Abstract
Near-isogenic lines (NILs) are valuable genetic resources for many crop species, including soybean (Glycine max). The development of new molecular platforms promises to accelerate the mapping of genetic introgressions in these materials. Here, we compare some existing and emerging methodologies for genetic introgression mapping: single-feature polymorphism analysis, Illumina GoldenGate single nucleotide polymorphism (SNP) genotyping, and de novo SNP discovery via RNA-Seq analysis of next-generation sequence data. We used these methods to map the introgressed regions in an iron-inefficient soybean NIL and found that the three mapping approaches are complementary when utilized in combination. The comparative RNA-Seq approach offers several additional advantages, including the greatest mapping resolution, marker depth, and de novo marker utility for downstream fine-mapping analysis. We applied the comparative RNA-Seq method to map genetic introgressions in an additional pair of NILs exhibiting differential seed protein content. Furthermore, we attempted to optimize the comparative RNA-Seq approach by assessing the impact of sequence depth, SNP identification methodology, and post hoc analyses on SNP discovery rates. We conclude that the comparative RNA-Seq approach can be optimized with sufficient sampling and by utilizing a post hoc correction accounting for gene density variation that controls for false discoveries.
Near-isogenic lines (NILs) are valuable genetic resources for the identification of genomic regions and alleles responsible for trait variation. This is particularly true within the soybean (Glycine max) community, where NILs can be utilized to map the genomic regions responsible for the phenotypic variation of numerous traits, including seed composition, nutrient deficiency tolerance, maturity, and several others (Bernard et al., 1991).
Historically, the mapping of NIL introgression sites has relied on a wide range of electrophoresis-based molecular tools, including isozyme, RFLP, amplified fragment length polymorphism, and simple sequence repeat (SSR) analyses (Muehlbauer et al., 1989, 1991; Molnar et al., 2003; Nichols et al., 2006). More recently, automated genotyping technologies have accelerated the efficiency of genetic mapping. Such methods, including single feature polymorphisms (SFP) analysis of microarray data and single nucleotide polymorphism (SNP)-based genotyping methods, have been successfully applied to the mapping of soybean NILs and other mapping populations (Hyten et al., 2008; Kaczorowski et al., 2008; Bolon et al., 2010). However, the mapping resolution of all of these platforms is limited by the location and depth of informative markers available for a given species. Additionally, many of the markers will not be polymorphic for the specific set of genotypes utilized in a NIL introgression study.
The recent sequencing of the soybean genome (Schmutz et al., 2010) and recent advances in next-generation sequencing (NGS) technologies have the potential to overcome some of these limitations. Comparative NGS analyses of NILs with their respective parental lines offers the possibility of identifying SNP polymorphisms that are unique to each NIL-parent group. Furthermore, comparative NGS analyses offer a potentially greater marker depth than previous mapping methods. Direct RNA sequencing (RNA-Seq) via NGS allows for these goals to be accomplished at a lower cost, as the sequence coverage per SNP will be enriched within transcribed regions, thereby reducing the total amount of sequence required to confidently identify true polymorphisms.
In this study, we have attempted to map the introgression loci of the soybean NIL IsoClark (PI 547430) relative to its recurrent parent Clark (PI 548533). Previous studies of Clark and IsoClark NIL have characterized the differences between these lines at multiple levels of resolution, including morphological and transcriptional differences (O’Rourke et al., 2007a, 2007b, 2009). Compared with Clark, IsoClark is an iron-inefficient line, putatively caused by the introgression of iron-inefficient genetic material from the donor line T203. Iron deficiency chlorosis remains a problem of great economic importance for soybean growers (Hansen et al., 2003). Therefore, the Clark-T203-IsoClark family represents a soybean NIL family of both scientific and economic importance. Here, we have examined several genotyping technologies to improve the mapping of T203 introgression sites in IsoClark. Furthermore, we have applied our RNA-Seq-based methods toward mapping the introgression of two additional soybean NILs exhibiting seed composition differences (Nichols et al., 2006). We have compared some of the existing (Affymetrix SFP and Illumina GoldenGate) and emerging (Illumina NGS) technologies for soybean introgression mapping and speculate on what methods and analytical tools will be most useful in the postgenomic era.
RESULTS
Introgression Mapping Using Affymetrix SFPs
Affymetrix SFP analysis was used to identify putative T203 introgressions in the NIL genotype IsoClark. SFPs between Clark and IsoClark were considered indicative of potential T203 introgression sites. We compared 10-d and 14-d root transcripts from Clark and IsoClark, each grown hydroponically in iron-sufficient and iron-limiting conditions (see “Materials and Methods”). This analysis identified four obvious SFP clusters in the IsoClark genome, on chromosomes 3, 5, 8, and 16 (Table I; Fig. 1). Based on these analyses, it appears that the T203 introgression on chromosome 3 is the largest of the four. Eleven additional SFPs were identified outside of these clusters and were scattered throughout the genome (Fig. 1). These SFPs were inferred to be false positives unless validated by additional genotyping platforms.
Table I. Introgression mapping comparison.
