Abstract
Late-Embryogenesis Abundant (LEA) proteins accumulate to high levels during the last stages of seed development, when desiccation tolerance is acquired, and in vegetative and reproductive tissues under water deficit, leading to the hypothesis that these proteins play a role in the adaptation of plants to this stress condition. In this work, we obtained the accumulation patterns of the Arabidopsis (Arabidopsis thaliana) group 4 LEA proteins during different developmental stages and plant organs in response to water deficit. We demonstrate that overexpression of a representative member of this group of proteins confers tolerance to severe drought in Arabidopsis plants. Moreover, we show that deficiency of LEA proteins in this group leads to susceptible phenotypes upon water limitation, during germination, or in mature plants after recovery from severe dehydration. Upon recovery from this stress condition, mutant plants showed a reduced number of floral and axillary buds when compared with wild-type plants. The lack of these proteins also correlates with a reduced seed production under optimal irrigation, supporting a role in fruit and/or seed development. A bioinformatic analysis of group 4 LEA proteins from many plant genera showed that there are two subgroups, originated through ancient gene duplication and a subsequent functional specialization. This study represents, to our knowledge, the first genetic evidence showing that one of the LEA protein groups is directly involved in the adaptive response of higher plants to water deficit, and it provides data indicating that the function of these proteins is not redundant to that of the other LEA proteins.
Water deficit is a common environmental condition that leads to various responses that help in the adaptation or adjustment of an organism to this stress, considered one of the most important environmental stresses influencing plant productivity (Bray, 1997; Morison et al., 2008). The adverse effects of this environmental stress need to be counteracted, mainly because of the increasing soil desertification in cultivated and uncultivated regions. This fact demands that plants tolerate drying periods and elevated salt concentrations in the soil, which may be accompanied by extreme temperatures. Also, an interest in understanding the mechanisms by which plants sense and respond to these environmental cues accounts for the most important reasons to study in detail the responses that have been selected in plants to cope with water deficit.
The acquisition of desiccation tolerance during late stages of seed development is correlated with the induction of a set of small, highly hydrophilic proteins called Late-Embryogenesis Abundant (LEA) proteins (Dure et al., 1989). These proteins are ubiquitous in plants, and although there are several classifications, we will follow that of Battaglia et al. (2008), where they are classified into seven groups on the basis of sequence similarity. Analysis of the protein sequences in these groups from different plant species defined distinctive motifs within groups (Dure, 1993; Battaglia et al., 2008). The number of members is different for each LEA protein group and varies according to the plant species. Most LEA proteins are hydrophilins, a set of proteins characterized by their biased amino acid composition, richness in Gly and other small and/or charged residues, and high hydrophilicity index (Garay-Arroyo et al., 2000). This amino acid composition promotes their flexible structure in solution, existing mainly as random coils, with the exception of the hydrophobic or atypical LEA proteins (Singh et al., 2005). Moreover, hydrophilic LEA proteins from groups 2, 3, and 4 show a prevalence of typical spectroscopic patterns of intrinsically unstructured proteins, with the occurrence of transitions from intrinsically unstructured proteins to ordered conformations in the presence of helix-promoting solvents or air drying (McCubbin et al., 1985; Russouw et al., 1995; Eom et al., 1996; Lisse et al., 1996; Ismail et al., 1999; Wolkers et al., 2001; Soulages et al., 2002, 2003; Goyal et al., 2003; Shih et al., 2004; Tolleter et al., 2007). Their high content of water-interacting residues facilitates the scavenging of water molecules, which is of special importance during developmental stages where a programmed desiccation of tissues takes place, as in the dry seed (Dure et al., 1989), or when cells experience changes in their water status (Colmenero-Flores et al., 1999). Remarkably, there is also an elevated induction in the expression of these proteins in vegetative tissues after exposure to water deficit in basically all plants that have been analyzed. In recent years, proteins with similar characteristics and expression patterns have also been detected to be induced in response to osmotic stress in bacteria and yeast (Stacy and Aalen, 1998; Garay-Arroyo et al., 2000), algae (Honjoh et al., 1995, 2000; Tanaka et al., 2004), nematodes (Solomon et al., 2000; Browne et al., 2004), rotifers (Tunnacliffe et al., 2005), and arthropods (Menze et al., 2009).
One of the hypotheses regarding their function is that these proteins may act as protectors of macromolecules and/or some cellular structures during water deficit, by preferentially interacting with the available water molecules and providing a hydration shell to protect “target” integrity and function (Bray, 1997; Garay-Arroyo et al., 2000; Hoekstra et al., 2001). The use of an in vitro dehydration assay, in which the activity of malate dehydrogenase and lactate dehydrogenase was measured in the presence or absence of a hydrophilic protein, showed that plant hydrophilins (LEA proteins from groups 2, 3, and 4) and hydrophilins from Saccharomyces cerevisiae and Escherichia coli were able to protect these enzymatic activities under low water availability conditions (Reyes et al., 2005). Similarly, in vitro assays using the same or other enzymes have been used to assess the protective capacities of LEA proteins under dehydration and cold (Honjoh et al., 2000; Hara et al., 2001; Bravo et al., 2003; Goyal et al., 2005; Grelet et al., 2005; Nakayama et al., 2007; Reyes et al., 2008). In some of these assays, the ratio of LEA protein to enzyme was 1:1, suggesting that the LEA protein protective activity is not only due to the formation of a preferential hydration shell but also to an additional effect probably related to a direct interaction with their targets (Reyes et al., 2005, 2008).
There are many reports of LEA proteins expressed in transgenic plants under the control of regulated or constitutive promoters, showing tolerant phenotypes under drought, high salinity, or freezing stress (Xu et al., 1996; Sivamani et al., 2000; NDong et al., 2002; Chandra Babu et al., 2004; Puhakainen et al., 2004; Fu et al., 2007; Lal et al., 2007; Xiao et al., 2007; Dalal et al., 2009). Also, the heterologous expression in bacteria and yeast of some LEA proteins confers salt and freezing tolerance (Imai et al., 1996; Zhang et al., 2000; Liu and Zheng, 2005). However, this “gain-of-function” approach does not necessarily reflect their direct participation in the plant adjustment or adaptation to these stress conditions but rather their potential to confer tolerance when ectopically expressed. In contrast, the results of a “loss-of-function” approach will lead to a direct indication of the participation of a particular gene within this process. Even though there is a large extent of information regarding the different properties of LEA proteins, our knowledge concerning their role in plant adaptation to water-limiting conditions is insufficient.
In this work, we focus on the study of the group 4 LEA proteins of Arabidopsis (Arabidopsis thaliana). With only three genes in the genome (AtLEA4-1, AtLEA4-2, and AtLEA4-5), the AtLEA4 group is one of the smallest groups in Arabidopsis (Battaglia et al., 2008; Hundertmark and Hincha, 2008), which makes it accessible for a loss-of-function analysis. The LEA4 proteins are characterized by a high content of A, T, and G amino acid residues, the latter highly represented in unstructured proteins. They have a conserved N-terminal domain of 70 to 80 residues, predicted to form amphipathic α-helices, and a less conserved C-terminal region with variable size and random coil structure (Dure, 1993). Like other LEA proteins, the LEA4 group is highly accumulated in all embryo tissues of dry seeds (Roberts et al., 1993). Recently, Wise (2002) performed a bioinformatics analysis and questioned the existence of a group 4 of LEA proteins as a distinct group of LEA proteins from group 3. The algorithm used the overrepresentation/underrepresentation of particular amino acids within small motifs in the protein, giving rise to a different classification for these proteins (Wise, 2003). In support of the original classification proposed by Dure et al. (1989) and because of the high sequence conservation within this group in plants, in this work, we present genetic and functional evidence that group 4 of LEA proteins is indeed a distinct group conserved in the plant kingdom. The results reported here show that overexpression of one of the AtLEA4 proteins in Arabidopsis leads to a tolerant phenotype compared with their wild-type counterparts in their capability to endure severe water deficit and that the reduction in the accumulation levels of these proteins leads to plants more sensitive to water-limiting conditions than their wild-type genotypes. Altogether, these data constitute, to our knowledge, the first direct evidence indicating that LEA4 proteins are involved in the adaptive response of vascular plants to withstand water deficit.
RESULTS
The AtLEA4 Group Is Differentially Expressed during Embryogenesis and in Response to Water Deficit Treatments
To gain insight into the function of LEA proteins in the adaptation of vascular plants to water deficit, we carried out a functional analysis of the Arabidopsis LEA4 protein family, because it is one of the three LEA protein groups with fewer members (two for group 1, three for group 4, and three for group 6; Battaglia et al., 2008; Hundertmark and Hincha, 2008). These data were confirmed with a BLASTp analysis using a reported LEA4 homolog from cotton (Gossypium hirsutum) as query (LEA D-113; National Center for Biotechnology Information [NCBI] accession no. M19406); it retrieved three proteins that conformed the Arabidopsis LEA4 family, encoded in loci At1g32560, At2g35300, and At5g06760. In reference to their chromosomal locations, we named the corresponding proteins AtLEA4-1, AtLEA4-2, and AtLEA4-5 (Supplemental Fig. S1). As predicted by Dure (1993), the proteins in this group were conserved in the N-terminal portion (AtLEA4-1, 1–78; AtLEA4-2, 1–74; AtLEA4-5, 1–76), for which α-helix and “coiled-coil” structures were predicted to form in silico (Lupas et al., 1991; McGuffin et al., 2000). In contrast, their C-terminal region (AtLEA4-1, 79–134; AtLEA4-2, 75–97; AtLEA4-5, 77–158 [corresponding to a length of 56, 23, and 82 amino acids, respectively]) showed a putative random coil structure (Supplemental Fig. S1). These proteins were predicted to be basic proteins (pI = 8.65–9.67) with molecular masses of 14.9, 10.5, and 16.2 kD for AtLEA4-1, AtLEA4-2, and AtLEA4-5, respectively.
The presence and abundance of a LEA transcript and the corresponding protein during a developmental stage or in response to stress in a plant organ can provide information about their sensitivity to different types of environmental adverse conditions, which can be useful in the elucidation of their function. Hence, we analyzed the LEA4 group transcript and protein accumulation patterns during embryogenesis and in seedlings of Arabidopsis plants grown under optimal irrigation and in plants subjected to water deficit treatments. The results from reverse transcription (RT)-PCR experiments using total RNA showed that, in agreement with available microarray data (Schmid et al., 2005; Winter et al., 2007; Hruz et al., 2008), transcripts of the AtLEA4 family could be detected in flowers and during embryo development, but the highest abundance was detected at the dry seed stage. After seed germination, their transcript levels showed a significant reduction (Fig. 1A).
Figure 1.
Transcript and protein accumulation patterns of the AtLEA4 gene family during embryogenesis, germination, and seedling establishment. A, Semiquantitative RT-PCR analysis of AtLEA4 transcripts using total RNA (2 μg) from flowers and buds, developing siliques, seeds, and seedlings. ACTIN2 (ACT2) transcript was used as a loading control. B, Western-blot analysis using specific antibodies against each of the AtLEA4 proteins and protein extracts (15 μg) from the same stages of development described in A. Molecular mass markers are indicated (MW). Arrowheads indicate higher molecular mass bands, which were recognized specifically by the corresponding antibodies. Reversible stain with Ponceau red after transfer is shown as a loading reference. These results are representative of five independent experiments.
