZmLtpII.12 and ZmWBC11a genes, and we even observed a weak, nonsignificant decrease of the GUS activity with regard to the basal level. These results clearly demonstrated the capacity of OCL1 to activate the transcription of two genes of the lipid group (Table I) in the maize kernel.
In order to provide further arguments for a direct interaction between OCL1 and the regulatory region of ZmWBC11a, we mutated the L1 box 5′-TAAATG(C/T)A-3′ to 5′-TAAGGG(C/T)A-3′, thereby introducing the same mutation previously used in the PDF1 promoter to demonstrate loss of binding of the HD-ZIP IV factor ATML1 in Arabidopsis (Abe et al., 2001). We observed that in the presence of the mutated L1 box, OCL1 lost the capacity of transactivate the transcription of ZmWBC11a (Fig. 4C). This result demonstrated that a native L1 box was required for the transactivation by OCL1 and suggested direct binding of OCL1 to the L1 box in intron 1 of ZmWBC11a.
DISCUSSION
Transcription of 14 Genes Is Altered in Plants Overexpressing OCL1
The identification and molecular characterization of 14 direct or indirect target genes of the HD-ZIP IV transcription factor OCL1 showed that half of them encode proteins known to be involved in the biosynthesis or transport of cuticular waxes in maize, fitting well with the preferential or specific expression of OCL1 in the epidermal cell layer of various plant organs. Plants overexpressing OCL1 do not only show up-regulation of seven genes with suggestive lipid-related annotations but also alterations in the wax composition of juvenile leaves. The annotations of the remaining seven genes were less informative and could not be readily linked to specific biological processes. The predicted localization of four gene products in the plasma membrane or cell wall hints at a role related to the cell envelope, which is not incompatible with epidermis-specific modifications. Two genes belong to the large gene families of TPR proteins and P450 cytochromes, which have been implicated in very diverse biological processes (Small and Peeters, 2000; Schuler and Werck-Reichhart, 2003); unfortunately, the position of the proteins encoded by OCL1 target genes is far from members with established functions in phylogenetic trees. The defense-related function of many P450 cytochromes provides a possible link with the last target gene Igl, which has previously been shown to be involved in the tritrophic defense of maize against herbivory (Frey et al., 2000).
Among the 14 genes with expression changes in OCL1-OE plants, eight genes showed an opposite trend in OCL1-RNAi knockdown lines, further supporting the hypothesis of a critical role of OCL1 in the transcriptional regulation of these target genes. The fact that a decrease in OCL1 mRNA levels by 50% does not affect the expression level of the remaining six genes can possibly be explained either by a more complex or a less sensitive chain of events between OCL1 expression levels and target gene transcription.
OCL1 is certainly not the only regulatory protein influencing the transcription of its target genes, and it likely interacts with different proteins or via different regulatory cascades in different parts of the maize plant. These conclusions are based on the fact that the up- or down-regulation by OCL1 in seedlings is not always observed in other organs of OCL1-OE plants and that the expression profiles of OCL1 and its target genes in the different organs of the maize plant overlap but do not coincide. A concrete example for independent regulation of a target gene by two different regulatory pathways is the regulation of Igl by volicitin (Frey et al., 2004) and OCL1. Igl expression was lower in OCL1-OE than in wild-type leaves, yet a treatment with volicitin increased Igl expression by a similar factor in both materials (data not shown).
OCL1 Regulates Target Genes Involved in Lipid Metabolism or Transport
Our data provide further evidence for the hypothesis that HD-ZIP IV transcription factors play important regulatory roles in the differentiation or maintenance of the epidermis in general and cuticle-related lipid metabolism and transport in particular. This hypothesis is based on the L1-specific expression pattern of most HD-ZIP IV family members on the one hand (Ariel et al., 2007) and on transcriptome data comparing epidermal cells with underlying tissues in Arabidopsis (Suh et al., 2005) and maize (Nakazono et al., 2003) on the other hand, where several HD-ZIP IV genes were found strongly up-regulated in epidermal tissues, just like genes involved in lipid metabolism and transport.
Here, we demonstrate the causal relationship between the overexpression of OCL1 and the up-regulation of genes coding for a nonspecific, type 2 lipid transfer protein (nsLTPII), an AtCXE18-like carboxylesterase, a SEC14/PITP, three ABC transporters of the WBC11/ABCG11 clade, and a FAR.
