Abstract
To accommodate fluctuating nutrient levels in the soil, plants modulate their metabolism and root development via signaling mechanisms that rapidly reprogram the plant transcriptome. In the case of nitrate, over 1,000 genes are induced or repressed within minutes of nitrate exposure. To identify cis-regulatory elements that mediate these responses, an enhancer screen was performed in transgenic Arabidopsis (Arabidopsis thaliana) plants. A 1.8-kb promoter fragment from the nitrate reductase gene NIA1 was identified that acts as a nitrate enhancer when fused to a 35S minimal promoter. Enhancer activity was localized to a 180-bp fragment, and this activity could be enhanced by the addition of a 131-bp fragment from the nitrite reductase promoter. A promoter construct containing the 180- and 131-bp fragments was also induced by nitrite and repressed by ammonium, indicating that it was responsive to multiple nitrogen signals. To identify specific regulatory elements within the 180-bp NIA1 fragment, a transient expression system using agroinfiltration of Nicotiana benthamiana was developed. Deletion analysis identified three elements corresponding to predicted binding motifs for homeodomain/E-box, Myb, and Alfin1 transcription factors. A fully active promoter showing nitrate and nitrite enhancer activity equivalent to that of the wild-type 180-bp fragment could be built from these three elements if the spacing between the homeodomain/E-box and Myb-Alfin1 sites was equivalent to that of the native promoter. These findings were validated in transgenic Arabidopsis plants and identify a cis-regulatory module containing three elements that comprise a nitrate enhancer in the NIA1 promoter.
Nitrate has two functions in plants; it serves as a nutrient and as a signal. Plants undergo many physiological and developmental changes when they are exposed to nitrate. One of the fastest responses is reprogramming of the plant transcriptome. When plants are exposed to nitrate, genes in the nitrate assimilation pathway (NRT, NIA, NiR) are induced within minutes (Wang et al., 2003, 2007; Scheible et al., 2004). Other genes, which are involved in carbon and energy metabolism that support nitrate assimilation, are also rapidly induced (Stitt, 1999; Wang et al., 2000, 2003, 2004; Stitt et al., 2002; Scheible et al., 2004; Fritz et al., 2006). Transcriptome analyses have shown that over 1,500 genes are rapidly induced or repressed by nitrate and that the processes of pentose phosphate oxidation, glycolysis, trehalose synthesis, nitrogen, and amino acid metabolism are most affected (Wang et al., 2003, 2004, 2007; Scheible et al., 2004; Gutierrez et al., 2007). These rapid transcriptome responses provide the basis for the longer-term responses that direct root growth, development, and architecture, root-to-shoot ratios, and germination rates (Forde, 2002; Alboresi et al., 2005; Filleur et al., 2005; Forde and Walch-Liu, 2009).
Even though nitrate responses have been reported for plants for over 40 years, the first biochemical response being reported in 1957 (Tang and Wu, 1957), the regulatory genes that mediate these responses are just now being identified. So far, several potential transcription factors, two kinases and a transceptor have been linked to nitrate regulation (Krouk et al., 2010). The ANR1 MADS-box transcription factor, which is induced by nitrogen deprivation, controls lateral root branching in response to nitrate (Zhang and Forde, 1998; Gan et al., 2005). The NIN-like gene NLP7, encoding a potential bZIP DNA-binding protein, was identified through its sequence similarity to the nitrate regulatory gene NIT2 in Chlamydomonas (Camargo et al., 2007) and functions in nitrate induction of several nitrate assimilatory genes (Castaings et al., 2009). Three members of the lateral organ boundary domain gene family (LBD37, 38, and 39) were identified as nitrate-inducible genes and shown to be repressors of anthocyanin biosynthetic and nitrate assimilatory genes (Rubin et al., 2009). Two kinase genes CIPK8 and CIPK23 were identified as nitrate-inducible genes. Mutations in CIPK8 result in reduced gene induction at high but not low nitrate concentrations (Hu et al., 2009) while mutations in CIPK23 increase gene induction at low nitrate concentrations, indicating that these kinases selectively target different phases (i.e. high-affinity versus low-affinity phases) of the nitrate response. Interestingly, CIPK23 phosphorylates the nitrate transporter NRT1.1 (CHL1; Tsay et al., 1993), which has been shown to act as a nitrate regulator/sensor (Ho et al., 2009; Wang et al., 2009; Krouk et al., 2010).
Additional genes have been identified that are involved in overall nitrogen regulation or starvation. A Dof transcription factor improves nitrogen use efficiency at low nitrogen (Yanagisawa et al., 2004), and the master clock control gene CCA1 regulates organic nitrogen metabolism (Gutierrez et al., 2008). Microarray analysis of specific cell types in roots uncovered a miRNA (microRNA167 from Arabidopsis [Arabidopsis thaliana]) that mediates cell-specific control of root development in response to nitrogen (Gifford et al., 2008). A genetic screen for mutants impaired in nitrogen starvation responses identified the NLA gene, which encodes a RING-type ubiquitin ligase needed for adaptive responses to nitrogen but not phosphorus deprivation and for repressing senescence (Peng et al., 2007).