Comparison of Clark-IsoClark polymorphism rates at larger introgression sites using three different genotyping platforms. The number of SNPs identified from the RNA-Seq data depends on the SNP-calling algorithm and the number of libraries compared (single-library comparisons versus four-library comparisons). The RNA-Seq method 1 and method 2 analyses protocols are described in “Materials and Methods.” N/A, Not applicable.
| Chromosome | Position | Approximate Size | SFP | GoldenGate | RNA-Seq Single Library (Method 1) | RNA-Seq Four Library (Method 1) | RNA-Seq Single Library (Method 2) | RNA-Seq Four Library (Method 2) |
| Mb | Mb | |||||||
| Gm03 | 36.3–45.8 | 9.2 | 13 | 15 | 105 | 120 | 102 | 204 |
| Gm04 | 44.7–45.6 | 1 | 0 | 1 | 7 | 21 | 6 | 23 |
| Gm05 | 38.2–39.5 | 1 | 4 | 2 | 15 | 24 | 17 | 34 |
| Gm08 | 2.0–3.5 | 1.5 | 0 | 4 | 1 | 2 | 1 | 14 |
| Gm08 | 43.8–47.0 | 3.2 | 7 | 1 | 0 | 8 | 2 | 17 |
| Gm13 | 35.5–35.9 | 0.5 | 1 | 1 | 10 | 24 | 7 | 23 |
| Gm16 | 30.4–31.9 | 1.5 | 6 | 3 | 21 | 48 | 23 | 50 |
| Other | N/A | N/A | 10 | 0 | 13 | 14 | 97 | 104 |
| Total | N/A | N/A | 41 | 27 | 172 | 261 | 255 | 469 |
Figure 1.
Chromosomal positions of Affymetrix SFPs and GoldenGate SNPs identified between Clark and IsoClark. Chromosomes are labeled at the top according to number, and centromere positions are shown as white circles. Red lines indicate the physical map positions of Affymetrix SFPs, and blue lines indicate the physical map positions of GoldenGate SNPs. Genomic regions coincident for both SFPs and SNPs are indicated with yellow boxes, and genomic regions exhibiting only GoldenGate SNPs are indicated with white boxes.
Introgression Mapping Using the Illumina GoldenGate Platform
The Illumina GoldenGate genotyping platform was used to identify putative T203 introgressions in IsoClark. SNPs between Clark and IsoClark were considered indicative of potential T203 introgression sites. This analysis identified seven loci that were polymorphic between Clark and IsoClark (Table I; Fig. 1). Four of these seven loci had been previously identified as likely introgressions based on SFP analysis. One of the remaining loci, on chromosome 13, colocalized with a solo SFP. The two remaining loci, near the top of chromosome 8 and toward the bottom portion of chromosome 4, did not colocalize with any previously identified SFP (Table I; Fig. 1).
Introgression Mapping Using Two SNP Calling Methods on a Single Library of Illumina RNA-Seq Data
Illumina RNA-Seq data were used to identify putative T203 introgressions in IsoClark. SNPs identified de novo between Clark and IsoClark transcripts were mapped to the soybean reference genome, and their genomic positions were considered as potential T203 introgression sites. Altogether, RNA-Seq SNP discovery was performed in four ways: single-library comparisons using method 1 (see description below and in “Materials and Methods”), four-library comparisons using method 1, single-library comparisons using method 2 (see description below and in “Materials and Methods”), and four-library comparisons using method 2. This approach allowed us to compare the sensitivity and accuracy of RNA-Seq SNP discovery across different analytical methods and sequence depths.
For the single-library comparisons, Illumina NGS was performed on the RNA isolated from the 10-d iron-limiting root samples, resulting in 30,897,337 short-read sequences. These sequences were then aligned to the soybean genome (Glyma1.01 genome assembly) to identify SNPs between Clark and IsoClark in protein-coding regions. SNPs were considered indicative of T203 genomic introgression sites. Two methods were used to identify SNPs and to gain a measure of confidence in the SNPs determined by each method.
Method 1 used the program SOAP2 (Li et al., 2009b) to align the short-read sequences to the soybean genome (Schmutz et al., 2010). SNPs were then identified from the SOAP2 alignment using the program SOAPsnp (Li et al., 2009a). Only unique alignments were considered. SNPs were screened for a minimum base-call quality score of 10 and average quality score of 20. For further filtering requirements, see “Materials and Methods.”
Method 2 used the program GSNAP (Wu and Nacu, 2010) to align the short-read sequences to the soybean genome. GSNAP can handle short-read sequences that fall over splice junctions. All mismatches from the best alignment for a read were tallied in a database, and a reporting script required the potential SNP to meet the following criteria: a minimum of two unique alignments, average quality score of 20, and a minimum of 80% of the reads uniquely aligned to the position calling the SNP within a sample. For further filtering requirements, see “Materials and Methods.”
There were 172 SNPs identified by method 1 (Supplemental Table S1) and 255 SNPs identified using Method 2 (Supplemental Table S2) when applied to the 10-d root RNA-Seq single-library comparison. The putative introgression sites previously identified by SFP and GoldenGate SNP analyses accounted for 159 of the 172 SNPs identified using method 1 and 158 of the 255 SNPs identified using method 2 (Table I). Thus, the larger introgression sites identified using SFP and GoldenGate analyses were generally confirmed by the SNPs called by each method, particularly the sites on chromosomes 3, 5, and 16 (Table I). Introgression sites on chromosomes 4 and 13, which were tentatively identified by SFP and/or GoldenGate analyses, were strongly confirmed by the RNA-Seq data (Table I). Surprisingly, the two chromosome 8 introgression sites identified by SFP and/or GoldenGate analyses were not strongly supported by the RNA-Seq SNP data obtained from this single-library comparison (Table I).
Introgression Mapping Using SNP Data from Multiple Illumina RNA-Seq Libraries
To determine if the quantity of the short-read sequence data for identifying introgression sites was limiting sensitivity, we analyzed eight additional Illumina RNA-Seq data sets using method 1 and method 2, four from Clark and four from IsoClark plants grown for 19 d, after which the plants were exposed to iron-sufficient and iron-limiting conditions for 24 h. We refer to this comparison as the “four-library comparison.” The 19-d root and leaf data set contained 91,303,822 short-read sequences. Therefore, this experiment included four times the number of experimental conditions and approximately three times the number of short-read sequences than were used in the RNA-Seq single-library comparison described in the previous section.
