Abstract
Mass spectrometry allows the elucidation of molecular details of the interaction domains of the individual components in macromolecular complexes subsequent to cross-linking of the individual components. Here, we applied chemical and UV cross-linking combined with tandem mass-spectrometric analysis to identify contact sites of the nuclear import adaptor snurportin 1 to the small ribonucleoprotein particle U1 snRNP in addition to the known interaction of m3G cap and snurportin 1. We were able to define previously unknown sites of protein–protein and protein–RNA interactions on the molecular level within U1 snRNP. We show that snurportin 1 interacts with its central m3G-cap-binding domain with Sm proteins and with its extreme C-terminus with stem-loop III of U1 snRNA. The crosslinking data support the idea of a larger interaction area between snurportin 1 and U snRNPs and the contact sites identified prove useful for modeling the spatial arrangement of snurportin 1 domains when bound to U1 snRNP. Moreover, this suggests a functional nuclear import complex that assembles around the m3G cap and the Sm proteins only when the Sm proteins are bound and arranged in the proper orientation to the cognate Sm site in U snRNA.
INTRODUCTION
Owing to the recently achieved improvements in protein-analytical techniques—in particular, in mass-spectrometric (MS) analysis—cross-linking is one method of choice for the direct identification of interacting components and their regions of interaction. The latter proves particularly useful in the absence of high-resolution overall structures, and when only partial structure information or the structure of only one component is available.
In this manner, various macromolecular interactions have been elucidated, including the spatial rearrangement of protein complexes (1–4), the tertiary folding of RNA (5), the stoichiometry and protein–protein interactions within larger macromolecular assemblies such as the exosome complex from yeast (6) and protein–DNA (7–10) and protein–RNA (11–14) interactions.
Here we used MS to investigate the quaternary structure of the U1 small nuclear ribonucleoprotein (U1 snRNP) complex together with the bound nuclear import factor snurportin 1 (SPN1) after protein–protein and protein–RNA cross-linking. U snRNPs (U1, U2, U4/U6.U5) are the key molecules in eukaryotic pre-mRNA splicing. They associate with numerous other spliceosomal proteins in a stepwise manner on the pre-mRNA. U snRNPs consist of a uridine-rich small nuclear RNA (U snRNA) and a set of particle-specific proteins. Common to all U snRNPs is a set of seven small, evolutionarily conserved proteins, the so-called Sm proteins (B/B′, D1, D2, D3, E, F and G). These seven proteins share a common conserved fold (15,16) through which they form a heptameric ring-like structure (17,18). This Sm ring is tightly associated with the conserved Sm site motif (PuAU4-6GPu) of the U snRNAs (19) and generate the so-called Sm core (20,21).
In eukaryotic splicing, the U1 snRNP is crucial for recognition of the 5′ splice site (SS) of pre-mRNA, which is achieved by base pairing of the 5′-end U1 snRNA with the 5′ SS. The base pairing is strengthened by additional protein–RNA interactions between the 5′ SS and members of the serine–arginine (SR) protein family and the U1 snRNP-specific protein C and (22). Due to its moderate protein complexity, the U1 snRNP is the best characterized snRNP particle in terms of the cell biological function of the various RNA stem-loops, protein–RNA interaction at the stem-loops, and its overall structure. Very recently, Pomeranz Krummel et al. (23) published the three-dimensional structure of partly truncated U1 snRNPs, derived from crystallization of reconstituted particles. The overall structure reveals the location of the 5′-end of the U1 snRNA, the Sm ring on the U1 snRNA and the U1-specific proteins U1-A, U1-70K and U1-C. Notably, the determination of the structures—with a resolution of only >5 Å—made use of protein–protein and protein–RNA cross-linking data from previous biochemical and structural studies to fit the U1-specific proteins [e.g. the U1-70K, (23)] and the Sm proteins (22,24) into the overall structure.