Western-blot experiments showed that AtLEA4-1 protein accumulated abundantly in flowers and immature siliques (Fig. 1B). Unexpectedly, in dry seeds, during stratification and germination, the 14.9-kD AtLEA4-1 protein was undetectable. Instead, a protein with an apparent higher molecular mass (AtLEA4-1-L) was specifically recognized by immunopurified LEA4-1 antibodies in dry or stratified seed protein extracts. The accumulation level of AtLEA4-1-L protein was higher in dry than in stratified and in germinating seedlings. Although in the experiment shown in Figure 1B, the AtLEA4-1 protein was not detected in germinating seedlings (24 h after incubation at 25°C), longer exposures allowed its detection at low levels (data not shown). Once the seeds have germinated, the AtLEA4-1-L protein disappears and the 14.9-kD LEA4-1 was detected again starting at 2 d after germination (DAG; Fig. 1B). Regarding AtLEA4-2 and AtLEA4-5 proteins, they showed high accumulation levels during late embryogenesis and in dry seeds, followed by a gradual reduction in germinating seedlings (Fig. 1B). Western-blot experiments using specific antibodies raised against AtLEA4-2 did not recognize a protein with the expected molecular mass (10.5 kD); instead, in all cases, they specifically detected a protein with a higher molecular mass (approximately 30 kD; Fig. 1B). Similar results were obtained using antibodies from different sources. The facts that this band was not detected when the AtLEA4-2 transcript was silenced by an artificial microRNA (a-miR; see below), and that this detection was competed with an AtLEA4-2 peptide (data not shown), demonstrated the specificity of this antibody.
AtLEA4 group transcript and protein levels were also determined in 2-week-old seedlings incubated in liquid Murashige and Skoog (MS) medium containing 100 μm abscisic acid (ABA), 25% (w/v) polyethylene glycol 8000 (PEG), or NaCl (100, 150, or 200 mm) during 12 h. Results in Figure 2A show that all three genes were responsive to water deficit imposed by PEG treatment; upon ABA and NaCl treatments, the AtLEA4-5 gene was shown to be the most responsive. Increasing concentrations of NaCl led to a gradual increase in the accumulation of AtLEA4-5 transcripts, whereas in the cases of AtLEA4-1 and AtLEA4-2, the accumulation of their transcripts was hardly detected (Fig. 2A). Regarding their protein levels, in agreement with their transcript accumulation patterns, the three proteins of the family accumulated upon PEG treatment. However, in response to ABA and NaCl treatments, some differences were observed between their transcript and protein accumulation patterns (Fig. 2B). Even though the AtLEA4-1 and AtLEA4-2 transcripts were barely detected upon NaCl treatments, their responsiveness to this stress condition was evident by the accumulation of their proteins (Fig. 2B). This was not the case in response to ABA, where the accumulation levels of AtLEA4-1 and AtLEA4-2 proteins did not change upon ABA treatment (Fig. 2B). The fact that the AtLEA4-1 protein was detected in spite of the low levels of its transcript (Fig. 2) suggested the participation of posttranscriptional control mechanisms modulating the levels of this protein. As in the western-blot experiments described above, the AtLEA4-2 protein was detected with an apparent higher molecular mass. Similar to AtLEA4-1 and AtLEA4-2 proteins, AtLEA4-5 protein accumulated in response to increasing NaCl concentrations; however, its accumulation levels were lower than those observed in response to ABA, even though AtLEA4-5 transcript accumulated to similar levels as upon salt treatments. The conspicuous contrast between transcript and protein accumulation patterns in response to ABA again suggested the involvement of a posttranscriptional control for the adjustment of protein levels. Overall, these results demonstrated that the three members of the LEA4 family were differentially expressed under normal developmental stages and upon stress treatments, suggesting that a functional diversification occurred in the course of evolution.
Figure 2.
Transcript and protein expression patterns of the AtLEA4 gene family from seedlings after 12-h treatments with ABA or abiotic stress. A, Northern-blot analysis using specific probes for each AtLEA4 transcript and total RNA (20 μg) from 2-week-old plants grown in vitro. Hybridization with 28S rRNA was used as a loading control. B, Western-blot analysis using specific antibodies for each AtLEA4 protein and total extracts (15 μg) from the same treatments as in A. Molecular mass markers are indicated (MW), and arrowheads show higher molecular mass bands, which were specifically recognized by the corresponding antibodies. Reversible stain with Ponceau red was used as a loading reference. These results are representative of three independent experiments.
AtLEA4-5 Constitutive Expression Leads to Tolerance to Severe Drought in Arabidopsis Adult Plants
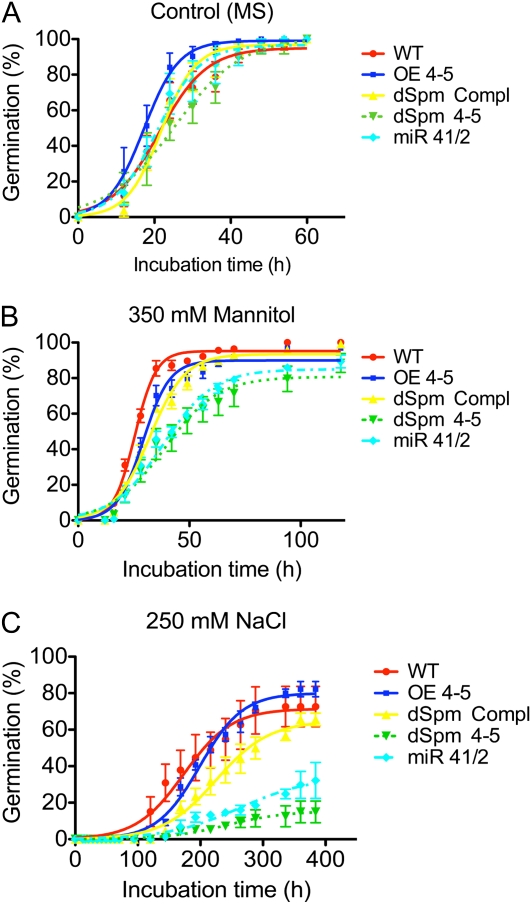
Arabidopsis plants were transformed by floral dipping using an Agrobacterium tumefaciens strain carrying a 35S::AtLEA4-5::NOS fusion, which led to the constitutive expression of the AtLEA4-5 gene (Fig. 3A). We selected the AtLEA4-5 gene for overexpression analysis because it showed the strongest response to water deficit treatments, as determined by its transcript and protein accumulation patterns among all three family members. Once the overexpression of the protein of interest was verified under control and drought conditions (Fig. 3, A and B), five independent homozygous transgenic Arabidopsis lines were used to analyze their phenotype under optimal and limiting irrigation (for details, see “Materials and Methods”). These lines overexpressed the AtLEA4-5 protein under both growth conditions, showing higher accumulation levels than the wild type even upon water deficit treatment (Fig. 3, A and B). The phenotypic analysis of these overexpression lines showed that many characteristics throughout development were similar to those shown by the wild type (see Fig. 8 below; data not shown). To evaluate the contribution of the AtLEA4-5 protein to the tolerance of Arabidopsis plants to low water availability conditions, the selected transgenic lines were subjected to water deficit during germination and in the adult stage. During germination, the effects of high concentrations of NaCl (250 mm) and of high osmolarity imposed by mannitol (350 mm) were determined by monitoring germination (radicle emergence) up to 16 d. The transgenic homozygous lines overexpressing AtLEA4-5 protein did not show a significantly improved ability to germinate under these conditions, because their germination rate was similar to that observed for wild-type seeds (OE 4-5; see Fig. 5 below).
Figure 3.
Phenotypic analysis of 35S::AtLEA4-5::NOS adult plants under dehydration and after recovery from drought. A, Western-blot analysis using antibodies against AtLEA4-5 and total protein extracts (15 μg) from the wild type (WT) and homozygous independent transgenic lines (lines 2–4, 6, and 7) under optimum irrigation. B, Western blot as in A using drought-treated plants in the adult stage. Molecular mass markers (MW) are indicated. Reversible stain after transfer with Ponceau red is shown as a loading reference. C, RWC of adult plants under irrigation or subjected to drought. One-way ANOVA showed significant differences between groups for drought-treated plants (P = 0.0065). Bars indicate means ± se (n = 3). D, Whole plant biomass under irrigation or after 10 d of recovery from severe drought. Significant differences were found between groups in the plant biomass recovered after stress (P = 0.0007). Bars indicate means ± se (n = 3). E, Number of floral and axillary buds per plant under irrigation (n = 3) and after recovery from drought (n = 9). Bars indicate means ± se. Significant differences between groups were found under irrigation (P < 0.0001) and after recovery from stress (P = 0.0033). Different letters above each bar indicate statistically significant differences using Dunnett’s posttests (P < 0.05).
Figure 8.
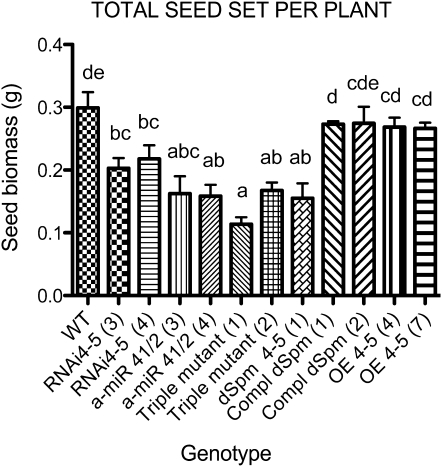
Total seed production of AtLEA4 single (RNAi 4-5, dSpm 4-5), double (a-miR 4-1/2), and triple mutants grown under optimal irrigation. Plants were germinated in vitro and transplanted to develop and set seeds under optimum irrigation conditions. Seeds from the wild type (WT) and two independent homozygous lines from each construct used in this study were harvested throughout the productive cycle until senescence. The transgenic lines overexpressing the AtLEA4-5 gene in the dSpm mutant (dSpm 4-5) and wild-type backgrounds (OE 4-5) are shown as controls. Bars indicate means ± se (n = 5). The numbers in parentheses show the lines selected from each construct (as indicated in Figs. 4 and 7). Significant differences among genotypes were determined by one-way ANOVA (P < 0.0001) statistical analysis. Different letters show significant differences between groups as indicated by Dunnett’s posttests (P < 0.05).
Figure 5.
Accumulated germination percentage of the wild type (WT) and AtLEA4 transgenic lines under optimal growth conditions or under stress. Germination was quantified by radicle emergence using seeds of homozygous lines plated on standard MS medium (control [A]), MS medium with 0.35 m mannitol (osmotic stress [B]), or MS medium with 0.25 m NaCl (ionic + osmotic stress [C]). Transgenic lines used were the 35S::AtLEA4-5::NOS construct in the wild-type background (OE 4-5) and the 35S::AtLEA4-5::NOS construct in the dSpm mutant background (dSpm Compl). Also, insertion mutant in the AtLEA4-5 gene (dSpm 4-5) and double mutant in AtLEA4-1 and AtLEA4-2 genes silenced with an a-miR construct (miR 4-1/2) were analyzed. Seeds were stratified for 3 d and incubated in a growth chamber at 25°C for the indicated times. Error bars indicate se of three replicates (n = 300), which were fit to a sigmoidal dose-response curve. Significant differences between genotypes were found in three parameters of the curve fit (steepness of the curve, Y value at the top plateau, and X value when the response is halfway between bottom and top) at P > 0.0011 (A), P < 0.0001 (B), and P < 0.0001 (C). [See online article for color version of this figure.]