Even though several transcription factors have been identified, little is known about the cis-acting regulatory elements in nitrate-regulated promoters. In early work, several mutant analyses defined minimal nitrate-responsive promoters and identified a 12-bp motif that was conserved among nitrate-regulated promoters; however, no sequence with enhancer function, i.e. capable of conferring nitrate regulation to a heterologous promoter such as the 35S minimal promoter, was reported (Lin et al., 1994; Hwang et al., 1997; Dorbe et al., 1998; Sivasankar et al., 1998). More recently, a 150-bp fragment from the AtNRT2.1 promoter was shown to have nitrate enhancer function in Arabidopsis plants treated with 0.3 mm nitrate for 7 d compared with plants treated with 0.3 mm ammonium (Girin et al., 2007). The 150-bp fragment contains several predicted transcription factor binding sites that may serve as cis-acting elements. Most recently, a 43-bp enhancer fragment in the NiR promoter was found to confer strong nitrate enhancer function when quadruplicated then fused to a 35S minimal promoter (Konishi and Yanagisawa, 2010). The promoter construct was tested in cotyledons of 3- to 4-d-old transgenic Arabidopsis plants treated with nitrate for 4 h. Further work showed that the 43-bp fragment contained a conserved sequence [tGACcCTT-N(10)-AAGagctc] that was critical for nitrate inducibility and that this fragment did not confer Gln repression.
One reason for the slow progress in identifying specific elements within nitrate enhancers is that no transient system has been reported in higher plants for rapid testing of nitrate-responsive promoters. Experiments to date have been done with transgenic plants and take months to perform. A transient system would greatly expedite the identification and analysis of nitrate enhancer elements (NEEs). In this article, we report the development of a transient system to assay nitrate enhancer function. We describe the discovery of a NIA1 enhancer fragment and then apply the transient system to identify elements within the NIA1 enhancer. What we found was a complex of three elements that comprise a cis-regulatory module that mediates nitrate enhancer function.
RESULTS
Identification of a Nitrate Enhancer Fragment in the NIA1 Promoter
Our search for NEEs began with a screen for enhancer fragments from the nitrate reductase gene NIA1 (At1g77760). Fragments from promoter and transcribed regions of NIA1 were cloned into a vector containing a 35S minimal promoter fused to GUS. Constructs were transformed into Arabidopsis plants using Agrobacterium tumefaciens and tested for nitrate-inducible GUS expression. Transgenic plants were grown for 10 d on ammonium succinate as the sole nitrogen source then treated with 10 mm KNO3 for 24 h before assaying for GUS activity using histochemical staining. Out of four NIA1 fragments tested, only one (the 1.8-kb NIA1-3 fragment, which is located in the distal promoter region 5′ to the start of transcription) showed nitrate enhancer activity (Supplemental Fig. S1). The other fragments showed baseline activity similar to that of the 35S minimal promoter alone.
The 1.8-kb NIA1-3 DNA was subdivided into smaller fragments (Supplemental Fig. S2) and tested again with the 35S minimal promoter in transgenic Arabidopsis plants. Enhancer function was localized to a 640-bp fragment (NIA1-14). NIA1-14 was subdivided further into 150- to 200-bp overlapping fragments and then tested; only one fragment (NIA1-14e, 180 bp) showed nitrate enhancer activity (Supplemental Figs. S3 and S4A).
Construction and Analysis of a Strong Nitrate-Inducible Promoter: NRP
To quantify the nitrate induction levels for each of the positive clones described above, transgenic plants were analyzed using the quantitative GUS assay as described in “Materials and Methods.” Constructs containing the largest fragment, NIA1-3 (1.8 kb), showed 3.0-fold induction (averaged for three independent transgenic lines) while the NIA1-14 (630 bp) and NIA1-14e (180 bp) fragments showed 2.3-fold and 1.9-fold induction, respectively (Fig. 1; data for each independent transgenic line are shown in Supplemental Fig. S5). These results showed that reducing the size of the enhancer DNA reduced the level of induction. Efforts to subdivide NIA1-14e into smaller fragments, which included testing overlapping 30-bp fragments in triplicate, were unsuccessful in further localizing the enhancer activity.
Figure 1.
Nitrate enhancer analysis of NIA1 promoter fragments in Arabidopsis transgenic lines. Transgenic seedlings grown on agarose plates with ammonium and no nitrate were transferred to plates containing 10 mm nitrate or 10 mm chloride for 24 h then assayed for promoter (GUS) activity as described in “Materials and Methods.” The averages of three independent transgenic lines are shown (error bars show se, GUS activity shown as nmol/mg protein h). Each GUS assay was performed in triplicate for each transgenic line (data for individual lines are shown in Supplemental Fig. S5). The fold of nitrate induction is indicated above each bar. The diagrams under the histogram show the promoter constructs used for these experiments.