In higher eukaryotes, with the exception of U6, the biogenesis of all other U snRNPs requires a cytoplasmic maturation step in which the Sm proteins are assembled with the help of the ‘survival motor neuron’ complex [SMN complex, (20,21,25–29)] at the Sm site. Moreover, the monomethylated 5′ cap of the U snRNA (which was added co-transcriptionally in the nucleus) is converted into a trimethylguanosine (TMG) cap (m3G cap) by the action of hypermethylase trimethylguanosine synthetase 1 (Tgs1) (30,31). The TMG is one element of a nuclear localization signal within the U snRNP core that is required for the import of the assembled U snRNPs into the nucleus (32). It is specifically recognized and bound by the U snRNP import adaptor SPN1, which bridges the interaction to the actual import receptor importin-β (33). It is suggested that importin-β in complex with Sm proteins bound to U1 snRNA and SMN generates an import-competent snRNP complex that enters the nucleus (34). After import into the nucleus the complex dissociates, and the U1 snRNP becomes fully assembled and can participate in eukaryotic pre-mRNA splicing through recognition of the 5′ SS in pre-mRNAs (35).
SPN1 comprises three distinct domains: an N-terminal importin β-binding domain, a central m3G-cap-binding domain, and C-terminal residues so far found to be involved only in binding the export receptor CRM1 [(32,36,37); see also Figure 6]. To date, the only known interaction between SPN1 and U1 snRNP was the binding of the 5′-TMG cap of the U snRNA to SPN1 (32,37,38). Recent crystallographic and cross-linking studies have shown that the central domain of SPN1, encompassing residues 97–300, binds the TMG, and it has therefore been termed TMG-binding domain (32,37–39). The TMG-binding domain harbors two tryptophane residues, namely 107 and 276, which are in direct contact with the TMG cap (37). The N-terminal part of SPN 1 (positions 1–65) is important for the interaction with the import factor importin-β (40–43) as well as for the binding of SPN1 to the export receptor CRM1 (36). Additional biochemical and structural analysis demonstrated that for binding of the m3G cap the N-terminal part and the 60 C-terminal residues could be omitted (32,37). Interestingly, despite the fact that the N-terminal domain of SPN1 interacts with CRM1 and that in the co-crystal structure of SPN1 bound to CRM1 the majority of the C-terminus is non-structured, the extreme C-terminal 12 residues of SPN1 have been shown to interact with CRM1 too and are thus structured. Whether these C-terminal residues are also involved in the interaction with other elements of U snRNPs is not known. Furthermore, it is unknown whether SPN1 makes additional contacts with core components of U1 snRNPs (Sm proteins and U1 snRNA) or is merely in close proximity to them.
In this study, we are able to extend the structural model of U1 snRNP by mass-spectrometric mapping of the interaction regions between the import factor SPN1 and the U1 snRNA and the Sm proteins B and D3 as well as the interaction sites between the Sm protein D2 to U1 70K protein. The combination of protein–protein and protein–RNA cross-linking in native U1 snRNP bound to SPN1 and the detection of interacting regions between the components by mass spectrometry provides yet another example of the analytical power of mass spectrometry in structural research and in the elucidation of the spatial arrangement of components within large molecular assemblies. Furthermore, protein–protein cross-linking by using heterobifunctional reagents has turned out to be highly specific in mapping the spatial arrangement of proteins within a complex and also identifies such domains in proteins that were hitherto not known to be involved in protein–protein interactions.
MATERIALS AND METHODS
Purification of native U1 snRNA, U1 snRNPs and recombinant SPN1, reconstitution of [U1 snRNP–SPN1]
U1 snRNPs were purified from HeLa nuclear extract with mAb H20 (44) by standard procedures (45). Native U1 snRNA was isolated from U1 snRNPs after treatment of U1 snRNPs with proteinase K (final concentration 0.5 mg/ml) in the presence of 1% (v/v) SDS for 90 min at 37°C. RNA was extracted by chloroform/phenol treatment and precipitated from the aqueous phase with ethanol. Pellets were washed with 80% ethanol and dried in a SpeedVac. The sample was purified by urea-gel electrophoresis. Bands containing U1 snRNA were excised, and RNA was eluted with TNES buffer (20 mM Tris–HCl, pH 8.0; 300 mM NaCl; 5 mM EDTA; 0.1% SDS) and further extracted with chloroform/phenol as described above. SPN1 was purified as described by Strasser et al. (39).