Plant nsLTPs are small, soluble proteins that facilitate the transfer of fatty acids, phospholipids, glycolipids, or steroids between membranes. They are encoded by gene families with 49 and 52 members in Arabidopsis and rice, respectively (Boutrot et al., 2008). The lipid-binding capacities of the proteins and the epidermis-specific expression of many nsLtp genes are well documented (Kader, 1996). Roles in two distinct biological processes, defense and cuticle biosynthesis, have been demonstrated. On the one hand, overexpression of barley (Hordeum vulgare) LTP2 enhances tolerance to Pseudomonas syringae in Arabidopsis (Molina and Garcia-Olmedo, 1997), and defective induced resistance1 mutants lack systemic acquired resistance after attack by Pseudomonas (Maldonado et al., 2002). On the other hand, certain Ltp genes can be induced by the presence of cutin monomers (Kim et al., 2008), and mutant plant lines lacking LTPG1 show a dramatic reduction of C29 alkanes (Debono et al., 2009). The two biological roles may involve a common molecular mechanism, since the ltpG1 mutant also shows enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola (Lee et al., 2009). The OCL1 target ZmLtpII.12 clusters far from Ltp genes with established biological functions in a phylogenetic tree, and its closest characterized neighbors are TaLTP2 and HvLTP2. While no precise role has been attributed to the latter genes, there is converging evidence that ZmLtpII.12 and consequently OCL1 are involved in lipid transfer for cuticle biosynthesis and/or plant defense.
The second OCL1 target coding for a carboxylesterase close to AtCXE18 is also linked to both lipid metabolism and plant defense. While the biochemical function of AtCXE and related carboxylesterases (EC 3.1.1.1) is to hydrolyze esters of short-chain fatty acids (Cummins et al., 2007), a majority of carboxylesterase genes have been associated with functions in plant defense (Marshall et al., 2003).
The third lipid-related OCL1 target contains a SEC14/PITP domain named after the yeast mutant sec14 perturbed in endosome trafficking and distinct trans-Golgi export pathways (Curwin et al., 2009). PITPs catalyze phosphatidylinositol and phosphatidylcholine transfer in vitro, and PITP deficiencies are known to be responsible for several diseases in mammals (Bankaitis et al., 2005). For example, lack of α-TOCOPHEROL TRANSFER PROTEIN causes vitamin E deficiency due to an impaired transport of α-tocopherol (Manor and Morley, 2007).
Three further OCL1 target genes involved in lipid transport are ZmWBC11a, ZmWBC11b, and ZmWBC11c, coding for ABC transporters of the WBC subfamily (also called ABCG subfamily), which is specialized in the ATP-dependent translocation of steroids and other lipids in animals (Velamakanni et al., 2007). In the plant kingdom, mutant analysis has identified CER5 (WBC12) and WBC11 as key components of the cuticular lipid export pathway (Pighin et al., 2004; Bird et al., 2007). The mutants cer5 and wbc11 present a decrease of total cuticular wax load and varying effects on wax composition; in addition, wbc11 presents a decrease in cutin load. While BLAST searches with entire protein sequences identified AtWBC11 as the closest relative of ZmWBC11a, a phylogenetic tree based on conserved blocks of maize, rice, and Arabidopsis sequences revealed a more complex picture (Supplemental Fig. S1). The orthologs of AtWBC11 seem to be ZmWBC11d and ZmWBC11e, which are not regulated by OCL1, while the three OCL1-controlled maize proteins ZmWBC11a, ZmWBC11b, and ZmWBC11c fell into a sister clade containing only maize and rice sequences. Nevertheless, the phylogenetic closeness to WBC11 and CER5 together with the changes in cuticular wax load observed in OCL1-OE plants strengthen the hypothesis that ZmWBC11a, ZmWBC11b, and ZmWBC11c are part of the cuticular lipid export pathway.