To continue with our search for specific enhancer elements, a search was made for DNA sequences that could augment the enhancer activity of the 180-bp NIA1 fragment. We tested several fragments near the start of transcription of the Arabidopsis NiR gene because several reports showed that NiR promoter constructs were nitrate inducible in transgenic plants (Wilkinson, 1992; Rastogi et al., 1993, 1997; Neininger et al., 1994; Sander et al., 1995; Dorbe et al., 1998). A 475-bp NiR fragment showing nitrate induction in transgenic Arabidopsis plants was divided into two fragments (upstream 260 bp and downstream 215 bp fragments relative to the start of transcription) and inserted between the 180-bp NIA fragment and the 35S minimal promoter (Supplemental Fig. S6). The 260-bp NiR fragment strongly boosted the nitrate enhancer activity of the 180-bp NIA1 fragment while the 215-bp fragment showed no effect. The 260-bp NiR fragment was not serving simply as a spacer as 260 bp of Escherichia coli DNA showed little effect. The 260- and 215-bp NiR fragments alone (with just the 35 minimal promoter) had little to no activity. The 260-bp NiR fragment was subdivided into three overlapping fragments and tested with the 180-bp NIA1 fragment. The most 3′ fragment (131-bp R) showed the strongest effect while the other two (130-bp L and M) showed little to no activity. We subsequently used the tripartite promoter, containing the 180-bp NIA1-14e, 131-bp R NiR, and 35S minimal fragments and called NRP for nitrate-regulated promoter, for subsequent experiments.
Using the quantitative GUS assay, we found that the NRP-GUS construct showed 14-fold induction, which was much higher than the 1.9-fold induction found for the 180-bp NIA1 fragment alone (Fig. 1). A nitrate titration was performed with the NRP-GUS construct by analyzing GUS mRNA levels in roots after treating transgenic plants with nitrate at various concentrations for 20 min (Fig. 2). Nitrate inductions of 2-fold or more were observed at 100 μm nitrate concentrations or greater, and the dose response was similar to the nitrate induction observed for the endogenous NIA1 gene.
Figure 2.
Nitrate induction of NRP-GUS mRNA in transgenic lines. NRP-GUS transgenic plants (line R56-3) were grown on agarose plates with nitrate-free growth media containing 2.5 mm ammonium succinate for 9 d under continuous light. The plates were then flooded with 15 mL of fresh medium plus KNO3 or KCl for 20 min at concentrations as indicated. Roots were collected for total RNA preparation at end of treatments. GUS (A) and NIA1 (B) mRNA levels were assayed by real-time quantitative PCR (Wang et al., 2007) with clathrin (At4g24550) as the reference gene. Experiments were in triplicate, and error bars show se.
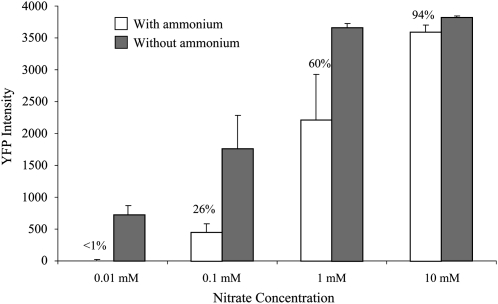
Another feature of many nitrate-induced genes is feedback repression (Girin et al., 2007). The NRP construct was tested for ammonium repression by treating transgenic plants with nitrate in the presence or absence of ammonium. Transgenic Arabidopsis plants containing an NRP-YFP construct were grown on ammonium as the sole nitrogen source then treated with nitrate for 16 h at various concentrations of nitrate in the presence or absence of 5 mm ammonium (Fig. 3). At low nitrate concentrations, the presence of ammonium strongly reduced the amount of NRP-YFP expression (99% inhibition at 0.01 mm nitrate and 74% inhibition at 0.1 mm nitrate). At higher nitrate concentrations, ammonium had much less effect (only 40% inhibition at 1 mm nitrate and 6% inhibition at 10 mm nitrate). These results show that the enhancer fragments in NFP retain sensitivity to ammonium repression, which is most apparent at low nitrate levels.
Figure 3.
Ammonium repression of NRP promoter. NRP-YFP transgenic plants were grown on vertical agarose plates for 4 d with 2.5 mm ammonium succinate as the sole nitrogen source. The seedlings were then transferred to fresh plates with media containing KNO3 at the indicated concentrations with or without 2.5 mm ammonium succinate. Seedlings transferred to plates without nitrate in the presence or absence of 2.5 mm ammonium succinate were used as controls. Seedlings were incubated under continuous light for 16 h then YFP fluorescence was determined by fluorescence microscopy with ImageJ software (National Center for Biotechnology Information). Each data point represent an average of six readings with se as indicated. The numbers above the white bars show the percent of fluorescence intensity in nitrate-treated plants treated with ammonium compared with nitrate-treated plants without ammonium.
Development of a Transient Expression System to Assay Nitrate Enhancers
To accelerate the identification of enhancer elements, a transient expression system was developed using agroinfiltration of Nicotiana benthamiana. This system has been used to study protein expression (Kopertekh and Schiemann, 2005; Joensuu et al., 2010), gene silencing (Waterhouse and Helliwell, 2003; de Felippes and Weigel, 2010), and protein-protein interactions (Ohad et al., 2007). It has also been used for promoter analysis (Yang et al., 2000). We adapted it for our nitrate-regulated promoters by growing plants on nitrate-free media with ammonium as the sole nitrogen source before agroinfiltration then irrigating plants with 10 mm KNO3 to induce gene expression. KNO3 was replaced with KCl for control plants. Leaves were assayed for promoter (GUS) activity 3 to 4 d after infiltration.