Reconstituted [U1 snRNP–SPN1] was obtained by incubating 1.7 nmol recombinant SPN1 with 0.25 nmol native U1 snRNPs for 10 min on ice and then loading the mixture onto a Superdex 200 PC column (PC 3.2/30) running in SE buffer (20 mM Hepes–KOH, pH 7.9; 150 mM NaCl; 1.5 mM MgCl2) at a flow rate of 40 µl/min. The maximum sample volume per run was 50 µl. Fractions containing [U1 snRNP–SPN1] complexes were subsequently used for protein–protein cross-linking (see below). Alternatively, for protein–RNA cross-linking, [U1 snRNA–SPN1] binary complex was obtained by incubating 1.2 nmol (∼60 µg) recombinant SPN1 with 0.4 nmol (70 µg) native U1 snRNPs for 30 min on ice, and directly subjecting the complex to UV irradiation (see below). For large-scale cross-linking of SPN1 to native U1 snRNA, 5 nmol of U1 snRNA was incubated with ∼12 nmol recombinant SPN1 and processed as described below.
UV cross-linking and ESI and MALDI mass-spectrometric analysis of cross-linked peptide–oligonucleotides
Reconstituted [U1 snRNP–SPN1] and/or native U1 snRNA reconstituted with SPN1 were cross-linked at a concentration of 0.1–0.2 µg/µl according to Kühn-Hölsken et al. (46) in a volume of 1 ml (analytical scale) or 12.5 ml (preparative scale). UV cross-linking was carried out on ice for 5 min at a wavelength of 254 nm and the sample was further processed (precipitation with ethanol, trypsin digestion and purification by size-exclusion chromatography) as described by Kühn-Hölsken et al. (46). RNA-containing fractions were hydrolyzed with RNase A and T1, and subjected to LC-coupled tandem mass-spectrometric analysis on a hybrid triple quadrupole/linear ion-trap mass spectrometer (4000 QTRAP LC/MS/MS System, Applied Biosystems/MDS Analytical Technologies) coupled to a Tempo 1D nano LC System (Applied Biosystems/MDS Analytical Technologies). Chromatography, initial detection of the cross-links in negative ion mode by precursor-ion scanning and subsequent verification and structural analysis of peptide–RNA oligonucleotide cross-links in positive ion mode by Multiple Reaction Monitoring (MRM)-triggered product ion-scanning experiments were performed exactly as described recently (11).
MALDI-MS/MS analysis of peptide–RNA cross-links was performed after semi-preparative purification of cross-links derived from UV-irradiated [U1 snRNA–SPN1] on a microbore liquid chromatography system (SMART, GE Healthcare) as described by Kühn-Hölsken et al. (46). Purified fractions that contained cross-linked peptide–RNA oligonucleotides were mixed with DHB (10 mg/ml) as matrix and analyzed on a MALDI-ToF/ToF mass spectrometer (4800 MALDI-TOF/TOF Analyzer, Applied Biosystems/MDS Analytical Technologies) under standard conditions in positive-ion mode.
Enrichment of cross-linked peptide–RNA oligonucleotides by immobilized metal-affinity chromatography (IMAC) was performed after treatment of cross-linked [U1 snRNA–SPN1] complexes with trypsin and RNases A and T1 and separation of the products on a capillary LC system (140C microgradient system, Applied Biosystems). Capillary LC separation and subsequent IMAC enrichment and elution of bound species were carried out exactly according to Kühn-Hölsken et al. (14). Enriched species were analyzed by MALDI-ToF mass spectrometry (Reflex IV, Bruker) using DHB as matrix.
Sulfo–MBS cross-linking and MALDI-mass spectrometric analysis of cross-linked peptides
Cross-linking of reconstituted and purified [U1 snRNP–SPN1] complexes was performed with 5 µM sulfo-MBS (Pierce) for 30 min at room temperature (47). The cross-linking reaction was stopped by adding DTT and Gly-HCl to a final concentration of 10 and 5 mM, respectively, for 30 min at 37°C. Cross-linked samples were precipitated with 5 volumes of acetone, washed with 80% ethanol and dried in a SpeedVac. Pellets were dissolved in SDS sample buffer (with DTT) and loaded onto SDS-polyacrylamide gels. After electrophoresis, Coomassie-stained bands were excised, and cross-linked proteins were digested with trypsin according to Shevchenko et al. (48), except that no DTT/iodoacetamide treatment was performed. Peptides were extracted and separated on C18 reversed phase (RP) column (Nucleosil 100-5 C18, 200 × 0.075 mm, 5 µm, 100 Å, Macherey-Nagel) under standard conditions using acetonitrile (ACN) in water with 0.1% (v/v) trifluoroacetic acid as eluent, with a gradient from 7.5 to 37.5% ACN in 0.1% formic acid over 60 min; the flow rate was 250 µl/min. Eluting species were mixed on-line with α-cyano-4-hydroxycinnamic acid (CHCA, 10 mg/ml). 15-second fractions were collected with a Probot microfraction collector (LC Packings) on a stainless steel MALDI target and analyzed by using a MALDI-ToF/ToF mass spectrometer (4800 MALDI-TOF/TOF Analyzer, Applied Biosystems/MDS Analytical Technologies).
