SUMMARY
The Ras signaling pathway allows cells to translate external cues into diverse biological responses. Depending on context and the threshold reached, Ras signaling can promote growth, proliferation, differentiation, or cell survival. Failure to maintain precise control of Ras can have adverse physiological consequences. Indeed, excess Ras signaling disrupts developmental patterning and causes developmental disorders [1–2], while in mature tissues, it can lead to cancer [3–5]. We identify Rabex-5 as a new component of Ras signaling crucial for achieving proper pathway outputs in multiple contexts in vivo. We show that Drosophila Rabex-5 restricts Ras signaling to establish organism size, wing vein pattern, and eye-versus-antennal fate. Rabex-5 has both Rab5 GEF activity that regulates endocytic trafficking [6] and also ubiquitin ligase activity [7–8]. Surprisingly, over-expression studies demonstrate that Rabex-5 ubiquitin ligase activity, not its Rab5 GEF activity, is required to restrict wing vein specification and to suppress the eye phenotypes of oncogenic Ras expression. Furthermore, genetic interaction experiments indicate that Rabex-5 acts at the step of Ras, and tissue culture studies show that Rabex-5 promotes Ras ubiquitination. Together, these findings reveal a new mechanism for attenuating Ras signaling in vivo and suggest an important role for Rabex-5-mediated Ras ubiquitination in pathway homeostasis.
Key words/phrases: Rabex-5, RabGEF1, Ras, ERK, A20, ubiquitin ligase, tumor suppressors, oncogenes, overgrowth
RESULTS AND DISCUSSION
Rabex-5 loss causes increased size
We characterized the gene Rabex-5 (also called RabGEF1, annotated CG9139, Fig. S1) in Drosophila. Rabex-5 was first identified as a Rab5 guanine nucleotide exchange factor (GEF) with a highly conserved VPS9 domain [6]. Studies of Rab5 and other VPS proteins indicate that they play a role in growth control [9–10]. We used gal4/UAS expression [11] of an inverted repeat allele of Rabex-5, Rabex-5IR [12], to perform tissue-specific RNAi, and we generated deletion alleles Rabex-5ex9, Rabex-5ex40 and Rabex-5ex42 (Fig. S1, Supplemental Experimental Procedures).
Deletion allele homozygotes, heterozygotes with one copy of Rabex-5ex9, Rabex-5ex40 or Rabex-5ex42 over the large deletion Df(3L)BSC250, and flies undergoing strong, constitutive Rabex-5 RNAi died as giant larvae or giant pupae often containing melanotic tumors (Fig. 1A–B; not shown). Using RNAi conditions to reduce but not eliminate Rabex-5 resulted in viable adults whose length, weight, and wing size increased by over 10% (Fig. 1C–E).
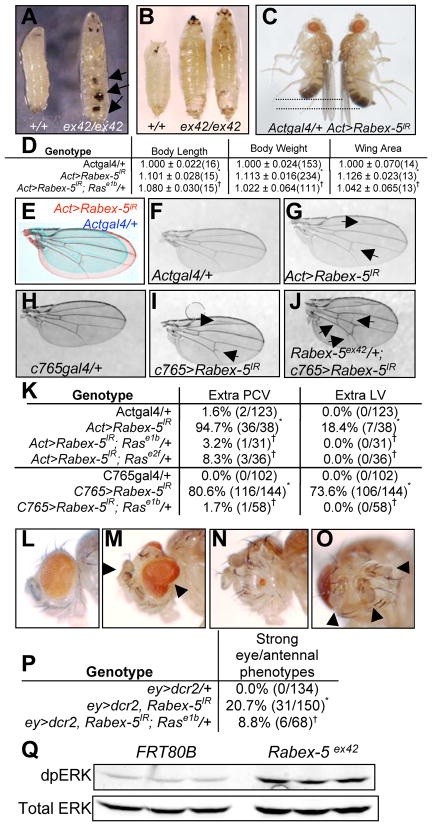
Figure 1. Loss of Rabex-5 causes increased size, ectopic wing veins, and eye/antennal phenotypes.
