Abstract
The ATPase cycle of the heat shock protein 70 (HSP70) is largely dependent on the ability of its nucleotide binding domain (NBD), also called ATPase domain, to undergo structural changes between its open and closed conformations. We present here a combined study of the Hsp70 NBD sequence, structure and dynamic features to identify the residues that play a crucial role in mediating the allosteric signaling properties of the ATPase domain. Specifically, we identify the residues involved in the shortest-path communications of the domain modeled as a network of nodes (residues) and links (equilibrium interactions). By comparing the calculations on both closed and open conformation of Hsp70 NBD, we identified a subset of central residues located at the interface between the two lobes of the NBD near the nucleotide binding site, which form a putative communication pathway invariant to structural changes. Two pairs of residues forming contacts at the interface in the closed conformation of the NBD are observed to no longer interact in the open conformation, suggesting that these specific interactions may play a switch role in establishing the transition of the NBD between the two functional forms. Sequence co-evolution analysis and collective dynamics analysis with elastic network model further confirm the key roles of these residues in Hsp70 NBD dynamics and functions.
1. Introduction
Heat shock proteins (HSPs), also known as molecular chaperones, are ATP-regulated machines that perform several housekeeping activities in the cell: they assist in folding newly synthesized peptides, or unfolding and refolding partially folded or misfolded proteins, they regulate the intracellular trafficking of proteins, facilitate, in particular the recognition of those to be degraded by the proteasome, and most importantly, assist in the correct folding, and prevent the aggregation, of the proteins denatured in response to heat and other environmental stresses [1,2].
Hsp70 is one of the most ubiquitous members of the HSP family. It exists in almost all organisms [3]. It is composed of two domains: the ATPase domain, also referred as nucleotide binding domain, NBD [4], and the substrate binding domain (SBD) [5]. The two domains regulate the activity of each other via allosteric communication: ATP hydrolysis at the NBD increases the substrate binding affinity of the SBD, thus lowering the substrate exchange rate of the latter; on the other hand, the replacement of the ADP produced upon ATP hydrolysis by a new ATP (nucleotide exchange) lowers the binding affinity of the SBD thus enhancing the release and exchange of substrates [3].
The regulation of substrate binding affinity during the ATPase cycle is a crucial aspect of the chaperone activity of Hsp70, and notably, of other HSP family members [6]. The ATPase domain undergoes conformational changes between open and closed forms during the ATPase cycle, which correspond to different nucleotide binding states. The open conformation has been observed in the presence of ATP [7,8] and in the complexes formed with a class of co-chaperones called nucleotide exchange factors (NEFs) [9-11]. NEFs are known to assist nucleotide exchange by stabilizing the open form. Nucleotide exchange efficiency is viewed to be largely dependent on the conformational change to an open state.
In the present study, we examined the type of conformational changes occurring in the ATPase domain, and their influence on inter-residue communication pathways. Our previous examination of another ATP-regulated allosteric machine, the bacterial chaperonin GroEL, showed that the structure has access to intrinsically favored collective dynamics, on the one hand, and to well-defined signal transduction pathways that transmit allosteric effects away from the ATP binding site, on the other [12]. Redistribution of on-pathway interactions during the most cooperative (global) modes of motion of the chaperonin has been proposed to be a mechanism of allosteric regulation [13]. Toward gaining insights into the dynamic aspects of allosteric regulation, this time in the Hsp70 ATPase domain, we adopted here a multi-pronged approach: First, we identified a number of key residues distinguished by their central role in so far as the allosteric signal transduction across the molecule is concerned. This approach takes account of all atom-atom contacts, which are mapped into a low resolution, residue-level representation of internal interactions. A number of residues lining the cleft between the two lobes of the NBD appear to modulate the opening and closing of the cleft. Second, we analyzed the sequence conservation and co-evolution patterns of these residues. Third, we examined their collective dynamics using a simple elastic network model, the Gaussian network model (GNM) [14,15].
Notably, the presently identified “central” residues belong to two groups in terms of their evolutionary patterns. Residues in the first group are highly conserved across Hsp70 NBD sequence homologs. These residues exhibit little, if any, movement in the global modes predicted by the GNM, serving as hinge centers near the nucleotide binding site. The second group comprises co-evolving residue pairs. These residue pairs tend to concertedly make and break contacts upon closing or opening of the nucleotide-binding cleft. Our results indicate that (i) the GNM-predicted global modes of the Hsp70 ATPase domain entail alterations in inter-residue contact topology, which in turn facilitate nucleotide binding or release; (ii) conserved residues participate in hinge centers in the global modes thus playing a role in maintaining the native contact topology; whereas sequentially variable but correlated residues exhibit a moderate mobility essential to enable functional changes in conformation.