When Nicotiana leaves were infiltrated with agrobacteria containing the vector alone (just the 35S minimal promoter fused to GUS), very low levels of GUS activity were observed in nitrate- and chloride-treated leaves (Fig. 4, vector). Infiltration with a full-length 35S promoter fused to GUS showed very high, unregulated activity (Fig. 4, 35S). In contrast, the NRP tripartite promoter, which showed strong nitrate induction in transgenic Arabidopsis plants, also showed strong nitrate induction in the transient system (Fig. 4, NRP). Thus, the Nicotiana system can be used to assay nitrate-responsive promoters.
Figure 4.
Testing the N. benthamiana transient expression system. Leaves of Nicotiana were infiltrated with agrobacteria containing the indicated promoter constructs fused to GUS then assayed 3 to 4 d later for GUS activity as described in “Materials and Methods.” Plants were irrigated with either 10 mm KCl or 10 mm KNO3 30 min before infiltration and 24 h before harvest. Experiments were done in triplicate with error bars showing se. Numbers above the bars show fold induction. Labels are as follows: Vector, 35S minimal promoter; 35S, 35S full-length promoter; NRP, tripartite promoter containing the NIA1 180 bp, NiR 131 bp, and 35S minimal promoter fragments.
Three NEEs Identified in the 180-bp Nitrate Enhancer Fragment
The search for NEEs in the 180-bp NIA1-14e fragment began with a deletion analysis using the Nicotiana transient expression system. NIA1-14e deletion fragments were tested in the context of the tripartite promoter; that is, they replaced the wild-type 180-bp NIA1 fragment in the NRP-GUS construct. A series of 3′ deletions of NIA1-14e showed that 71 bp could be deleted with no loss of nitrate induction but that a 105-bp deletion eliminated the response (Fig. 5, 3d1–3d4). At the 5′ end, even the first deletion reduced nitrate induction as follows: Deletion of 19 bp (Fig. 5, 5d1) reduced induction by 40%; deletion of 43 bp (Fig. 5, 5d2) reduced it by 60%. These results show that the 3′ end of NIA1-14e is dispensable and that a 109-bp subfragment of NIA1-14e (3d3) is sufficient for full activity.
Figure 5.
Deletion analysis of the NIA1 180-bp enhancer fragment. Deletions of the NIA1-14e fragment are shown to the left of the histogram. Deletion end points are shown with arrows, and remaining nucleotides are shown with solid black line. The histogram shows GUS activities after nitrate and chloride treatments with fold inductions shown to the right of each bar. Experiments were done in triplicate with error bars showing se.
Next, 4-bp deletions were made at four sites in the 109-bp fragment that correspond to potential regulatory motifs (i.e. predicted transcription factor binding sites based on the AthaMap analysis software [www.athamap.de]; Fig. 6A). A deletion in the 5′ HVH21 site reduced the nitrate response by 26% relative to the wild-type 109-bp fragment of NIA1-14e (Fig. 6C, -hvh21). A deletion in the Myb-1 site (-myb1) had little effect while a deletion in the Myb-2 (-myb2) or Alfin1 (-alfin) sites reduced the response by 100% and 75%, respectively. These results indicate that the Myb-2 and Alfin1 sites are critical for enhancer function while the 5′ HVH21 site plays a contributing role.
Figure 6.
Identification of enhancer elements in the 109-bp NIA1 enhancer fragment. A, Sequence of the 109-bp fragment showing sites of transcription factor binding sites as predicated by AthaMap. The 4-bp deletions that were tested are underlined. B, Location of the HVH21 and Myb-2+Alfin1 fragments. C, GUS activity for each construct using the Nicotiana transient system for nitrate- and chloride-treated plants with fold induction shown above each bar. Experiments were performed in triplicate with ses shown. wt, Wild type.
If the above conclusion is correct, then a fragment containing these three sites should reconstruct full nitrate induction comparable to that of the 109-bp or 180-bp NIA1-14e fragments. First, a fragment containing only the Myb-2 and Alfin1 sites was tested (Fig. 6B, Myb2+Alfin1); very little nitrate response was observed (Fig. 6C, +myb2+-alfin). When the HVH21 site was added to the Myb-2+Alfin1 fragment (Fig. 6C, +HVH21), activity was restored; however, the activity depended on the distance between the HVH21 sequence and the Myb-2+Alfin1 fragment. Spacers shorter than the native length gave reduced induction: A 7-bp spacer (Fig. 6C, 7 sp) restored only 53% of the nitrate induction, and a 16-bp spacer (Fig. 6C, 16 sp) restored only 43% of the induction compared with the wild-type 109-bp fragment (sequences of constructs are given in Supplemental Fig. S7). Other spacers (23 and 26 bp) gave even less activity. However, a 44-bp spacer (Fig. 6C, 44 sp, the same length found in the native DNA) showed the highest level of induction (140% of the 109-bp fragment). These results show that the combination of the HVH21, Myb-2, and Alfin1 sites with sufficient spacing between the HVH21 and the Myb-2-Alfin1 sites is sufficient for full nitrate enhancer function.
Validation in Transgenic Arabidopsis Plants
Several key constructs tested in the transient system described above were transformed and tested in transgenic Arabidopsis plants. The original tripartite NRP construct showed 14-fold induction (Fig. 7, NRP; see also Fig. 1). The Alfin1 single mutant showed almost no activity (Fig. 7, -Alfin1). The Myb-2 and HVH21 double mutant showed no activity (Fig. 7, -HVH21-Myb2). For the reconstructed fragment containing the HVH21 site fused to Myb-2+Alfin1 with a 44-bp spacer (same length as the native sequence), full induction was restored (Fig. 7, +HVH21+Myb2+Alfin1). Thus, the results in transgenic Arabidopsis plants validate the presence of three NEEs in the NIA1-14e fragment of the NIA1 promoter.