A-B) FRT80B control, left. Homozygous Rabex-5 deletion (ex42/ex42) resulted in giant larvae (A, right) and pupae (B, right) often containing melanotic masses (arrows, A). C) Constitutive Rabex-5 RNAi (Act>Rabex-5IR, right) increased adult size compared to Actgal4/+ controls (left, males shown). D) Table of Act>Rabex-5IR phenotypes. Length, weight, wing area normalized to 1.000 for controls and shown as a ratio to controls ± standard deviation; number of samples in parentheses. E) Overlay of Act>Rabex-5IR (red) and control Actgal4/+ (blue) wings. F) Control Actgal4/+ wing. G) Act>Rabex-5IR wings contain extra PCV (lower arrow) and extra LV (upper arrow). H) Control c765gal4/+ wing. I) Rabex-5 RNAi in the wing at 30°C using c765gal4 caused extra veins (arrows) and was enhanced (J) by deletion allele Rabex-5ex42. K) Table of wing phenotypes. Wings with ectopic veins over total wings in parentheses. L) Control ey>dcr2 eye. M–O) Strong Rabex-5 RNAi in the early eye (ey>dcr2, Rabex-5IR) produced (M) overgrown eyes, (N) small eyes, (O) extra antennae (overhead view of N) and other abnormalities (Fig. S2). P) Table of eye results. *statistically significant difference compared to control (p<0.05); †statistically significant suppression by Ras alleles (p<0.05). Images and quantitation in D–O from females; quantitation in P of males. Q) Western blot of larvae homozygous for control FRT80B or Rabex-5ex42. dpERK (upper panel) and total ERK (lower panel) staining showed 3.9 ± 0.23 (standard deviation) fold increased ERK activation in mutant larvae compared to controls. Additional controls in Fig. S1.
Loss of Rabex-5 causes ectopic wing veins and eye/antennal effects
Constitutive or wing-specific Rabex-5 RNAi resulted in extra posterior crossveins (PCV) and extra longitudinal veins (LV) (Fig. 1G, 1I–K, Fig. S1) compared to controls (Fig. 1F, 1H). Removing one copy of Rabex-5 enhanced Rabex-5 RNAi in the wing (Fig. 1J), and homozygous Rabex-5ex42 mutant wing tissue also formed extra veins (Fig. S1). Thus, it is unlikely this phenotype resulted from RNAi off-target effects.
Strong Rabex-5 RNAi in the early eye led to 10–25% of eyes with severe phenotypes including overgrown eyes, outgrowths, small or lost eyes, and extra antennae (Fig. 1M–P; Fig. S1) that were not observed in controls (Fig. 1L). Similar phenotypes were seen in eyes containing primarily Rabex-5ex42 mutant tissue (Fig. S1).
Rabex-5 mutant larvae exhibit increased ERK activity
These phenotypes are consistent with failure to regulate Ras. Increased Ras signaling causes melanotic tumors [13] as seen upon loss of Rabex-5. Ras activity is required for wing vein specification and in excess causes ectopic veins [14–15] as seen upon Rabex-5 reduction. Ras is also important in the eye. Increased Ras signaling early in eye development induces hyperplastic growth [16] while exceeding certain thresholds may promote antennal fates instead of eye fates [17]. Eye phenotypes from Rabex-5 loss are consistent with Ras activity near the threshold between driving hyperplasia and promoting an eye-to-antenna switch. To evaluate Ras signaling, we assessed dually phosphorylated ERK (dpERK), a downstream Ras effector. Western analysis showed 2.5 to 5-fold increased ERK activity in mutant larvae compared to controls when normalized for total ERK (Fig. 1Q, Fig. S1). Previous in vitro studies reported that Rabex-5 loss in mast cells in culture extended the duration of ERK activation when triggered by aggregation of FcεR1 receptors or by stem cell factor [18–19], consistent with our in vivo findings.
