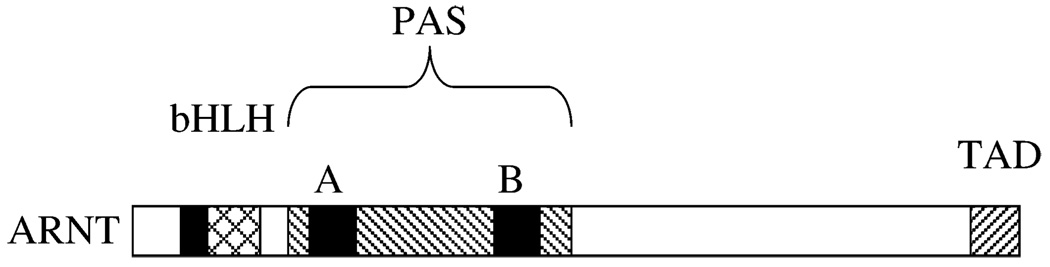
The aryl hydrocarbon receptor nuclear translocator (ARNT) is a member of the bHLH PAS (basic helix–loop–helix Period/ARNT/Single-minded) family of transcription factors. It is the obligatory dimerization partner for many other members of this family, including the aryl hydrocarbon receptor (AHR) and hypoxia-inducible factors 1α and 2α (HIF-1/2α). Agonists for AHR include a variety of environmentally important toxicants, including 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), and polycyclic aromatic hydrocarbons (PAHs) such as 3-methylcholanthrene (3MC) and benzo[a]pyrene (BAP). The mechanism of transcriptional activation by AHR is best understood for the CYP1A1 gene. After binding ligand, AHR translocates into the nucleus and dimerizes with ARNT. The AHR/ARNT heterodimer then binds to specific regulatory sequences, termed xenobiotic response elements (XREs), in the enhancer regions of CYP1A1 (and other responsive genes). Transcriptional coactivators the associated with the AHR/ARNT dimer as it resides on the enhancer region. These coactivators in turn direct the recruitment of RNA polymerase II (pol II) and the other general transcription factors to the promoter regions of genes, resulting in the initiation of transcription of those genes. Transcriptional activation of genes (as yet not fully defined) in this fashion is believed to underlie many of the toxic effects of TCDD, including its potent carcinogenicity. Metabolism of PAHs by CYP1A1 and other enzymes that are upregulated via AHR/ARNT critically impact the carcinogenic activities of these compounds.
HIF-1/2α activity is regulated not via a small molecule ligand, but indirectly, via oxygen tension. Briefly, HIF-1/2α is proteolytically degraded very rapidly under normoxic conditions. Under reduced oxygen conditions, it is stabilized, translocates into the nucleus, and dimerizes with ARNT. The HIF-1/2α/ARNT heterodimer then binds hypoxia response elements in responsive genes, thereby activating these genes in a manner similar to that described above for AHR/ARNT. HIF-1/2 represents the “master regulator” of the hypoxic response, modulating the expression of a variety of genes important in a number of pathological conditions including cancer, heart disease, cerebrovascular disease, and chronic obstructive pulmonary disease. ARNT is therefore an essential participant in the physiologic response to two important environmental insults: chemical toxicants and hypoxia. ARNT also participates in several developmental pathways via dimerization with other partner proteins not discussed above.
The domain structure of ARNT is shown in Figure 1. The bHLH motif is located within its amino-terminal region. Further toward the carboxy-terminus is the PAS region, which contains two approximately fifty amino acid degenerate repeat segments, PAS-A and PAS-B. Dimerization of ARNT with AHR and with HIF-1α requires both the HLH and PAS regions. The basic region contacts DNA, although there is some evidence that part of the PAS region also participates in DNA binding. ARNT harbors a transactivation domain at its carboxyl terminus.
FIG. 1.
Domain structure of ARNT
In 1996, Yoshiaki Fujii-Kuriyama’s group reported the cloning of a second ARNT gene, ARNT2. Twelve of the thirteen amino acids in the basic regions of ARNT2 and ARNT are identical, including all amino acids known to contact DNA. The amino acid sequence over the HLH and PAS regions is 80% identical between the two proteins, although the carboxy-terminal halves of the two proteins exhibit less amino acid identity (Hirose et al., 1996). ARNT2 has a more limited tissue distribution than ARNT. In their original publication, Hirose and coworkers concluded that ARNT2 dimerizes with AHR as efficiently as does ARNT, and that the AHR/ARNT2 dimer so formed can activate XRE-dependent gene transcription. In the current issue of Toxicological Sciences, Dougherty and Pollenz (2008) revisit this issue and come to different conclusions.