2. Materials and Methods
2.1. Structural Data
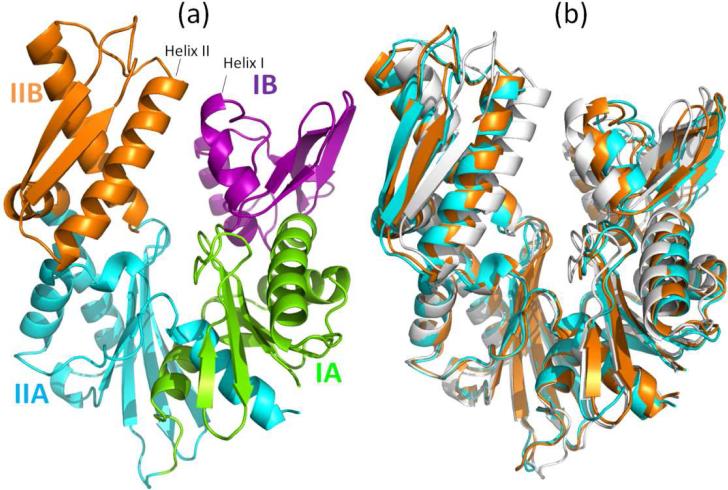
The Hsp70 ATPase domain consists of two lobes [4]. Lobe I consists of subdomains IA and IB, and lobe II, of subdomains IIA and IIB (Figure 1a). We used the structures of the bovine homolog of Hsp70 (Hsc70) (PDB id: 1hpm [16]) for the closed form of the NBD. For the open form, we considered two structures of the same species complexed with mammalian NEFs: a complex with BAG, and another with Sse1, with respective PDB identifiers of 1hx1 [9] and 3c7n [17]. The structural alignment in Figure 1(b) shows that there is a global change in the relative positions of subdomains IB and IIB, as the structure undergoes a conformational change between closed and open forms.
Figure 1.
(a) Subdomains of Hsp70 ATPase domain: IA (green; residues 1-39 and 116-188), IB (purple; residues 40-115), IIA (cyan; residues 189-228 and 307-C-terminus) and IIB (orange; residues 229-306). (b) Ribbon diagram of the superimposed closed and open conformations. The closed form (white) is a structure observed in the absence of nucleotide (PDB id: 1hpm) [16]. Two open forms are shown, both observed in the complexes formed with NEFs: In cyan is the structure from the complex with the Sse1 (PDB id: 3c7n) [17]; and in orange is that assumed when complexed with BAG (PDB id: 1hx1) [9]. The NEFs (Sse1 and BAG) are not shown here. The three structures have been aligned using the Kabsch algorithm ([21] as implemented in PyMol).
2.2. Sequence Data
We started from the multiple sequence alignment (MSA) of 4839 sequences retrieved from the PFAM database [18] for Hsp70 family (PFAM id: PF00012). This family includes a wide range of subfamilies, some of which have biological functions not represented by the canonical Hsp70 (e.g., the Hsp110 subfamily). We refined the MSA using the consensus sequence of the ATPase domain (380 residues) in the bovine cytosolic homolog of Hsp70. The refinement consists of the following steps: (i) iterative implementation of the Smith-Waterman algorithm (SW) for pairwise alignment [19] using the consensus sequence against each sequence in the MSA, and elimination of those below a threshold SW score (of 300) so as to retain the closest orthologs to the human (Hsc70) and bacterial (DnaK) chaperones, (ii) removal of the MSA columns that correspond to insertions with respect to the consensus sequence, thus restricting the number of amino acids to 380, and (iii) elimination of the sequences that contain more than 10 gaps. The refinement resulted in 1627 sequences, which has been subjected to sequence conservation and correlation analyses. Conserved residues were identified with the WEBLOGO web server [20], and correlated residues by mutual information (MI) analysis.
2.3. Identification of Central Residues
We adopted the approach proposed by Nussinov and coworkers [22] to identify the central residues of the ATPase domain. These are the residues that exhibit a high probability of participating in shortest-path communication when all such paths between all residue pairs are examined. The protein is modeled as a network to this aim, each node representing a residue. Nodes corresponding to residues A and B are connected if at least one atom of A is within 4.5 Å distance from an atom of B. In previous studies, such contact-based network models have been pointed out to exhibit properties of small-world networks [23,24]. We utilized as metric the characteristic path length (L) --- a global property of the network defined as the average shortest path length between all pairs of nodes. The shortest path between each pair of nodes is computed using the Dijkstra's algorithm. The centrality of residue k is measured by the difference ΔLk=Lhyp(k) - L in characteristic path length with respect to the original network, obtained for the hypothetical network where node k and all connected edges have been removed. If there is a significant increase in the characteristic path length due to removal of residue k, then residue k is considered to be a “central residue” in establishing internode communication. Central residues are hypothesized to play a role in allosteric signal transduction. Indeed, Nussinov and coworkers [22] have applied the method to seven distinct families, to find results consistent with experimental data. It should be noted however, that the identification of central residues may be sensitive to missing residues. In addition, the test set of proteins used in previous work for establishing the centrality contains globular proteins exclusively. The applicability of the method to other proteins remains to be established.