The RNA-Seq four-library comparison of the 19-d samples identified 261 SNPs with method 1 (Supplemental Table S3) and 469 SNPs with method 2 (Supplemental Table S4). The method 1 SNPs appeared primarily in the larger introgression sites, with only 14 located outside of these regions. The method 2 SNPs were found outside of the larger introgressions at a substantially higher frequency (Table I); the locations of these SNPs were scattered across the genome. Both method 1 and method 2 identified the two introgression sites on chromosome 8 that were essentially missed by the single-library comparison (Table I). However, the method 2 analysis identified these sites at a much higher frequency.
Without a substantial accumulation of SNPs in one region in the genome or the coincidental overlap of Affymetrix SFPs or Illumina GoldenGate polymorphisms with the NGS SNPs, it may be difficult to distinguish between a site of introgression and an RNA-Seq false-positive SNP call. This problem is further confounded by variations in gene density along each chromosome. In order to identify all or nearly all of the prominent T203 introgression sites, a statistical method for distinguishing between introgression sites and false positives randomly scattered across the genome was required.
Accounting for Gene Density Increases the Sensitivity of Introgression Mapping
The RNA-Seq data identified SNPs based on short-read sequences taken from protein-coding regions. To account for gene density and to provide a statistical measure of SNP clustering, an algorithm for SNP clustering utilizing a “bootstrap method” was developed.
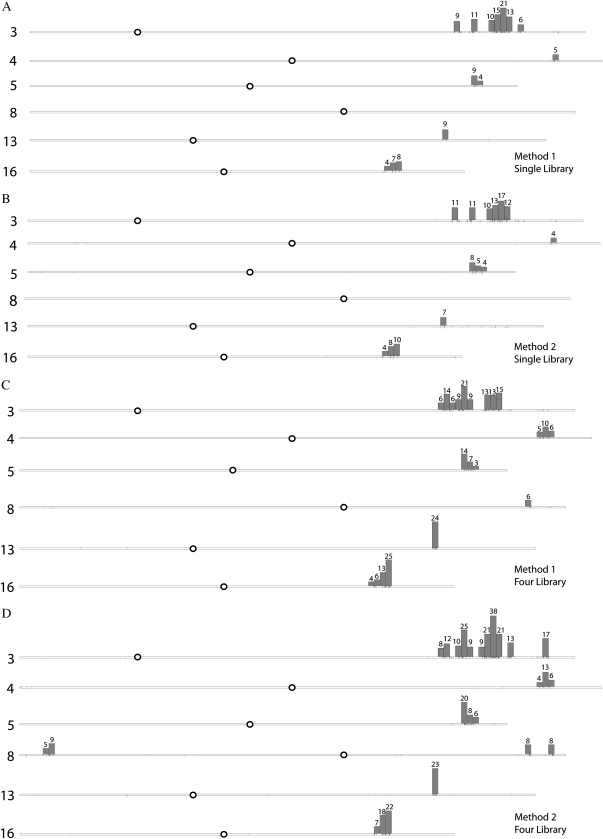
The simulated density of SNPs that might be found within a chromosomal interval by random chance was determined by choosing genes at random with replacement. For example, if 204 SNPs were identified on chromosome 3, then the positions of 204 genes from chromosome 3 were chosen at random with replacement. The position of the gene was estimated by averaging the start and end coordinates. This process was repeated 1,000 times to obtain an estimate of the mean SNP density and sd for a given interval. Intervals in the genome that contained a significantly higher density of SNPs than would be expected at random were inferred to be introgressed. An interval was considered to contain a significantly higher density of SNPs if there were three or more SNPs in the interval and the number of SNPs was greater than 3 sd above the mean SNP density expected by random chance for a given interval. When the bootstrap method was applied to SNPs identified using method 1 and method 2 on the RNA-Seq single-library comparison (10-d root), significant intervals were identified on chromosomes 3, 4, 5, 13, and 16 (Fig. 2, A and B). SNPs identified using method 1 and method 2 from the RNA-Seq four-library comparison (19-d root and leaf) revealed the same introgression sites and additional sites on chromosome 8 (Fig. 2, C and D). The introgression sites identified in the bootstrap method are conservative estimates of the full introgression site but account for 80% of the single-library SNPs and 93% of the four-library SNPs identified in Table I.
Figure 2.
Significant intervals of SNP clustering between the Clark and IsoClark lines were found on six chromosomes, 3, 4, 5, 8, 13, and 16, as determined from the bootstrap method. Chromosomes are labeled on the left according to number, and centromere positions are shown as white circles. Vertical boxes indicate 500,000-nucleotide intervals. The number of SNPs found in each interval is indicated above the interval. A, Clustering of SNPs obtained from the 10-d root data using method 1 on the single-library comparison. B, Clustering of SNPs obtained from the 10-d root data using method 2 on the single-library comparison. C, Clustering of SNPs obtained from the 19-d root and leaf data using method 1 on the four-library comparison. D, Clustering of SNPs obtained from the 19-d root and leaf data using method 2 on the four-library comparison.
These data suggest that the quantity and coverage of short-read sequences present in the RNA-Seq four-library comparison may alone be sufficient to identify the same introgression sites as were determined from a combination of SFP, GoldenGate, and RNA-Seq single-library comparison. More SNPs pass through the filtering criteria with the increased number of reads from the RNA-Seq four-library comparison. Additionally, the sensitivity of the RNA-Seq four-library comparison is aided by the sampling of RNA from different tissue types, ensuring that a more comprehensive set of transcripts (and genome space) was surveyed as compared with the single-library comparison.
Application of the Advanced NGS Introgression Mapping on a Second NIL Pair
To further validate our method for determined introgression sites, we performed the method 2 analysis followed by the bootstrapping post hoc method on an additional set of two NILs, HiPro and LoPro. The two NILs, derived by introgressing Glycine soja into a soybean background (see “Materials and Methods”), exhibit differential seed protein content (Nichols et al., 2006; Bolon et al., 2010). In this case, we were interested in identifying differential introgression patterns between the two lines; therefore, the RNA-Seq SNP comparison was performed directly between HiPro and LoPro, rather than between the NILs and the soybean recurrent parent.