Figure 7.
Validation of promoter constructs in transgenic Arabidopsis. Transgenic seedlings grown on agarose plates with ammonium and no nitrate were transferred to plates containing 10 mm nitrate or 10 mm chloride for 24 h. GUS activity (nmol/mg protein h) was determined for three independent transgenic lines then averaged (error bars show se). Each GUS assay was performed in triplicate for each transgenic line (data for individual lines shown in Supplemental Fig. S8). The fold of nitrate induction is indicated above each bar. Labels are as follows: -Alfin1, 4-bp deletion in Alfin1 site; -HVH21-Myb2, 4-bp deletions in HVH21 and Myb-2 sites; +HVH21+Myb2+Alfin1, HVH21 fragment fused to Myb-2+Alfin1 fragment with 44-bp spacer; NRP, tripartite promoter; WT, wild type. The first three constructs used the 109-bp 3d3 fragment.
Analysis of the Nitrite Response for the 180-bp Enhancer
Nitrite also serves as a potent signal to rapidly induce and repress nitrate-regulated genes including NIA1 and NiR (Wang et al., 2007). We tested transgenic plants containing various NRP constructs to determine if the NRP promoter responded to nitrite, and if a nitrite response was dependent on the same enhancer elements required for nitrate induction.
NRP-GUS transgenic plants were grown with ammonium as the sole nitrogen source, treated with 1 mm KNO2 (using 1 mm KCl as control) for 30 min, then GUS mRNA levels in roots were measured (Fig. 8). The NRP-GUS construct showed 16-fold induction by nitrite, which is similar to the level induced by nitrate. Nitrite inductions of modified NRP constructs with either the Alfin1 site or both the HVH21 and Myb-2 sites mutated were reduced 9- and 6-fold, respectively. The reconstructed enhancer fragment containing all three sites with a spacer of native length between the HVH21 and Myb-2 sites showed 11-fold induction by nitrite, almost as much as the original NRP promoter. For a control, mRNA levels for the endogenous NiR gene were determined, and they showed similar induction levels in all four sets of transgenics plants. These results show that the NRP construct is strongly induced by nitrite, and the same three enhancer elements needed for nitrate induction are also needed for nitrite induction.
Figure 8.
Nitrite response of NRP-GUS constructs in transgenic plants. Plants were grown on vertical agarose plates with nitrate-free growth media containing 2.5 mm ammonium succinate for 7 d under continuous light. Plates were then flooded with 15 mL of fresh medium plus 1 mm KNO2 or KCl for 30 min. Roots were collected at end of treatment for total RNA preparation. Messenger RNA levels of GUS (A) and NiR (B) were assayed by real-time quantitative PCR with clathrin (At4g24550) as the reference gene. Two independent transgenic lines for each construct were used. Experiments were in triplicates, and error bars show se. Labels are as described in Figure 7 legend.
DISCUSSION
Our results identify a 180-bp fragment in the NIA1 promoter that has nitrate enhancer function and locate three cis-regulatory elements within the 180-bp fragment that account for the enhancer activity. These regulatory elements act synergistically to form a cis-regulatory module (as defined in Priest et al., 2009) that mediates nitrate induction. Together with the elements identified by Konishi et al. in the 43-bp fragment of NiR (Konishi and Yanagisawa, 2010), these sequence motifs provide insights into the identity of NEEs in plants.
The three regulatory elements in the 180-bp NIA1 fragment are candidate transcription factor binding sites. They were initially identified as potential binding sites by the AthaMap software. The first motif, a potential HVH21 site, contains a TGAC consensus sequence that binds homeodomain proteins of the knotted class 1 type (Krusell et al., 1997). This site also overlaps with an E-box, which has a consensus sequence CANNTG and binds factors in the bHLH family of factors (Toledo-Ortiz et al., 2003). The E-box element in the 180-bp fragment (CAAGTG) is very similar to the canonical E-box sequence: CACGTG. Interestingly, there are homeodomain factors that bind E-boxes (Aigner et al., 2007); thus, this site has the potential to bind either bHLH or homeodomain factors. The two Myb sites predicted by AthaMap correspond to motifs that contain the consensus sequence C-G/C-GTT-G/A, which was originally described as a site that binds GA-induced Myb proteins from barley (Hordeum vulgare; Gubler et al., 1999). Even though the two GA-Myb sites have identical GTTG core sequences, the first one shows no activity while the second is essential. The Alfin1 site has a core consensus sequence of C/A-CAC, which was first shown to bind a novel zinc-finger protein in alfalfa (Medicago sativa; Bastola et al., 1998).