2.4. Gaussian Network Model (GNM)
We used the Gaussian network model (GNM) [14,15] for analyzing the equilibrium dynamics of the ATPase domain. Details on the method can be found in our previous studies. In summary, the structure is modeled as an elastic network, the position of each node being identified by that of each α-carbon; and pairs of nodes (Cα) within a cutoff distance of 7.3 Å are connected by elastic springs to account for the tendency of the structure to maintain its inter-residue contact topology under native state conditions. Knowledge of inter-residue contact distribution permits us to construct the Kirchhoff matrix Γ, also known as Laplacian in graph theory. Eigenvalue decomposition of Γ, or its inverse, C = Γ-1,
| (1) |
yields information on the spectrum of collective modes (fluctuations) intrinsically accessible to the structure under equilibrium conditions. Here λk and uk represent the kth eigenvalue and eigenvector, respectively of Γ, the summation is performed over all N-1 nonzero eigenmodes, for a protein of N residues/nodes and λk-1Ck is the contribution of the kth mode to C. The diagonal elements of Ck represent the normalized square fluctuations of the N residues, also called the mobility profile, induced by mode k, and the off-diagonal elements scale with the cross-correlations between residue fluctuations. Each mode contribution is weighted by the inverse eigenvalue (which is proportional to the square root of the mode frequency) such that the lowest frequency modes make the largest contribution to C. We focus here on the modes in the low frequency regime, as the major determinants of potentially functional movements, and examine the weighted average mobility profiles Σkλk-1Ck/Σkλk-1 for 1 ≤ k ≤10.
2.5. Mutual Information (MI)
We adopted the mutual information (MI) content as a measure of the degree of co-evolution between residue pairs [25,26]. Accordingly, each of the N columns in the MSA generated for a protein of N residues is considered as a discrete random variable Xi (1 ≤ i ≤ N) that takes on one of the 20 amino-acid types with some probability. The MI associated with the random variables Xi and Yj corresponding to the ith and jth positions is defined as
| (2) |
where P(Xi ; Yj) is the joint probability of observing amino acid type X at position i, and Y at position j; P(Xi) and P(Yj) are the marginal/singlet probabilities for amino acids of types X and Y at the two respective positions. Eq (2) permits us to evaluate the N × N MI matrix I with elements Iij = I(Xi , Yj). Iij varies in the range [0, Imax], with the lower and upper limits corresponding to uncorrelated and most correlated pairs of residues. The MI is a classical concept from information theory; however, like other methods based on sequence information, the performance of the MI metric also relies on the quality of MSA.
3. Results and Discussion
3.1. Centrality Profile of Different Conformations of NBD
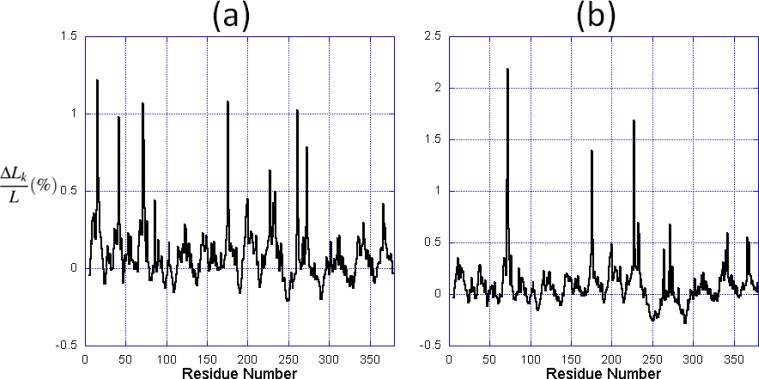
We calculated the centrality profile of all the three structures, the NBD in the closed state and the two others, in the open state. The results are shown in Figure 2 for the unbound (panel (a) and Sse1-bound (panel (b)) of the ATPase domain. The centrality profile for the BAG-bound form exhibits patterns similar to those observed in panel (b). From the characteristic path length and the RMSD calculated from structural alignment, we infer that the lobes of the Sse1-bound NBD are further apart than the BAG-bound form.
Figure 2.
The centrality profile for NBD residues. The profiles are calculated for (a) the closed form, and (b) the open form in the Sse1-bound NBD. The abscissa represents the fractional change ΔLk/L in characteristic path length compared to the original network, maxima referring to nodes that have a strong impact on communication efficiency, if removed.