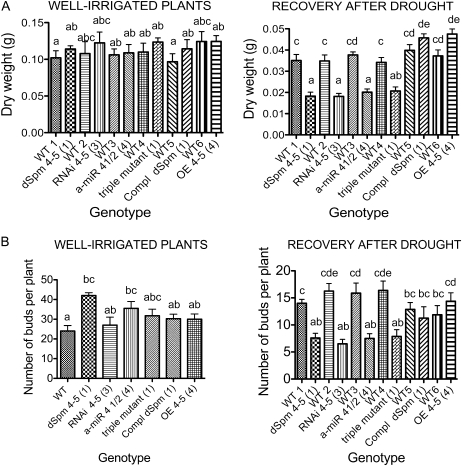
In the adult stage, two different experiments were carried out growing the plants in a substrate with low water retention. In order to evaluate the impact of AtLEA4-5 overexpression on the production of buds after recovery from severe dehydration, plants at the flowering stage were subjected to water deficit by halting irrigation during 14 d, when the substrate water potential (Ψsubstrate) was approximately −6.45 ± 0.57 MPa (Supplemental Fig. S2), after which plants were rehydrated and allowed to recover during 10 d; at this point, axillary and floral buds were counted (for details, see “Materials and Methods”). As shown in Figure 3, C and D, and Supplemental Figure S2, the transgenic lines overexpressing AtLEA4-5 protein showed a better recovery from this severe dehydration treatment as compared with wild-type plants. Homozygous AtLEA4-5-overproducing lines showed not only recovery of vegetative tissues (e.g. rosette leaves) but also a higher number of axillary and floral buds, contrasting with wild-type plants that, even though they were able to recover some of their rosette leaves, were incompetent in bud maintenance (Fig. 3E; Supplemental Fig. S2; Supplemental Table S1). Total biomass accumulation recorded at the end of the recovery period showed that four of the five 35S::AtLEA4-5::NOS lines presented a significantly higher biomass when compared with wild-type plants (Fig. 3D).
In an independent experiment, wild-type and AtLEA4-5-overexpressing plants at the flowering stage were subjected to water deficit by halting irrigation during 10 d, when Ψsubstrate was approximately −4.62 ± 0.62 MPa; at this point, complete plants were harvested to determine relative water content (RWC; for details, see “Materials and Methods”). The results show that plants overexpressing AtLEA4-5 protein exhibited a higher RWC upon water deficit when compared with wild-type plants (Fig. 3C). These data support the conclusion that the overproduction of AtLEA4-5 protein confers tolerance to water-limiting conditions to Arabidopsis plants, as determined by RWC, biomass accumulation, and bud maintenance after recovery from severe dehydration.
The AtLEA4-5 Insertion Mutant and Posttranscriptional Gene Silencing Mutants in AtLEA4-1 and AtLEA4-2 Genes Show Sensitive Phenotypes in Response to Osmotic Stress during Germination and to Drought Treatments in Adult Plants
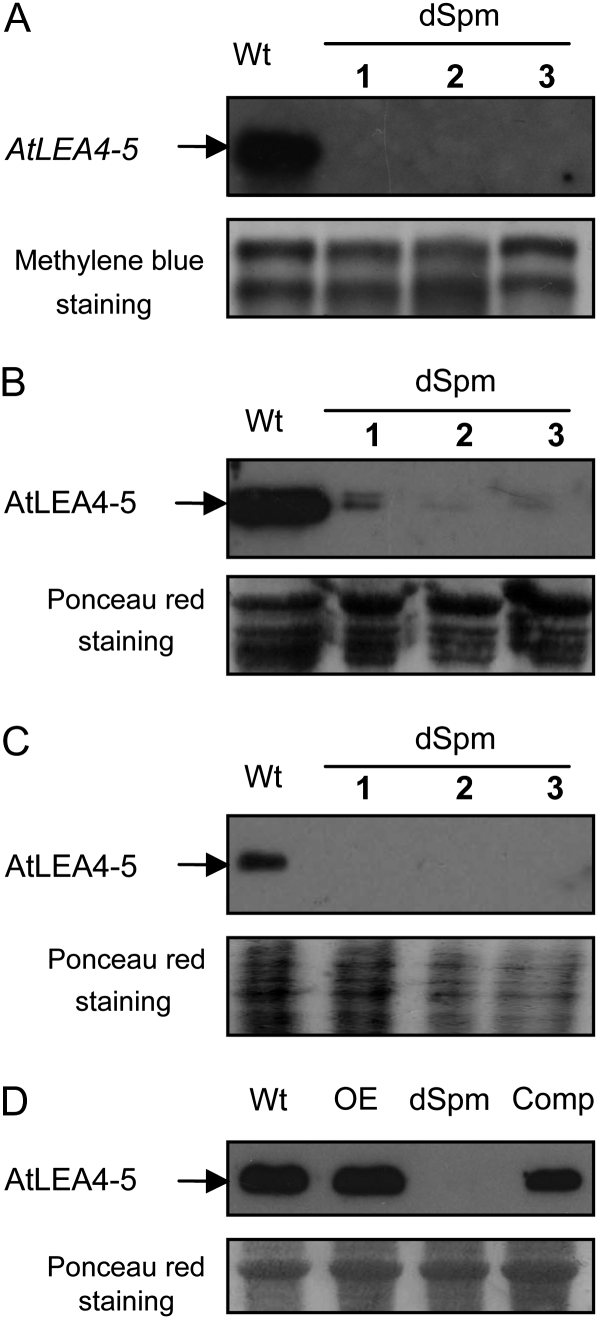
In this work, we analyzed the phenotype of mutants affecting the three members of the LEA4 gene family in response to water deficit conditions. A nonautonomous Suppressor-mutator transposon insertion (dSpm, Tissier et al., 1999) in the promoter of AtLEA4-5 was obtained from the European Arabidopsis Stock Centre (accession no. N122943). Southern-blot experiments confirmed the dSpm transposon insertion site in the AtLEA4-5 gene promoter as described in the European Arabidopsis Stock Centre database (Supplemental Fig. S3). To investigate whether this mutation affects the production of the corresponding protein, northern- and western-blot analyses were carried out. The results showed that this insertion led to a severe reduction in the levels of AtLEA4-5 transcript and protein when detected in dry seeds and roots from adult plants grown under water deficit as opposed to wild-type plants (Fig. 4, A–C).
Figure 4.
Reduction in the expression levels of AtLEA4-5 transcript and its corresponding protein in the transposon insertion mutant (dSpm). A, Northern-blot analysis using a specific probe for AtLEA4-5 and total RNA (10 μg) from dry seeds of wild-type (Wt) and dSpm mutant plants. Reversible stain with methylene blue after transfer was used as a loading reference. B, Western-blot analysis using specific antibodies against AtLEA4-5 and total protein extracts (10 μg) from dry seeds of wild-type and dSpm mutant plants. C, Western-blot analysis using protein extracts from roots under dehydration (5 μg) of wild-type and dSpm mutant plants. D, Western-blot analysis using total protein extracts (10 μg) from adult plants under dehydration, showing wild-type, homozygous transgenic lines with constitutive expression of AtLEA4-5 protein in the wild-type background (OE), transposon insertion in the AtLEA4-5 gene (dSpm), and its complementation with a 35S::AtLEA4-5::NOS construct (Comp). For B to D, reversible staining with Ponceau red after transfer was used as a loading reference.
This mutant was used to analyze its phenotype during germination in media with or without NaCl (250 mm) or mannitol (350 mm) as well as in adult plants subjected to dehydration-rehydration treatments. As shown in Figure 5, mutant seeds were unable to withstand the stress treatments, because a significant reduction in their germination rate compared with that of wild-type seeds was detected, thus indicating that AtLEA4-5 protein was necessary for optimal germination efficiency under water deficit. To evaluate the participation of the AtLEA4-5 protein in the ability of the plant to maintain the production of floral and axillary buds under water limitation, plants were grown under optimal irrigation conditions until bolting, and at this point they were subjected to a dehydration treatment until the Ψsubstrate was approximately −4.62 ± 0.62 MPa (note that the dehydration treatment in this experiment was less severe than that used with overexpressing lines); subsequently, plants were rehydrated during a 6-d recovery period (for details, see “Materials and Methods”). After this time, axillary and floral buds were counted and then plants were harvested to determine total biomass accumulation. Results in Figure 6 show that significant differences were detected in biomass accumulation after recovery from drought between mutant and wild-type plants and that the production of buds in the AtLEA4-5 mutants was affected by the dehydration-rehydration treatment when compared with wild-type plants. In this case, wild-type plants recovered some buds, in contrast to the phenotype of wild-type plants during the characterization of 35S::AtLEA4-5::NOS, where the dehydration treatment was more severe (Supplemental Fig. S2, B and D). The lower severity of the dehydration-rehydration treatments for the experiments involving mutants of the AtLEA4 genes was necessary to be able to monitor the susceptibility of the mutant lines in comparison with wild-type plants, which did not survive upon severe dehydration. To verify that the phenotype detected in the mutant plants was due to the absence of a AtLEA4-5 functional gene, complementation experiments were carried out by transforming the dSpm transposon insertion mutant with the 35S::AtLEA4-5::NOS construct (Fig. 4D). The phenotypic analysis of the transformed mutants performed in the T3 generation showed that the overexpressed AtLEA4-5 gene was able to complement the AtLEA4-5::dSpm mutant during germination under low water availability conditions (Fig. 5) and in adult plants after dehydration-rehydration treatments (Fig. 6). In this type of experiment, where the dehydration was not as severe as in the previous one used for the characterization of the AtLEA4-5-overexpressing lines (Columbia [Col] background; Fig. 3), there were no significant differences with the AtLEA4-5 gene complementation lines (Col, AtLEA4-5::dSpm background) and wild-type plants (Figs. 5 and 6).
Figure 6.
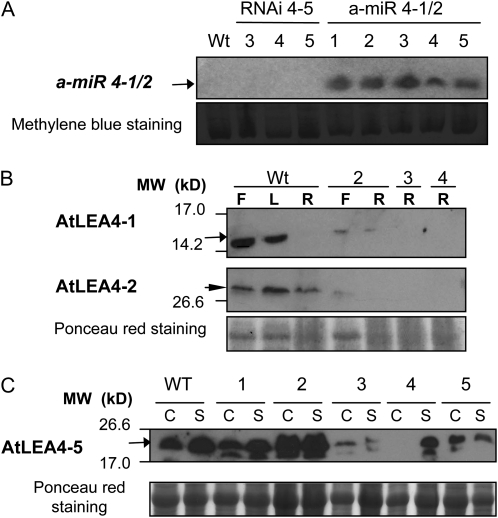
Phenotypic analysis of adult plants with altered accumulation levels of the AtLEA4 protein family. Seedlings grown in vitro for 2 weeks were transplanted to a low-water-retention substrate and kept under optimum irrigation with nutrient solution until flowering under greenhouse conditions. Wild type (WT) and homozygous lines were grown in the same pot. Dehydration was followed by loss of water from the substrate, and pots were rotated in the tray every 2 d to maintain uniform water loss during drought treatment. A, Biomass of whole plants under control conditions (well-irrigated plants) or after 6 d of recovery from stress. One-way ANOVA was applied for each treatment to compare the performance of the different lines. This analysis showed significant differences between lines after recovery from drought (P < 0.0001). Bars indicate means ± se (n = 8). B, Number of axillary and floral buds per plant under optimum irrigation (n = 4) or after 6 d of recovery from stress (n = 8). Significant differences between groups were found using one-way ANOVA under control conditions (well-irrigated plants; P = 0.0095) and after recovery from stress (P < 0.0001). Bars indicate means ± se. Different letters show significant differences between bars (P < 0.05) as indicated by Tukey’s posttests. Homozygous lines were used in all experiments: transposon insertion in the AtLEA4-5 gene (dSpm 4-5), posttranscriptional gene silencing single mutant with RNAi-directed silencing of the AtLEA4-5 gene (RNAi 4-5), posttranscriptional gene silencing double mutants in AtLEA4-1 and AtLEA4-2 genes using an a-miR (a-miR 4-1/2), and the resulting F2 crosses from mutants of RNAi with a-miR (triple mutant). This figure also shows data from lines ectopically overexpressing AtLEA4-5 protein (35S::AtLEA4-5::NOS) in the wild type (OE 4-5) or in the AtLEA4-5::dSpm mutant background (Compl dSpm). The numbers in parentheses indicate the homozygous line used for the phenotypic analysis as shown in Figures 4 and 7.