Plants Overexpressing OCL1 Show Changes in Cuticular Wax Composition
A direct link between OCL1 and cuticle biosynthesis was established by the observation that the C24 to C28 fatty alcohol contents were significantly increased in the leaf blade and ester contents were systemically affected in the sheath and blade of OCL1-OE leaves. Since the decarbonylation pathway, which is responsible for the synthesis of aldehydes and alkanes, appears less affected, it seems that OCL1 expression principally affects the acyl reduction pathway. The qualitative shift in the composition of epicuticular waxes could possibly be explained by the up-regulation of the last lipid-related OCL1 target ZmFAR1, coding for a fatty acid reductase. Just like four other ZmFAR genes not regulated by OCL1, ZmFAR1 is related to CER4 (Supplemental Fig. S3). The phenotype of the OCL1-OE plants is somewhat complementary to the Arabidopsis cer4 mutant, which does not accumulate C24 to C28 primary alcohols and contains intermediate levels of C30 primary alcohols (Rowland et al., 2006). More detailed comparisons between ZmFAR1 and CER4, which may accept a narrower range of substrates, are interesting but difficult, because the phylogenetic tree does not allow the identification of orthologuous gene pairs and rather establishes the existence of orthologous groups, and because the composition of cuticular waxes is quite different between maize and Arabidopsis leaves, the major compounds being C32 alcohols and aldehydes in the former but C29 and C31 alkanes in the latter case. The same limitations are valid for comparisons between the phenotypes of OCL1-OE plants and the Arabidopsis wbc11 mutant, which is characterized by a significant decrease in C29 alkanes and C26 to C28 primary alcohols (Bird et al., 2007). In addition, in the WBC11 subfamily characterized by obligatory dimerization (Kusuhara and Sugiyama, 2007), the possible formation of heterodimers between ZmWBC11a, ZmWBC11b, and ZmWBC11c would further multiply the hypotheses.
A defect in fatty acid reduction has been reported in the maize double mutant gl5/gl20, blocked in the production of primary alcohols and showing a high accumulation of aldehydes (Bianchi et al., 1978). While gl5 has been mapped on chromosome 4 in BIN 4.03 between markers pdi1 and umc2211, no map position is available for the duplicate locus gl20 (http://www.maizegdb.org). However, neither OCL1 itself nor any of the five ZmFAR genes mapped close to gl5, leaving the possibility that one of them represents gl20.
Transcriptional Activation Requires the OCL1 Activation Domain and an L1 Box in ZmWBC11a
With ZmWBC11a, at least one of the 14 OCL1 target genes seems to be directly activated by OCL1, since the transactivation of the iWBC11a-GUS reporter construct by OCL1 depends both on the presence of the activation domain in OCL1 and an intact L1 box in the 347-bp fragment of ZmWBC11a driving the GUS reporter gene. While our transactivation assays are no formal proof of physical interaction, the hypothesis of OCL1 binding to the ZmWBC11a L1 box is further substantiated by previous gel-shift assays or DNaseI footprints, which established in vitro physical interaction between HD-ZIP proteins and double-stranded oligonucleotides (19–21 bp) containing an L1 box for ATML1 (Abe et al., 2001), PDF2 (Abe et al., 2003), and GL2 (Ohashi et al., 2003). It is also noteworthy that the Arabidopsis AtWBC11 contained two adjacent L1 boxes in its promoter, the shift between intron and promoter likely being the consequence of quite different intron/exon structures of AtWBC11 and ZmWBC11a. Taking into account that OCL1, ATML1/PDF2, and GL2 belong to different clades of the HD-ZIP IV family and that Helianthus annuus homeodomain protein1 (HAHR1) interacts with an L1L box (Tron et al., 2001), our data lend further evidence to the hypothesis that the interaction between HD-ZIP proteins and an L1 or L1L box is not restricted to certain family members or to Arabidopsis but is a widespread phenomenon in the family and across species.
The transactivation of a 2,906-bp upstream fragment of ZmLtpII.12 depended also on the presence of the activation domain in OCL1 but did not involve an L1 box, suggesting either the need for at least one additional regulatory protein in the signaling cascade between OCL1 and the ZmLtpII.12 regulatory region or OCL1 binding to alternative sites, which may be variants of the L1 or L1L box. Preliminary results of serial promoter deletions indicate that more than 800 bp upstream of the ATG are needed for transactivation by OCL1 (data not shown). Further deletion analysis and sequence comparisons with upstream regions and introns of the other 12 OCL1 target genes may lead to the identification of a novel cis-element recognized by an HD-ZIP IV protein and/or a yet unknown intermediary protein.