Comparison of panels (a) and (b) shows that the two profiles exhibit similar features (i.e., peaks and minima at the same regions), while the relative heights of the peaks vary. In particular, the peaks near residues located at the inter-lobe interface, that is residues 257-276 (helix II) and residues 10-60, are suppressed in the open form. In contrast, some residues located at the nucleotide binding pocket (e.g., Arg342 and Asp366) are increased in the open form (note that the ordinate scales are different in the two panels). The increase in centrality suggests that they assume an enhanced role in establishing the communication away from the active site in the open form.
We consider the top ranking (top 2%, or equivalently, top 8 residues) residues in the centrality profile in each case and refer to them as the central residues in the following text. Among them four are distinguished as central residues in all of three structures, regardless of the open or closed state of the NBD: Lys71, Arg72, Glu175 and His227; in contrast, the other four residues vary with the conformation (Table 1).
Table 1.
Central residues in the closed and two open conformations of Hsp70 NBD.
| PDB id | L(Å) | Binding NEF | conformation | Central residues(*) | |
|---|---|---|---|---|---|
| shared by all | Specific to examined structures | ||||
| 1hpm | 14.06 | None | closed | Lys71 Arg72 Glu175 His227 | Tyr15 Tyr41 Arg261 Arg272 |
| 1hx1 | 14.67 | BAG | open | Ala60 Arg261 Arg342 Asp366 | |
| 3c7n | 15.26 | Sse1 | open | Leu73 Asp232 Lys271 Arg342 | |
(*) fully conserved residues are written in boldface (see Fig. 4a). Note that Leu73 and Asp232 are also highly conserved.
3.2. Central Residues Have Different Structural and Sequence Variation Characters
We examined the position of the central residues on the structure (Figure 3), and performed sequence analyses to examine their conservation profile and co-evolutionary properties (Figure 4).
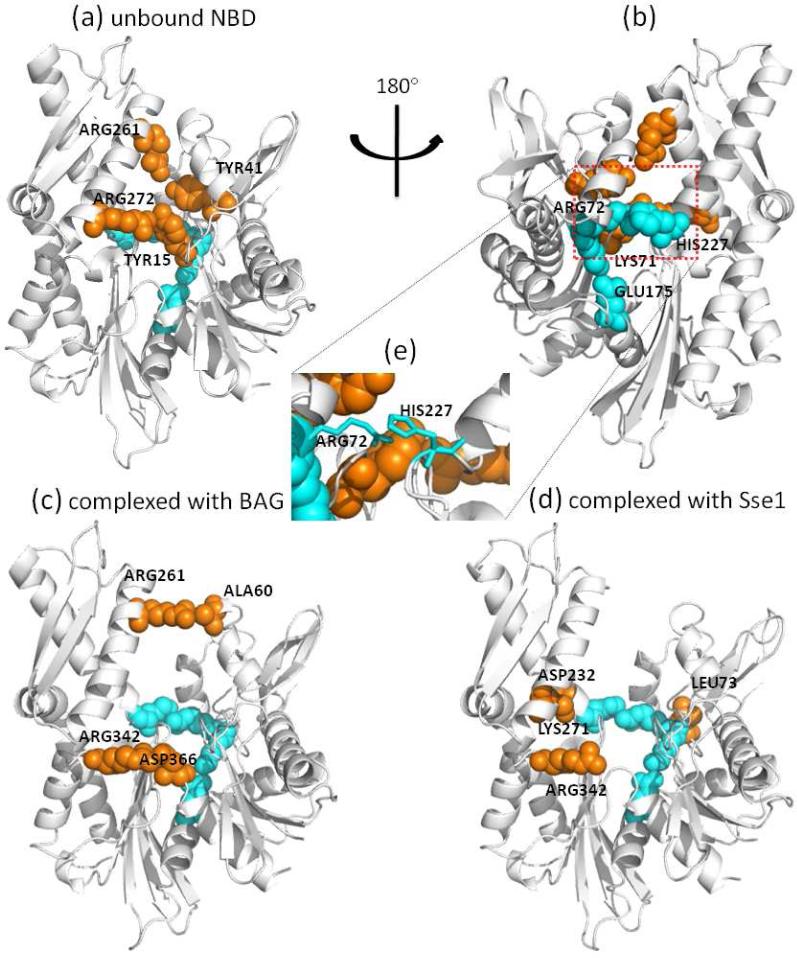
Figure 3.
Position of Hsp70 NBD central residues. Panels (a) and (b) display the positions on the NBD closed form, (c) on the NBD open form bound to BAG and (d) on NBD open form bound to Sse1. Panel (e) highlights the interaction of Arg72 and His227. The conserved central residues are colored cyan; other central residues are colored orange. Panel (b) is the rotated view of panel (a), and the conserved central residues are only labeled in panel (b). The diagram was generated with PyMOL. (http://pymol.sourceforge.net).