RESULTS
Protein–protein contacts between SPN1 and U1 snRNP
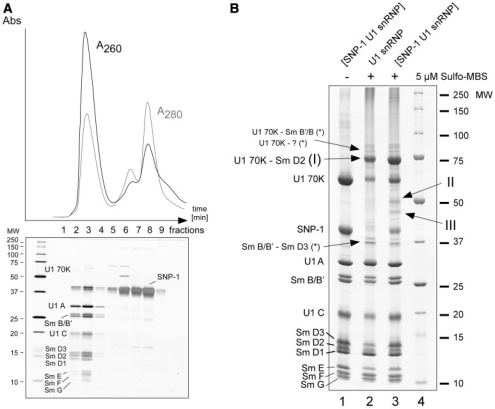
To investigate in detail how SPN1 interacts with native U1 snRNP, we first purified [U1 snRNP–SPN1] complex by size exclusion chromatography (SE) after incubation of purified native U1 snRNPs with recombinant SPN1 (Figure 1A). The respective SDS-PAGE analysis of the corresponding SE fraction showed that SPN1 forms a stable complex with U1 snRNP that elutes in fraction 2 and 3 and can be clearly separated from non-bound SPN1 (fractions 4–9, Figure 1A). Moreover, the SDS-PAGE analysis with silver staining of the proteins, although not strictly quantitative, revealed that SPN1 is bound to U1 snRNP probably in an equimolar manner (Figure 1A, lower panel). Fractions containing the complex were incubated with the cross-linking reagent sulfo-MBS and after precipitation of the thus probed complex, SPN1 and U1 snRNP proteins are separated by SDS-PAGE (Figure 1B). The chosen cross-linking reagent sulfo-MBS is a water-soluble heterobifunctional cross-linker consisting of an N-hydroxysuccinimide (NHS) ester and a maleimide group that links amines (e.g. ε-Lys) and sulfhydryl groups (e.g. Cys), thereby generating a covalent spacer of ∼7.3 Å between the cross-linked species. Sulfo-MBS has been used successfully in earlier studies to demonstrate the close proximity of the U1-specific proteins SmD3 to SmB/B′, 70K to SmD2 and U1-70K to protein C of the U1 snRNP (48).
Figure 1.
(A) Purification of the [U1snRNP-SPN1] complex. SPN1 and U1 snRNPs mixed in a 7 to 1 molar ratio were incubated on ice for 10 minutes prior to removal of excess of unbound SPN1 by size exclusion chromatography (SE). The fractions obtained were analyzed by SDS–PAGE and visualized by Silver staining. Upper Panel: Chromatogram of the SE elution. Lower panel: Proteins in the respective fractions were separated by SDS–PAGE. Fractions 2 and 3 containing the [U1snRNP-SPN1] complex were used for subsequent cross-linking experiments. Of note, U1 70K protein escapes detection by silver staining (45). (B) Protein–protein cross-linking of reconstituted [SPN1–U1 snRNP] complexes using sulfo-MBS. Sulfo-MBS-induced protein–protein cross-links were analyzed by SDS-PAGE and subsequently stained with Coomassie. Lane 1, non-cross-linked [U1 snRNP-SPN1]; lane 2, cross-linked [U1 snRNP] using 5 µM sulfo-MBS; lane 3, cross-linked [U1 snRNP-SPN1] using 5 µM sulfo-MBS. The positions of the individual U1 snRNPs proteins are indicated. Previously identified U1 70K–SmD2 (marked with I), U1 70K-Sm B/B′, and SmB–SmD3 cross-linked species are indicated (*). A potential cross-link of U1 70K with presumably U1 C protein is indicated as well. Arrows II and III mark additional putative protein–protein cross-links that were not observed in previous studies (48) and may thus be expected to be specific for SPN1. Lane 4, molecular-weight marker (MW).