Size, eye, and wing phenotypes are Ras-dependent
Ras mutations dominantly suppressed the extra wing veins caused by constitutive and wing-specific Rabex-5 RNAi (Fig. 1K, Fig. S1), reduced the percentage of flies with severe eye phenotypes upon Rabex-5 RNAi (Fig. 1P), and suppressed the increased body size, body length, and wing area of constitutive Rabex-5 RNAi (Fig. 1D, Fig. S1). Body weight of all flies and wing area in female flies were no longer statistically different from controls. Mutations in Egfr, sos, Raf, MEK, and ERK dominantly suppressed the extra PCV RNAi phenotype (Fig. S1). Mutation in ERK also dominantly suppressed homozygous Rabex-5ex42 phenotypes (Fig. S1).
These data indicate that Rabex-5 restricts Ras signaling through ERK to achieve appropriate organism size, to specify wing vein pattern, and to establish eye versus antennal fate in Drosophila.
Loss of Rabex-5 enhances activated Ras and Egfr but not Raf
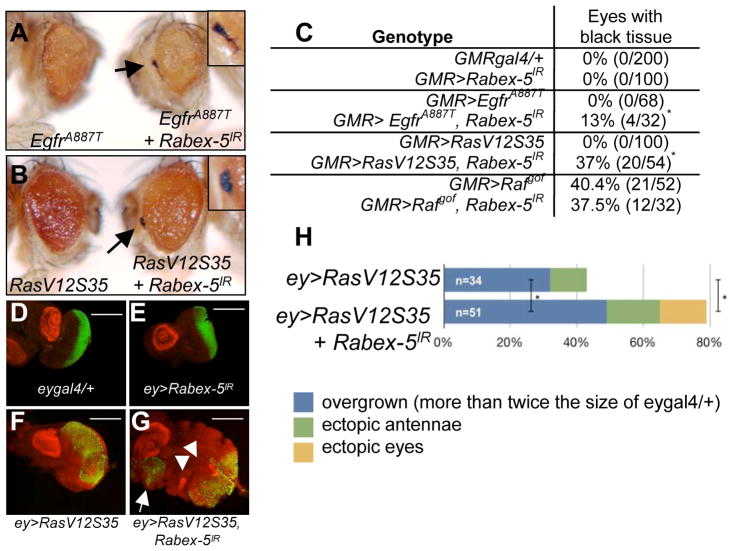
If Rabex-5 restricts Ras signaling, mutations activating the pathway in an otherwise wild-type background may be somewhat inhibited by endogenous Rabex-5. Therefore, loss of Rabex-5 might exacerbate the consequence of expressing activated pathway components. Activated Egfr (EgfrA887T), RasV12S35 (oncogenic Ras signaling primarily through ERK), and activated Raf (Rafgof) [20] (details in Supplemental Experimental Procedures) expression in differentiating eye cells each caused cause rough eyes of reduced size (EgfrA887T, Fig. 2A, Fig. S2; RasV12S35, Fig. 2B, Fig. S2; Rafgof, not shown). As these phenotypes worsened, we observed black tissue and lethality (Fig. S2). Loss of Rabex-5 significantly enhanced the number of eyes with black tissue and significantly increased lethality of EgfrA887T or RasV12S35 expression (Fig. 2A–C, Fig. S2). Rabex-5 RNAi in differentiating eye cells resulted in no obvious abnormalities (Fig. S2), so phenotypic enhancement was unlikely due to additive effects of combining two visible phenotypes. Unlike its enhancement of EgfrA887T and RasV12S35 phenotypes, reducing Rabex-5 in differentiating eye cells did not enhance the phenotype of Rafgof expression (Fig. 2C; not shown).
Figure 2. Loss of Rabex-5 enhances activated Egfr and oncogenic Ras.