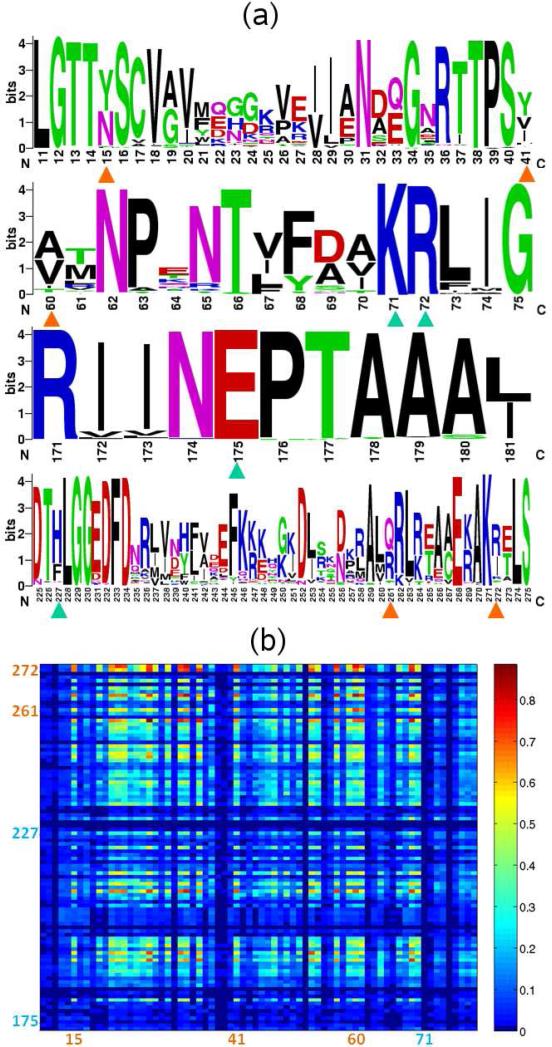
Figure 4.
Sequence analysis of central residues. (a) Sequence logo of the SCRs (marked with cyan triangles) and other central residues (marked with orange triangles) identified in the closed and open (BAG-complexed) ATPase domain. (b) Mutual information map between residues 173-274 and 10-80, which includes the central residues located at the lobe interface in the closed state. The SCRs’ residue numbers are written in cyan, whereas others are written in orange.
The four residues that are invariant to conformational changes are colored cyan in Figure 3, and are labeled in Figure 3b. Interestingly, these (sequentially separated) residues appear to form a (spatially contiguous) communication path across the lobes, starting from His227 and ending at Glu175. Indeed, Lys71 and Glu175 serve as catalytic residues [27,28] and are believed to regulate a proline switch that regulates the inter-domain allosteric interactions. Studies by Johnson and McKay also show that mutations of Lys71 and Glu175 impede the functional conformational change of ATPase domain, which appears to block the signal transduction between subdomains IIB and IA [29]. His227 is located at the calcium binding site and its mutation significantly weakens the catalytic activity [30,31]. Since the central residues are found to be mediating allosteric communications in a variety of protein families [22], we propose these residues to play not only a catalytic role, but also a signaling role in communicating the nucleotide exchange events to the other regions of the NBD, including for example the interface with the substrate-binding domain.
These four residues are also highly conserved. The sequence logo [20] shown in Figure 4a indicates that residues Lys71, Arg72 and Glu175 are fully conserved. His227, although not conserved, can only be substituted by phenylalanine, although histidine probability is much higher, suggesting that a large aromatic group may be functional at this position. The interaction of Arg72 and His227, as can be seen from Figure 3e, can be viewed as a highly conserved amino-aromatic interaction [32], which is presumably maintained when histidine is replaced by phenylalanine. So even though His227 tolerates a mutation to phenylalanine, its interaction with Arg72 is conserved. In the following text we will refer to these 4 residues as the shared central residues (SCR).
The other central residues also exhibit patterns relevant to functional changes in NBD conformation. In the closed form, these residues (Tyr15, Tyr41, Arg261 and Arg272) are distributed along the cleft formed by lobes I and II to form two closely interacting pairs: Arg272---Tyr15 and Arg261---Tyr41. These pairs serve as two bridges that connect subdomain IIB with IA (Arg272---Tyr15) and with IB (Arg261---Tyr41). Bukau and coworkers [33] have shown that the salt bridges formed between helix I and helix II, labeled in Figure 1a, affect the nucleotide exchange of NBD. We speculate that among the residues located on these two helices, these two pairs arginines and tyrosines, also involved in amino-aromatic interactions, play a key role in controlling the subdomain closure and opening that in turn ensure efficient nucleotide stabilization or release, respectively. Moreover, since the central residues are supposed to be the most “indispensable” residues in establishing the shortest-path communications, the two pairs we identified might be the “anchors” that maintain the closed conformation of NBD. Indeed, this conjecture is reinforced by the collective dynamics of the NBD in the next section.