Dougherty and Pollenz introduced ARNT2 into an ARNT-deficient (and ARNT2-deficient) mutant of Hepa-1 cells, and found that the protein failed to cooperate with AHR in TCDD induction of the endogenous Cyp1a1 gene. In a recent paper, Fujii-Kuriyama’s group (Sekine et al., 2006) revise their original opinion, and come to the same conclusion as Dougherty and Pollenz. Like Dougherty and Pollenz, Sekine and coworkers tested the effect of ARNT2 on the endogenous CYP1A1 gene, while in the original paper, Hirose and coworkers investigated the effect of ARNT2 on a transiently transfected artificial XRE-driven reporter gene. It is likely that the reporter gene assay lead to confounding results because of the abnormally high copy number of the reporter gene in the transfected cells and/or because of the artificial, nonchromatin state of the reporter gene. It therefore seems safe to assume that ARNT2 is indeed incapable of cooperating with AHR in the activation of XRE-dependent genes (at least with regard to CYP1A1 and in these cells). The important question then becomes—at which step(s) in the AHR transcription pathway is ARNT2 defective? Of considerable interest in this regard, Sekine and coworkers demonstrate that ARNT2 functions nearly as efficiently as ARNT in the hypoxic induction of gene expression. ARNT2 is therefore not a “dead” protein (which is also indicated by the fact that ARNT2 knockout mice die perinatally).
Dougherty and Pollenz set out to address the above question. In the meticulously quantitative fashion characteristic of the Pollenz laboratory, they demonstrate that ARNT2 dimerizes in vitro with AHR as efficiently as ARNT, and that the AHR/ARNT2 dimer binds the XRE DNA segment with an efficiency equal to that of AHR/ARNT. Thus, ARNT2 is not inherently defective in these two steps of the AHR induction pathway. In contrast, Sekine and coworkers reported that ARNT2, unlike ARNT, is incapable of dimerizing with AHR. One difference between the experimental designs of the two research groups is that Dougherty and Pollenz used the high affinity AHR ligand, TCDD, in their experiments, while Sekine and coworkers used the lower affinity ligand, 3MC. Dougherty and Pollenz therefore tested whether the use of different ligands might explain the difference in results obtained. They found that 3MC (and BAP) was indeed less effective than TCDD at inducing AHR/ARNT2 dimerization as well as the subsequent association of the dimer with an XRE, while the compounds were as effective as one another at inducing these activities for AHR and ARNT. These observations suggest the interesting possibility that the ligands induce slightly different conformations of AHR. Nevertheless, the differences observed by the use of differing ligands in these experiments are unlikely to be of sufficient magnitude to fully explain the contrasting results obtained by the two research groups with regard to the ability of ARNT2 to dimerize with AHR and bind the XRE; another factor is likely to contribute to the different results. Unlike the in vitro experiments of Dougherty and Pollenz, Sekine and coworkers assessed the dimerization of AHR and ARNT2 within cultured cells, and this seems likely to represent a major reason for the differences in results obtained. Indeed, Dougherty and Pollenz also obtained some evidence suggesting that, within the cell, ARNT2 is incapable of dimerizing with AHR and/or binding the XRE in concert with AHR. Some aspect of cell structure or physiology therefore appears to preclude dimerization of AHR with ARNT2 or the subsequent association of the resultant dimer with the Cyp1a1 gene. One possibility is that there exists a protein in mammalian cells that inhibits dimerization of ARNT2 with AHR, which is not present in the extracts used to generate the ARNT2 and AHR proteins for the in vitro assays. The above observations are eerily reminiscent of those made by our laboratory a number of years ago on a protein repressor expressed in certain cultured cells that inhibits CYP1A1 induction. We found that the AHR/ARNT dimer functioned normally with regard to dimerization and XRE binding when assays were performed with cellular extracts of these cells, but that the dimer was clearly defective in binding XREs in the enhancer of the endogenous Cyp1a1 gene when binding was assayed in vivo in nondisrupted cells. We hypothesized that these conflicting results might be explained by lability of the repressor or to an inability to recover the repressor in cellular extracts (Watson et al., 1992).
