Abstract
TILLING (for Targeting Induced Local Lesions IN Genomes) is a well-established method for identifying plants carrying point mutations in genes of interest. A traditional TILLING project requires a significant investment of time and resources to establish the mutant population and screening infrastructure. Here, we describe a modified TILLING procedure that substantially reduces the investment needed to perform mutation screening. Our motivation for developing iTILLING was to make it practical for individual laboratories to rapidly perform mutation screens using specialized genetic backgrounds. With iTILLING, M2 seeds are collected in bulk from the mutagenized population of plants, greatly reducing the labor needed to manage the mutant lines. Growth of the M2 seedlings for mutation screening, tissue collection, and DNA extraction are all performed in 96-well format. Mutations are then identified using high-resolution melt-curve analysis of gene-specific polymerase chain reaction products. Individual plants carrying mutations of interest are transferred from the 96-well growth plates to soil. One scientist can complete an iTILLING screen in less than 4 months. As a proof-of-principle test, we applied iTILLING to Arabidopsis (Arabidopsis thaliana) plants that were homozygous for the mekk1-1 (for MAPK/ERK kinase kinase 1) mutation and also carried a MEKK1 rescue construct. The goal of our screen was to identify mutations in the closely linked MEKK2 and MEKK3 loci. We obtained five mutations in MEKK2 and seven mutations in MEKK3, all located within 20 kb of the mekk1-1 T-DNA insertion. Using repeated iterations of the iTILLING process, mutations in three or more tandemly duplicated genes could be generated.
The process of reverse genetics has been widely used by plant biologists to study gene function. In Arabidopsis (Arabidopsis thaliana), three approaches that have been used to generate populations of plants for reverse genetic analysis are insertional mutagenesis (Wisman et al., 1998; Alonso et al., 2003), fast neutron mutagenesis to induce deletions (Li et al., 2001), and chemical mutagenesis to induce point mutations (McCallum et al., 2000). In order to find individual plants carrying point mutations of interest, a process called TILLING (for Targeting Induced Local Lesions IN Genomes) was developed whereby genes are screened for mutations using a PCR-based assay (McCallum et al., 2000). Although originally developed for use with Arabidopsis, the TILLING process has been subsequently applied to a wide range of plants, including barley (Hordeum vulgare; Caldwell et al., 2004), Brassica napus (Wang et al., 2008), Brassica oleracea (Himelblau et al., 2009), Brassica rapa (Stephenson et al., 2010), Lotus japonicus (Perry et al., 2009), maize (Zea mays; Till et al., 2004), Medicago truncatula (Le Signor et al., 2009), oat (Avena sativa; Chawade et al., 2010), pea (Pisum sativum; Triques et al., 2007), potato (Solanum tuberosum; Elias et al., 2009), rice (Oryza sativa; Till et al., 2007), sorghum (Sorghum bicolor; Xin et al., 2008), soybean (Glycine max; Cooper et al., 2008), tomato (Solanum lycopersicum ; Gady et al., 2009), and wheat (Triticum aestivum; Dong et al., 2009). TILLING has also been used in Drosophila (Winkler et al., 2005), zebrafish (Wienholds et al., 2003), and Caenorhabditis elegans (Gilchrist et al., 2006).
The chemical mutagen most commonly used to create the mutant populations used for TILLING is ethyl methanesulfonate (EMS). When working with plants, seeds are soaked in EMS to induce mutations throughout the genome. Mutagenized seeds are then planted on soil, and the resulting plants are grown to maturity to produce M2 seeds, which are collected from the plants individually or in small pools. Next, M2 seed samples from each individual plant are germinated and grown to produce tissue from which DNA can be extracted. The resulting large collection of ordered DNA samples and the corresponding M2 seeds constitute the infrastructure of a TILLING population. PCR-based screening can then be used to find individual plants in the population carrying mutations in genes of interest (McCallum et al., 2000). Once established, this type of TILLING infrastructure can serve the needs of an entire research community through a fee-for-service screening operation (Colbert et al., 2001; Martín et al., 2009).
Several different strategies have been developed for identifying the mutations present in a TILLING population, but all of them involve detecting heteroduplex PCR products. A heteroduplex is formed when a mixture of wild-type and mutant PCR products are melted and reannealed, resulting in DNA duplexes that contain a single-base mismatch. TILLING was originally described using denaturing HPLC to identify mutations based on the differential retention times of heteroduplexes and homoduplexes in the chromatography column (McCallum et al., 2000). TILLING has since been modified so that endonucleases are used to cleave PCR products containing a heteroduplex. Cleavage products are then separated via gel electrophoresis to identify banding patterns indicative of mutations (Colbert et al., 2001).
More recently, high-resolution melting analysis of PCR products has been used to identify heteroduplexes when performing TILLING (Dong et al., 2009; Gady et al., 2009). High-resolution melting analysis was originally developed for use in clinical settings to identify known single-nucleotide polymorphisms and small insertions/deletions potentially linked to genetic diseases (Erali et al., 2008). With high-resolution melting, the mismatch in a heteroduplex is visualized as a melting event that occurs more rapidly or at a lower temperature than the corresponding homoduplex. Montgomery et al. (2007) demonstrated that mutation scanning with high-resolution melting is a robust technique with greater than 95% sensitivity in distinguishing heteroduplexes from homoduplexes. It has also been observed that the sensitivity with which mutations in PCR products can be identified using DNA melting analysis depends on the resolution of the instrumentation used for collecting the melt-curve data (Zhou et al., 2005; Herrmann et al., 2006).
Although traditional TILLING is a high-throughput method for mutation screening, the establishment of the initial screening population and the corresponding ordered DNA samples requires a substantial up-front investment of time and money. Because of this situation, TILLING resources are available for only two genetic backgrounds in Arabidopsis: wild-type Columbia-0 and Landsberg erecta (Greene et al., 2003; Martín et al., 2009). If a scientist is interested in identifying mutations in a more specialized genetic background, the costs associated with establishing a TILLING population can be prohibitive. Therefore, we were interested in determining if a modified version of the TILLING process could be developed that would substantially reduce the investment of time and resources necessary to perform mutation screening. The individualized TILLING procedure, or iTILLING, which we describe in this paper provides one solution to this challenge.