Interestingly, residues at these four positions also tend to co-evolve as may be seen from the MI map in Figure 4b. By examining the sequence logo (Figure 4a), we found that the variation of amino acids at these residues primarily arises from the difference between the Hsp70 mammalian homolog Hsc70 and the Hsp70 bacterial homolog Dnak. The interactions between the two lobes of DnaK, as well as its interaction with NEF (GrpE in this case) primarily consist of hydrophobic contacts; whereas in Hsc70, there is a prominence of electrostatic interactions. The co-evolution of these central residues is in line with the specificity of their interactions in different organisms.
In the BAG-bound NBD, which assumes a less open conformation between the two NEF-bound structures, there still remains a contacting residue pair between the tips of subdomains IB and IIB (Arg261---Ala60, see Figure 3c), but this interaction can hardly account for the interface between the lobes. On the other hand, Arg342 and Asp366 are both conserved and form the nucleotide binding pocket. Their interactions are crucial for maintaining the conformation of the active site. In the Sse1-bound NBD, because subdomains IB and IIB have undergone a rotation, Asp232 becomes in contact with Lys227, which implies a putative extension of the SCR to subdomain IIB. Similarly, Leu73 extends SCR to subdomain IB. Lys271 and Arg342 are both conserved residues at the active site, and mutagenesis study showed that Arg342 is crucial for sulfogalactolipid recognition [34].
3.3. Global Dynamics of ATPase Domain and Role of Central Residues
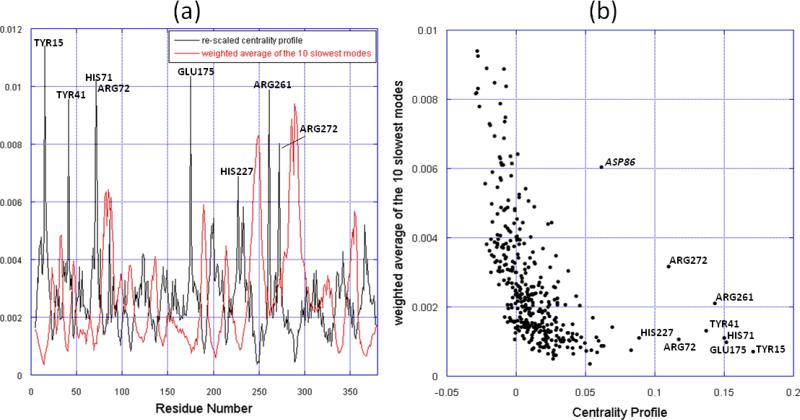
As suggested in [22], the central residues generally relate to the system fragility; that is, these residues ought to remain “stable” to maintain the biological function of the molecule. From the sequence perspective, this requires sequence conservation; from the structural dynamics perspective, one might expect to see little variations, if any, in their spatial positions. In order to critically examine their dynamical characters, we examined the equilibrium dynamics of the ATPase domain using the GNM. We focused in particular on the low frequency end of the spectrum of modes, given that these modes are usually highly cooperative and relevant to function [35,36]. We compared the centrality profile and the mobility profile resulting from the weighted average of the 10 slowest modes of the closed-form NBD (Figure 5). Strikingly, the mobility profile (which represents the normalized distribution of square fluctuations in residue positions driven by these modes) exhibits minima at the peaks of the centrality profile, and vice versa. Minima in the mobility profile represent sites that act as hinges (or anchors) in the collective modes. Notably, all the central residues coincide with minima (Figure 5a), which is indicative of their key role in the global motions of the NBD. Arg261 and Arg272 are of particular interest: first, their mobility is higher than that of other central residues, suggesting a lower energy barrier for them to dissociate from lobe I to facilitate the cleft opening; second, helix II as the linkage between two most mobile regions of NBD, is implicated in functional motions.
Figure 5.
Comparison of the slowest modes and the centrality profile. (a) Mobility profile resulting from the weighted average of the 10 slowest modes and the re-scaled centrality profile of the NBD closed form. The centrality profile is re-scaled for visual comparison. The peaks corresponding to central residues are labeled, in addition to another outlier, Asp86, distinguished by its high mobility. (b) Mobility versus centrality for all residues in the NBD closed form. The points corresponding to central residues are labeled. The ordinate and abscissa values are taken from the two curves in panel a, for each residue.
Overall, the centrality profile and the slow modes curve are negatively correlated, which can be observed in Figure 5b. Figure 5a indicates the correspondence between the peaks of one curve and the valleys of the other, in most cases. In Figure 5b, the residues with high centrality (≥ 0.05) are characterized with low mobility, except for Asp86 (labeled in italic in Figure 5b). Indeed, Asp86 is located in an exposed helix that accounts for the rotation of subdomain IB and forms a salt bridge with Arg72, which in turn is one of the shared central residues presently identified. It appears that the salt bridge between Asp86 and Arg72 is critical to the motion of the exposed helix. On the other hand, the residues with negative centrality are usually located at the ends or tips of the structure, consistent with their high mobility.