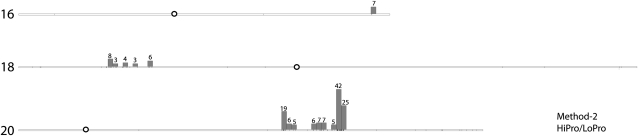
Twenty-eight libraries taken from a variety of tissues and seed developmental stages were included in the RNA-Seq SNP analysis. These data included 97,637,480 short-read sequences. Within this seed protein NIL data, 387 SNPs were identified (Supplemental Table S5). Approximately 40% (153 out of the 387 SNPs) were located within genomic regions determined to be significant based on our bootstrap algorithm. The remaining SNPs in each experiment were randomly scattered across the genome. Our method was able to easily identify the well-known introgressed region on chromosome 20 (Nichols et al., 2006; Bolon et al., 2010). It also identified regions on chromosome 16 and chromosome 18 that were previously unknown (Fig. 3). SNP GoldenGate analysis on HiPro and LoPro validated all three of these introgressions (data not shown).
Figure 3.
Significant clusters of SNPs for the seed protein lines were found on three chromosomes, 16, 18, and 20, as determined from the bootstrap method. Chromosomes are labeled on the left according to number, and centromere positions are shown as white circles. Vertical boxes indicate 500,000-nucleotide intervals. SNPs were identified via method 2. The number of SNPs found in each interval is indicated above the interval. SNPs were clustered from the seed protein RNA-Seq data that contained 14 libraries for each NIL.
Introgression Validation
The IsoClark introgression sites on chromosomes 4, 5, 13, and 16 were confirmed through resequencing by PCR amplification of Clark, IsoClark, and T203 DNA (the introgression on chromosome 3 is well established and did not require further validation). Additionally, candidate introgressions were also validated with SSR markers. SSR markers BARCSOYSSR_04_1282, BARCSOYSSR_04_1286, BARCSOYSSR_04_1297, and BARCSOYSSR_04_1299 were polymorphic between Clark and IsoClark on chromosome 4. Similarly, SSR markers Sat_217 and Sat_271 were polymorphic on chromosome 5. SSR marker Satt228 was polymorphic on chromosome 8 (nucleotide position 45,272,500). SSR marker Satt490 was polymorphic on chromosome 13. SSR markers BARCSOYSSR_16_1047, BARCSOYSSR_16_1057, BARCSOYSSR_16_1059, and BARCSOYSSR_16_1070 were polymorphic on chromosome 16. All markers and positions were developed by Song et al. (2004, 2010) and are available on Soybase (http://soybase.org). Only the predicted introgression between 2.0 and 3.5 Mb on chromosome 8 was not confirmed through resequencing or SSR markers due to problematic primers or lack of SSR markers in that region. This region, however, has additional support from Illumina GoldenGate SNP data. A similar introgression validation was performed for the HiPro and LoPro NILs. Resequencing by PCR amplification confirmed the candidate introgression on chromosome 16 but was unable to confirm the introgression on chromosome 18 (the introgression on chromosome 20 is well established and did not require further validation). However, all three of these introgressions have been validated by GoldenGate SNP data (see previous section).
DISCUSSION
Comparison of SFP, SNP GoldenGate, and NGS RNA-Seq for Genetic Introgression Mapping
The Affymetrix SFP and Illumina GoldenGate SNP methodologies are established as genetic mapping approaches that are far more efficient than electrophoresis-based methods for genome-wide mapping applications (Hyten et al., 2008; Kaczorowski et al., 2008; Bolon et al., 2010). In our introgression mapping for the IsoClark NIL, the SFP and GoldenGate platforms primarily identified an overlapping set of putative introgression sites (Fig. 1). The GoldenGate platform identified seven introgression sites, which were validated in subsequent experiments, indicating that this platform is robust for introgression mapping. The SFP analysis identified five of these seven sites. However, the SFP analysis also identified 10 polymorphic markers outside of these larger introgressions, some of which are believed to be false positives.
The SFP and GoldenGate SNP mapping approaches are relatively efficient and inexpensive. However, in our analyses, the Affymetrix SFP and GoldenGate platforms identified a relatively small number of polymorphic markers (Table I). The low number of markers limits our ability to resolve the introgression boundaries and leaves open the possibility of missing smaller introgressions altogether.
The RNA-Seq methodologies clearly identified a much greater number of polymorphic loci within the known introgression sites (Table I). The increased marker coverage allowed us to identify the introgression boundaries at a higher resolution. The two introgressions on chromosome 8, however, were exceptional in this regard. The introgression at positions 2.0 to 3.5 Mb was easily identified by the GoldenGate approach, and the introgression at positions 43.9 to 47.0 Mb was easily identified by the SFP approach. It is unclear what properties of the chromosome 8 introgressions caused this phenomenon; the gene content and transcription levels are both relatively high in these regions (Libault et al., 2010; Schmutz et al., 2010; Severin et al., 2010). The other five larger introgressions were most clearly identified by the RNA-Seq approach, regardless of which of the four RNA-Seq analyses was considered.
Importantly, the RNA-Seq approach offers two important benefits that standardized mapping platforms do not. First, the SNP markers identified via RNA-Seq are specific to the genetic materials of interest. By contrast, the soybean GoldenGate SNP panel is derived from different genetic materials than was used in our study; many of the 1,536 SNPs would be nonpolymorphic between our original parents, Clark and T203, and therefore would be uninformative for this study. The RNA-Seq data, however, identify SNPs that are necessarily polymorphic between our genetic materials of interest. The SNPs identified de novo by RNA-Seq can be directly used for fine-mapping on subsequent generations of this material using a custom SNP genotyping platform, like the MassARRAY (Sequenom) or SNPlex (Applied Biosystems) platform (Ding and Jin, 2009). Second, the RNA-Seq data may be mined for transcriptional differences or genetic alterations between Clark and IsoClark that may identify candidate genes that drive the differential iron susceptibility observed between the lines. The Affymetrix data will also allow for the analysis of transcript differences; however, the RNA-Seq data provide a larger sampling of transcripts and also permit the possible identification of frame-shift or nonsense mutations within introgressed loci.