A–C) Activated Egfr, EgfrA887T (left, A) or RasV12S35 (left, B) expressed in differentiating eye cells caused small, rough eyes. A–B) Rabex-5 RNAi (right) enhanced GMR>EgfrA887T (A) and GMR>RasV12S35 (B), evident by appearance of black tissue (arrows; enlarged in insets) and lethality (Fig. S2). C) Table quantifying effects of Rabex-5 loss on EgfrA887T, RasV12S35, and Rafgof. Female eyes are shown and quantified. In Rafgof eyes, Rabex-5 reduction did not statistically significantly change the frequency of black tissue compared to control (p>0.05). (D–G) ELAV (green) stains presumptive photoreceptors; Distal-less (red) stains presumptive antennae in larval discs. D) eygal4/+ control. E) ey>Rabex-5IR control. F) Larval disc expressing RasV12S35 in the early eye. G) RasV12S35 discs with reduced Rabex-5 showed increased overgrowth, extra antennae (arrowheads), and ectopic eyes (arrow). Scale bar, 200μm. H) Graph quantifying phenotypes in D–E. *statistically significant difference compared to control (p<0.05). Additional controls and quantitation in Fig. S2.
Expressing RasV12S35 in the early eye results in eye overgrowth and other abnormalities; loss of Rabex-5 dramatically enhanced these phenotypes (Fig. 2G–H, Fig. S2), notably the overgrowth, extra antennae, and ectopic eye phenotypes in larval eye discs (Fig. 2G–H, Fig. S2) compared to control discs (Fig. 2D–F).
The ubiquitin ligase domain of Rabex-5 restricts wing vein formation
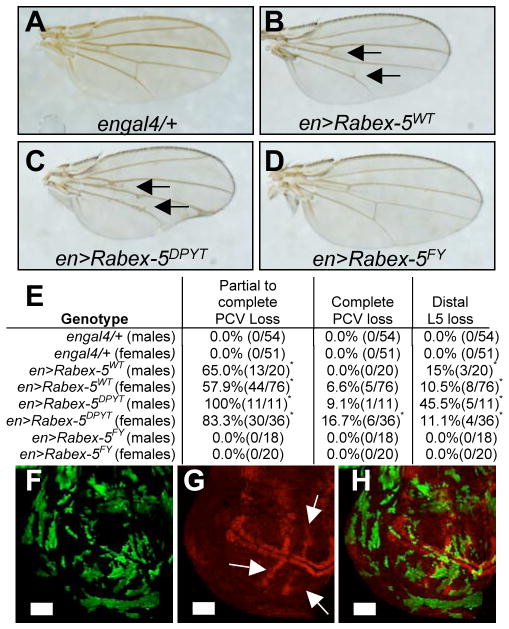
Rabex-5 displays GEF activity for Rab5 [8]. Rab5 regulates endocytosis and can influence receptor trafficking, so Rabex-5 could inhibit Ras via Rab5. In addition, Rabex-5 has a ubiquitin ligase domain [9–10]. We engineered Drosophila transgenic lines to express myc-tagged wild-type and mutant forms of Rabex-5. Rabex-5DPYT was designed to inactivate Rab5 GEF activity and Rabex-5FY to inactivate ubiquitin ligase activity within full-length protein (Supplemental Experimental Procedures). Both Rabex-5WT and Rabex-5DPYT expressed in the posterior wing resulted in loss of crossveins and loss of the distal part of longitudinal vein L5 (Fig. 3B–C, 3E, Fig. S3) compared to a control wing (Fig. 3A). In contrast, Rabex-5FY did not alter wing vein pattern (Fig. 3D–E). These results indicate that the ubiquitin ligase domain restricts wing vein formation. Clones of Rabex-5DPYT in wing discs showed loss of dpERK staining (Fig. 3F–H) establishing that wing vein loss likely resulted from decreased Ras signaling.
Figure 3. Rabex-5 regulates wing vein specification via its ubiquitin ligase domain.