Regulation of Cuticle Biosynthesis
Cuticular wax formation is known to be tightly regulated in response to both developmental and environmental cues. Several transcription factors regulating the activity of genes involved in the synthesis of the cuticle have recently been identified. In Arabidopsis, lines overexpressing WAX INDUCER1/SHINE1 (WIN1/SHN1; Aharoni et al., 2004; Broun et al., 2004) or the closely related AP2/EREBP family members SHN2 or SHN3 (Aharoni et al., 2004) trigger wax production, enhance drought tolerance, and modulate cuticular permeability. In addition, WIN1/SHN1 overexpression also increased cutin production by the induction of cutin biosynthesis genes (Kannangara et al., 2007). Increased cuticular wax accumulation and enhanced drought tolerance were also observed by the overexpression of M. truncatula WAX PRODUCTION1 (WXP1), belonging to a different clade of the AP2/EREBP family, in Medicago sativa (Zhang et al., 2005) or Arabidopsis (Zhang et al., 2007), where the paralogous WXP2 had similar effects. The overexpression of AtMYB41, an R2R3 MYB transcription factor, led to an increased leaf epidermal permeability and modulated the expression of genes involved in lipid and cuticle metabolism (Cominelli et al., 2008). MYB30, a Myb-domain transcription factor that is induced during incompatible interactions between Arabidopsis and several bacterial pathogens (Vailleau et al., 2002), appears to positively regulate the accumulation of alkanes in cuticular waxes (Raffaele et al., 2008).
A possible link between HD-ZIP IV transcription factors and cuticle biosynthesis has previously been suggested based on coordinated up-regulation of HD-ZIP IV genes and genes involved in cuticle biosynthesis in the epidermal layer of Arabidopsis (Suh et al., 2005) and maize (Nakazono et al., 2003). While no cuticle defect has been described in any of the 16 HD-ZIP IV mutants in Arabidopsis, a point mutation in a tomato HD-ZIP IV gene was very recently identified as the likely cause for cutin defects of the tomato fruit in the cd2 mutant (Isaacson et al., 2009). Phylogenetic analyses show that OCL1 falls in the same clade as CD2 and forms an orthologous group with ANL2, HDG1, HDG7, and HDG6/FLOWERING WAGENINGEN from Arabidopsis.
Our data on the regulation of lipid-related genes by OCL1 and alterations of the cuticle in OCL1-OE plants reinforce the hypothesis that, in addition to the above-cited members of the AP2/EREBP and MYB families, transcription factors of the HD-ZIP IV family contribute to the transcriptional regulation of cuticle biosynthesis. The presence of the START domain, which is involved in lipid binding and transport in animals (Ponting and Aravind, 1999), opens the way to the very speculative hypothesis that the activation of lipid or cuticle biosynthetic pathways by HD-ZIP IV proteins may depend on the sensing of regulatory lipids or metabolic intermediates via the START domain.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The maize (Zea mays) inbred line A188 (Gerdes and Tracy, 1993) and transgenic A188 plants overexpressing OCL1 were grown in a greenhouse fulfilling French S2 safety standards for the culture of transgenic plants with a 16-h illumination period (100 W m−2) at 24°C/19°C (day/night) and without control of the relative humidity. Seeds were germinated in 0.2 L of Favorit MP Godets substrate (Eriterre) and were transferred at 21 DAS to 10 L of Favorit Argile TM substrate (Eriterre) supplemented with 4 g L−1 Osmocote Exact hi-end 15+9+12 fertilizer (Scotts). All plants were propagated by hand pollination.
T-DNA Construct and Plant Transformation
The plasmid used for the production of OCL1-OE and OCL1-RNAi plants contained the backbone of vector pSB11 (Ishida et al., 1996) and a Basta resistance cassette. For the OCL1-OE construct, the OCL1 coding sequence was amplified with primers A10-6HIS5′ and A10-6HIS3′ and placed under the control of the CsVMV promoter. For the OCL1-RNAi construct, the inverted 350-bp OCL1 fragments (amplified with primers OCL1-RNAi-5′ and OCL1-RNAi-3′) were separated by the rice (Oryza sativa) Tubulin intron and placed under the control of a rice Actin promoter followed by a rice Actin intron. Primer sequences are given in Supplemental Table S4.
Agrobacterium tumefaciens-mediated transformation of maize inbred line A188 was based on a published protocol (Ishida et al., 2007). Among the six independent transformation events, the two with strongest OCL1 expression (K2 and K3) were used in this study.
Microarray Analysis
The Genoplante maize microarray contained 58,752 oligonucleotides of 70 bases spotted on glass slides. The subtending unigene set had been established by clustering the Genoplante maize EST data (http://urgi.versailles.inra.fr/data/gnpSeq/genoplante_data.php) with all publicly available EST data. Hybridization was carried out as described (Zeidler et al., 2004). Experiments were done in biological triplicate with in vitro-amplified total RNA of 18-DAS maize seedlings from wild-type and OCL1-OE plants. While a dye swap of the Cy5- or Cy3-labeled probes was performed, only the Cy5 data were exploited. Quantile normalization of the raw data was carried out using Spotfire software. The criteria for the inclusion of a gene in the list of differentially expressed genes were a logR > 2 or < −2 and P < 0.01.