3.4. Summary of Different Types of Central Residues
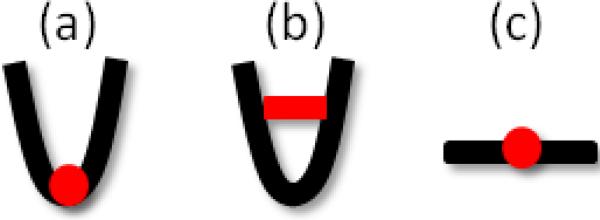
Here is an overview of the central residues based on our findings in this study (illustrated in Figure 6), and it remains to be seen how these results apply to central residues in other families of proteins. We can group the central residues into three categories depending on their location on the structure and/or their role in the structural dynamics:
The hinge point
bridging point at the interface near the cleft (or contact interface)
stretched linker (or long helix/loop stretching out)
Figure 6.
Three scenarios for the central residue's location on the structure.
In the first case, the central residues (e.g., SCR) connect two parts, at least one of which is highly mobile. These residues mediate the communications between different parts of the molecule, and transmit the information necessary for the proper functioning of the molecule. Perturbations at these residues are most likely to impede function. These residues are also highly conserved and serve as hinge points not only with respect to the two structural elements that are directly connected, but in the global dynamics of the entire NBD. In the second case, the central residues serve as linkages at the interface between substructures that have intrinsically access to alternative (e.g. open and closed) conformations. They act as the “anchoring point” of the interface, and can be the determinants of the motions of the moving parts. These residues are more exposed to the environment and more tolerant to mutations compared to the first case. Yet, their important role is signaled by correlated mutations that take place which presumably aim at restoring the key role (that of locking the closed form in this case). For residues in the third category, although we did not observe any such residue in this study, they have been observed in other systems. For example, the inter-domain linker between the NBD and SBD of the Hsp70 possess such residues, which evidently play a key role in establishing the allosteric communication between the two domains [37].
3.5. Comparison with results from other methods
We note that similar studies have been conducted for identifying allosteric residues and communication pathways, including the work of Tang and coworkers [38] who developed the AlloPathFinder package, the statistical coupling analysis (SCA) of Ranganathan and coworkers [39], or the pair-to-pair correlations analysis of Eyal et al. [40]. The performance of different methods for detecting sequence correlations has been comparatively studied by Fodor and Aldrich [41] and by Eyal et al. [42]. Calculations performed here for Hsp70 with AlloPathFinder indicated a number of pathways starting from His227 to Lys71/Glu175 which are composed of a series of residues deeply buried at the interface of subdomains IA and IIA (e.g, His227→Leu228→Leu200→Val337→Ala179→Glu175→Lys71). When the destination was set to Arg72, however, AlloPathFinder identified the pathway established by the contact between His227 and Arg72. The difference in the identified pathway(s) may be attributed to the fact that AlloPathFinder employs evolutionary information for weighting the individual steps along the communication paths, whereas our present approach for identifying pathways is exclusively based on topology. On the other hand, the application of SCA on Hsp70 indicates that Lys71 and Arg72 take part in a given cluster of highly coupled residues, while Glu175 belongs to another cluster, and His227does not belong to any such cluster (private communication with Dr. Lila Gierasch).
4. Conclusion
We presented here a computational study of the ATPase domain of Hsp70 to identify and analyze the residues that are crucial for efficient transduction of signals within this domain. This particular domain serves as a signal transduction module not only in molecular chaperones but many other proteins, as well, and understanding the position and role of key residues in establishing allosteric communication is a topic of broad interest. We identified a subset of central residues across sub-domains, which form a communication pathway invariant to structural change between open and closed forms. We also identified two pairs of interacting residues bridging the lobes in the closed conformation but no longer interacting in the open conformation.
The analysis of sequence correlations and collective dynamics assisted in assessing the key role of these residues in mediating domain movements relevant to function, supporting the functional character of central residues in allosteric systems..The findings independently obtained by centrality profile based on graph-theoretical methods, GNM based on statistical mechanical principles, and sequence analysis provide complementary perspectives on the allosteric potential, intrinsic dynamics and sequence evolution properties, respectively, of the examined system. These three pieces of data have been advantageously combined here to extract a uniform picture of the structural and dynamic aspects of Hsp70 NBD function. Further investigation of the detailed mechanism of transition between the NBD closed and open conformations and its coupling to the SBD, using the adaptive ENM [43], holds promise toward gaining deeper insights about, and further establishment of, the functional significance of particular residues and interactions inferred from the present study.