Because no mutants were available for the AtLEA4-1 and AtLEA4-2 genes, an a-miR construct was used for their posttranscriptional silencing. The high homology between AtLEA4-1 and AtLEA4-2 genes allowed the design of an a-miR (a-miR 4-1/2) able to target both genes for silencing using a region of maximal nucleotide identity between both transcripts (Supplemental Fig. S4). The precursor of a-miR 4-1/2 was expressed under the control of the 35S promoter to favor an efficient silencing of the genes of interest. Those lines showing the highest a-miR accumulation were selected to validate the expected silencing. The mature a-miR 4-1/2 was detected by small RNA northern blot from various T3 lines using 2-week-old plants grown under optimal conditions (Fig. 7A). The results from western-blot analyses using protein extracts from adult plants of the selected lines subjected to dehydration indicated that the designed a-miR was functional and able to silence the expression of both genes (Fig. 7B). Once confirmed that the silencing was functional, these lines were phenotypically characterized, applying the same treatments described above for the AtLEA4-5 insertion mutant. For the germination assays, the major effect was observed when seeds were germinated in the presence of NaCl (250 mm), where the silenced AtLEA4-1 and AtLEA4-2 mutant seeds showed 20% germination compared with 70% for wild-type seeds. When mannitol (350 mm) was used, no differences in the final germination percentage were detected; however, significant differences were observed between the silenced mutants and the wild-type germination rates (Fig. 5). An increased susceptibility to dehydration was also detected in experiments using adult plants subjected to dehydration-rehydration, where mutants showed a significant reduction in the amount of buds when compared with wild-type plants (Fig. 6; Supplemental Table S2).
Figure 7.
Posttranscriptional gene silencing of AtLEA4 genes using an a-miR (a-miR 4-1/2) to silence AtLEA4-1 and AtLEA4-2 or RNAi to silence AtLEA4-5 transcripts (RNAi 4-5). A, Northern blot using antisense probe of a-miR 4-1/2 and RNA (20 μg) from homozygous transgenic seedlings grown under optimal conditions to confirm the constitutive expression of mature a-miR 4-1/2. Wild type (Wt) and RNAi 4-5 lines (lanes 3–5) were used as controls. Reversible staining with methylene blue after transfer was used as a loading reference. B, Western blot using specific antibodies for AtLEA4-1 and AtLEA4-2 proteins and total protein extracts (10 μg) from adult plants grown under drought to show their accumulation levels in the wild type and in homozygous silenced plants (lanes 2–4). F, Flower; L, leaf; R, root. The arrow shows the proteins migrating with the expected molecular mass (MW) for the corresponding monomer size, and the arrowhead shows the higher molecular mass band specifically detected with AtLEA4-2 antibodies. The selected lines for further phenotypic analysis were lines 3 and 4. C, Western blot using antibodies against AtLEA4-5 protein and total protein extracts (10 μg) from homozygous RNAi 4-5 seedlings (lanes 1–5) grown in vitro for 2 weeks and immersed in liquid MS medium, where they were treated for 8 h without (C, for control) or with 25% PEG solution (S, for stress), showing different silencing levels. The selected lines for further phenotypic analyses were those showing the lower AtLEA4-5 protein accumulation (lanes 3–5). Reversible staining with Ponceau red after transfer was used as a loading reference in B and C.
To construct a triple mutant affecting the production of the three members of the AtLEA4 group and because both mutants, dSpm insertion in AtLEA4-5 and a-miR 4-1/2, are resistant to BASTA, we generated a different mutant affecting the expression of AtLEA4-5, containing kanamycin as the selection marker. To this end, we obtained a silencing mutant using RNA interference (RNAi) against AtLEA4-5 by the expression of the AtLEA4-5 open reading frame (ORF) sequence arranged as an inverted repeat, under the control of the 35S promoter (RNAi 4-5). Some of the transgenic lines containing this construct showed lower levels of expression than wild-type plants under the tested conditions (mutant lines 3–5 [Fig. 7C; Supplemental Fig. S5]); however, because silencing was stable just in T1 and T2 generations, all experiments with these lines were carried out using T2 homozygous plants. In this generation, we selected independent transgenic lines showing different levels of reduction in the AtLEA4-5 protein accumulation (lines 3–5; Fig. 7C), which were phenotypically characterized prior to the generation of the triple mutant. Analyses were carried out using Arabidopsis adult plants subjected to dehydration treatment until Ψsubstrate = −4.62 ± 0.62 was achieved under greenhouse conditions. The results from these analyses showed that RNAi 4-5-silenced lines accumulated less biomass, recovered fewer buds after 6 d of plant rehydration, and produced a lower total seed number per plant than wild-type plants (Supplemental Table S1; Supplemental Fig. S5). Homozygous T2 RNAi 4-5-silenced plants (mutant line 3 in Fig. 7C) were crossed with homozygous T2 a-miR 4-1/2-silenced plants (mutant lines 3 and 4 in Fig. 7B), choosing those lines where the silencing of the corresponding transcripts was more efficient. From the products of this cross containing both silencing constructs (a-miR 4-1/2 and RNAi 4-5), we selected those lines that showed the lower protein levels in the F2 generation for further characterization (Supplemental Fig. S6). Two independent lines were subjected to dehydration-rehydration treatments as those applied for the phenotypical analyses described above. The results showed a significantly lower dry biomass and lower recovery of buds after dehydration-rehydration in those plants affected in the production of the group 4 LEA proteins compared with wild-type plants grown in the same pot (Fig. 6). As for the RNAi 4-5-silenced plants, the silencing was functional only in the F1 and F2 generations: some homozygous F3 lines showed wild-type protein levels as well as wild-type phenotype upon dehydration-rehydration treatments (data not shown). Because RNA interference was not effective in reducing AtLEA4-5 protein accumulation in seeds, none of the lines containing the RNAi 4-5 construct were used for phenotypical characterization during germination.
Because the three protein members of LEA group 4 accumulated to high levels in dry seeds, we analyzed the effects that their lower levels could have on seed production under optimal growth conditions. To this end, individual seedlings of the different lines used in this study were grown in pots containing soil (Metromix) under optimal irrigation until senescence. Seeds were collected from each plant, and the dry biomass of seeds was determined. The comparison of their seed biomass showed that the absence of AtLEA4-5 protein, the low levels of AtLEA4-1 and AtLEA4-2 proteins, or of the complete protein family led to a lower seed biomass than in wild-type plants even when grown during their complete life cycle under optimal growth conditions (Fig. 8). Higher AtLEA4-5 protein levels obtained when AtLEA4-5 was overexpressed did not confer any advantage on this phenotype when compared with wild-type plants (Fig. 8).
Structural and Phylogenetic Analysis of Group 4 LEA Proteins Points to an Early Gene Duplication That Gave Rise to Two Distinct Subgroups with Arguably Divergent Functions
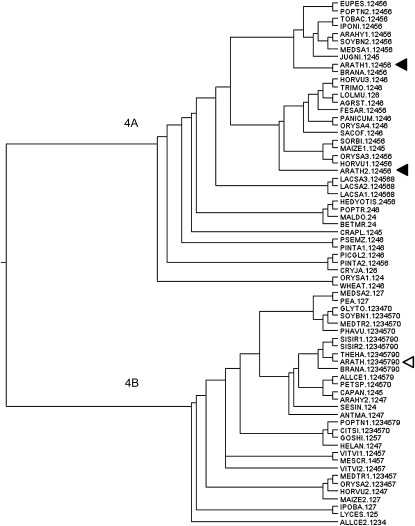
In a previous work, we showed that group 4 LEA proteins can be defined based on sequence similarity (Battaglia et al., 2008). In that work, we report the identification of two subgroups, where the proteins differed in the length of their C termini. Five sequence motifs were identified, three of which were common to all proteins, while the other two were present only in the longer proteins. In this work, we report a more thorough analysis, starting with a collection of 74 LEA group 4 proteins from angiosperms, gymnosperms, and bryophyta. A multiple sequence alignment showed a broadly conserved N-terminal region and a much more variable C-terminal region, where sequence repetitions and rearrangements were common (data not shown). Given that such variability confounds the creation of a correct multiple alignment for the C-terminal region, to describe the motif structure of the group 4 LEA proteins we used MEME, a motif discovery tool that does not rely on alignments (Bailey and Gribskov, 1998). Ten motifs were discovered. MEME is sensitive to the starting collection of sequences, so the new motifs do not match precisely those reported previously. Thus, we will not refer to the previous nomenclature. Motifs 1, 2, and 4 are almost universally distributed, being present in 70, 74, and 66 of the 77 proteins, respectively. The actual prevalence of motif 4 could be higher, because, being the left-most motif, it could easily be lost during cDNA construction. Motifs 6 and 7, although distinct, must be divergent variants of the same motif, because they are related in sequence, occupy equivalent positions in the proteins, and are mutually exclusive: proteins have either motif 6 (35 proteins) or motif 7 (32 proteins). Motif 5 is also very common; it is present in 46 proteins, where it usually lies near the C terminus, and sometimes it is present more than once per protein. Motifs 3, 9, and 10 appear in 13, seven, and 12 proteins, respectively. When present, they appear in proteins that have motif 7 and never in association with those that have motif 6 (Supplemental Table S3; Supplemental Fig. S7). Motif 8 is only present in the three proteins from lettuce (Lactuca sativa). It might be related to motif 3 because it has a similar size, occupies a similar position in the proteins, and, like motif 3, is only present in proteins that carry motif 7. However, motifs 3 and 8 are not similar in sequence.
The Pfam (listed as LEA_1, PFAM 03760) alignment that defines this family encompasses motifs 4, 1, 2, and either motif 6 or 7, in that order. We called this region of the proteins “the main conserved block” (MCB). Because there are no gaps between the four motifs and the region has the same length in all proteins, the MCB could be easily aligned. We used the MCB to reconstruct the phylogeny of the family. Although none of several methods achieved high reliability for all branches (something we impute to having many sequences and relatively few alignment columns), the resulting best trees from all strategies were very similar and are generally consistent with the species phylogeny, except for a deep branching, which probably indicates a very ancient gene duplication (a representative tree is shown in Fig. 9). The deepest divide was found between proteins having motif 6 and those having motif 7, showing a bootstrap value of 62/100 with neighbor joining and 80/100 with Fitch, but bootstrap values became 99/100 if we ignored the wheat (Triticum aestivum) LEA, which has a very poor motif 6 (MAST e-value = 0.00037; for the sequences used and motif presence, see Supplemental Table S3). This divide was also the most reliable with a maximum likelihood approach (Proml; bootstrap was 34/100, and 96/100 if the wheat LEA was ignored). The divide was not an artifact of the great difference in sequence between motifs 6 and 7, because the same divide and similar bootstrap values were obtained using motif 2 alone instead of the whole MCB. In general, motif 2 was informative of the evolutionary history of the complete proteins, indicating that it represents the defining core of group 4 LEA proteins and that throughout evolution the acquisition and loss of the other motifs has not been rampant. The presence of proteins with either motif 6 or motif 7, which allowed us to predict the associated “accessory motifs,” together with the finding that the deepest dichotomy in the phylogenetic tree was clearly attributable to the presence of either of these two motifs indicate that group 4 LEA proteins should be divided in two subgroups, 4A and 4B, according to the occurrence in a protein of motif 6 or motif 7. Because both subgroups were found within and outside the angiosperms (4A exists in several conifers, while 4B was present in the moss Physcomitrella patens), the gene duplication must have predated the evolution of seed plants. Furthermore, the side-to-side persistence of both subgroups, after hundreds of millions of years, in broadly distributed taxons suggests that the duplication gave rise to functional divergence in such a way that one subgroup cannot be substituted for the other. This acquisition of specialized subfunctions would explain why the expression of the two Arabidopsis proteins from subgroup 4A (AtLEA4-1 and AtLEA4-2) overlapped only partially with the expression of the one from subgroup 4B (AtLEA4-5).
Figure 9.