RESULTS
Overview of the iTILLING Approach
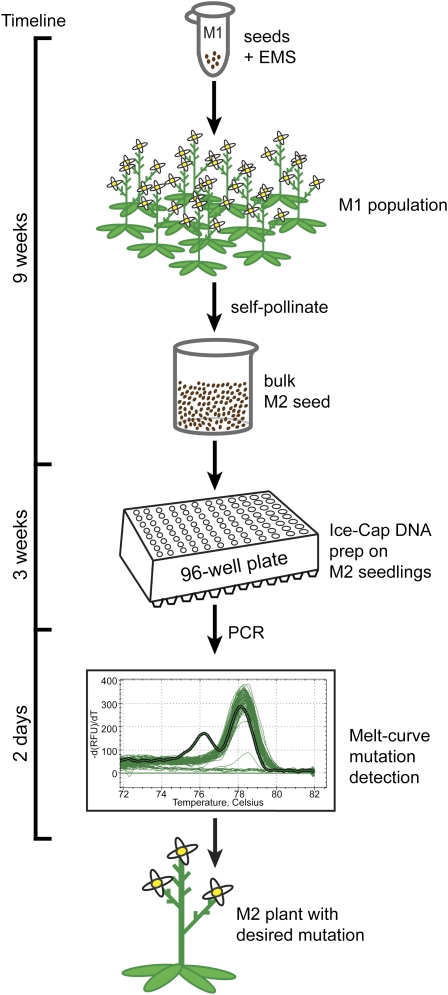
Like traditional TILLING, the process of iTILLING begins with the use of the chemical mutagen EMS to induce mutations throughout the genome (Fig. 1). Greater than 99% of the mutations induced by EMS treatment in Arabidopsis have been shown to be G-to-A single-base changes (Greene et al., 2003). The EMS-mutagenized seeds are sown on soil, where they germinate and give rise to M1 plants. These M1 plants self-pollinate and yield M2 seeds. Unlike traditional TILLING, which requires individual plant manipulations to collect the M2 seeds, the M2 seeds from the entire population are harvested in bulk with iTILLING. At this point in traditional TILLING, M2 plants are grown and high-quality DNA samples are prepared from small pools of plants and cataloged for long-term storage. With iTILLING, no cataloging of M2 individuals or DNA samples is required. Instead, M2 plants are screened for mutations as they are growing on 96-well plates. The rare individuals carrying a mutation of interest are ultimately moved off the 96-well plate and into soil, where they grow to maturity and produce M3 seeds.
Figure 1.
Work flow of iTILLING. Seeds are treated with EMS to produce the M1 population from which M2 seeds are collected in bulk. M2 seedlings are grown on 96-well Ice-Cap plates, and tissue samples are collected 96 at a time. PCR and melt-curve analysis are done to identify heteroduplex products indicative of a mutation. -dRFU/dT, Negative change in relative fluorescence units over the change in temperature. Plants carrying a desired mutation are transplanted from the 96-well plate to soil. The work flow from initial mutagenesis to the identification of mutations of interest is less than 4 months. [See online article for color version of this figure.]
iTILLING uses a previously described method called Ice-Cap to screen M2 seedlings in a high-throughput fashion on 96-well plates (Krysan, 2004; Clark and Krysan, 2007). When using the Ice-Cap system, seedlings are grown on agar plugs on 96-well spin column plates. Roots from each seedling grow down through the agar, onto a 96-well plate filled with water. The seedlings are approximately 3 weeks old when the roots reach the bottom of the second plate. The lower plate containing root tissue is frozen and separated from the seedling plate, resulting in the collection of 96 tissue samples. The plate containing root tissue is then sealed and rapidly agitated, causing a ball bearing present in each well to disrupt the tissue. After centrifugation of the plate, genomic DNA in the supernatant fluid is used as a PCR template to perform mutation screening of the intact M2 seedlings growing on the upper agar plate. High-resolution melting of PCR products is used to scan the amplicons for mutations. Mutation screening can begin as soon as the tissue is collected. If necessary, plants in Ice-Cap blocks can be stored in the dark at 4°C for at least 4 weeks before a seedling of interest is transplanted with no loss of viability (data not shown). Any M2 plant growing in an Ice-Cap well and carrying a mutation of interest can be easily removed from the 96-well plate to finish its life cycle in soil and produce seeds. Ice-Cap has been shown to be effective for use with Arabidopsis and rice, and it is expected that it could also be used with other plant species that have seeds small enough to fit into the wells of a 96-well plate (Krysan, 2004).
Using High-Resolution Melting to Detect Mutations
Mutations are detected in traditional TILLING with PCR amplification of pooled DNA, enzymatic cleavage of any heteroduplex products, and gel electrophoresis to separate the digested products. In contrast, iTILLING is a gel-free system that uses high-resolution melt-curve analysis of PCR products to reveal mutations of interest. High-resolution melting has recently been shown to be an effective alternative to enzymatic cleavage for mutation detection in TILLING populations (Dong et al., 2009; Gady et al., 2009).
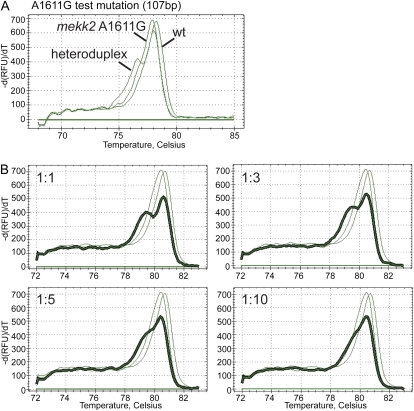
Gady et al. (2009) used a LightScanner System (Idaho Technology) high-resolution melting machine for their TILLING screen, whereas we utilized the CFX96 PCR Detection System from Bio-Rad Laboratories. Because our Bio-Rad machine has a lower resolution than the LightScanner System, we wanted to determine if a single-base change could be discerned using our melting capabilities. To generate a mutation that could be used as a test case for this purpose, we amplified a region of the Arabidopsis MEKK2 (for MAPK/ERK kinase kinase 2) gene (At2g08480) to generate PCR-induced mutations. The resulting PCR products were ligated into the pNEB193 vector and transformed into Escherichia coli, and individual clones were sequenced to identify mutations. The base change A1611G was identified and used for subsequent testing of the high-resolution melting system. Numbering of the mutation refers to the full-length genomic sequence of MEKK2, where the first base of the start codon corresponds to base 1.