We noted two primary drawbacks to the RNA-Seq approach. First, this technology is currently more expensive than using standardized platforms. This problem should be mitigated in the near future, as NGS is expected to become more affordable and accessible. Second, this approach targets mRNA transcripts; therefore, our marker depth is necessarily biased for gene-rich regions. Although we have applied a bootstrapping method to correct for gene density biases, severely gene-poor regions may not be represented in our analyses. Additionally, exonic regions tend to have more highly conserved sequences than noncoding regions. Introgression mapping could be improved if the NGS technology was used directly on DNA rather than RNA. With current technology, this would provide better genomic coverage but may not provide the sequence depth required for confident SNP identification at a reasonable cost. A more cost-effective strategy would be to perform comparative NGS on reduced representation genomic DNA libraries (Van Tassell et al., 2008; Fu et al., 2010; Hyten et al., 2010a). As sequencing technologies improve and the cost per library decreases, the limitations of sequencing depth and read length will no longer be an issue.
Altogether, our data indicate that the RNA-Seq approach offers the greatest depth and resolution for mapping most genomic introgressions; however, the SFP and GoldenGate approaches were more efficient for mapping certain introgressions. The combination of SFP, GoldenGate, and RNA-Seq data does not necessarily ensure that we have identified all the introgressed loci in these NILs. For example, when we combined the unique SNPs identified using method 1 and method 2, we noted that a cluster of four SNPs was identified within an approximately 480-kb interval on IsoClark chromosome 2 (positions 42.35–42.83 Mb). Intuitively, it would appear that these SNPs may define a genetic introgression; however, this region was not identified as significant by our bootstrap analyses using each method (1 and 2) individually. Using the methods described here, introgressions greater than 0.5 to 1.0 Mb can be efficiently mapped with relatively high resolution, assuming that there is an adequate level of sequence polymorphism between the parental lines. However, it may be difficult to identify introgressions that are small, located within gene-poor regions, or located within regions of low diversity between parental lines. Identification of such introgressions, such as the putative introgression on IsoClark chromosome 2, may require “manual” rather than automated analytical approaches, along with sufficient validation.
Optimizing NGS RNA-Seq for Genetic Introgression Mapping
We tested the impact of three different factors on RNA-Seq introgression mapping: (1) sequence depth; (2) SNP identification methodology; and (3) post hoc analysis accounting for gene density.
Clearly, the RNA-Seq method is more effective for introgression mapping when the sequence depth and tissue sampling range are expanded. Our data indicate that our four-library comparison with different tissue types and treatments identified greater than 1.5 times more SNPs than a single-library comparison (Table I). Consequently, introgression sites that were either poorly identified or not identified in the single-library analysis (namely, the two introgressions on chromosome 8) were more confidently identified in the four-library comparison.
We applied two different SNP identification methodologies to the RNA-Seq data, generically called method 1 and method 2 (see “Materials and Methods”). The two methods were each applied to the RNA-Seq single-library and four-library comparisons of Clark and IsoClark. These two identification methods appeared to offer an interesting tradeoff in benefits. Method 1 appeared to be the more conservative approach, as it identified fewer SNPs. However, only 5% to 7% of the SNPs were located outside of the putative larger introgression regions identified by SFP and GoldenGate genotyping; it is unclear what proportion of these SNPs represent false-positive calls. Method 2 appeared to be the more liberal method, identifying far more SNPs than method 1 (Table I). A high percentage of SNPs fell outside of the putative larger introgressions (approximately 22%–38%), indicating that this method may foster a higher rate of false discoveries. However, method 2 was more effective at identifying recalcitrant introgressions, primarily the two chromosome 8 introgressions. It is worth noting that the differential SNP discovery rates of method 1 and method 2 are not necessarily a function of the algorithms used (Li et al., 2009a, 2009b; Wu and Nacu, 2010) but are also influenced by the stringency of the identification parameters. Thus, either method could be performed with greater or reduced stringency, as needed by the user.
The post hoc bootstrap method was used to distinguish true introgressions from false discoveries by accounting for regional SNP clustering rates and gene density differences across the genome. This method proved most valuable when applied to the method 2 SNP calls, as this was the more permissive identification method and presumably identified a higher relative rate of false positives. The bootstrap method, when applied to the method 2 four-library comparison SNPs, identified all of the seven larger introgressions, including the recalcitrant introgressions on chromosome 8.
The data analyses presented here covered a range of tissues and conditions and were performed on a well-studied organism with a set of high-quality predicted gene models. Our analyses indicate several regions of introgression that have been confirmed for two different NILs. However, had the number of expressed genes been significantly lower than what is found in our data sets, it may have been prudent to only use expressed genes, rather than every gene predicted in the genome, when accounting for gene density with our bootstrap method.
CONCLUSION
In this report, we show that SFP, Illumina GoldenGate, and RNA-Seq are complementary methods for identifying genetic introgressions in NILs. We show that the depth of coverage of SNPs identified from NGS RNA-Seq technology in combination with a bootstrapping method is an effective tool for identifying introgression sites. As new NGS technologies arise (Eid et al., 2009; Rusk, 2009) and become more affordable, NGS of genomic DNA at greater depth will become feasible for mapping purposes.