One of the advances that made it possible to identify the three regulatory elements in the 180-bp NIA1 fragment was the inclusion of the 131-bp NiR fragment in the NRP construct. Even though the 180-bp fragment acted as a nitrate enhancer on its own (with only the 35S minimal promoter), its activity was increased almost 10-fold by the NiR fragment. An important clue about how the 131-bp fragment may be functioning has come from the work of Konishi and Yanagisawa (2010). They identified the sequence tGACcCTT-N(10)-AAGagct as a nitrate-responsive cis-element. This sequence is present at the 3′ end of the 131-bp NiR fragment used for our NRP constructs. Konishi and Yanagisawa proposed that a novel transcription factor binds to this site, but one can also find an HVH21 core sequence (TGAC) present in this element. Other analyses of the NiR promoter do not shed light on our 131-bp fragment; for example, it does not overlap with −230 to −200 region of the spinach (Spinacia oleracea) NiR promoter identified by Rastogi et al. as being important for nitrate induction (Rastogi et al., 1997).
One question that arises from our findings is how prevalent are HVH21/E-box, GA-Myb, and Alfin1 motifs in nitrate-regulated genes. Previous analysis has shown that the E-box site is not only present but is overrepresented in promoters of nitrate-regulated genes (Nero et al., 2009). Analysis of sequences previously shown to be involved in nitrate regulation revealed that a potential Alfin1 site is present in the Arabidopsis NRT2.1 150-bp enhancer fragment (Girin et al., 2007), a potential HVH21 site in the 43-bp NiR enhancer fragment (Konishi and Yanagisawa, 2010), and a potential Alfin1 site (in addition to the HVH21 site) in the 131-bp NiR fragment of NRP. We also scanned sequences of highly nitrate-induced genes of Arabidopsis and found examples where all three motifs were present in close proximity in the same order as that found in NIA1 (Supplemental Fig. S9). These regions contain potential nitrate cis-regulatory modules, but further work will be necessary to verify the functionality of these regions.
The NRP tripartite promoter described above for our analysis of cis-regulatory elements was also used in a genetic screen to identify nitrate regulatory mutants (Wang et al., 2009). Trans-acting mutants were identified that failed to show induction of the NRP-GUS construct in the presence of nitrate. Two lines were characterized and shown to have mutations at the nrt1.1 and nlp7 loci. NRT1.1 encodes a nitrate transporter (Tsay et al., 1993) and sensor (Remans et al., 2006; Walch-Liu and Forde, 2008; Ho et al., 2009; Wang et al., 2009) and NLP7 encodes a transcription factor (Castaings et al., 2009). These results show that the NRT1.1 and NLP7 signaling pathway(s) utilize the cis-regulatory elements in NRP to mediate nitrate induction. Our findings also show that these elements mediate ammonium repression and nitrite induction demonstrating that they are multifunctional.
It is our hope that the development of the transient expression system in N. benthamiana and the NRP tripartite construct will facilitate identification of other nitrate enhancer fragments and elements in the future. The transient system facilitates rapid analysis of nitrate responses for promoter constructs compared with several months needed for transgenic plants. The validation of the N. benthamiana results in Arabidopsis is encouraging and suggests that results from the transient system will be applicable to transgenic plants. The development of the tripartite promoter addresses a problem several labs, including our own, have reported for analyses of nitrate-regulated promoters. First, activity of nitrate-regulated promoters and enhancer fragments is greatly diminished as one reduces the size of the fragments. Second, our general enhancer screens are not identifying fragments that must contain enhancer elements. For example, our initial enhancer screen, using only the 35S minimal promoter, showed enhancer activity for the upstream NIA1-3 fragment but not for the more proximal NIA1-1 fragment, which must contain enhancer elements because it includes DNA shown in previous work to be sufficient for nitrate induction (Lin et al., 1994; Hwang et al., 1997). The 35S minimal promoter alone does not appear to be sensitive enough to detect enhancer activity from these proximal regions. The 131-bp NiR fragment used in the NRP constructs provides a possible solution as it greatly increases the sensitivity of the system to the NIA1 upstream nitrate enhancers. If the 131-bp NiR fragment acts similarly for other nitrate enhancers, it will help locate other fragments and elements that the 35S minimal promoter alone would miss.
MATERIALS AND METHODS
Transgenic Lines
Transgenic Arabidopsis (Arabidopsis thaliana) plants were produced by the floral-dip procedure using 4-week-old plants and Agrobacterium tumefaciens cultures containing the appropriate constructs (Bechtold et al., 1993). Seeds from treated plants were collected and screened for kanamycin resistance.
Growth and Treatment Conditions
For nitrate treatments of Arabidopsis, plants were grown for 10 to 14 d under 24 h light on agarose plates with nitrate-free growth media containing 2.5 mm ammonium succinate as described (Wang et al., 2003, 2004). Seedlings were transferred for 16 to 24 h to fresh growth media with either 10 mm NH4NO3 as the nitrogen source (replacing ammonium succinate) or 10 mm KCl. Seedlings were then collected for GUS assay. Each GUS assay contained 25 seedlings, and was performed in triplicate for each transgenic line.
For the transient system, Nicotiana benthamiana seeds were germinated in soil then seedlings were transferred to perlite at the two- to three-leaf stage. Plants were grown for three weeks at 25°C to 27°C with 16 h light and irrigated with modified hydroponic solution containing ammonium and no nitrate [10 mm KPO4, pH 6.5, 2.5 mm (NH4)2SO4, 2 mm MgSO4, 1 mm CaCl2, 0.1 mm FeNa2EDTA, and micronutrients: 50 μm H3BO3, 12 μm MnSO4, 1 μm ZnCl2, 1 μm CuSO4, and 0.2 μm Na2MoO4; Wang et al., 2003]. Plants were infiltrated with agrobacteria and irrigated with 10 mm KNO3 or KCl as described below.