A) Control engal4/+ wing. B) Expressing Rabex-5WT or (C) Rabex-5DPYT using engal4 resulted in crossvein loss (arrows). D) Expressing Rabex-5FY did not alter wing vein pattern. Female wings are shown. (E) Table of results from A–D. *statistically significant difference for transgenes compared to control (p<0.05). F–H) Wing clones expressing Rabex-5DPYT (green) show loss of dpERK staining (red in G–H, arrows in G). H) Merge of F, G. Scale bar, 50μm. Additional controls in Fig. S3.
Rabex-5 ubiquitin ligase domain blocks activated Ras in the eye
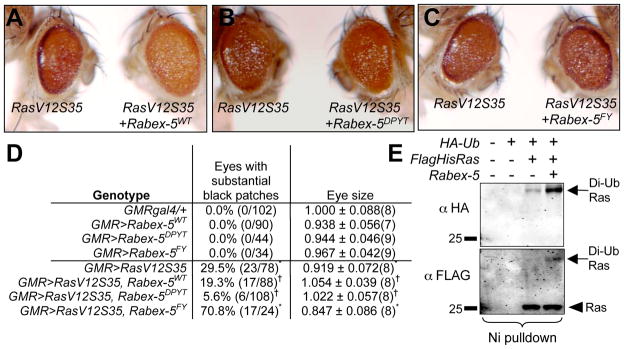
If Rabex-5 inhibits Ras, it could antagonize oncogenic Ras. To assay suppression, we expressed RasV12S35 in differentiating eye cells using conditions resulting in black tissue in 30% of eyes. Rabex-5WT and Rabex-5DPYT each reduced roughness, eye size, and black tissue phenotypes of RasV12S35 expressing eyes (Fig. 4A–B, 4D, Fig. S4). Change in roughness was subtle; eyes were scored double-blind by other lab members to confirm suppression in an unbiased manner. By quantitation, Rabex-5WT and Rabex-5DPYT statistically significantly suppressed the reduced eye size and black tissue phenotypes compared to RasV12S35 eyes (Fig. 4D, Fig. S4). Rescued eyes were statistically no different in size from controls. Rabex-5FY did not suppress RasV12S35 phenotypes (Fig. 4C–D, Fig. S4). Consistent with Fig. 2C, Rabex-5WT did not suppress appearance of black tissue in Rafgof eyes (Fig. S4). Enhancement of RasV12S35 by Rabex-5 loss (Fig. 2) and its suppression by the Rabex-5 ubiquitin ligase domain (Fig. 4) together with failure of Rabex-5 to suppress the Rafgof phenotype and its loss to enhance Rafgof (Fig. 2, Fig. 4, not shown) indicates that Rabex-5 restricts Ras signaling through its ubiquitin ligase domain most likely at the step of Ras.
Figure 4. Rabex-5 ubiquitin ligase domain suppresses oncogenic Ras.
A–C) Co-expressing Rabex-5WT (A, RasV12S35+Rabex-5WT, right) or Rabex-5DPYT (B, RasV12S35+Rabex-5DPYT, right) in differentiating eye cells suppressed RasV12S35 (left in A–C). C) Co-expressing Rabex-5FY (RasV12S35+Rabex-5FY, right) did not suppress RasV12S35. D) Summary table. Eye size was normalized to 1.000 for GMRgal4/+ controls and shown as a ratio to controls ± standard deviation. *statistically significant difference for Rabex-5 and/or RasV12S35 transgene expression compared to GMRgal4/+ control (p<0.05).† statistically significant suppression of GMR>RasV12S35 (p<0.05). L) Nickel pulldown of lysates from cells expressing tagged Ras, Ub, and Rabex-5. Rabex-5 expression increased Ras ubiquitination (right lane). Anti-HA (upper panel), anti-FLAG (lower panel) staining; di-ubiquitinated Ras, arrows; unconjugated Ras, arrowhead. Additional controls in Fig. S4.