MATERIALS AND METHODS
Plant Materials
Two pairs of soybean (Glycine max) NILs were used in this study: (1) a NIL line selected for differential iron deficiency chlorosis susceptibility; and (2) a NIL pair selected for differential seed protein. The iron-efficient parent line Clark (PI 548533) and the iron-inefficient NIL IsoClark (PI 547430) have been extensively described in previous studies (O’Rourke et al., 2007a, 2007b, 2009). The IsoClark NIL was derived from crossing Clark with iron-inefficient T203 (PI 54619), followed by five subsequent backcrosses to Clark. Subsequent self-mating yielded the iron-inefficient NIL IsoClark.
The seed protein NIL pair was derived from introgressing Glycine soja (PI468916) into soybean (A81-356022) and has been described previously (Nichols et al., 2006; Bolon et al., 2010). The BC5F5 plant P-C609-45-2-2 was heterozygous for the LG I protein quantitative trait locus (QTL) introgression from G. soja. The derived BC5F6 NILs segregated for the LG I protein QTL introgression. The BC5F6 line LD04-15154 (HiPro) maintained the introgression and the corresponding high seed protein phenotype. The BC5F6 line LD04-15146 (LoPro) segregated out the QTL introgression and exhibited the low seed protein phenotype.
RNA Sampling of Clark and IsoClark Root Tissues from Iron-Sufficient and Iron-Limiting Conditions (10 d and 14 d)
Clark and IsoClark were grown in hydroponic conditions as described by O’Rourke et al. (2009). Both genotypes were exposed to two different hydroponic treatments, iron sufficient [100 μm Fe(NO3)3] and iron limiting [50 μm Fe(NO3)3]. Roots were collected and flash frozen in liquid nitrogen following 10 and 14 d of growth. [The iron-limiting 14-d sample was switched to a 100 μm Fe(NO3)3 treatment at day 12.] RNA samples were purified from both Clark and IsoClark root tissues using the TRIzol method (Invitrogen) and DNase treated with the Ambion DNA-free kit according to the manufacturer’s instructions (Applied Biosystems/Ambion). The samples were then further purified using the RNeasy mini kit (Qiagen). These RNA samples are referred to as the “10-d root” and “14-d root” samples, respectively.
RNA Sampling of Clark and IsoClark Tissues following Iron Shock (19-d Root and Leaf)
Clark, IsoClark, and T203 seeds were germinated using germination paper soaked in water for 6 d in a growth chamber set at 27°C. Plants were grown in hydroponic conditions as described by O’Rourke et al. (2009) in the greenhouse for 13 d, which coincided with the fully open first trifoliate. At this time, the plants were placed in either iron-sufficient or iron-deficient conditions. Briefly, the plant roots were rinsed in six buckets of water for 15 s minimum in each bucket and then returned to a fresh hydroponic bucket either sufficient in iron [100 μm Fe(NO3)3·9H2O] or deficient in iron [50 μm Fe(NO3)3·9H2O].
Plants were grown for 24 h in their new iron environment, where the trifoliates, trifoliolates, and roots were harvested, placed in individual tubes and flash frozen in liquid nitrogen, and stored at –80°C. Total RNA was isolated using the RNeasy mini kit (Qiagen) following the Qiagen protocol for everything except the final elution step, which was extended by 5 min to optimize RNA concentration. Quality was checked using a NanoDrop Spectrophotometer (Thermo Scientific). These RNA samples from root and leaf in iron-sufficient and iron-deficient conditions are referred to as the “19-d” samples.
RNA Sampling of the Seed Protein NIL
Seeds from NILs generated from soybean (A81-356022) and G. soja (PI468916) specific for the LG I seed protein QTL were grown in growth chambers to mimic Illinois field growing conditions, as described by Bolon et al. (2010). Briefly, 14 tissues that included seven stages in seed development were harvested from the two NILs: HiPro (LD0-15154) and LoPro (LD0-15146), with high and low seed protein phenotypes. RNA was extracted as described by Bolon et al. (2010). These RNA samples are referred to as the “HiPro” and “LoPro” NIL samples.
SFP Analysis
The 10-d and 14-d root RNA samples were labeled and hybridized to the Affymetrix GeneChip Soybean Genome Array according to the manufacturer’s instructions. Three biological replicates for each genotype and treatment were collected and hybridized. All data are accessible at http://www.ncbi.nlm.nih.gov/geo/ under accession number GSE22227.
SFPs between Clark and IsoClark were identified based on the Affymetrix data as described previously (Xu et al., 2009). SFPs between Clark and IsoClark identified at any of the four levels of comparison (10-d iron sufficient, 10-d iron limiting, 14-d iron sufficient, or 14-d iron limiting) were included in the downstream SFP analyses. The Affymetrix SFP probe sets were mapped back to the Williams 82 soybean genome reference sequence (Schmutz et al., 2010). T203 genomic introgressions into IsoClark were inferred based on SFP colocalization clusters.
Illumina GoldenGate Mapping
Clark, IsoClark, and T203 DNA samples were purified using the Qiagen DNeasy method according to the manufacturer. These DNA samples were genotyped using the Illumina GoldenGate Universal Soy Linkage Panel (USLP 1.0) of 1,536 SNP loci for soybean, as described previously (Hyten et al., 2010b).
Illumina NGS of RNA
The Illumina NGS platform was used to identify SNPs between Clark and IsoClark. The three Clark and IsoClark RNA biological replicates from the 10-d root tissues in the iron-limiting condition were pooled within each genotype and submitted for NGS analysis. Similarly, the 19-d root and leaf RNA samples grown in iron-sufficient and iron-deficient conditions were each pooled among three biological replicates within each genotype and submitted for NGS analysis. Therefore, eight different pooled samples from the 19-d study were sequenced, consisting of Clark root and leaf in stressed and unstressed conditions and IsoClark root and leaf in stressed and unstressed conditions.