qRT-PCR
Approximately 100 mg of fresh tissue was quick frozen in liquid nitrogen and ground to powder with mortar and pestle. Total RNA was extracted with 1 mL of TRIzol reagent according to the instructions of the supplier (Invitrogen). After ethanol precipitation, the RNA was resuspended in 30 μL of RNase-free water and treated with RNase-free DNase. The DNase was inactivated according to the instructions of the supplier (Ambion). Approximately 5 μg of total RNA were reverse transcribed using random hexamers (Amersham Biosciences) and reverse transcriptase without RNaseH activity (Fermentas) in a final volume of 20 μL. A total of 2.5 × 105 copies of GeneAmplimer pAW109 RNA (Applied Biosystems) were added to the RT reaction.
The cDNA was diluted 50 times, and 2 μL was used in a volume of 20 μL containing 10 μL of Platinum SYBR Green qPCR SuperMix UDG according to the instructions of the supplier (Invitrogen) to carry out qPCR on a DNA Engine Opticon 2 (Bio-Rad). Dilution series (2n with n = 0–7) of a mixture of all cDNAs within a comparison were used to fix the threshold cycle (CT). Gene expression levels relative to the 18S rRNA reference gene were calculated by the ΔΔCT method (Schmittgen and Livak, 2008). The primers used are listed in Supplemental Table S4.
Sequence Analysis
The cDNA sequences corresponding to the 70mers present on the microarray were established by BLASTN individual EST sequences or full-length cDNA sequences at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/). Consensus sequences were obtained using VectorNTI ContigExpress software (Invitrogen) and regularly updated. Genomic sequences were obtained by BLASTN of the cDNA sequence against the high-throughput genomic sequences database at NCBI. Deduced amino acid sequences were annotated by BLASTP against the Arabidopsis (Arabidopsis thaliana) genome at NCBI and screened for known, conserved domains using the CDS database.
Wax Composition Analysis
Maize leaf 4 sheath and blade were isolated, and their surfaces were measured by image analysis of a scan (leaf sheath was considered a cylinder for determining total surface). Cuticular waxes were extracted by immersing tissues for 30 s in 20 mL of chloroform containing 20 μg of docosane as the internal standard. Extracts were dried under a gentle stream of nitrogen, dissolved into 150 μL of BSTFA-TMCS [for N,O-bis(trimethylsilyl)trifluoroacetamide):trimethylchlorosilane (99:1)], and derivatized at 85°C for 1 h. Surplus BSTFA-TMCS was evaporated under nitrogen, and samples were dissolved in 200 μL of hexane for analysis using an Agilent 6850 gas chromatograph and helium as the carrier gas (1.5 mL min−1). The gas chromatograph was programmed with an initial temperature of 80°C for 1 min and increased at 15°C min−1 to 260°C, held for 10 min at 200°C, increased again at 5°C min−1 to 320°C, and held for 15 min at 320°C. Qualitative analyses were performed using an HP-5MS column (30 m × 0.25 mm × 0.25 μm) and an Agilent 5975 mass spectrometric detector (70 eV, mass-to-charge ratio of 50–750). Quantitative analyses were performed using an HP-1 column (30 m × 0.32 mm × 0.25 μm) and a flame ionization detector. Quantification was based on peak areas and the internal standard docosane.
Laser-Capture Microdissection and RT-PCR
From the region of maximum width of fully expanded leaf 4, 1-cm2 sections were fixed in acetone and paraffin embedded as described (Ohtsu et al., 2007). Epidermal and mesophyll subepidermal cells were microdissected from 10-μm sections using the Arcturus XT infrared laser-capture microdissection system with the following settings for epidermal/mesophyll cells, respectively: laser spot size, 10/20 μm; laser pulse duration, 20/30 ms; and laser power, 50/70 mW. About 5,000 epidermal cells (predominantly adaxial) and 2,000 mesophyll cells were collected and RNA extracted with the PicoPure RNA isolation kit (Arcturus). RNA samples were treated with DNase I (Qiagen) and amplified (two rounds) with the TargetAmpTM 2-Round aRNA Amplification kit 2.0 (Epicentre Biotechnologies). RT and PCR were carried out as described above, including a control experiment without reverse transcriptase. Primer sequences are given in Supplemental Table S4.