Rabex-5 promotes Ras ubiquitination
The importance of a functional ubiquitin ligase domain for Ras inhibition suggests that ubiquitination of one or more substrates mediates effects on Ras signaling. Previously, we and others showed Ras ubiquitination reduces Ras signaling to ERK [21–22]. In S2 cells, Rabex-5 promoted Ras ubiquitination via its ligase domain (Fig. 4E, Fig. S4) but not Rab5 ubiquitination, showing specificity for Ras (Fig. S4). F25 Y26 mutation impairs ubiquitin binding [7], so Rabex-5 could bind ubiquitinated Ras to block downstream signaling, or Rabex-5 may affect Ras by targeting other substrates. However, the simplest model is that Rabex-5 constrains Ras signaling by direct Ras ubiquitination. In fact, mammalian Rabex-5 promotes Ras ubiquitination in a purified system [23].
CONCLUSIONS
Our findings represent the first evidence that Rabex-5 attenuates Ras signaling in vivo. We show that inhibition occurs at the step of Ras possibly by direct Ras ubiquitination, and such inhibition plays a critical role in control of size and tissue fate in Drosophila. Mammalian Rabex-5 promotes ubiquitination of HRas and NRas [23] suggesting that Rabex-5 restriction of Ras is highly conserved. Interestingly, we did not see supernumary R7 cells upon Rabex-5 loss (not shown), indicating that Rabex-5 antagonizes Ras in some but not all developmental contexts. Germline mutations increasing Ras signaling occur in “Rasopathies” [1–2] which are associated with patterning defects making Rabex-5 an attractive candidate to evaluate in developmental disorders. Indeed, Rabex-5 wing vein phenotypes resemble those seen in a Drosophila model of Noonan Syndrome [24]. Somatic mutations causing higher Ras activity are common in cancer [3–5]. Rabex-5 overgrowth phenotypes resemble those of mutation in many Drosophila tumor suppressors [25–26]. Thus, our study suggests that Rabex-5 ubiquitin ligase activity could serve as a novel means of tumor suppression.
EXPERIMENTAL PROCEDURES
Student unpaired t-tests compared length, weight, wing area (Fig. 1D) and eye size (Fig. 4D). Chi-square or Fisher’s exact tests compared PCV, LV, or eye phenotypes (Fig. 1K, 1P, 2C, 2H, 3E, 4D). S2 cells cultured at 25°C were transfected with pIE1–4-Flag-6HisRas, Actin-Gal4, UAS-HA-Ubiquitin and UAS-myc-Rabex-5 using Cellfectin (Invitrogen). Ubiquitin conjugates were isolated from cell lysates as described [21]. Flies were raised on standard media at 25°C unless otherwise stated. Larvae were dissected, stained, and imaged as before [21]. Extracts were prepared from individual larvae and Westerns were visualized with the Li-Cor Odyssey as before [21]. For genotypes and additional details, see Supplemental Experimental Procedures.
Highlights.