References
- 1.Hartl FU, Hayer-Hartl M. Science. 2002;295:1852. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Hayer-Hartl M. Nat Struct Mol Biol. 2009;16:574. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B, Horwich AL. Cell. 1998;92:351. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KM, Luca-Flaherty C, McKay DB. Nature. 1990;346:623. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhu XT, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Science. 1996;272:1606. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Nature. 2006;440:1013. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya A, Kurochkin AV, Yip GN, Zhang Y, Bertelsen EB, Zuiderweg ER. J. Mol. Biol. 2009;388:475. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8471. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Science. 2001;291:1553. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 10.Polier S, Dragovic Z, Hartl FU, Bracher A. Cell. 2008;133:1068. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 11.Harrison CJ, HayerHartl M, DiLiberto M, Hartl FU, Kuriyan J. Science. 1997;276:431. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 12.Chennubhotla C, Bahar I. Molecular Systems Biology. 2006;2:36. doi: 10.1038/msb4100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chennubhotla C, Yang Z, Bahar I. Mol. Biosyst. 2008;4:287. doi: 10.1039/b717819k. [DOI] [PubMed] [Google Scholar]
- 14.Yang LW, Rader AJ, Liu X, Jursa CJ, Chen SC, Karimi HA, Bahar I. Nucleic Acids Research. 2006;34:W24. doi: 10.1093/nar/gkl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahar I, Atilgan AR, Erman B. Folding & Design. 1997;2:173. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 16.Wilbanks SM, McKay DB. J. Biol. Chem. 1995;270:2251. doi: 10.1074/jbc.270.5.2251. [DOI] [PubMed] [Google Scholar]
- 17.Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, Jin S, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R. Molecular Cell. 2008;31:232. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer ELL, Bateman A. Nucleic Acids Research. 2006;34:D247. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith TF, Waterman MS. J. Mol. Biol. 1981;147:195. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 20.Crooks GE, Hon G, Chandonia JM, Brenner SE. Genome Res. 2004;14:1188. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch W. Acta Crystallographica Section A. 1976;32:922. [Google Scholar]
- 22.del Sol A, Fujihashi H, Amoros D, Nussinov R. Molecular Systems Biology. 2006;2:2006.0019. doi: 10.1038/msb4100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watts DJ, Strogatz SH. Nature. 1998;393:440. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 24.Greene LH, Higman VA. J. Mol. Biol. 2003;334:781. doi: 10.1016/j.jmb.2003.08.061. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Eyal E, Bahar I. Bioinformatics. 2008;24:1243. doi: 10.1093/bioinformatics/btn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke ND. Protein Science. 1995;4:2269. doi: 10.1002/pro.5560041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien MC, Flaherty KM, McKay DB. J. Biol. Chem. 1996;271:15874. doi: 10.1074/jbc.271.27.15874. [DOI] [PubMed] [Google Scholar]
- 28.Vogel M, Bukau B, Mayer MP. Molecular Cell. 2006;21:359. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 29.Johnson ER, McKay DB. Biochemistry. 1999;38:10823. doi: 10.1021/bi990816g. [DOI] [PubMed] [Google Scholar]
- 30.Sriram M, Osipiuk J, Freeman B, Morimoto R, Joachimiak A. Structure. 1997;5:403. doi: 10.1016/s0969-2126(97)00197-4. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Yano M, Washida H, Kido H. Biochem. J. 2004;378:793. doi: 10.1042/BJ20031680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burley SK, Petsko GA. FEBS Lett. 1986;203:139. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- 33.Brehmer D, Rudiger S, Gassler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B. Nature Structural Biology. 2001;8:427. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- 34.Mamelak D, Lingwood C. J. Biol. Chem. 2001;276:449. doi: 10.1074/jbc.M006732200. [DOI] [PubMed] [Google Scholar]
- 35.Bahar I, Atilgan AR, Demirel MC, Erman B. Physical Review Letters. 1998;80:2733. [Google Scholar]
- 36.Bahar I, Rader AJ. Curr. Opin. Struct. Biol. 2005;15:586. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Molecular Cell. 2007;26:27. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang S, Liao JC, Dunn AR, Altman RB, Spudich JA, Schmidt JP. J. Mol. Biol. 2007;373:1361. doi: 10.1016/j.jmb.2007.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suel GM, Lockless SW, Wall MA, Ranganathan R. Nature Structural Biology. 2003;10:59. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 40.Eyal E, Pietrokovski S, Bahar I. Bioinformatics. 2007;23:1837. doi: 10.1093/bioinformatics/btm256. [DOI] [PubMed] [Google Scholar]
- 41.Fodor AA, Aldrich RW. J. Biol. Chem. 2004;279:19046. doi: 10.1074/jbc.M402560200. [DOI] [PubMed] [Google Scholar]
- 42.Eyal E, Frenkel-Morgenstern M, Sobolev V, Pietrokovski S. Proteins. 2007;67:142. doi: 10.1002/prot.21223. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, Majek P, Bahar I. PLoS Comput. Biol. 2009;5:e1000360. doi: 10.1371/journal.pcbi.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]