Consensus phylogenetic tree of LEA4 protein sequences from plants. The phylogenetic tree was obtained from analyses with several programs from the Phylip suite of phylogenetic programs. Bootstrapped data sets were obtained with Seqboot using 100 repetitions. The reconstruction from 74 proteins and translated EST sequences showed two conserved subgroups that diverged before the appearance of vascular plants. AtLEA4-1 and AtLEA4-2 (black arrowheads) belong to subgroup 4A, whereas AtLEA4-5 (white arrowhead) belongs to subgroup 4B. The numbers (1–0) next to each taxon indicate the presence of motifs, but their arrangement within the protein sequence is shown in Supplemental Table S3, where motif 10 corresponds to “0” in this figure.
DISCUSSION
The high conservation of LEA protein families in the plant kingdom and the high correlation between their abundance and water deficit conditions denote a relevant role for these proteins in this environment. Even though circumstantial evidence has suggested the participation of these proteins in the adaptation of higher plants to low water availability, no direct genetic evidence in this regard had been reported. In this work, we analyzed the group 4 LEA genes and provide data showing that they participate in the adaptive response to this environmental stress in Arabidopsis plants throughout their life cycle.
Even though there is some information regarding the transcript accumulation patterns for the group 4 LEA genes in Arabidopsis (Delseny et al., 2001; Hoth et al., 2002; Seki et al., 2002; Oono et al., 2003; Schmid et al., 2005; Winter et al., 2007; Hruz et al., 2008), we were interested in knowing their accumulation under the growth conditions and developmental stages relevant for this work, as well as having information on the correlation between the abundance of a transcript and its corresponding protein. Our results and those reported previously agree in that the highest accumulation for all group 4 LEA transcripts occurs at the desiccation tolerance acquisition stage during seed development and up to 2-DAG seedlings. Also, our results concur that AtLEA4-5 transcript is the one that reaches the highest abundance in most stress conditions tested, in particular in response to hyperosmotic, drought, and ABA treatments, when compared with the other two members of the family. The two genes that showed the highest similarity, AtLEA4-1 and AtLEA4-2, also showed a high correlation in their transcript accumulation patterns, suggesting similar functions, in agreement with our phylogenetic analysis, which localized both genes in the same clade or subgroup (see below).
Transcript levels contrasted with protein abundance in mature flowers and during seedling development in the case of AtLEA4-1, where protein levels are similar to those accumulated in seeds, even though low transcript abundance was detected. Interestingly, we found that, in dried and stratified seeds, AtLEA4-1 protein migrated with an apparent higher molecular mass, suggesting that under severe dehydration this protein experiences a posttranslational modification or conformational changes. Unexpectedly, in the case of AtLEA4-2, we were unable to detect the protein in the predicted molecular mass (10.5 kD); instead, we specifically detected a protein with a higher molecular mass, whose accumulation pattern correlates with that obtained for its transcript. A possible explanation for this observation is that, in contrast to AtLEA4-5 and AtLEA4-1, secondary structure prediction analysis suggests the lowest percentage of random coil (20% of the protein) for AtLEA4-2, indicating that the higher structural order in this protein could favor the formation of homo- or hetero-oligomers. In some cases (Figs. 1B and 2B), instead of one band, two bands were detected, suggesting the formation of two types of oligomers due to preferential interactions. Notice that all the members of the AtLEA4 protein family are predicted to form coiled-coil structures (Lupas et al., 1991) involved in protein-protein interactions, reinforcing a hypothesis where these proteins may interact with each other and/or with other protein partners in the cell. In the case of AtLEA4-2, most of its residues (80%) are predicted to be involved in the formation of coiled-coils, supporting the existence of strong interactions between AtLEA4-2 monomers.
The analysis of the protein accumulation patterns for the different members of this family suggests the participation of posttranscriptional control mechanisms to modulate protein levels, as has been suggested for other LEA proteins (Colmenero-Flores et al., 1999). One example of this situation is evident in the case of the AtLEA4-5 transcript/protein upon ABA and salt treatments (Fig. 2), where transcript accumulation levels are similar for both conditions whereas protein levels in response to ABA are much higher than in response to NaCl, indicating that ABA may be involved in a putative posttranscriptional regulation of this transcript.
To gain insight into the role of this LEA gene family in plant adaptation to water deficit conditions, we carried out a phenotypic analysis of Arabidopsis plants deficient in the gene products of this family. Arabidopsis mutants unable to synthesize AtLEA4-5 protein showed that it plays a relevant role in the adjustment of the plant to hyperosmotic (350 mm mannitol) and high-salt (250 mm NaCl) conditions during germination, where they showed lower germination rate and efficiency. The absence of this protein also affected the ability of Arabidopsis plants to recover from the drought treatments applied to plants in the adult stage. Mutant plants not only showed a significantly lower biomass accumulation but also a reduced capacity to produce axillary and floral buds when compared with wild-type plants. In agreement with this, the deficiency of AtLEA4-5 led to low seed production upon water limitation (see Supplemental Fig. S6). A similar effect was observed in those plants where AtLEA4-1 and AtLEA4-2 were silenced using a specific a-miR, indicating that there is no redundancy in their participation under these adverse environments. This interpretation was further supported by the results obtained from plants where the three genes of the family were silenced (RNAi 4-5 × a-miR 4-1/2). These plants with undetectable or low levels of the three proteins showed a lower reduction in biomass accumulation and also in the production of axillary and floral buds in response to drought treatments.
The participation of this gene family in the plant response to water limitation was also supported by the results obtained from the overexpression of the AtLEA4-5 gene, which showed a higher production of axillary and floral buds compared with wild-type plants, when plants were subjected to severe dehydration-rehydration treatments. An additional observation from these experiments was that plants overproducing AtLEA4-5 protein showed a higher ability to restore their tissues after rehydration (see Supplemental Fig. S6 and Supplemental Table S1). These results indicate that the overproduction of AtLEA4-5 protein is able to confer a higher tolerance to severe drought treatment (greater than 85% substrate water loss).
Interestingly, we found that even under optimal irrigation conditions, the seed biomass from the different mutants (dSpm 4-5, a-miR 4-1/2, RNAi 4-5; triple silenced) was lower than that of wild-type plants, maybe due to a protective role of these proteins on the plant machinery needed for optimal fruit and/or seed development. Also, it should be considered the presumed protective role that these proteins may have on the meristematic regions or primordia (in agreement with their effect on the number of floral and axillary buds under water deficit), which would be relevant for the formation of inflorescences and consequently in the total number of seeds. Because the determination of seed size by thousand seed weight (data not shown) indicates that none of the mutants affect this phenotype, the possibility that inefficient seed filling is responsible for lower seed biomass in the different mutants could be discarded. Plants overproducing AtLEA4-5 did not show a higher seed biomass under optimal irrigation, suggesting that during normal seed development, higher levels than those present in wild-type seeds are not required for successful seed production.
The in silico analysis of the known group 4 LEA proteins confirmed the proposal by Battaglia et al. (2008) that group 4 LEA proteins can be defined through sequence conservation, which is a much stronger criterion than amino acid properties (Wise, 2003), and that the group is formed by two subgroups, each characterized by different sequence motifs. Motifs 1, 2, 4, and 6/7 are so broadly distributed that they could be the signature of the group. However, only 52 out of the 77 sequences included in this work have all four motifs. If we were to choose a single motif as signature of the group, that would probably be motif 2, not only because it is present in most sequences but also because its phylogeny is informative of the phylogeny of the complete proteins. By superimposing the NCBI taxonomy of the organisms on the phylogeny of group 4 LEA proteins, we concluded that the two subgroups originated from a very early duplication that predated the branching of monocots and dicots, because the two types of proteins can be found in both taxons. Outside the flowering plants, subgroup 4A is represented by all five sequences from moss, while subgroup 4B is represented by the six ESTs from conifers. It is tempting to suggest that 4B is older than 4A, because 4B is present in vascular and nonvascular plants while 4A proteins are absent from the completely sequenced genome of Physcomitrella. However, 4A sequences could be found in nonvascular plants as more species are examined. On the other hand, it is significant that many genera have members from both subgroups. These include Allium, Hordeum, Oryza, and Zea among monocots (MAIZE1 lacks motif 6, but a phylogeny based on motif 2 places it in subgroup 4A) and Arachis, Medicago, Glycine, Brassica, Arabidopsis, Ipomoea, and Populus among the dicots. This observation strongly suggests that the initial duplication gave rise to functional divergence or subfunctionalization. A reasonable hypothesis is that, in protecting other proteins or cellular structures, both subgroups have similar mechanisms but different targets, a prediction supported by both subgroups sharing a common core (formed by motifs 4, 1, and 2 and either motif 6 or 7, two motifs that are clearly related) but differing in the presence of additional motifs that could be involved in specificity. In the case of Arabidopsis, the two group 4A proteins (AtLEA4-1 and AtLEA4-2) have similar patterns of expression that differ from that of the group 4B protein (AtLEA4-5). Our experimental observations correlate well with the conclusion from the bioinformatic analysis, because dissimilar patterns are what would be expected for proteins whose functions have diverged.
Since the first recognition of several families of these hydrophilic and flexible proteins in cotton embryos by Dure et al. (1989), the focus of the research on LEA proteins has been mostly directed to the constitutive or regulated expression in plants, to test their protective effect on the survival of vegetative tissues during the first stages in development (for a recent review, see Battaglia et al., 2008). For group 4 LEA proteins, few examples of functional analysis have been reported; that is the case of the overexpression of a close homolog to AtLEA4-5 from Brassica napus (BnLEA4-1), which led to increased tolerance in seedlings (Dalal et al., 2009). Regarding a loss-of-function approach, only one study has been reported for a gene in this family in Arachis hypogaea, where transient virus-induced silencing of the homolog gene in tobacco (Nicotiana tabacum) plants led to an apparent enhanced susceptibility to low moisture in vegetative tissues (Senthil-Kumar and Udayakumar, 2006). In contrast to these studies and others (for a recent review, see Battaglia et al., 2008), in this work, we addressed the role of a complete LEA gene family on the adaptation of higher plants to drought and desiccation using a genetic approach, where we correlated the presence and abundance of members of this family with the plant adjustment to stress in most developmental stages of their life cycle. We also showed that their specific induction and high abundance is necessary, but not sufficient, to adapt to drought stress, suggesting that each LEA gene has evolved to help in the adaptive process of higher plants depending on the developmental stage or particular tissues as well as stress type and severity. These results also support the idea that there is no functional redundancy among the different LEA protein groups. Altogether, the genetic and phylogenetic evidence presented in this work strongly supports the essential role that group 4 LEA proteins play in the adaptive process to water deficit in higher plants. Furthermore, we include a phenotypical analysis that considers the impact of drought on Arabidopsis plants at the adult stage, focusing on the survival and/or recovery of reproductive organs. This type of analysis allowed us to assign to these proteins a role in the protection of reproductive organs, a property relevant for the offspring rather than for the vegetative tissues, and consequently important from the evolutionary and agronomic points of view.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana Col) seeds were germinated in MS 1×: MS salt mixture (4.3 g L−1; Caisson Laboratories), 1% Suc (Research Organics), and 0.5 g L−1 MES (Research Organics). Seeds were surface sterilized with absolute ethanol for 2 min, washed with a 40% sodium hypochlorite + 0.02% Triton X-100 (Sigma) solution for 8 min, and rinsed thoroughly with distilled water five times. Seeds were sown in petri dishes containing solid MS medium, stratified for 4 d at 4°C in darkness, and transferred to a culture room at 25°C, 60% to 65% relative humidity, and 16-h/8-h photoperiod with white light at 60 to 80 μE m−2 s−1 intensity, until used as indicated.