Rabex-5 loss causes Ras-dependent increased size, extra wing veins, and eye effects
The Rabex-5 ubiquitin ligase domain restricts Ras signaling in vivo
Rabex-5 promotes Ras ubiquitination
Supplementary Material
Acknowledgments
We thank D. Bar-Sagi, R. Cagan, M. Mlodzik, S. Aaronson, A. Jenny, Z.Q. Pan, M. O’Connell, K. Sadler-Edepli and their labs; S. Wong, R. Blair, E. DeMoll for assistance. Rase1b, Rase2f, UAS EgfrA887T, and UAS Rafgof were from the Bloomington Stock Center; Rabex-5IR from the VDRC. rlEMS698 and UAS RasV12S35 provided by M. Mlodzik. CMP is a Kimmel Scholar; this work was supported by NIH/NCI CA140451. Confocal imaging was done at the MSSM-Microscopy Shared Resource Facility (NIH-NCI shared resources grant 5R24 CA095823-04, NSF Major Research Instrumentation grant DBI-9724504, NIH shared instrumentation grant 1 S10 RR0 9145-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelb BD, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS-mitogen activated protein kinase signal transduction. Hum Mol Genet. 2006;15:R220–226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- 3.Young A, Lyons J, Miller AL, Phan VT, Alarcón IR, McCormick F. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 4.Duursma AM, Agami R. Ras interference as cancer therapy. Semin Cancer Biol. 2003;13:267–273. doi: 10.1016/s1044-579x(03)00040-3. [DOI] [PubMed] [Google Scholar]
- 5.Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Horiuchi H, Lippe R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattera R, Tsai YC, Weissman AM, Bonifacino JS. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J Biol Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- 9.Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7:1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- 10.Vaccari T, Rusten TE, Menut L, Nezis IP, Brech A, Stenmark H, Bilder D. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J Cell Sci. 2009;122:2413–2423. doi: 10.1242/jcs.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 12.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 13.Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, Hultmark D. A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci U S A. 2004;101:14192–14197. doi: 10.1073/pnas.0403789101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunner D, Oellers N, Szabad J, Biggs WH, 3rd, Zipursky SL, Hafen E. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 15.Sawamoto K, Okano H, Kobayakawa Y, Hayashi S, Mikoshiba K, Tanimura T. The function of argos in regulating cell fate decisions during Drosophila eye and wing vein development. Dev Biol. 1994;164:267–276. doi: 10.1006/dbio.1994.1197. [DOI] [PubMed] [Google Scholar]
- 16.Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001;104:687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- 18.Tam SY, Tsai M, Snouwaert JN, Kalesnikoff J, Scherrer D, Nakae S, Chatterjea D, Bouley DM, Galli SJ. RabGEF1 is a negative regulator of mast cell activation and skin inflammation. Nat Immunol. 2004;5:844–852. doi: 10.1038/ni1093. [DOI] [PubMed] [Google Scholar]
- 19.Kalesnikoff J, Rios EJ, Chen CC, Nakae S, Zabel BA, Butcher EC, Tsai M, Tam SY, Galli SJ. RabGEF1 regulates stem cell factor/c-Kit-mediated signaling events and biological responses in mast cells. Proc Natl Acad Sci U S A. 2006;103:2659–2664. doi: 10.1073/pnas.0511191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Raf acts downstream of the EGF receptor to determine dorsoventral polarity during Drosophila oogenesis. Genes Dec. 1994;8:629–639. doi: 10.1101/gad.8.5.629. [DOI] [PubMed] [Google Scholar]
- 21.Yan H, Chin M-L, Horvath EA, Kane EA, Pfleger CM. Impairment of ubiquitylation by mutation in Drosophila E1 promotes both cell-autonomous and non-cell-autonomous Ras-ERK activation in vivo. J Cell Sci. 2009;122:1461–1470. doi: 10.1242/jcs.042267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21:679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Lubkov V, Taylor LJ, Bar-Sagi D. Feedback Regulation of Ras Signaling by Rabex-5-mediated Ubiquitination. Curr Biol. 2010 doi: 10.1016/j.cub.2010.06.051. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oishi K, Gaengel K, Krishnamoorthy S, Kamiya K, Kim IK, Ying H, Weber U, Perkins LA, Tartaglia M, Mlodzik M, Pick L, Gelb BD. Transgenic Drosophila models of Noonan syndrome causing PTPN11 gain-of-function mutations. Hum Mol Genet. 2006;15:543–553. doi: 10.1093/hmg/ddi471. [DOI] [PubMed] [Google Scholar]
- 25.Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu Rev Genet. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- 26.Read RD, Bach EA, Cagan RL. Drosophila C-terminal Src kinase negatively regulates organ growth and cell proliferation through inhibition of the Src, Jun N-terminal kinase, and STAT pathways. Mol Cell Biol. 2004;24:6676–6689. doi: 10.1128/MCB.24.15.6676-6689.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.