To evaluate our ability to detect the mekk2 (A1611G) mutation using the Bio-Rad CFX96 instrument, we amplified a 107-bp product from plasmid DNA template of the mekk2 (A1611G) mutant by itself, wild-type MEKK2 by itself, and a 1:1 mixture of these two templates. As shown in Figure 2A, the heteroduplex was distinguishable from both homoduplexes. Additionally, a dilution series using the mekk2 (A1611G) polymorphism was used to evaluate the sensitivity of our melt-curve assay. PCR products from reactions performed using template dilutions of 1:1, 1:3, 1:5, and 1:10 mutant-to-wild-type DNA were analyzed using the Bio-Rad CFX96 machine. Analysis of the dilution series indicated that a mutant allele in a dilution of up to 1:10 mutant-to-wild-type DNA could be distinguished from either wild-type or mutant homoduplexes (Fig. 2B).
Figure 2.
Use of high-resolution melting for mutation detection. A, Melt curves of PCR products amplified from wild-type (wt) DNA, mekk2 (A1611G) mutant DNA, and a 1:1 mix of wild-type and mutant DNA. A heteroduplex is formed when a wild-type DNA strand anneals to a mutant strand. B, Melt curves of PCR products amplified using mixtures of wild-type and mekk2 (A1611G) mutant template DNA. The ratios of mutant to wild-type DNA used in the reactions were 1:1, 1:3, 1:5, and 1:10. Thick lines represent the melt peaks of the heteroduplex products. Thin lines represent the melt peaks of the mekk2 (A1611G) homoduplex (left peak) and the wild-type homoduplex (right peak). -dRFU/dT, Negative change in relative fluorescence units over the change in temperature. [See online article for color version of this figure.]
Using a very high-resolution melting system, single-nucleotide polymorphism heteroduplexes in amplicons of up to 1,000 bp can be visualized (Montgomery et al., 2007). For our iTILLING screen, we chose to use amplicons in the size range of 95 to 125 bp. At less than 95 bp, we found that the fluorescence signal from the PCR product was not reliably above the background fluorescence level. In amplicons of more than 125 bp, a single base change was not sufficient to cause robust differential melting of heteroduplex and homoduplex PCR products (data not shown).
Applying iTILLING to the MEKK Gene Cluster
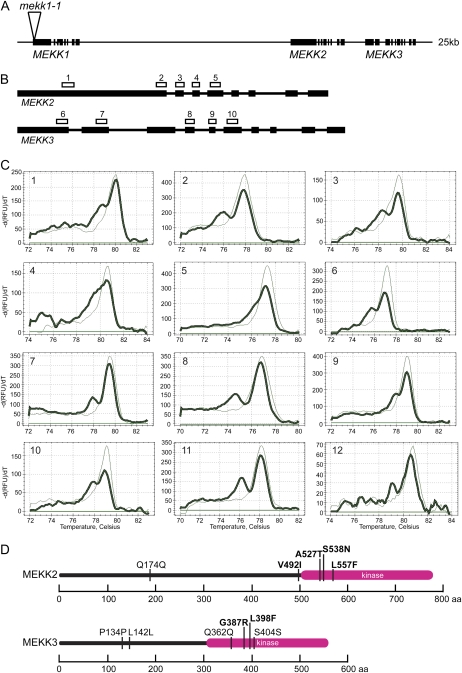
Mutations in the MEKK1 gene have been described previously (Ichimura et al., 2006; Nakagami et al., 2006; Su et al., 2007; Suarez-Rodriguez et al., 2007). These studies have indicated that MEKK1 plays a role in biotic and abiotic stress responses. Plants homozygous for a mutation in MEKK1 are strongly dwarfed and display constitutive defense responses (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007). Mutations in the two closely related family members MEKK2 and MEKK3 have not been described; however, mekk2 and mekk3 T-DNA insertional alleles do not display any obvious abnormal phenotypes under standard growth conditions (Salk_150039 and WiscDsLox 472D11; data not shown). These three MEKK genes are tandemly located on chromosome 4 in an approximately 25-kb region (Fig. 3A). Because of the low recombination rate expected for such tightly linked loci, the creation of double- or triple-mutant plants using T-DNA insertions within the MEKK gene cluster would be difficult to achieve. As an alternative, we applied iTILLING to plants homozygous for the T-DNA mutation mekk1-1 that also carried an unlinked homozygous MEKK1 rescue construct. Because the mekk1-1 T-DNA mutation is homozygous, any base change identified in MEKK2 or MEKK3 will be in cis-configuration with the mekk1-1 mutation. As plants homozygously mutant for mekk1-1 are strongly dwarfed (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007), the presence of the wild-type MEKK1 rescue construct in our mutagenesis population ensures that the M1 plants are phenotypically normal (Suarez-Rodriguez et al., 2007).
Figure 3.
Detection of mutations in the MEKK gene cluster. A, MEKK1, MEKK2, and MEKK3 are located in a 25-kb region of chromosome IV. Exons are shown as thick black bars. The mekk1-1 T-DNA insertion is shown as a triangle in MEKK1. B, Gene structures of MEKK2 and MEKK3 are shown from start to stop codons. Exons are shown as thick black bars. The locations of amplicons 1 through 10 used for mutation screening in MEKK2 and MEKK3 are shown as white boxes above each gene. C, Melt curves for mutations 1 through 12 identified via iTILLING in MEKK2 and MEKK3. Thick lines represent the melt curves of the heteroduplex products. Thin lines represent the melt curve of a wild-type homoduplex. -dRFU/dT, Negative change in relative fluorescence units over the change in temperature. D, Location of the mutations detected in MEKK2 and MEKK3. The protein sequences are represented as a horizontal line. The kinase domains of the proteins are indicated by the thicker line. Mutations causing amino acid changes are highlighted in boldface letters. aa, Amino acids. [See online article for color version of this figure.]
Seeds collected from a mekk1-1/mekk1-1; PMEKK1:MEKK1/PMEKK1:MEKK1 plant were mutagenized using 0.2% EMS. We evaluated the mutation frequency in the resulting M1 population using the method described by Mednik (1988) and observed a value of P = 0.75. A population of approximately 10,000 M1 plants treated with 0.2% EMS was grown to maturity in soil, and the resulting M2 seeds were collected in bulk. M2 seeds were then plated in Ice-Cap blocks for seedling growth and root tissue collection. The high-throughput Ice-Cap method was used to collect root tissues samples 96 at a time, and DNA was extracted from each plate. Eighty-one plates of M2 seedlings were grown and sampled in this manner, including 69 plates with one seedling per well and 12 plates with two seedlings per well. When screening one seedling per well, we expected to only be able to identify plants that were heterozygous for a mutation of interest. The two-seedling-per-well strategy is described in more detail later in this paper.