For the expression pattern analyses of group 4 LEA transcripts and proteins from embryogenesis to seedling establishment, wild-type seeds were germinated in MS 1×, and after 10 d of growth in vitro they were transferred to 3- × 3- × 3-inch pots containing Metromix 200 (Hummert International) and kept under optimal irrigation with nutrient solution [5 mm KNO3, 2.5 mm KH2PO4, pH 5.6, 2 mm MgSO4, 2 mm Ca(NO3)2, 50 μm Fe-EDTA, and micronutrients solution (70 μm H3BO3, 14 μm MnCl2, 0.5 μm CuSO4, 1 μm ZnSO4, 0.2 μm NaMoO4, 10 μm NaCl, and 0.01 μm CoCl2)] until adult stage. To follow embryogenesis from early stages through the end of seed development, flowers were marked at the day of anthesis and siliques were collected at intervals until dry seeds were obtained, as follows: buds and flowers at anthesis, 1 to 5 d post anthesis (DPA), 6 to 10 DPA, 11 to 15 DPA, 16 to 20 DPA, and dry seeds. Seeds were sown in MS 1× and collected at intervals during stratification (3 d at 4°C), germination (radicle emergence), and seedling establishment at 2 DAG, 5 DAG, 8 DAG, and 11 DAG. Plants were maintained in a growth room at 21°C, relative humidity of 70%, and 16-h/8-h photoperiod with white light at 60 μE m−2 s−1 intensity.
Water Deficit Treatments
The expression pattern analyses of group 4 LEA transcripts and proteins were carried out at different developmental stages as indicated. Some experiments were performed using 2-week-old seedlings grown in vitro in MS 1× and subjected to water deficit treatments by transferring them to petri dishes with 15 mL of liquid MS supplemented with PEG-8000 (25% [w/v]; Research Organics) or NaCl (100–200 mm). Also, a treatment using 100 μm ABA (Sigma) was included. Seedlings were incubated in a culture room at 25°C for 12 h with constant shaking. Germination experiments were carried out in MS medium supplied with NaCl (100–250 mm; J.T. Baker) or 350 mm mannitol (Sigma).
Phenotypic analyses were carried out during germination or in adult plants as described in the “Results” section. Germination experiments were conducted under optimal conditions (MS 1×) or under stress by adding 100 to 250 mm NaCl (J.T. Baker) or 350 mm mannitol (Sigma). Dehydration and rehydration experiments were carried out in adult plants at their reproductive stage, for which 10- to 12-d seedlings (germinated in vitro in MS 1×) were transplanted to 3- × 3- × 3-inch pots with Turface (Hummert International), an inert clay substrate. Since the substrate is inert and has low water retention, plants were kept under field capacity (Ψsubstrate = −0.455 ± 0.70 MPa) using nutrient solution and distilled water and maintained like that until the beginning of the drought experiments (or until the end of the experiment in the case of control plants). Because of the low water retention of this type of substrate, withholding watering leads to a faster and more severe water deficit than in standard soil such as Metromix 200. Drought was imposed by halting irrigation after flowering, when plants were between 4 and 6 weeks old (dehydration-rehydration treatment). Rotation of the pots within each tray during the course of the drought experiments was performed to allow even water loss in all pots. For the phenotypical characterization of the loss-of-function mutants, a mild drought treatment was applied by withholding watering from 10 to 12 d under light greenhouse conditions and controlled temperature until water loss in each pot led to Ψsubstrate = −4.617 ± 0.619 MPa. A more severe stress was applied when AtLEA4-5-overexpressing transgenic lines were characterized, where the substrate water loss was followed up to 14 d after the irrigation was stopped, when Ψsubstrate = −6.447 ± 0.574 MPa. The variation of days to achieve the required water loss from the substrate depended on the relative humidity of the season; therefore, we conducted drought experiments only in the dry season of the year, where the relative humidity is below 50% (March-April-May). Pots were then rehydrated with nutritive solution and left to recover 6 to 10 d; after that period, we determined the percentage of survival by the number of plants showing recovery of vegetative tissues (rosette leaves) and the number of buds per plant (floral and axillary).
Total plant biomass was weighed separately after drying whole plants in paper bags for 48 h at 80°C. Sample tissues were frozen in liquid nitrogen and kept at −70°C until their use for RNA and/or protein extraction.
Ψsubstrate was determined with a Dew Point microvoltmeter (n = 10 ± sd), following the manufacturer’s instruction manual (model HR-33T; Wescor).
RWC during drought experiments was calculated as RWC = (FW − DW)/(TW − DW), where FW is the fresh weight of adult plants, using leaf discs from rosette leaves of adult plants or whole adult plants, TW is the turgid weight after floating for 8 to 24 h in distillate water, and DW is the dry weight after oven drying in paper bags at 80°C for 48 h (Tezara et al., 2002).
Statistical Analyses
The germination experiments included three replicates (plates) with 100 seeds per replicate. The data were accumulated over time and fit to sigmoidal dose-response curves with variable slope [Y = bottom + {(top − bottom)/1 + 10(LogEC50-X)*Hillslope}], also called four-parameter logistic equation. Bottom is the Y value at the bottom plateau (constrained to zero); top is the Y value at the top plateau; LogEC50 is the X value when the response is halfway between bottom and top; Hillslope describes the steepness of the curve. The null hypothesis was that two curve-fit parameters (Hillslope and LogEC50) from each data set were the same. For the water deficit treatments, the experimental design included three replicates (pots) with four to six plants in each replicate. The dehydration-rehydration experiments were analyzed by one-way ANOVA, and significant differences between groups were searched through Dunnett’s or Tukey’s multiple comparisons posttests. Bartlett’s test for equal variances showed no significant differences (P < 0.05) within each group, so one-way ANOVA could be applied in all cases. The null hypothesis was that there were no significant differences in the means between groups. All the statistical analyses and curve fitting were performed using Prism 5 for Mac OS X (GraphPad Software).
Vectors, Bacterial Strains, and Plant Transformation
AtLEA4-5 Constructs
The AtLEA4-5 ORF (477 bp) was cloned into pBluescript KS+ (ampicillinR; Stratagene) from a cDNA library (leaf, flowers, and siliques) using specific primers, which add NcoI and SalI restriction sites (5′-AAACCATGGAGTCGATGAAAGAAAC-3′ and 5′-GCGGTCGACCCGTTTATCCAGTATATCC-3′). This cDNA was used as a template for random labeling of PCR fragments to make a specific probe for hybridization in northern-blot experiments. The vector was digested with NcoI/SalI to subclone into pBin35S-nos binary vector (kanamycinR; Bevan, 1984) to overexpress the corresponding protein in bacteria and plants. This cDNA was also cloned into pHannibal RNAi vector (ampicillinR; Commonwealth Scientific and Industrial Research Organization vectors) as inverted repeats separated by an intron (XhoI-KpnI fragment for sense orientation and BamHI-XbaI fragment for antisense orientation) in order to be transcribed as a stem-loop precursor and processed by the plant’s RNAi machinery to silence the endogenous mRNAs of this gene. A 4-kb NotI fragment containing the RNAi construction for AtLEA4-5 was subcloned into pArt27 binary vector (kanamycinR, spectinomycinR; Gleave, 1992). From this transformation, T3 homozygous lines were obtained; however, RNAi silencing was neither observed in dry seeds nor in vegetative tissues during dehydration. For this reason, homozygous RNAi-silenced T2 lines, where silencing was evident, were used for their phenotypic characterization. To select those transgenic lines with the highest decrease in AtLEA4-5 protein levels compared with the wild type, seedlings were subjected to osmotic stress using 25% PEG; from this screening, three lines were selected for phenotypic analysis.
For AtLEA4-5 antibody production, the AtLEA4-5 ORF was subcloned as a NcoI/SalI fragment into pTRC99A vector (ampicillinR; Amann et al., 1988) to induce the overproduction of the native recombinant protein in Escherichia coli (see “Antibodies Preparation” below).
AtLEA4-1 and AtLEA4-2 Constructs
The AtLEA4-1 ORF (406 bp) was isolated from cDNA from osmotically stressed Arabidopsis plants using specific primers 5′-AATCCATGGAATCGGCGAAACAGATAAGCGATATGGC-3′ and 5′-GCCGTCGACCGGATTAGTAGTGATGATGATTATGAT-3′. The AtLEA4-2 ORF (294 bp) was isolated from genomic DNA, because the gene has no intron, using specific primers 5′-GCGGGATCCCTCGAGATGCAGTCGGCGAAGGAAAAG-3′ and 5′-CGATCTAGAGGTACCGTTTTAGATCTGTCCCGGCGG-3′. Both ORFs were cloned into entry vector pCRII-TOPO (ampicillinR, kanamycinR; Invitrogen) to obtain the PCR fragments used as probes in northern-blot experiments. They were also cloned into pENTR/SD/D-TOPO (kanamycinR; Invitrogen) using Gateway technology (Invitrogen); the AtLEA4-1 and AtLEA4-2 glutathione S-transferase (GST) translational fusions obtained after recombination with destination vector pDEST24 (ampicillinR; Invitrogen) were used for antibody production.
To overcome the lack of available mutants showing a significant decrease of AtLEA4-1 and AtLEA4-2 transcripts, an a-miR (4-1/2) was designed to posttranscriptionally silence the corresponding transcripts using a single construct able to trigger the silencing of both genes (Niu et al., 2006). Specific primers were designed to introduce PCR-based point mutations into a microRNA precursor (ath-miR159a) to allow the production of a-miR 4-1/2 that specifically triggers the silencing of the desired genes instead of its original target (Niu et al., 2006). Primer sequences were as follows: forward, 5′-ATAGATCTTGATCTGACGATGGAAGGACATGGCCAGATCGGTCAAACATG-3′, and reverse, 5′-TTGACCCGGGATGGACATGGCCAGTACAGCCAAAGAAG-3′. Mutated precursor was cloned into pENTR/SD/D-TOPO and recombined with binary vector pK2GW7 (spectinomycinR, kanamycinR; Karimi et al., 2002). Analysis of the transgenic plants containing the a-miR 4-1/2 precursor showed a constitutive expression of the mature a-miR 4-1/2 (5′-UUUGGCUGUACUGGCCAUGUC-3′) until the T3 generation. The silencing of the AtLEA4-1 and AtLEA4-2 transcripts was determined by detection of the corresponding transcripts and/or proteins. Those lines that showed reduced or no accumulation of the AtLEA4-1 and AtLEA4-2 proteins were selected for further characterization.
Agrobacterium Transformation
Electrocompetent Agrobacterium tumefaciens C58/pGV2260 (rifampicinR, ampicillinR) cells were electroporated with each construct, and selective marker-resistant clones were verified by colony PCR using specific gene primers. Isolated colonies were cultured to transform wild-type Arabidopsis (Col) plants by the floral dipping method (Clough and Bent, 1998). T0 seeds were plated in MS medium with kanamycin sulfate (50 mg mL−1; Sigma) and/or BASTA (glufosinate-ammonium, 50–15 μg mL−1 [Finale; Aventis]) until resistant plants were distinguished, transplanted to soil, and self-fertilized to obtain T1 progeny. Insertion number was estimated in independent lines, germinating 200 T1 seeds and selecting those with a 3:1 resistant:sensitive segregation ratio. T2 and/or T3 lines were generated to obtain homozygous lines from plants transformed with each construct. The selected homozygous lines were confirmed to be 100% resistant to kanamycin (OE 4-5, RNAi 4-5), BASTA (dSpm 4-5, a-miR 4-1/2), or kanamycin + BASTA (triple mutant, complementation of dSpm 4-5 mutant).
Transgene expression levels were verified in homozygous plants by RT-PCR and northern blot using specific primers for each transcript; protein levels were also confirmed by western blot using specific polyclonal antibodies for each protein of the group. Phenotypic characterization experiments were conducted in MS 1× (without kanamycin/BASTA) using only those transgenic lines that had been previously confirmed to be homozygous for each construct.
Genomic DNA Extraction and Southern-Blot Experiments
For PCR experiments, quick minipreparations of genomic DNA from Arabidopsis were obtained as described by Edwards et al. (1991). Genomic DNA was obtained by large-scale extractions as described by Taylor et al. (1993). Southern blotting was carried out using genomic DNA (40 μg) from wild-type and mutant homozygous lines from the European Arabidopsis Stock Centre (N122943), followed by digestion with ClaI, HindIII, and BamHI and separation by electrophoresis. Transfer and hybridization were carried out following standard protocols under stringent conditions. AtLEA4-5 probe was obtained by random labeling with [α32P]dCTP (3,000 Ci mmol−1) an 800-bp PCR product using specific primers for the AtLEA4-5 gene. After washing in high-stringency conditions, the membrane was exposed to Kodak film using an intensifying white screen (Amersham).