RNA-Seq data acquisition from Illumina sequencing methods was carried out by the National Center for Genome Resources. These techniques along with RNA-Seq data analysis methods for the seed protein NILs have been described by Severin et al. (2010). Briefly, poly(A)-containing RNA isolated from total RNA was converted to cDNA. Illumina adapters were added by ligation and size selected by electrophoresis for approximately 500-bp fragments. The purified DNA libraries were PCR amplified for 15 cycles and assessed by Nanodrop ND-1000 for quality and quantity before loading onto an Illumina flow cell. Short reads of 36 bp were obtained and processed through image analysis, base-calling quality filtering, and per base confidence scores. Sequence reads were then aligned to the 8× soybean genome sequence assembly.
NGS SNP Discovery Using Method 1
Software SOAP2 (Li et al., 2009b) and SOAPsnp (Li et al., 2009a) were used for SNP discovery between Clark and IsoClark genotypes using an RNA-Seq single-library comparison (the 10-d iron-limiting root samples). A customized pipeline was developed for this analysis. Briefly, 15,260,698 36-base read sequences of Clark and 15,636,639 reads of IsoClark from Illumina sequencing were aligned to the soybean genome sequence (Schmutz et al., 2010) using SOAP2. Only the unique alignment hits were selected by setting the program parameter r = 0. All position loci of the alignment files were screened by SOAPsnp for SNPs and pair compared between Clark and IsoClark. The potential SNPs were selected using the criteria of minimum base-call quality of 10, average quality of 20, and minimum best hits of four. The SNP was not allowed to be an ambiguous base (e.g. SNP ≠ “N”).
In order to plot the SNP alignment, all short-read sequences that encompassed the SNP positions were extracted from the original Illumina read files. For each SNP, the short reads of Clark and IsoClark and the 68-base genomic sequences that encompass the SNPs in the middle were aligned by using emma of the EMBOSS suite (Rice et al., 2000). The aligned sequences were plotted using the EMBOSS prettyplot program.
The locations of the SNPs discovered were extracted, and an R script was created for mapping these SNPs onto soybean chromosomes using a 1,000-base window size. The protocol described is referred to as the RNA-Seq method 1.
To determine the difference in sensitivity between an RNA-Seq single-library comparison and a four-library comparison, method 1 was also applied to the four 19-d samples of Clark and IsoClark: root under iron-sufficient conditions, root under iron-limiting conditions, leaf under iron-sufficient conditions, and leaf under iron-limiting conditions. The pooled 19-d samples contained 32,030,175 36-base read sequences in Clark and 59,273,647 read sequences in IsoClark.
NGS SNP Discovery Using Method 2
For comparison, the software GSNAP (Wu and Nacu, 2010) was also used for SNP discovery between Clark and IsoClark genotypes on the RNA-Seq single-library comparison (the same 10-d iron-limiting root samples used for method 1) and the 19-d four-library comparison. Briefly, Clark and IsoClark reads from Illumina sequencing were each aligned to the soybean genome sequence using GSNAP. The alignment program was set to allow for alignment over a splice junction. Alignments of short-read sequences without at least 34 matches were not considered. The following requirements were also needed for a SNP to be called: a minimum of two unique alignments calling the SNP, average base-call quality of 20, and minimum of 80% of the reads uniquely aligned to the position calling the SNP. SNPs were further screened for a minimum short-read coverage of four and a difference in allelic frequency between the NILs of 50%. The protocol described is referred to as the RNA-Seq method 2.
Statistical Significance and Visualization of SNP Clusters
A specific number of SNPs were found on each chromosome using method 1 and method 2. To determine which regions on the chromosome had a significantly higher density of SNPs than might be found by random chance, the same number of SNPs found on each chromosome was simulated using a bootstrapping protocol (Supplemental File S1). Since the sequence used to identify SNPs was taken from protein-coding regions, the locations of the simulated SNPs were generated from the average position of each gene on the chromosome chosen at random. Each chromosome was divided into 500,000-nucleotide tandem intervals, resulting in a total of 1,908 intervals analyzed across the 20 chromosomes. The average number of simulated SNPs and sd within each 500,000-nucleotide interval was determined from 1,000 simulations. SNPs in an interval were considered significant if the number of SNPs was greater than 3 sd above the simulated SNPs in the interval and the total SNP count in the interval was three or more. Once the intervals with significant SNP clustering were determined, these regions were plotted onto a scaled version of each chromosome using the rectangle-drawing function in R. The protocol described is referred to as the bootstrap method.
Laboratory Confirmation of Genomic Introgressions Identified in Silico
Genomic regions identified as candidate introgressions were identified, and a small portion of the sequence in the region was extracted from Soybase (http://soybase.org). Primers were used to PCR amplify Clark, IsoClark, and T203 DNA. PCRs were conducted using a touchdown method starting with a 60°C annealing temperature and decreasing by 0.5°C each cycle for 29 cycles. Choice Taq (Denville Scientific) was used, and PCRs were at concentrations according to the manufacturer’s protocol.
PCR products were cleaned using an exonuclease 1 and shrimp alkaline phosphatase method. Cleaned PCR products were used in a cycle sequencing reaction. The sequencing protocol was adapted from the Applied Biosystems BigDye Terminator version 3.1 Cycle Sequencing kit. Sequencing was done on an Applied Biosystems 3730xl 96-capillary 50-cm array DNA analyzer. Sequence was end trimmed using Applied Biosystems Sequence Analysis version 5.2. Sequence ends were trimmed until fewer than four of 20 bases had quality scores less than 20. The sequences generated from each primer pair were aligned using Sequencher version 4.9 (Gene Codes Corporation).
Additionally, SSR markers were chosen from Soybase (http://soybase.org) in candidate regions of introgression in the IsoClark line. PCRs were conducted using a touchdown method starting with a 60°C annealing temperature and decreasing by 0.5°C each cycle for 29 cycles. Choice Taq (Denville Scientific) was used, and PCR was at concentrations according to the manufacturer’s protocol. Bromphenol blue loading dye was added to the PCR and loaded onto a 6% polyacrylamide gel run at 250 V for 2.5 h. Bands were visualized at 312 λ using a grayscale digital camera (Scion Corporation). The lowest band was scored and compared with 10- and 100-bp ladders.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. SNPs identified by method 1 on the single-library comparison.