In order to screen for mutations in the M2 seedlings, five amplicons in MEKK2 and five in MEKK3 were selected for PCR amplification and melt-curve analysis (Table I; Fig. 3B). These amplicons were designed to maximize the probability of identifying stop codons. We also chose amplicons in regions of the genes encoding the highly conserved ATP-binding regions of MEKK2 and MEKK3 (Fig. 3B). Because of the high level of amino acid conservation in this domain of the proteins, point mutations in this region are likely to have a deleterious effect on protein function. Using these 10 PCR amplicons, approximately 8,000 plants were screened for mutations in a total of 650 bp of target sequence across the two genes. Putative mutations identified in the primary screen were further tested by reamplification and melting of the PCR amplicon, followed by DNA sequencing to determine the authenticity and location of the mutation. For each valid mutation identified, putative mutations from an average of 1.8 plants were tested by DNA sequencing. M2 plants carrying verified mutations were transplanted from the 96-well plate to soil in order to produce M3 seeds. In all cases, transplantation to soil did not harm the plants, and seeds were successfully collected from all of the mutant lines.
Table I. PCR primers used for mutation screening.
| Amplicon | Primer | Sequence (5′–3′) | Amplicon Size |
| bp | |||
| 1 | F | GAGTTGATGATGGAGAAATAGA | |
| R | AACGCTCATGCTCCATAC | 111 | |
| 2 | F | TCCAATACCTCACCGATTTG | |
| R | TGAAATGGCTTCATACACAGAG | 108 | |
| 3 | F | GCTGAATGTAGAGATGGGGACT | |
| R | AACTTCCATACCCCCTCA | 107 | |
| 4 | F | CCTTAGCCACATGTTTTCATT | |
| R | CGGGCCATACCTTGTCTGT | 103 | |
| 5 | F | TTGTATATTTTTCTTGAGCTTG | |
| R | GAAGATATTTCAAACCGTCA | 124 | |
| 6 | F | TATCCATTATTTCGCCGAGT | |
| R | CCCTGAACTGCCTTCATACA | 113 | |
| 7 | F | CGTGCCAAGTTGATTGAAAA | |
| R | GGCTTCATACACAGAGGCAAA | 111 | |
| 8 | F | AGAGATGGGGACTTCTTTGCT | |
| R | TGACTTTCATACCCCCTCAA | 99 | |
| 9 | F | TTTATTTTTCTACAGGAGATTG | |
| R | GAAAGAGAGAACAAGACTACAC | 100 | |
| 10 | F | CATCTTTCTTGAGCTTGTAACC | |
| R | ATAATTCAACCCAGCAAGAA | 115 |
A total of 12 mutations were identified in our screen (Table II). The corresponding melt curves are shown in Figure 3C. Because we found two individual plants carrying the same mekk3 (C1212T) mutation, a total of 11 unique base changes were identified. In addition, we found one plant that had mutations in both MEKK2 and MEKK3, although the mutation in MEKK2 was silent. We identified one mutation per 415 kb of sequence screened, or one mutation approximately every seven Ice-Cap blocks. Both missense and silent mutations were identified, and these were located across the length of the regions scanned (Fig. 3D). No nonsense mutations were identified in our screen. Based on the specific coding sequences present in the amplicons screened in our study, 13.5% of the possible G/C-to-A/T mutations in our survey would be expected to produce a stop codon. χ2 analysis indicates that our observation of zero nonsense mutations in a sample of 12 total mutations does not represent a statistically significant dearth of nonsense mutations. In addition, the absence of nonsense mutations in our screen is not likely to be due to the lethality of such mutations, since T-DNA insertional alleles of mekk2 and mekk3 have been identified and found to cause no obvious abnormal phenotype (data not shown). χ2 analysis of our data also shows that the rate of silent mutations identified in our screen is not significantly different from the rate that would be expected given random mutation of the G/C base pairs present in the sequences targeted by our screen. The heteroduplex melt curves for the 12 mutations identified varied in size and shape (Fig. 3C). It is likely that differences in amplicon sequence contribute to this variance.
Table II. Mutations identified in MEKK2 and MEKK3 using iTILLING.
| Mutation | Gene | het/hmza | Amino Acid Effectb |
| 1 | MEKK2 | hmz | A527T |
| 2 | MEKK2 | het | L557F |
| 3 | MEKK2 | het | S538N |
| 4 | MEKK2 | het | V492I |
| 5 | MEKK2 | het | Q174Q |
| 6 | MEKK3 | het | G387R |
| 7 | MEKK3 | het | L398F |
| 8 | MEKK3 | hmz | L142L |
| 9 | MEKK3 | hmz | S404S |
| 10 | MEKK3 | het | S404S |
| 11 | MEKK3 | het | Q362Q |
| 12 | MEKK3 | het | P134P |
Indicates whether the mutation was initially identified in the heterozygous (het) or homozygous (hmz) state.
Indicates the amino acid affected by each mutation and its position in the protein. Amino acid changes are noted in boldface.
Screening Plants Grown Two per Well with iTILLING
Because induced mutations are heterozygous in the M1 generation, individuals in the M2 population can be heterozygous or homozygous for any given mutation. When growing and screening plants one per well using the 96-well Ice-Cap platform, only heterozygous plants can be detected during melt-curve analysis of PCR products. The ability to identify homozygous mutations in the M2 population, therefore, could increase our mutation detection rate by 50%. When screening seedlings individually, however, we cannot identify the plants with homozygous lesions because they do not produce a heteroduplex PCR product. In order to identify homozygous mutants, a wild-type DNA template must be present during PCR amplification in addition to the mutant template provided by the homozygous plant. One strategy for providing this wild-type template to each sample would be to grow seedlings two per well on the Ice-Cap plates. If two individuals from an M2 population were grown together in one well, the likelihood of the two seedlings carrying the same mutation at any locus would be very low. Because of this situation, each plant could act as a wild-type control for the other. To determine if this strategy would work in practice, we directly tested if growing two seedlings per well could be used to identify homozygous mutations.