Transient Expression Assays
All the constructs were transformed into A. tumefaciens strains C1C58. Individual agrobacteria colonies, grown for 20 h in 5-mL Luria broth containing 50 μg/mL rifampicin, 25 μg/mL gentamycine, 5 μg/mL tetracycline, were used to inoculate a 50-mL culture (Luria broth, 20 μm acetosyringone, 10 mm MES, pH 5.7, 5 μg/mL tetracycline), which was grown for 16 to 20 h at 28°C (Llave et al., 2000). Bacteria were pelleted, resuspended in infiltration medium (10 mm MgCl2, 10 mm MES, pH 5.7, 150 μm acetosyringone) to an OD600 of 0.5, then incubated at room temperature for 3 h (Llave et al., 2000).
After the initial 3 weeks of growth on nitrate-free hydroponic solution, N. benthamiana plants were irrigated with hydroponic solution containing either 10 mm KCl or 10 mm KNO3 30 min before infiltration. The third leaf form the top was then infiltrated with agrobacteria using a 1-mL syringe (without needle) by injecting 0.2 mL of the agrobacteria solution into leaf. Plants were again irrigated with hydroponic solution containing 10 mm KCl or KNO3 2 to 3 d after infiltration, then the third leaf from the top was collected for GUS assays 24 h after the final irrigation.
Quantitative GUS Assays
For transgenic Arabidopsis plants, 25 seedlings were ground in liquid nitrogen, and soluble proteins were extracted with GUS buffer (100 mm KPO4, pH 7.8, 2 mm EDTA, 5% glycerol, 2 mm dithiothreitol). GUS assays were performed as described (Beaud et al., 2005). Reactions were initiated by mixing 50 μL of protein extract with 120 μL of 1 mm p-nitrophenyl-β-d-glucuronide (Sigma-Aldrich). Reactions were incubated at 37°C for 1 to 4 h (GUS activity stayed linear for up to 16 h) then stopped by adding 800 μL 125 mm Na2CO3. OD415 was then measured with a spectrophotometer. Standard curves were made using p-nitrophenol (0–0.5 mm).
For the transient Nicotiana system, leaves were harvested and frozen in −80°C 3 to 4 d post infiltration. Frozen leaf tissue (0.1 g) was pulverized then suspended into 1 mL GUS extraction buffer (0.1 m KPO4, pH 7.8, 2 mm EDTA, 5% glycerol, 2 mm dithiothreitol). Glucuronidase activity was assayed in a GUS assay buffer (50 mm Na PO4, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 10 mm 2-mercaptoethanol, and 1 mm p-nitrophenyl β-d-glucuronide) as described (Jefferson et al., 1986).
DNA Constructs
Promoter constructs were introduced into the HindIII and PstI sites of the binary vector pDW294, which contains a cauliflower mosaic virus 35S minimal promoter upstream of the coding region for GUS as described (Busch et al., 1999).
Transcription factor binding sites were predicted using AthaMap (www.athamap.de; Bülow et al., 2009). Sequence of the NIA1-NiR components of the NRP tripartite promoter is provided at GenBank as number GQ374175.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GQ374175.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nitrate enhancer screen for the NIA1 gene.
Supplemental Figure S2. Enhancer analysis of subfragments of NIA1-3.
Supplemental Figure S3. Enhancer analysis of NIA1-14.
Supplemental Figure S4. Sequences of NRP enhancer fragments.
Supplemental Figure S5. Nitrate responses for NIA1 fragments in individual transgenic lines.
Supplemental Figure S6. Analysis of NiR fragments in transgenic plants.
Supplemental Figure S7. Sequences of NIA1-14e fragments used in promoter analysis.
Supplemental Figure S8. Nitrate responses for mutant NRP constructs in individual transgenic lines.
Supplemental Figure S9. Potential cis-regulatory modules in selected nitrate-responsive genes.
Supplementary Material
Acknowledgments
We wish to thank Detlef Weigel for contributing plasmid pDW294 and Abraham Tang for assistance with the nitrite treatments.