RNA Extraction and Northern-Blot Experiments
Total RNA was extracted from flowers, immature siliques, and dry seeds according to Vicient and Delseny (1999). Total RNA from control and drought-stressed plants was extracted with Trizol (Invitrogen) according to the manufacturer’s instructions. Small RNA-enriched preparations were also obtained with Trizol reagent, omitting the washing step with 75% ethanol. Northern-blot hybridizations were performed according to Sambrook et al. (1989). RNA (20 μg) from each sample was separated on formaldehyde-agarose gels, equilibrated with 10× SSC, and transferred to nitrocellulose Hybond N+ (Amersham) membranes. Hybridizations were carried out following standard protocols. Probes were prepared by random labeling of PCR products from the corresponding gene fragments with [α-32P]dCTP (3,000 Ci mmol−1) and purified with Ilustra Microspin G-25 columns (GE Healthcare). After washing in high-stringency conditions, membranes were used to expose Kodak x-ray films from 1 to 3 d at −70°C. For microRNA detection, total RNA samples were resolved on 15% acrylamide-8 m urea-Tris-borate/EDTA gels, blotted under semidry conditions to nitrocellulose membranes, and prehybridized with Ultra-Hyb Oligo solution (Ambion). Hybridizations were performed at 42°C overnight, and after two washes with 2× SSC, 0.1% SDS for 30 min, membranes were exposed to Kodak films. Antisense probes were labeled with 15 pmol of [γ-32P]ATP (3,000 Ci mmol−1) and purified with mini Quick Spin columns (Roche).
RT-PCR
For semiquantitative RT-PCR experiments, 1 μg of DNase I (1 unit μL−1; Invitrogen)-treated total RNA was used. cDNA was synthesized with oligo(dT) (1 μg μL−1) with the sequence 24[T]TVN, where V = A/G/C and N = A/G/C/T, and SuperScript III reverse transcriptase (Invitrogen) following the manufacturer’s instructions. Polyadenylated sequences were amplified using Platinum Taq DNA polymerase (Invitrogen) and specific primers to amplify the ORF of each gene. AtLEA4-1 was amplified with primers 5′-AATCCATGGAATCGGCGAAACAGATAAGCGATATGGC-3′ and 5′-GCCGTCGACCGGATTAGTAGTGATGATGATTATGAT-3′. AtLEA4-2 was amplified with primers 5′-GCGGGATCCCTCGAGATGCAGTCGGCGAAGGAAAAG-3′ and 5′-CGATCTAGAGGTACCGTTTTAGATCTGTCCCGGCGG-3′. For AtLEA4-5 amplification, primers were 5′-GTAGGATCCCTCGAGATGCAGTCGATGAAAGAAACAGC-3′ and 5′-TCGTCTAGAGGTACCCCGTTTATCCAGTATATCCCCC-3′. For the semiquantitative PCR, the reaction mixture contained 0.8 μL of the cDNA preparation, 200 μm deoxyribonucleotide triphosphates, 1.5 mm MgCl2, 0.2 μm of each primer, 2 μL of 10× PCR buffer, and 1 unit of Platinum Taq DNA Polymerase. PCR conditions were 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 58°C to 63°C (depending on the pair of oligonucleotides used) for 30 s, 70°C for 30 s, and 70°C for 5 min. The ACTIN2 gene, used as the normalizing standard, was amplified with the oligonucleotide primers 5′-GTATGTTGCCATTCAGGCCGTTCTTTCTCT-3′ and 5′-CGACCCGCAAGATCAAGACGAAGGA-3′.
Protein Extraction and Western Blot
Total proteins from flowers, immature siliques, dry seeds, and vegetative organs were extracted as described by Hurkman and Tanaka (1986) with some modifications. Frozen tissues were homogenized in extraction buffer:phenol at 1:3 proportion (0.7 m Suc, 0.5 m Tris, 30 mm HCl, 2% β-mercaptoethanol, and 12 mg mL−1 polyvinylpolypyrrolidone). Organic phase was extracted again with the same buffer:phenol proportion without polyvinylpolypyrrolidone and precipitated with 3 volumes of 0.1 m sodium acetate in methanol. Pellet was washed once with cold 80% acetone and solubilized in a buffer containing 20 mm Tris, pH 8.0, 50 mm NaCl, and 0.1% SDS. Proteins were quantified using Bradford assay with Bio-Rad dye reagent. Total proteins (10–15 μg) were separated on 16.5% polyacrylamide gels in 0.1 m Tris, 0.1 m Tricine, and 0.1% SDS (Schägger and von Jagow, 1987). Proteins were transferred to nitrocellulose membranes (Hybond C; Amersham), washed with phosphate-buffered saline (PBS) 1×, blocked with 5% skim milk (in PBS 1×), and probed overnight with anti-AtLEA4-5 (1:2,000), anti-AtLEA4-2 (1:2,000), or anti-AtLEA4-1 (1:1,000) antibodies. Membranes were washed and incubated with secondary antibody (1:5,000; anti-rabbit horseradish peroxidase; Zymed) for 1 h at room temperature, washed with PBS 1×, and developed with peroxidase substrates from Supersignal West Pico (Pierce). Membranes were exposed to x-ray films (Kodak).
Antibody Preparation
AtLEA4-5 antibodies were produced using native recombinant protein, whereas antibodies against AtLEA4-1 and AtLEA4-2 were obtained using GST fusion proteins. Recombinant proteins were purified from E. coli after induction with isopropylthio-β-galactoside. For the purification of AtLEA4-5 native protein, pelleted cells were resuspended in ice-cold buffer containing 20 mm Tris, 50 mm NaCl, and 1 mm phenylmethylsulfonyl fluoride and lysed by sonication. After centrifugation, cleared supernatant was boiled 10 min and put in ice for 10 min. This step enriches boiling soluble proteins from bacteria, most of it being the recombinant protein (Jepson and Close, 1995). To obtain AtLEA4-1-GST and AtLEA4-2-GST fusion proteins, pelleted cells were resuspended in ice-cold PBS and sonicated three times. After addition of Triton X-100 (1% final concentration), cell debris were discarded and supernatant was incubated with glutathione-agarose beads (Sigma) and washed three times with cold PBS. Soluble fusion protein was obtained after boiling in 10% SDS.
For polyclonal antibody production, purified proteins were separated on polyacrylamide gels and stained with Coomassie Brilliant Blue, and the band corresponding to the protein of interest was eluted from the gel, equilibrated with distilled water and emulsified with complete Freund’s adjuvant (Gibco) at a 1:1 ratio, homogenized exhaustively, and used to immunize New Zealand female rabbits through multiple intradermal injections. Preimmune serum was previously taken. The titer and specificity of the antibodies were monitored through an inoculation protocol using dilutions of the purified recombinant proteins and protein extracts from dry seeds and from osmotically stressed plants. The IgG fraction was precipitated from serum with saturated ammonium sulfate and stored at 4°C with 0.02% sodium azide. To verify the specific recognition of the antibodies for their own antigen, antibodies for each of the three group members were cross-tested with the purified recombinant proteins by western blot. As a negative control, preimmune serum was also used to incubate with membranes. Immunopurified antibodies were obtained by affinity purification according to Lillie and Brown (1987). Custom rabbit-specific polyclonal antibodies against AtLEA4-2 were produced against a peptide corresponding to a nonconserved region between the members of this family (CSKEAQAKADLHQSK; GenScript). This antibody specifically recognized a GST-AtLEA4-2 fusion protein produced in bacteria. To verify the antibody specificity, competition experiments were carried out using increasing concentrations of the corresponding purified antigens (recombinant AtLEA4-5, AtLEA4-2_GST, and AtLEA4-2 peptides).
Bioinformatic Analyses
Homologs of AtLEA4-5 protein were searched in the NCBI EST database using BLAST algorithm (tBLASTn protein query versus EST database), selecting only ESTs corresponding to nearly complete ORFs for each protein. Additionally, we included five bryophyte sequences obtained from the COSMOSS database at www.cosmoss.org (a computational biology resources for the moss Physcomitrella patens; Lang et al., 2005). In all, 77 sequences were analyzed after translating them in silico (the complete list, including the “short names” used in this work, is shown in Supplemental Table S3).
The predicted hydrophilicity, molecular mass, and pI of the proteins of interest were verified using Kyte and Doolittle (1982) plots and Protean (a protein sequence analysis software; DNASTAR). Their predicted secondary structure was established using PSIPRED (a protein structure prediction server; McGuffin et al., 2000) and the COILS programs (for prediction of coiled-coil regions in proteins; Lupas et al., 1991).
Sequence motifs were inferred with MEME (Multiple Em for Motif Elicitation; Bailey and Gribskov, 1998; Bailey et al., 2006) from 72 group 4 LEA proteins (all but the five proteins from P. patens) using default parameters. The model used was “zoops” (zero or one per sequence). Ten motifs were obtained, and these were searched using the MEME-associated tool MAST (Motif Alignment and Search Tool; Bailey and Gribskov, 1998; Bailey et al., 2006) over the complete collection (this time including the moss proteins).
Multiple alignments were carried out with ClustalW (Larkin et al., 2007). Alignments were made with the whole proteins, with their MCB, which is the concatenation of motifs 4-1-2-6/7 found by MEME, or with motif 2 alone. All phylogenetic analyses were made with the Phylip suite of phylogenetic programs (Felsenstein, 2005). Bootstrapped data sets were obtained with Seqboot using 100 repetitions. Distance matrices were obtained with Protdist. Distance-based phylogenies were made in parallel with Neighbor and Fitch. Maximum likelihood-based phylogenies were built with Proml using the Jones-Taylor-Thornton model of amino acid change. Consensus trees were obtained with Consense. Trees were visualized and edited with ATV (A Tree Viewer; Zmasek and Eddy, 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence similarity between group 4 LEA proteins in Arabidopsis and prediction of coiled-coil regions in these proteins.
Supplemental Figure S2. Phenotypic analysis of plants overexpressing AtLEA4-5 protein.
Supplemental Figure S3. Description of the transposon insertion mutant (dSpm 4-5) in the AtLEA4-5 gene.
Supplemental Figure S4. Design of an a-miR to silence AtLEA4-1 and AtLEA4-2 genes and the construction of RNAi-triggered silencing for the AtLEA4-5 gene.
Supplemental Figure S5. Protein reduction levels in posttranscriptional gene silencing single, double, and triple mutants of the AtLEA4 gene family subjected to drought
Supplemental Figure S6. Phenotypic analysis of T2 homozygous plants expressing an RNAi to silence AtLEA4-5 transcript after recovery from dehydration
Supplemental Figure S7. Conserved motifs and their arrangement in the LEA4 protein sequences from plants.
Supplemental Table S1. Phenotypic analysis of T3 homozygous adult plants overexpressing AtLEA4-5 protein (35S::AtLEA4-5::NOS) during drought and after recovery from stress.
Supplemental Table S2. Phenotypic analysis of a T4 homozygous transposon insertion mutant in the AtLEA4-5 gene and of homozygous T2 posttranscriptional gene silencing mutants in the AtLEA4 gene family during drought and after recovery from stress.
Supplemental Table S3. Taxonomic classification of the sequences used for motif search, sequence number, and sequence name depicted in Figure 9.
Supplementary Material
Acknowledgments
We thank M. Rocha for critical reading of the manuscript and stimulating discussions and R.M. Solórzano and R.E. Quiroz for technical assistance. We are also grateful to the Oligonucleotides Synthesis, DNA Sequencing, Computer, and Animal Facilities of the Instituto de Biotecnología for providing us with the oligonucleotides, DNA sequences, technical assistance, and handling of rabbits, respectively. The European Arabidopsis Stock Centre kindly provided the seeds of the transposon insertion mutant in the AtLEA4-5 gene.