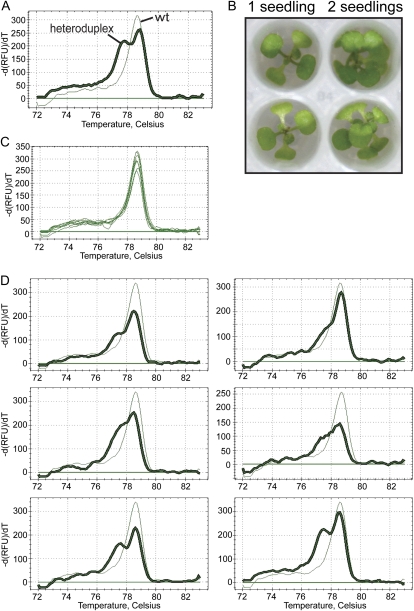
To begin, we performed a reconstruction experiment in which we planted one wild-type seed and one known homozygous mutant seed together in the wells of Ice-Cap plates. The homozygous mutant line chosen for this experiment was mekk3 (C1212T), which carries a mutation that is detected using PCR amplicon 10. As shown in Figure 4B, seedlings grown two per well appear healthy, indicating that the Ice-Cap plates have sufficient space to accommodate two seedlings. Root tissue collection and DNA extraction of the plates were performed as usual. It was expected that the DNA extracts from the wells with two seedlings would contain a mixture of both the wild-type and mutant genomes. Melt-curve analysis of PCR products targeting the mekk3 (C1212T) mutation was performed. Heteroduplexes were observed in 193 out of the 294 samples from seedlings grown two per well (70%; Fig. 4D). Although each heteroduplex in this experiment had the same sequence composition, the shape of the melt curves was variable (Fig. 4D). It seems likely that differences in the relative ratio of wild-type to mutant template in the different DNA extracts may explain the variability of the shape of the melt curves. This explanation is supported by the results observed in the dilution experiment shown in Figure 2B.
Figure 4.
Mutation detection for seedlings grown two per well using Ice-Cap. A, Characteristic melt curves from amplicon 10 from wild-type (wt) and heterozygous mekk3 (C1212T) plants. The thick line represents the melt curve of the heteroduplex product. The thin line represents the melt peak of the wild-type homoduplex. B, Top view of seedlings growing on a 96-well Ice-Cap plate. Wells on the left contain one seedling; wells on the right contain two seedlings. C, Melt curves of PCR products from amplicon 10 for eight wild-type seedlings. D, Melt curves from plants grown two per well on Ice-Cap plates with one wild-type seedling and one homozygous mekk3 (C1212T) mutant seedling per well. Thick lines represent the melt curves of the mekk3 (C1212T) heteroduplex products. Thin lines represent the melt curves of wild-type homoduplex products. -dRFU/dT, Negative change in relative fluorescence units over the change in temperature. [See online article for color version of this figure.]
When screening an M2 population with two seeds per well on the Ice-Cap plates, heteroduplexes are generated when a homozygous mutant seedling is paired with a wild-type seed (Fig. 4D). Heteroduplexes would also occur when a heterozygous mutant is paired with a wild-type seed. In the latter case, the EMS-induced base change is present on one chromosome out of the four present in that DNA extract. Based on our dilution experiment (Fig. 2B), up to a 1:10 dilution of the mutant allele with wild-type DNA can be detected with our melting capabilities. This led us to believe that mutation detection in heterozygous plants would be possible when growing plants two per well. In order to evaluate this possibility, we performed a second reconstruction experiment. Seeds were plated two per well: one seed was from a wild-type plant and the other was from a population that was segregating the mekk3 (C1212T) mutation. We observed heteroduplex melt curves for 51 out of the 134 wells tested (38%; data not shown). Seeds from a population that is segregating a known mutation can be wild type, heterozygous, or homozygous mutant at the locus of interest. The Mendelian segregation ratio predicts that 75% of the plants in the population should have at least one copy of the mutant allele. If our screening efficiency were 100%, we would have expected to observe heteroduplexes in approximately 75% of the wells for this experiment. The lower rate of heteroduplex detection could be due to differences in the relative contribution of the two seedlings to the final DNA extract, which could lead to a situation where the ratio of wild-type to mutant DNA drops below the detection limit of our melt-curve analysis. Reduced fitness of the mutant plants could potentially exacerbate this situation.
In the reconstruction experiment described above, we detected heteroduplexes in 38% of the wells of an Ice-Cap plate when growing seedlings from a segregating mutant population two per well with a wild-type companion. For comparison, if seedlings from a segregating mutant population were grown one per well, we would expect to be able to identify heteroduplexes in a maximum of 50% of the wells of an Ice-Cap plate, since only heterozygous individuals from the population would be detected. Initially, it may appear that the rate of mutation detection per plate is lower when analyzing two seedlings per well when compared with one seedling per well. It should be noted, however, that this reconstruction experiment involved plating one wild-type seed into each well. For a real mutation screen, both seedlings in each well would be from the mutant population. For this reason, one must double the observed rate of mutation detection found in the reconstruction experiment in order to estimate the rate with which mutations would be recovered using two-per-well screening. Based on this analysis, screening seedlings two per well would be expected to yield mutations at a rate near that found in screens performed with one seedling per well. In our reconstruction experiments, growing seedlings two per well had the effect of reducing the rate with which heterozygous mutants were detected. Since on average one would expect there to be twice as many heterozygous mutant plants as homozygous mutant plants in the population, improving the rate with which heterozygous mutants are detected from seedlings growing two per well would likely increase the overall efficiency of the mutation screen. The use of a higher resolution melt-curve instrument could be useful for this purpose. Three of the 12 mutations we identified in our iTILLING screen of MEKK2 and MEKK3 are homozygous lesions that were found by screening two seedlings per well.
DISCUSSION
TILLING has become well established as an effective method for identifying induced mutations in populations of mutagenized organisms (Comai and Henikoff, 2006). The most common implementation of the TILLING process is for one laboratory to establish a TILLING facility that serves as a public resource to provide the entire research community with access to the mutation screening infrastructure. This approach has proven to be effective for providing large numbers of users with access to mutant alleles in standard wild-type genetic backgrounds. The iTILLING procedure that we describe in this study is intended to serve as a complementary strategy to traditional TILLING. Where traditional TILLING excels at providing large numbers of users access to mutations in a standard genetic background, iTILLING is designed to provide a single laboratory access to mutations in a specialized genetic background by substantially reducing the required effort and costs. iTILLING is best suited for the identification of mutations at a relatively small number of loci due to the short-lived nature of the screening population.
When using iTILLING, M2 seeds can be collected in bulk from the M1 population. Bulking seeds saves considerable time over the cataloging of M2 seeds required in standard TILLING. With iTILLING, M2 seeds are grown and tissue samples are taken in the 96-well format of Ice-Cap (Krysan, 2004; Clark and Krysan, 2007). When a mutation is identified, the associated M2 plant is still living on a 96-well plate. Once the mutation has been confirmed, this plant is removed from the plate and transferred to soil, where it will grow to maturity and yield seeds. Plants that do not contain mutations of interest can be discarded. Since the M2 population is not cataloged, and since individual plants are screened and discarded, the population is ephemeral. With all processes running optimally, a single scientist could perform this screen in less than 4 months, from the initial mutagenesis through the identification of mutant seedlings. This time frame could increase with unforeseen technical difficulties.