References
- Aigner K, Descovich L, Mikula M, Sultan A, Dampier B, Bonne S, van Roy F, Mikulits W, Schreiber M, Brabletz T, et al. (2007) The transcription factor ZEB1 (deltaEF1) represses Plakophilin 3 during human cancer progression. FEBS Lett 581: 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN. (2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28: 500–512 [DOI] [PubMed] [Google Scholar]
- Bastola DR, Pethe VV, Winicov I. (1998) Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol Biol 38: 1123–1135 [DOI] [PubMed] [Google Scholar]
- Beaud D, Tailliez P, Anba-Mondoloni J. (2005) Genetic characterization of the beta-glucuronidase enzyme from a human intestinal bacterium, Ruminococcus gnavus. Microbiology 151: 2323–2330 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Bülow L, Engelmann S, Schindler M, Hehl R. (2009) AthaMap, integrating transcriptional and post-transcriptional data. Nucleic Acids Res 37: D983–D986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Camargo A, Llamas A, Schnell RA, Higuera JJ, Gonzalez-Ballester D, Lefebvre PA, Fernandez E, Galvan A. (2007) Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 19: 3491–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al. (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57: 426–435 [DOI] [PubMed] [Google Scholar]
- de Felippes FF, Weigel D. (2010) Transient assays for the analysis of miRNA processing and function. Methods Mol Biol 592: 255–264 [DOI] [PubMed] [Google Scholar]
- Dorbe MF, Truong HN, Crete P, Daniel-Vedele F. (1998) Deletion analysis of the tobacco Nii1 promoter in Arabidopsis thaliana. Plant Sci 139: 71–82 [Google Scholar]
- Filleur S, Walch-Liu P, Gan Y, Forde BG. (2005) Nitrate and glutamate sensing by plant roots. Biochem Soc Trans 33: 283–286 [DOI] [PubMed] [Google Scholar]
- Forde BG. (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Forde BG, Walch-Liu P. (2009) Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant Cell Environ 32: 682–693 [DOI] [PubMed] [Google Scholar]
- Fritz C, Palacios-Rojas N, Feil R, Stitt M. (2006) Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J 46: 533–548 [DOI] [PubMed] [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG. (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222: 730–742 [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T, Lejay L, Wirth J, Widiez T, Palenchar PM, Nazoa P, Touraine B, Gojon A, Lepetit M. (2007) Identification of a 150 bp cis-acting element of the AtNRT2.1 promoter involved in the regulation of gene expression by the N and C status of the plant. Plant Cell Environ 30: 1366–1380 [DOI] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Gifford ML, Poultney C, Wang R, Shasha DE, Coruzzi GM, Crawford NM. (2007) Insights into the genomic nitrate response using genetics and the Sungear Software System. J Exp Bot 58: 2359–2367 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. (2009) AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Hwang CF, Lin Y, D’Souza T, Cheng CL. (1997) Sequences necessary for nitrate-dependent transcription of Arabidopsis nitrate reductase genes. Plant Physiol 113: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsh D. (1986) beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA 83: 8447–8451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu JJ, Conley AJ, Lienemann M, Brandle JE, Linder MB, Menassa R. (2010) Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol 152: 622–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. (2010) Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J 63: 269–282 [DOI] [PubMed] [Google Scholar]
- Kopertekh L, Schiemann J. (2005) Agroinfiltration as a tool for transient expression of cre recombinase in vivo. Transgenic Res 14: 793–798 [DOI] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay YF. (2010) Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13: 265–272 [DOI] [PubMed] [Google Scholar]
- Krusell L, Rasmussen I, Gausing K. (1997) DNA binding sites recognised in vitro by a knotted class 1 homeodomain protein encoded by the hooded gene, k, in barley (Hordeum vulgare). FEBS Lett 408: 25–29 [DOI] [PubMed] [Google Scholar]
- Lin Y, Hwang CF, Brown JB, Cheng CL. (1994) 5′ proximal regions of Arabidopsis nitrate reductase genes direct nitrate-induced transcription in transgenic tobacco. Plant Physiol 106: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Carrington JC. (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA 97: 13401–13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neininger A, Back E, Bichler J, Schneiderbauer A, Mohr H. (1994) Deletion analysis of a nitrite-reductase promoter from spinach in transgenic tobacco. Planta 194: 186–192 [Google Scholar]
- Nero D, Krouk G, Tranchina D, Coruzzi GM. (2009) A system biology approach highlights a hormonal enhancer effect on regulation of genes in a nitrate responsive “biomodule”. BMC Syst Biol 3: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Shichrur K, Yalovsky S. (2007) The analysis of protein-protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol 145: 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ. (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Priest HD, Filichkin SA, Mockler TC. (2009) Cis-regulatory elements in plant cell signaling. Curr Opin Plant Biol 12: 643–649 [DOI] [PubMed] [Google Scholar]
- Rastogi R, Back E, Schneiderbauer A, Bowsher C, Moffatt B, Rothstein SJ. (1993) A 330 bp region of the spinach nitrite reductase gene promoter directs nitrate-inducible tissue specific expression in transgenic tobacco. Plant J 4: 317–326 [Google Scholar]
- Rastogi R, Bate N, Sivasankar S, Rothstein SJ. (1997) Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Plant Mol Biol 34: 465–476 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A. (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103: 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander L, Jensen PE, Back LF, Stummann BM, Henningsen KW. (1995) Structure and expression of a nitrite reductase gene from bean (Phaseolus vulgaris) and promoter analysis in transgenic tobacco. Plant Mol Biol 27: 165–177 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar S, Rastogi R, Jackman L, Oaks ASR. (1998) Analysis of cis-acting DNA elements mediating induction and repression of the spinach nitrite reductase gene. Planta 206: 66–71 [Google Scholar]
- Stitt M. (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Stitt M, Muller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A. (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Tang PS, Wu HY. (1957) Adaptive formation of nitrate reductase in rice seedlings. Nature 179: 1355–1356 [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. (1993) A herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG. (2008) Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J 54: 820–828 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Crawford N. (2007) Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol 145: 1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM. (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA. (2003) Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet 4: 29–38 [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ. (1992) Molecular and Genetic Characterization of the Nitrate Reductase Structural Genes (NIA1 and NIA2) of Arabidopsis thaliana. University of California at San Diego, La Jolla, CA, pp 198–204 [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T. (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101: 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YN, Li RG, Qi M. (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Zhang HM, Forde BG. (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.