References
- Amann E, Ochs B, Abel KJ. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69: 301–315 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. (1998) Combining evidence using P-values: application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias A. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148: 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo LA, Gallardo J, Navarrete A, Olave N, Martinez J, Alberdi M, Close TJ, Corcuera LJ. (2003) Cryoprotective activity of a cold-induced dehydrin purified from barley. Physiol Plant 118: 262–269 [Google Scholar]
- Bray EA. (1997) Plant responses to water deficit. Trends Plant Sci 2: 48–54 [Google Scholar]
- Browne JA, Dolan KM, Tyson T, Goyal K, Tunnacliffe A, Burnell AM. (2004) Dehydration-specific induction of hydrophilic protein genes in the anhydrobiotic nematode Aphelenchus avenae. Eukaryot Cell 3: 966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra Babu R, Zhang JS, Blum A, Ho T, Wu R, Nguyen HT. (2004) HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci 166: 855–862 [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colmenero-Flores JM, Moreno LP, Smith C, Covarrubias AA. (1999) PvLEA-18, a member of a new late-embryogenesis-abundant protein family that accumulates during water stress and in the growing regions of well-irrigated bean seedlings. Plant Physiol 120: 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal M, Tayal D, Chinnusamy V, Bansal KC. (2009) Abiotic stress and ABA-inducible group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotechnol 139: 137–145 [DOI] [PubMed] [Google Scholar]
- Delseny M, Bies-Ethève N, Carles C, Hull G, Vicient C, Raynal M, Grellet F, Aspart L. (2001) Late Embryogenesis Abundant (LEA) protein gene regulation during Arabidopsis seed maturation. J Plant Physiol 158: 419–427 [Google Scholar]
- Dure L. (1993) Structural motifs in LEA proteins. Close TJ, Bray EA, , Plant Responses to Cellular Dehydration during Environmental Stress. American Society of Plant Physiologists, Rockville, MD, pp 91–103 [Google Scholar]
- Dure L, Crouch M, Harada JJ, Ho T, Mundy J, Quatrano RS, Thomas TL, Sung ZR. (1989) Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol 12: 475–486 [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom J, Baker W, Kintanar A, Wurtele ES. (1996) The embryo-specific EMB-1 protein of Daucus carota is flexible and unstructured. Plant Sci 115: 17–24 [Google Scholar]
- Felsenstein J. (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle [Google Scholar]
- Fu D, Huang B, Xiao Y, Muthukrishnan S, Liang GH. (2007) Overexpression of barley HVA1 gene in creeping bentgrass for improving drought tolerance. Plant Cell Rep 26: 467–477 [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. (2000) Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem 275: 5668–5674 [DOI] [PubMed] [Google Scholar]
- Gleave AP. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goyal K, Tisi L, Baran A, Browne J, Burnell A, Zurdo J, Tunnacliffe A. (2003) Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J Biol Chem 278: 12977–12984 [DOI] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A. (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelet J, Benamar A, Teyssier E, Avelange-Macherel MH, Grunwald D, Macherel D. (2005) Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol 137: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Terashima S, Kuboi T. (2001) Characterization and cryoprotective activity of cold responsive dehydrin from Citrus unshiu. J Plant Physiol 58: 1333–1339 [Google Scholar]
- Hoekstra F, Golovina E, Buitink J. (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci 6: 431–438 [DOI] [PubMed] [Google Scholar]
- Honjoh K, Matsumoto H, Shimizu H, Ooyama K, Tanaka K, Oda Y, Takata R, Joh T, Suga K, Miyamoto T, et al. (2000) Cryoprotective activities of group 3 late embryogenesis abundant proteins from Chlorella vulgaris C-27. Biosci Biotechnol Biochem 64: 1656–1663 [DOI] [PubMed] [Google Scholar]
- Honjoh K, Yoshimoto M, Joh T, Kajiwara T, Miyamoto T, Hatano S. (1995) Isolation and characterization of hardening-induced proteins in Chlorella vulgaris C-27: identification of late embryogenesis abundant proteins. Plant Cell Physiol 36: 1421–1430 [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH. (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. (2008) LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9: 118–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkman WJ, Tanaka CK. (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol 81: 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai R, Chang L, Ohta A, Bray EA, Takagi M. (1996) A lea-class gene of tomato confers salt and freezing tolerance when expressed in Saccharomyces cerevisiae. Gene 170: 243–248 [DOI] [PubMed] [Google Scholar]
- Ismail AM, Hall AE, Close TJ. (1999) Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol 120: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson SG, Close TJ. (1995) Purification of a maize dehydrin protein expressed in Escherichia coli. Protein Expr Purif 6: 632–636 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lal S, Gulyani V, Khurana P. (2007) Overexpression of HVA1 gene from barley generates tolerance to salinity and water stress in transgenic mulberry (Morus indica). Transgenic Res 17: 651–663 [DOI] [PubMed] [Google Scholar]
- Lang D, Eisinger J, Reski R, Rensing S. (2005) Representation and high-quality annotation of the Physcomitrella patens transcriptome demonstrates a high proportion of proteins involved in metabolism among mosses. Plant Biol 7: 238–250 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. (1987) Artifactual immunofluorescent labelling in yeast, demonstrated by affinity purification of antibody. Yeast 3: 63–70 [DOI] [PubMed] [Google Scholar]
- Lisse T, Bartels D, Kalbitzer HR, Jaenicke R. (1996) The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineum displays no defined three-dimensional structure in its native state. Biol Chem 377: 555–561 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng Y. (2005) PM2, a group 3 LEA protein from soybean, and its 22-mer repeating region confer salt tolerance in Escherichia coli. Biochem Biophys Res Commun 331: 325–332 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- McCubbin WD, Kay CM, Lane B. (1985) Hydrodynamic and optical properties of the wheat germ Em protein. Can J Biochem Cell Biol 63: 803–811 [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Menze MA, Boswell L, Toner M, Hand SC. (2009) Occurrence of mitochondria-targeted Late Embryogenesis Abundant (LEA) gene in animals increases organelle resistance to water stress. J Biol Chem 284: 10714–10719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison JI, Baker NR, Mullineaux PM, Davies WJ. (2008) Improving water use in crop production. Philos Trans R Soc Lond B Biol Sci 363: 639–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Okawa K, Kakizaki T, Honma T, Itoh H, Inaba T. (2007) Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol 144: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NDong C, Danyluk J, Wilson KE, Pocock T, Huner NP, Sarhan F. (2002) Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins: molecular characterization and functional analyses. Plant Physiol 129: 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24: 1420–1428 [DOI] [PubMed] [Google Scholar]
- Oono Y, Seki M, Nanjo T, Narusaka M, Fujita M, Satoh R, Satou M, Sakurai T, Ishida J, Akiyama K, et al. (2003) Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca 7000 full-length cDNA microarray. Plant J 34: 868–887 [DOI] [PubMed] [Google Scholar]
- Puhakainen T, Hess MW, Makela P, Svensson J, Heino P, Palva ET. (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol 54: 743–753 [DOI] [PubMed] [Google Scholar]
- Reyes JL, Campos F, Wei H, Arora R, Yang Y, Karlson DT, Covarrubias AA. (2008) Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ 31: 1781–1790 [DOI] [PubMed] [Google Scholar]
- Reyes JL, Rodrigo MJ, Colmenero-Flores JM, Gil JV, Garay-Arroyo A, Campos F, Salamini F, Bartels D, Covarrubias AA. (2005) Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ 28: 709–718 [Google Scholar]
- Roberts JK, DeSimone NA, Lingle WL, Dure L., III (1993) Cellular concentrations and uniformity of cell-type accumulation of two LEA proteins in cotton embryos. Plant Cell 5: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russouw PS, Farrant J, Brandt W, Maeder D, Lindsey GG. (1995) Isolation and characterization of a heat-soluble protein from pea (Pisum sativum) embryos. Seed Sci Res 5: 137–144 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning: A Laboratory Manual Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schägger H, von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Udayakumar M. (2006) High-throughput virus-induced gene-silencing approach to assess the functional relevance of a moisture stress-induced cDNA homologous to Lea4. J Exp Bot 57: 2291–2302 [DOI] [PubMed] [Google Scholar]
- Shih MD, Lin SD, Hsieh JS, Tsou CH, Chow TY, Lin TP, Hsing YIC. (2004) Gene cloning and characterization of a soybean (Glycine max L.) LEA protein, GmPM16. Plant Mol Biol 56: 689–703 [DOI] [PubMed] [Google Scholar]
- Singh S, Cornilescu CC, Tyler RC, Cornilescu G, Tonelli M, Lee MS, Markley JL. (2005) Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein. Protein Sci 14: 2601–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivamani E, Bahieldin A, Wraith JM, Al-Niemi T, Dyer WE, Ho TD, Qu R. (2000) Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci 155: 1–9 [DOI] [PubMed] [Google Scholar]
- Solomon A, Salomon R, Paperna I, Glazer I. (2000) Desiccation stress of entomopathogenic nematodes induces the accumulation of a novel heat-stable protein. Parasitology 121: 409–416 [DOI] [PubMed] [Google Scholar]
- Soulages JL, Kim K, Arrese EL, Walters C, Cushman JC. (2003) Conformation of a group 2 late embryogenesis abundant protein from soybean: evidence of poly (l-proline)-type II structure. Plant Physiol 131: 963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulages JL, Kim K, Walters C, Cushman JC. (2002) Temperature-induced extended helix/random coil transitions in a group 1 late embryogenesis-abundant protein from soybean. Plant Physiol 128: 822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy RAP, Aalen RB. (1998) Identification of sequence homology between the internal hydrophilic repeated motifs of group 1 late-embryogenesis-abundant proteins in plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis. Planta 206: 476–478 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Ikeda K, Miyasaka H. (2004) Isolation of a new member of group 3 late embryogenesis abundant protein gene from a halotolerant green alga by a functional expression screening with cyanobacterial cells. FEMS Microbiol Lett 236: 41–45 [DOI] [PubMed] [Google Scholar]
- Taylor BH, Manhart JR, Amasino RM. (1993) Isolation and characterization of plant DNAs. Glick BR, Thompson JE, , Methods in Plant Molecular Biology and Biotechnology, Ed 2 CRC Press, Boca Raton, FL, pp 37–47 [Google Scholar]
- Tezara W, Mitchell V, Driscoll SP, Lawlor DW. (2002) Effects of water deficit and its interaction with CO2 supply on the biochemistry and physiology of photosynthesis in sunflower. J Exp Bot 53: 1781–1791 [DOI] [PubMed] [Google Scholar]
- Tissier A, Marillonnet S, Klimyuka V, Patela K, Torres MA, Murphy G, Jones J. (1999) Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Jaquinod M, Mangavel C, Passirani C, Saulnier P, Manon S, Teyssier E, Payet N, Avelange-Macherel MH, Macherel D. (2007) Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell 19: 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A, Lapinski J, McGee B. (2005) A putative LEA protein, but no trehalose, is present in anhydrobiotic bdelloid rotifers. Hydrobiologia 546: 315–321 [Google Scholar]
- Vicient CM, Delseny M. (1999) Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem 268: 412–413 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ. (2002) The POPPs: clustering and searching using peptide probability profiles. Bioinformatics (Suppl 1) 18: S38–S45 [DOI] [PubMed] [Google Scholar]
- Wise MJ. (2003) LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA. (2001) Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta 1544: 196–206 [DOI] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L. (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115: 35–46 [DOI] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T, Wu R. (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ohta A, Takagi M, Imai R. (2000) Expression of plant group 2 and group 3 lea genes in Saccharomyces cerevisiae revealed functional divergence among LEA proteins. J Biochem 127: 611–616 [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR. (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics 17: 383–384 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.