The main nonsalary cost associated with the iTILLING procedure is the cost of the consumables used to perform PCR. Using homemade Taq polymerase and 20-μL PCRs as described in this study, we calculated the cost per reaction to be approximately $0.05 US. This calculation includes the cost of the 96-well PCR plates. For the approximately 80,000 PCRs performed in this study, that corresponds to a total of approximately $4,000 US. Since we identified 12 mutations in our screen, the cost per mutation was approximately $333 US. By using a higher resolution melt-curve instrument, it should be possible to screen for mutations using larger PCR amplicons, which would in turn allow one to screen a given stretch of the genome using fewer PCRs. Doubling the size of the PCR amplicons used for screening would cut in half the total cost of mutation screening. For this reason, it should be possible to substantially reduce the cost per mutation by using more advanced melt-curve instrumentation.
While the publicly available Arabidopsis TILLING resources are in the Columbia-0 and Landsberg erecta genetic backgrounds (Colbert et al., 2001; Martín et al., 2009), iTILLING makes it practical for a laboratory to search for mutations in any specialized genetic background that may be of interest. As the hundreds of distinct Arabidopsis ecotypes available from stock centers become more widely used, iTILLING could be applied to search for induced mutations directly in any of these nonstandard ecotypes. iTILLING could also be used to screen for mutations in any complex genetic background, such as plants already possessing point mutations, insertional mutations, or transgenes.
Perhaps more importantly, iTILLING could be used to create multimutants in tandemly duplicated gene families. Sixty-five percent of genes in the Arabidopsis genome are in gene families of two or more members, and more than 1,500 of them are found in tandemly repeated clusters (Arabidopsis Genome Initiative, 2000). In a three-member gene cluster composed of genes A, B, and C, one could potentially create a plant carrying linked mutations in all three genes in less than 1 year. Starting with a plant carrying a homozygous T-DNA insertion in gene A, one round of iTILLING could be performed to search for mutations in gene B. Once a desired mutation was found, the plant would be moved to soil to set seed. These double mutant seeds carrying mutations in genes A and B could be used for a second round of iTILLING, in which mutagenized plants would be screened for mutations in gene C. If a second round of iTILLING were to be performed, then M3 seeds from a chosen M2 plant would be used for a second round of EMS mutagenesis. Due to the independent assortment of mutations in the previous generations, each M3 individual is expected to carry an average of 62.5% of the mutations present in the original M1 generation. Mutations that persist in the M3 population will have come through three generations of purifying selection. In addition, it has been shown that populations of Arabidopsis plants can be generated that carry a much higher density of EMS mutations than those used in our study (Martín et al., 2009). Thus, performing a second round of iTILLING using M3 seeds would likely be possible without the need for an intervening generation of back-crossing. In this way, iTILLING could be used to overcome the challenges posed by functional redundancy that are often confronted when performing genetic analysis of tandemly duplicated genes.
With traditional TILLING, mutation screening is performed using pooled DNA samples, typically containing approximately eight M2 plants per pool. iTILLING does not require pooling of DNA samples, but pooling could be applied. Multiple plants could be grown and sampled together on the 96-well plates, such as in the two-per-well growth strategy described in this study. Alternatively, plant tissue could be harvested and DNA samples prepared individually and then individual DNA extracts combined to form a pool, as in traditional TILLING. The main factor limiting the extent to which DNA extracts can be pooled is the sensitivity of the mutation detection platform. With the amplicon melting assay, it has been demonstrated that higher resolving power will allow detection of single-base-change mutations in more highly pooled samples as well as in amplicons of greater length (Reed and Wittwer, 2004; Montgomery et al., 2007; Gady et al., 2009). The dilution experiment shown in Figure 2B indicated that the Bio-Rad CFX96 instrument could identify mutant amplicons diluted up to 1:10 with the wild type. This ratio corresponds to pools of five seedlings, since heterozygous plants would contribute one mutant and one wild-type allele to the DNA extract. As reported by Gady et al. (2009), DNA pools of eight individuals can be effectively screened if a higher resolution melting instrument is used.
iTILLING is designed to allow a scientist to identify useful mutations in a gene or genes of interest. The aim of iTILLING is not to exhaustively catalog all of the mutations present in a mutagenized population. For this reason, we recommend collecting seeds from the entire M1 population en masse so that all the progeny from the approximately 10,000 parent plants are handled as a single sample. This strategy minimizes the chance that one will oversample the progeny of a given M1 parent during the PCR phase of the process. If seeds were collected, for example, from pools of 100 M1 parents, then a given 96-well plate would have a high probability of sampling one M1 parent more than one time. This situation would reduce the overall efficiency with which novel mutations are detected, since the same parental line would be screened multiple times. As stated above, the goal of iTILLING is to identify novel mutations in the most efficient manner possible, not to catalog all the mutations present in the population. The highest efficiency of novel mutation detection will be achieved when the total population is undersampled rather than oversampled. Screening a large pool of bulked M2 seeds reduces the likelihood of oversampling and also facilitates seed collection.
The density of mutations observed in our iTILLING population was one mutation per 415 kb screened. Our mutagenesis treatment was performed using 0.2% EMS (approximately 20 mm). For comparison, Greene et al. (2003) reported a rate of one mutation per 300 kb screened in a traditional TILLING project involving EMS treatments ranging from 20 to 45 mm (Till et al., 2003), while Martín et al. (2009) reported one mutation per 89 kb with treatments of 20 to 40 mm EMS. In addition, Jander et al. (2003) described an EMS-mutagenized Arabidopsis population that was screened for mutations in CSR1 based on the phenotype of acquired herbicide resistance. This population was treated with 20 mm EMS and reported to have one mutation per 180 kb (Jander et al., 2003). The rate with which we observed mutations in our iTILLING screen was lower than that of the other TILLING projects. This may be due to a lower rate of initial mutagenesis in our EMS treatment or to a reduced screening sensitivity of our mutation detection instrument. Use of a higher resolution instrument for performing melt-curve analysis could improve our ability to observe mutations as well as allow for an increase in the amplicon size used for mutation detection. By using larger amplicons, one would be able to screen more target sequence with fewer PCRs, thus lowering the cost and accelerating the speed with which mutations can be identified.