Transactivation Tests
The promoter regions of ZmLtpII.12 and the first intron of ZmWBC11a were amplified using specific primer pairs (Supplemental Table S4). After cloning into pCRII-Blunt-Topo (Invitrogen) and sequencing, they were fused with the GUS reporter gene, the endogenous ATG (ZmLtpII.12), or an in-frame ATG in exon 2 (ZmWBC11a), becoming the start codon of the GUS. Plasmid DNA prepared with the PureLink HiPure Plasmid Filter Midiprep kit (Invitrogen) was used in transient transformation of 15-DAP maize kernels by particle bombardment.
The 15-DAP maize kernels were surface sterilized by pulverization of Pursept-A (Poly-Labo). The pericarp was removed in a rectangular window on the adaxial side, exposing the embryo and part of the endosperm. The kernels were plasmolyzed for 4 h on Murashige and Skoog medium (4.3 g L−1 MS M0221 [Duchefa], 30 g L−1 Suc, 0.2 g L−1 Asn, 36.4 g L−1 sorbitol, 36.4 g L−1 mannitol, 1 mg L−1 2,4-dichlorophenoxyacetic acid, and 3 g L−1 Gelrite, pH 5.6) prior to bombardment.
Conditioned samples were transformed using a particle-inflow gun PDS-1000/He Biolistic Particle Delivery System (Bio-Rad). For each type of sample, the parameters were optimized according to Sanford et al. (1993). In the standard protocol, gold particles of 1 μm diameter (Bio-Rad) coated with 5 μg of plasmid DNA were propelled by helium gas under pressure (7,500 kPa) toward the samples, which were placed at 6 cm below the gun orifice. A partial vacuum (90 kPa) increased the speed of the particles. A 20-μm nylon mesh placed 3 cm above the targets protected the samples from the gas blast and dispersed the particles evenly onto them. The GUS assays were performed 48 h after transformation.
GUS Assays
Transiently transformed kernels were incubated in 5-bromo-4-chloro-3-indolyl-β-d-GlcUA for 24 h at 37°C according to Jefferson et al. (1986). Proteins were extracted with 500 μL of buffer (50 mm phosphate buffer, 10 mm EDTA, 0.1% sodium lauryl sarcosine, 0.1% Triton X-100, and 10 mm β-mercaptoethanol). The supernatant was used for quantification of GUS activity. Protein extracts were incubated with 2 mm 4-methylumbelliferyl β-d-glucuronide at 37°C during 2 h. The fluorescent product 4-methylumbelliferone (MU) was measured with a Fluoroskan II (Labsystems). Reference samples with known quantities of MU were used to determine the quantity of MU produced, which was expressed in μmol MU mg−1 protein min−1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of maize and Arabidopsis WBC proteins.
Supplemental Figure S2. Comparison of cuticle structure in wild-type and OCL1-OE leaves.
Supplemental Figure S3. Phylogenetic tree of maize and Arabidopsis FAR proteins.
Supplemental Table S1. Relative expression levels of confirmed OCL1 target genes in 18-DAS plantlets.
Supplemental Table S2. Expression of 11 OCL target genes in maize organs and during kernel development as determined by qRT-PCR.
Supplemental Table S3. Composition of cuticular waxes on juvenile leaves of OCL1-OE and OCL1-RNAi plants and their wild-type siblings.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Isabelle Anselme-Bertrand for her precious advice on electron microscopy. Isabelle Desbouchages, Alexis Lacroix, and Priscilla Angelot are acknowledged for maize culture, Hervé Leyral and Claudia Bardoux for the preparation of buffers and media, the Biogemma transcriptomics team for the hybridization and analysis of microarrays, Pierre Chambrier for advice on transient transformation, and Cédric Finet for help with phylogenetic trees. Monika Frey kindly provided us with volicitin.