The experiments of Dougherty and Pollenz illustrate the usefulness of performing an in vitro analysis of the components of the AHR transcription activation pathway, but also highlight the importance of performing such analysis in conjunction with in vivo assays. It will be important to fully resolve whether ARNT2 can participate in AHR action in living cells. Novel experimental approaches will be needed to achieve this goal. In this regard, Dougherty and Pollenz usefully identify several immortal cell lines that express ARNT2 (as well as ARNT). Chromatin immunoprecipitation assays would be useful to determine whether ARNT2 is recruited in a TCDD-dependent fashion to the endogenous CYP1A1 enhancer in these cells, although it should be pointed out that it has become increasingly clear that recruitment of the AHR/ARNT dimer to gene enhancers does not necessarily result in activation of gene transcription (Yang et al., 2008; R.T. Taylor, S. Beedanagari, and O. Hankinson, unpublished data). Ultimately, knockout mouse models will likely resolve this issue. ARNT and ARNT2 knockout mice die in utero and shortly after birth, respectively, but Arntfl/fl mice are available, and can be used to render the ARNT gene inactive in selected tissues (Tomita et al., 2000).
The question as to whether ARNT2 can interact with AHR in vivo is relevant not only to the conventional pathway in which the AHR/ARNT dimer activates gene transcription, but also potentially to other, newly discovered pathways involving this dimer. Thus, the liganded AHR/ARNT dimer has been shown to recruit the estrogen receptor to Estrogen Response Elements in estradiol-responsive genes and to either activate or attenuate transcription of these genes, depending on the absence or presence of estradiol (Ohtake et al., 2003). In conjunction with ARNT, liganded AHR directs the proteolytic degradation of the estrogen and androgen receptors through its activity as a component of a novel ubiquitin ligase complex (Ohtake et al., 2007). ARNT also acts a transcriptional coactivator for the estrogen receptor in the absence of AHR (Ruegg et al., 2008) and dimerizes with, and thereby mediates the effects of, a number of other bHLH PAS proteins involved in developmental processes. The question arises as to whether ARNT2 also participates in these pathways. Further elucidation of the role of this interesting protein is eagerly awaited.
ACKNOWLEDGMENTS
I thank Drs Erin K. Hsu and Feng Wang for critical reading of the manuscript.
FUNDING
National Institutes of Health grants (R01CA28868 and R01ES015384).
REFERENCES
- 1.Dougherty EJ, Pollenz RS. Analysis of AH receptor-ARNT and AH receptor-ARNT2 complexes in vitro and in cell culture. Toxicol. Sci. 2008 doi: 10.1093/toxsci/kfm300. [epub 2007 Dec 20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS factor (Arnt2) with close sequence similarity to the aryl hydrocarbon receptor nuclear translocator (Arnt) Mol. Cell. Biol. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 4.Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, et al. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature. 2003;423:545–550. doi: 10.1038/nature01606. [DOI] [PubMed] [Google Scholar]
- 5.Ruegg J, Swedenborg E, Wahlstrom D, Escande A, Balaguer P, Pettersson K, Pongratz I. The transcription factor aryl hydrocarbon receptor nuclear translocator functions as an estrogen receptor {beta}-selective coactivator, and its recruitment to alternative pathways mediates antiestrogenic effects of dioxin. Mol. Endocrinol. 2008;22:304–316. doi: 10.1210/me.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sekine H, Mimura J, Yamamoto M, Fujii-Kuriyama Y. Unique and overlapping transcriptional roles of arylhydrocarbon receptor nuclear translocator (Arnt) and Arnt2 in xenobiotic and hypoxic responses. J. Biol. Chem. 2006;281:37507–37516. doi: 10.1074/jbc.M606910200. [DOI] [PubMed] [Google Scholar]
- 7.Tomita S, Sinal CJ, Yim SH, Gonzalez FJ. Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Mol. Endocrinol. 2000;14:1674–1681. doi: 10.1210/mend.14.10.0533. [DOI] [PubMed] [Google Scholar]
- 8.Watson AJ, Weir-Brown KI, Bannister RM, Chu FF, Reisz-Porszasz S, Fujii-Kuriyama Y, Sogawa K, Hankinson O. Mechanism of action of a repressor of dioxin-dependent induction of Cyp1a1 gene transcription. Mol. Cell Biol. 1992;12:2115–2123. doi: 10.1128/mcb.12.5.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Solomon S, Fraser LR, Trombino AF, Liu D, Sonenshein GE, Hestermann EV, Sherr DH. Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in premalignant and malignant mammary tissue. Int. J. Cell. Biochem. 2008 doi: 10.1002/jcb.21630. [epub 2007 Dec 4] [DOI] [PubMed] [Google Scholar]