We have shown that iTILLING offers a new method for locating mutations in tandemly repeated genes. Other methods have been described that also allow for the identification of mutations in tandem genes, including selection for genomic deletions (Li et al., 2001), site-directed mutagenesis using zinc-finger nucleases (Lloyd et al., 2005), the use of a specialized Ds transposon system with Cre/Lox functionality (Zhang et al., 2003), and the selection of meiotic recombinants between closely linked mutations of interest (Jander and Barth, 2007). One potential benefit of iTILLING compared with many of these alternative methods would be the relative speed with which double- and triple-mutant lines could be developed via iTILLING for a given tandem gene pair. If one were working with larger arrays of duplicated genes, iTILLING may not be the most practical alternative. As with some of the approaches for identifying mutations in tandem genes described above, mutations identified using iTILLING can vary in their effect on the protein in question, and additional nontarget mutations may occur.
Next-generation sequencing technologies have greatly reduced the cost of determining the sequence of entire genomes in recent years, and it is expected that this trend of lower cost and greater coverage will continue (Metzker, 2010). Depending on the research budget that one has available, it may be possible to perform whole-genome sequencing on DNA samples collected using the iTILLING procedure described above. Indexed primers could, in theory, be used to track individual plants from the 96-well growth plates, allowing genome sequencing to be performed using pooled samples. In this way, the iTILLING platform could potentially be used to rapidly generate and characterize mutant populations at the whole-genome level.
iTILLING is not intended to take the place of traditional TILLING. Although the establishment of public TILLING facilities requires a large initial investment, the resulting populations can be screened repeatedly over many years by hundreds of scientists. By contrast, iTILLING involves an ephemeral mutant population that can be quickly and easily developed, but with no intention of maintaining it. Because of the ease with which an iTILLING population can be produced and screened, iTILLING provides a new strategy for performing personalized reverse genetics.
MATERIALS AND METHODS
Production of the MEKK2 Polymorphism
The mekk2 (A1611G) polymorphism was generated by PCR amplification of a region of MEKK2 from Arabidopsis (Arabidopsis thaliana) Columbia-0 genomic DNA using a mutant version of Taq polymerase (Kermekchiev et al., 2003) and the following primers: AscI-F, 5′-AAAAGGCGCGCCAGCCTCCTCCGGTGATGAAA-3′; PacI-R, 5′-AAAATTAATTAAGGTGGATTAGCAACCTACTTTTGTCTG-3′. After digestion with the restriction enzymes AscI and PacI, this product was ligated into the pNEB193 vector backbone and transformed into Escherichia coli. Sequencing of plasmids extracted from individual bacterial colonies identified the A1611G mutation, which is located within amplicon 3 shown in Table I. Numbering of the mutation refers to the full-length genomic sequence of MEKK2, where the first base of the start codon corresponds to base 1.
EMS Treatment
The Arabidopsis mekk1-1 homozygous mutant line (SALK_052557; Alonso et al., 2003) was rescued with the wild-type construct PMEKK1:MEKK1 as described (Suarez-Rodriguez et al., 2007). mekk1-1/mekk1-1; PMEKK1:MEKK1/PMEKK1:MEKK1 seeds were imbibed for 2 d at 4°C in the dark and then treated with 0.2% (v/v) EMS (Sigma-Aldrich) in water for 16 h at 22°C under low light, with shaking at 100 rpm. Treated seeds were rinsed 10 times with water and then planted on soil using 0.1% agar to dispense the seeds. EMS-mutagenized lines were grown at 24°C in long days of 16 h of light, 8 h of dark for 9 weeks. Plants were allowed to self-fertilize. M2 seeds were collected in bulk from an M1 population of approximately 10,000 individuals.
Ice-Cap and Plant Growth
Arabidopsis M2 seeds were grown using the Ice-Cap growth platform as described (Krysan, 2004; Clark and Krysan, 2007). Seeds were plated, germinated, and grown on 96-well plates containing an agar plug. Plants in Ice-Cap blocks were grown for 2 to 5 weeks in a continuous watering system in fluorescent light banks under constant light at a light intensity of 115 μmol m−2 s−1. Roots grew down through the agar onto a second 96-well plate filled with water. When roots reached the bottom of the second plate, the root tissue contained on the lower plate was captured by freezing the plates in a dry ice-ethanol bath. Root samples were separated from the seedlings by separating the plates, leaving viable plants on the upper plate. Twenty-five microliters of a solution of 500 mm Tris, pH 8, 50 mm EDTA, pH 8, was added to the root plate. The plates were sealed, and root tissue was pulverized with steel beads by shaking for 3.5 min at 350 rpm. Insoluble material was pelleted by centrifugation of the plates at 3,400 rpm for 10 min. The supernatant fluid was then diluted 1:2 in water. The genomic DNA from this dilution was used as a PCR template for amplification and mutation scanning. After root extraction, the Ice-Cap plates containing the seedlings still growing in agar plugs were tightly sealed and covered with foil for storage in the dark at 4°C for up to 4 weeks. Plants of interest were extracted from Ice-Cap wells and transplanted to soil with their associated agar plugs. Plants in soil were grown under constant light at 22°C.
PCR and High-Resolution Melting
Genomic DNA collected from plants grown on 96-well Ice-Cap plates was used as a template for PCR. PCRs with a final volume of 20 μL contained 0.2 μm of each PCR primer, 2.5 μm SYTO13 nucleotide-binding dye (Invitrogen), 0.2 mm of each deoxyribonucleotide triphosphate (Promega), 75 mm Tris-HCl, pH 9, 20 mm (NH4)2SO4, 3 mm MgCl2, 0.01% (v/v) Tween 20, DNA prepared from Ice-Cap, and a mutant form of Taq polymerase that has a reduced activity at room temperature (Kermekchiev et al., 2003). A liquid-handling robot (SciClone ALH3000; Caliper LifeSciences) was used to dispense 2 μL of the DNA template into each PCR. Primers used for PCR amplification are listed in Table I. PCR products amplified for mutation detection were melted using the CFX96 PCR Detection System from Bio-Rad Laboratories. The SYBR/FAM emission/detection channel (450–530 nm) was used to detect fluorescence of SYTO13 bound to the PCR products. The protocol used to melt PCR amplicons was as follows: 96°C for 30 s, 40°C for 15 s, ramp from 72°C to 83°C at 0.1°C per s, capturing fluorescence images at each temperature step. Putative mutations were identified by the occurrence of melt curves suggestive of a heteroduplex. Candidate mutations were further tested by performing an independent PCR and melt-curve analysis using a freshly isolated DNA extract. The location of each mutation was determined by dye-terminator DNA sequencing of the amplicon of interest.