References
- Abe M, Katsumata H, Komeda Y, Takahashi T. (2003) Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643 [DOI] [PubMed] [Google Scholar]
- Abe M, Takahashi T, Komeda Y. (2001) Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J 26: 487–494 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. (2007) The true story of the HD-Zip family. Trends Plant Sci 12: 419–426 [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Phillips S, Yanagisawa L, Li X, Routt S, Xie Z. (2005) Phosphatidylinositol transfer protein function in the yeast Saccharomyces cerevisiae. Adv Enzyme Regul 45: 155–170 [DOI] [PubMed] [Google Scholar]
- Bianchi G, Salamini F, Avato P. (1978) Glossy mutants of maize. 8. Accumulation of fatty aldehydes in surface waxes of gl5 maize seedlings. Biochem Genet 16: 1015–1021 [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Boutrot F, Chantret N, Gautier MF. (2008) Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 9: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. (2008) Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J 53: 53–64 [DOI] [PubMed] [Google Scholar]
- Costaglioli P, Joubes J, Garcia C, Stef M, Arveiler B, Lessire R, Garbay B. (2005) Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim Biophys Acta 1734: 247–258 [DOI] [PubMed] [Google Scholar]
- Cummins I, Landrum M, Steel PG, Edwards R. (2007) Structure activity studies with xenobiotic substrates using carboxylesterases isolated from Arabidopsis thaliana. Phytochemistry 68: 811–818 [DOI] [PubMed] [Google Scholar]
- Curwin AJ, Fairn GD, McMaster CR. (2009) Phospholipid transfer protein Sec14 is required for trafficking from endosomes and regulates distinct trans-Golgi export pathways. J Biol Chem 284: 7364–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debono A, Yeats TH, Rose JK, Bird D, Jetter R, Kunst L, Samuels L. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. (1996) The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J 10: 393–402 [DOI] [PubMed] [Google Scholar]
- Eigenbrode SD, Espelie KE. (1995) Effects of plant epicuticular lipids on insect herbivores. Annu Rev Entomol 40: 171–194 [Google Scholar]
- Frey M, Spiteller D, Boland W, Gierl A. (2004) Transcriptional activation of Igl, the gene for indole formation in Zea mays: a structure-activity study with elicitor-active N-acyl glutamines from insects. Phytochemistry 65: 1047–1055 [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Pare PW, Schmelz EA, Tumlinson JH, Gierl A. (2000) An herbivore elicitor activates the gene for indole emission in maize. Proc Natl Acad Sci USA 97: 14801–14806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, Niebel Fde C, Lescure N, Cullimore J. (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9: 233–242 [DOI] [PubMed] [Google Scholar]
- Gerdes JT, Tracy WF. (1993) Pedigree diversity within the Lancaster surecrop heterotic group of maize. Crop Sci 33: 334–337 [Google Scholar]
- Glover BJ. (2000) Differentiation in plant epidermal cells. J Exp Bot 51: 497–505 [DOI] [PubMed] [Google Scholar]
- Gray JE, Holroyd GH, van der Lee FM, Bahrami AR, Sijmons PC, Woodward FI, Schuch W, Hetherington AM. (2000) The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716 [DOI] [PubMed] [Google Scholar]
- Guan XY, Li QJ, Shan CM, Wang S, Mao YB, Wang LJ, Chen XY. (2008) The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol Plant 134: 174–182 [DOI] [PubMed] [Google Scholar]
- Guimil S, Dunand C. (2007) Cell growth and differentiation in Arabidopsis epidermal cells. J Exp Bot 58: 3829–3840 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Farbos I, Lagercrantz U, von Arnold S. (2001) PaHB1 is an evolutionary conserved HD-GL2 homeobox gene expressed in the protoderm during Norway spruce embryo development. Genesis 30: 220–230 [DOI] [PubMed] [Google Scholar]
- Ingram GC, Boisnard-Lorig C, Dumas C, Rogowsky PM. (2000) Expression patterns of genes encoding HD-ZipIV homeo domain proteins define specific domains in maize embryos and meristems. Plant J 22: 401–414 [DOI] [PubMed] [Google Scholar]
- Ingram GC, Magnard JL, Vergne P, Dumas C, Rogowsky PM. (1999) ZmOCL1, an HDGL2 family homeobox gene, is expressed in the outer cell layer throughout maize development. Plant Mol Biol 40: 343–354 [DOI] [PubMed] [Google Scholar]
- Isaacson T, Kosma DK, Matas AJ, Buda GJ, He Y, Yu B, Pravitasari A, Batteas JD, Stark RE, Jenks MA, et al. (2009) Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J 60: 363–377 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Hiei Y, Komari T. (2007) Agrobacterium-mediated transformation of maize. Nat Protoc 2: 1614–1621 [DOI] [PubMed] [Google Scholar]
- Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. (1996) High efficiency transformation of maize (Zea mays L) mediated by Agrobacterium tumefaciens. Nat Biotechnol 14: 745–750 [DOI] [PubMed] [Google Scholar]
- Ito M, Sentoku N, Nishimura A, Hong SK, Sato Y, Matsuoka M. (2002) Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J 29: 497–507 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsh D. (1986) Beta-glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA 83: 8447–8451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffree CE. (2006) The fine structure of the plant cuticle. Biology of the Plant Cuticle, Vol 23 Blackwell Publishing, Oxford, pp 11–110 [Google Scholar]