Supplemental Table S2. SNPs identified by method 2 on the single-library comparison.
Supplemental Table S3. SNPs identified by method 1 on the four-library comparison.
Supplemental Table S4. SNPs identified by method 2 on the four-library comparison.
Supplemental Table S5. SNPs identified by method 2 on the seed protein NIL pair.
Supplemental File S1. Script used to determine intervals of significant SNP density based on a bootstrap method.
Supplementary Material
Acknowledgments
We thank Nathan Weeks for valuable discussions and information technology support. We thank B.J. Haun and Eric Eischens for technical support. We also thank the BioMedical Genomics Center at the University of Minnesota for Affymetrix microarray support.
References
- Bernard RL, Nelson RL, Cremeens CR. (1991) USDA Soybean Genetic Collection: isoline collection. Soybean Genet Newsl 18: 27–57 [Google Scholar]
- Bolon YT, Joseph B, Cannon SB, Graham MA, Diers BW, Farmer AD, May GD, Muehlbauer GJ, Specht JE, Tu ZJ, et al. (2010) Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biol 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Jin S. (2009) High-throughput methods for SNP genotyping. Methods Mol Biol 578: 245–254 [DOI] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. (2009) Real-time DNA sequencing from single polymerase molecules. Science 323: 133–138 [DOI] [PubMed] [Google Scholar]
- Fu Y, Springer NM, Gerhardt DJ, Ying K, Yeh CT, Wu W, Swanson-Wagner R, D’Ascenzo M, Millard T, Freeberg L, et al. (2010) Repeat subtraction-mediated sequence capture from a complex genome. Plant J 62: 898–909 [DOI] [PubMed] [Google Scholar]
- Hansen NC, Schmitt MA, Anderson JE, Strock JS. (2003) Iron deficiency of soybean in the upper Midwest and associated soil properties. Agron J 95: 1595–1601 [Google Scholar]
- Hyten DL, Cannon SB, Song Q, Weeks N, Fickus EW, Shoemaker RC, Specht JE, Farmer AD, May GD, Cregan PB. (2010a) High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics 11: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Choi IY, Song QJ, Specht JE, Carter TE, Shoemaker RC, Hwang EY, Matukumalli LK, Cregan PB. (2010b) A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci 50: 960–968 [Google Scholar]
- Hyten DL, Song Q, Choi IY, Yoon MS, Specht JE, Matukumalli LK, Nelson RL, Shoemaker RC, Young ND, Cregan PB. (2008) High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor Appl Genet 116: 945–952 [DOI] [PubMed] [Google Scholar]
- Kaczorowski KA, Kim KS, Diers BW, Hudson ME. (2008) Microarray-based genetic mapping using soybean near-isogenic lines and generation of SNP markers in the Rag1 aphid resistance interval. Plant Genome 1: 89–98 [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J. (2009a) SNP detection for massively parallel whole-genome resequencing. Genome Res 19: 1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. (2009b) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Molnar SJ, Rai S, Charette M, Cober ER. (2003) Simple sequence repeat (SSR) markers linked to E1, E3, E4, and E7 maturity genes in soybean. Genome 46: 1024–1036 [DOI] [PubMed] [Google Scholar]
- Muehlbauer GJ, Specht JE, Staswick PE, Graef GL, Thomas-Compton MA. (1989) Application of the near-isogenic line gene mapping technique to isozyme markers. Crop Sci 29: 1548–1553 [Google Scholar]
- Muehlbauer GJ, Staswick PE, Specht JE, Graef G, Shoemaker RC, Keim P. (1991) RFLP mapping using near-isogenic lines in soybean, Glycine max (L.) Merr. Theor Appl Genet 81: 189–198 [DOI] [PubMed] [Google Scholar]
- Nichols DM, Glover KD, Carlson SR, Specht JE, Diers BW. (2006) Fine mapping of a seed protein QTL on soybean linkage group I and its correlated effects on agronomic traits. Crop Sci 46: 834–839 [Google Scholar]
- O’Rourke JA, Charlson DV, Gonzalez DO, Vodkin LO, Graham MA, Cianzio SR, Grusak MA, Shoemaker RC. (2007a) Microarray analysis of iron deficiency chlorosis in near-isogenic soybean lines. BMC Genomics 8: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JA, Graham MA, Vodkin L, Gonzalez DO, Cianzio SR, Shoemaker RC. (2007b) Recovering from iron deficiency chlorosis in near-isogenic soybeans: a microarray study. Plant Physiol Biochem 45: 287–292 [DOI] [PubMed] [Google Scholar]
- O’Rourke JA, Nelson RT, Grant D, Schmutz J, Grimwood J, Cannon S, Vance CP, Graham MA, Shoemaker RC. (2009) Integrating microarray analysis and the soybean genome to understand the soybeans iron deficiency response. BMC Genomics 10: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rusk N. (2009) Cheap third-generation sequencing. Nat Methods 6: 244–245 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the paleopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Jia G, Zhu Y, Grant D, Nelson RT, Hwang EY, Hyten DL, Cregan PB. (2010) Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci 50: 1950–1960 [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109: 122–128 [DOI] [PubMed] [Google Scholar]
- Van Tassell CP, Smith TP, Matukumalli LK, Taylor JF, Schnabel RD, Lawley CT, Haudenschild CD, Moore SS, Warren WC, Sonstegard TS. (2008) SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat Methods 5: 247–252 [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WW, Cho S, Yang SS, Bolon YT, Bilgic H, Jia H, Xiong Y, Muehlbauer GJ. (2009) Single feature polymorphism discovery by computing probe affinity shape powers. BMC Genet 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.