Acknowledgments
We thank Peter Jester for expert technical assistance with this project.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. (2004) A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.). Plant J 40: 143–150 [DOI] [PubMed] [Google Scholar]
- Chawade A, Sikora P, Bräutigam M, Larsson M, Vivekanand V, Nakash MA, Chen T, Olsson O. (2010) Development and characterization of an oat TILLING-population and identification of mutations in lignin and beta-glucan biosynthesis genes. BMC Plant Biol 10: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KA, Krysan PJ. (2007) Protocol: an improved high-throughput method for generating tissue samples in 96-well format for plant genotyping (Ice-Cap 2.0). Plant Methods 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S. (2001) High-throughput screening for induced point mutations. Plant Physiol 126: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Henikoff S. (2006) TILLING: practical single-nucleotide mutation discovery. Plant J 45: 684–694 [DOI] [PubMed] [Google Scholar]
- Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, Jamai A, El-Mellouki T, Liu S, Ritchie R, Nielsen N, et al. (2008) TILLING to detect induced mutations in soybean. BMC Plant Biol 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Vincent K, Sharp P. (2009) Simultaneous mutation detection of three homoeologous genes in wheat by high resolution melting analysis and mutation surveyor. BMC Plant Biol 9: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias R, Till BJ, Mba C, Al-Safadi B. (2009) Optimizing TILLING and Ecotilling techniques for potato (Solanum tuberosum L). BMC Res Notes 2: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erali M, Voelkerding KV, Wittwer CT. (2008) High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol 85: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gady ALF, Hermans FWK, Van de Wal MHBJ, van Loo EN, Visser RGF, Bachem CWB. (2009) Implementation of two high through-put techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Methods 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist EJ, O’Neil NJ, Rose AM, Zetka MC, Haughn GW. (2006) TILLING is an effective reverse genetics technique for Caenorhabditis elegans. BMC Genomics 7: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR, et al. (2003) Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KL. (2006) Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem 52: 494–503 [DOI] [PubMed] [Google Scholar]
- Himelblau E, Gilchrist EJ, Buono K, Bizzell C, Mentzer L, Vogelzang R, Osborn T, Amasino RM, Parkin IAP, Haughn GW. (2009) Forward and reverse genetics of rapid-cycling Brassica oleracea. Theor Appl Genet 118: 953–961 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. (2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Jander G, Baerson SR, Hudak JA, Gonzalez KA, Gruys KJ, Last RL. (2003) Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol 131: 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Barth C. (2007) Tandem gene arrays: a challenge for functional genomics. Trends Plant Sci 12: 203–210 [DOI] [PubMed] [Google Scholar]
- Kermekchiev MB, Tzekov A, Barnes WM. (2003) Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR. Nucleic Acids Res 31: 6139–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ. (2004) Ice-cap: a high-throughput method for capturing plant tissue samples for genotype analysis. Plant Physiol 135: 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Signor C, Savois V, Aubert G, Verdier J, Nicolas M, Pagny G, Moussy F, Sanchez M, Baker D, Clarke J, et al. (2009) Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol J 7: 430–441 [DOI] [PubMed] [Google Scholar]
- Li X, Song Y, Century K, Straight S, Ronald P, Dong X, Lassner M, Zhang Y. (2001) A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J 27: 235–242 [DOI] [PubMed] [Google Scholar]
- Metzker ML. (2010) Sequencing technologies: the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Lloyd A, Plaisier CL, Carroll D, Drews GN. (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA 102: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín B, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C. (2009) A high-density collection of EMS-induced mutations for TILLING in Landsberg erecta genetic background of Arabidopsis. BMC Plant Biol 9: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. (2000) Targeted screening for induced mutations. Nat Biotechnol 18: 455–457 [DOI] [PubMed] [Google Scholar]
- Mednik IG. (1988) On methods evaluating the frequencies of induced mutations in Arabidopsis based on embryo test data. Arabidopsis Inf Serv 26: 67–72 [Google Scholar]
- Montgomery J, Wittwer CT, Palais R, Zhou L. (2007) Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc 2: 59–66 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Soukupová H, Schikora A, Zársky V, Hirt H. (2006) A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Perry J, Brachmann A, Welham T, Binder A, Charpentier M, Groth M, Haage K, Markmann K, Wang TL, Parniske M. (2009) TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiol 151: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed GH, Wittwer CT. (2004) Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem 50: 1748–1754 [DOI] [PubMed] [Google Scholar]
- Stephenson P, Baker D, Girin T, Perez A, Amoah S, King GJ, Østergaard L. (2010) A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SH, Suarez-Rodriguez MC, Krysan P. (2007) Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity. FEBS Lett 581: 3171–3177 [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. (2007) MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Cooper J, Tai TH, Colowit P, Greene EA, Henikoff S, Comai L. (2007) Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, et al. (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13: 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Weil C, Springer N, Burtner C, Young K, Bowers E, Codomo CA, Enns LC, Odden AR, et al. (2004) Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triques K, Sturbois B, Gallais S, Dalmais M, Chauvin S, Clepet C, Aubourg S, Rameau C, Caboche M, Bendahmane A. (2007) Characterization of Arabidopsis thaliana mismatch specific endonucleases: application to mutation discovery by TILLING in pea. Plant J 51: 1116–1125 [DOI] [PubMed] [Google Scholar]
- Wang N, Wang Y, Tian F, King GJ, Zhang C, Long Y, Shi L, Meng J. (2008) A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol 180: 751–765 [DOI] [PubMed] [Google Scholar]
- Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RHA, Cuppen E. (2003) Efficient target-selected mutagenesis in zebrafish. Genome Res 13: 2700–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler S, Schwabedissen A, Backasch D, Bökel C, Seidel C, Bönisch S, Fürthauer M, Kuhrs A, Cobreros L, Brand M, et al. (2005) Target-selected mutant screen by TILLING in Drosophila. Genome Res 15: 718–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. (1998) Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA 95: 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J. (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Raina S, Li H, Li J, Dec E, Ma H, Huang H, Fedoroff NV. (2003) Resources for targeted insertional and deletional mutagenesis in Arabidopsis. Plant Mol Biol 53: 133–150 [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang L, Palais R, Pryor R, Wittwer CT. (2005) High-resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin Chem 51: 1770–1777 [DOI] [PubMed] [Google Scholar]