- Jenks MA, Eigenbrode SD, Lemieux B. (2002) Cuticular waxes of Arabidopsis. Somerville C, Meyerowitz E, eds, The Arabidopsis Book, Vol 34 American Society of Plant Biologists, Rockville, MD, pp 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader JC. (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Hofte H, Pauly M, Riechmann JL, Broun P. (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled AS, Vernoud V, Ingram GC, Perez P, Sarda X, Rogowsky PM. (2005) Engrailed-ZmOCL1 fusions cause a transient reduction of kernel size in maize. Plant Mol Biol 58: 123–139 [DOI] [PubMed] [Google Scholar]
- Kim TH, Park JH, Kim MC, Cho SH. (2008) Cutin monomer induces expression of the rice OsLTP5 lipid transfer protein gene. J Plant Physiol 165: 345–349 [DOI] [PubMed] [Google Scholar]
- Kubo H, Peeters AJ, Aarts MG, Pereira A, Koornneef M. (1999) ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Samuels AL. (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Kusuhara H, Sugiyama Y. (2007) ATP-binding cassette, subfamily G (ABCG family). Pflugers Arch 453: 735–744 [DOI] [PubMed] [Google Scholar]
- Lee SB, Go YS, Bae HJ, Park JH, Cho SH, Cho HJ, Lee DS, Park OK, Hwang I, Suh MC. (2009) Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol 150: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Porat R, Nadeau JA, O’Neill SD. (1996) Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Manor D, Morley S. (2007) The alpha-tocopherol transfer protein. Vitam Horm 76: 45–65 [DOI] [PubMed] [Google Scholar]
- Marshall SD, Putterill JJ, Plummer KM, Newcomb RD. (2003) The carboxylesterase gene family from Arabidopsis thaliana. J Mol Evol 57: 487–500 [DOI] [PubMed] [Google Scholar]
- Messing J, Dooner HK. (2006) Organization and variability of the maize genome. Curr Opin Plant Biol 9: 157–163 [DOI] [PubMed] [Google Scholar]
- Molina A, Garcia-Olmedo F. (1997) Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J 12: 669–675 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Burglin TR. (2006) MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol 140: 1142–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA. (2009) Stomatal development: new signals and fate determinants. Curr Opin Plant Biol 12: 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. (2006) Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol 141: 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS. (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. (2002) The biopolymers cutin and suberin. Somerville C, Meyerowitz E, eds, The Arabidopsis Book, Vol 34 American Society of Plant Biologists, Rockville, MD, pp 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T. (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Ohtsu K, Smith MB, Emrich SJ, Borsuk LA, Zhou R, Chen T, Zhang X, Timmermans MC, Beck J, Buckner B, et al. (2007) Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.). Plant J 52: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. (2008) Corn genomics pops wide open. Science 319: 1333. [DOI] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. (1999) START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 24: 130–132 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, Huard C, Blee E, Mongrand S, Domergue F, Roby D. (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Riederer M. (2006) Introduction: biology of the plant cuticle. Biology of the Plant Cuticle, Vol 23 Blackwell Publishing, Oxford, pp 1–10 [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L. (2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Sanford JC, Smith FD, Russell JA. (1993) Optimizing the biolistic process for different biological applications. Methods Enzymol 217: 483–509 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D. (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Shen B, Sinkevicius KW, Selinger DA, Tarczynski MC. (2006) The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol Biol 60: 377–387 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N. (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139: 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. (2009) The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J 60: 564–574 [DOI] [PubMed] [Google Scholar]
- Tron AE, Bertoncini CW, Palena CM, Chan RL, Gonzalez DH. (2001) Combinatorial interactions of two amino acids with a single base pair define target site specificity in plant dimeric homeodomain proteins. Nucleic Acids Res 29: 4866–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D. (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velamakanni S, Wei SL, Janvilisri T, van Veen HW. (2007) ABCG transporters: structure, substrate specificities and physiological roles. A brief overview. J Bioenerg Biomembr 39: 465–471 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Laigle G, Rozier F, Meeley RB, Perez P, Rogowsky PM. (2009) The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J 59: 883–894 [DOI] [PubMed] [Google Scholar]
- Zeidler M, Zhou Q, Sarda X, Yau CP, Chua NH. (2004) The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J 40: 355–365 [DOI] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY. (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42: 689–707 [DOI] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Sumner LW, Wang ZY. (2007) Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol Biol 64: 265–278 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.