In contrast to the experiment where sulfo-MBS was omitted in the cross-linking experiments (Figure 1B, lane 1), treatment of U1 snRNP and the [U1 snRNP–SPN1] complex resulted in several additional protein bands that represent potential cross-linked protein species (Figure 1B, compare lanes 2 and 3). Routine ESI-MS analysis revealed that the most prominent bands correspond to cross-linked U1-70K–D2, and the weaker ones to SmB/B′–SmD3, and U1 70K–Sm B/B′ as identified in earlier studies by western blot analysis (48) (data not shown). In the band migrating below the double cross-link band corresponding to U1 70K–Sm B/B′ we could only detect U1 70K by MS. This band might correspond to a U1 70K–U1 C cross-link as detected in earlier studies albeit with a different cross-linking reagent, i.e. DSB (49). The fact that we could not detect any other peptides suggests tentatively that the other cross-linked protein could be U1 C. It is known that U1 C produces only a very limited number of tryptic fragments and readily escapes detection by MS or is identified by 1 peptide only even when a large amount of U1 C is analyzed (50). In addition, two hitherto uncharacterized bands migrating at 40–50 kDa were detected in cross-linked [U1 snRNP–SPN1] (Figure 1B, lane 3, bands II and III) that are not observed in cross-linking studies using U1 snRNP only suggesting that these contain SPN1 cross-linked to U1 snRNP specific proteins. Routine ESI-MS analysis identified SNP1 and Sm proteins within these bands.
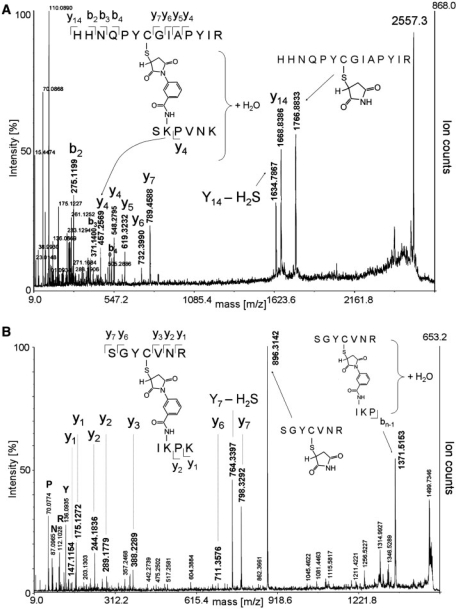
These together with the U1 70K–D2 band (Figure 1B, lane 1, band I) were analyzed in more detail by LC-offline MALDI MS/MS in order to identify the actual cross-linked peptide regions. Of note, the other weaker bands with cross-linked proteins were not further investigated mainly owing to the fact that their intensity was too weak to expect sequence information of the cross-linked protein regions of the corresponding proteins. To identify cross-linked peptides, the MS and MSMS spectra of those precursors that did not match with the peptide sequences of any U1 snRNP-specific protein and/or SPN1 (including all possible protein modifications) in the database search were inspected more closely. Potential peptide–peptide cross-links should display m/z values consistent with the masses of the cross-linked peptides and the mass increment of 199 Da caused by the cross-linker. We first annotated those MSMS spectra derived from the previously identified U1-70K–SmD2 cross-link (Figure 1B, lane 1, band I) for validation of cross-links. After trypsinization and elution of the peptides, we identified a precursor at m/z 2557.3 that matched precisely to the U1-70K tryptic fragment HHNQPYCGIAPYIR (1667.8 Da; positions 33–46 of U1-70K) cross-linked via sulfo-MBS (199.1 Da) to the Sm-D2 tryptic peptide SKPVNK (671.3 Da; positions 87–92) plus a water molecule (18.0 Da). Maleimides can add water through hydrolysis, thus being slowly converted to their cis-maleamic acid derivatives (51). Figure 2A shows the fragmentation pattern of this precursor. The product ion spectrum reveals a triplet of fragment ions resembling the fragmentation of peptides containing disulfide bridges, i.e. showing a characteristic loss of 34 Da (–H2S) (52). The fragment ion at m/z 1766.8 corresponds to the U1-70K tryptic peptide (HHNQPYCGIAPYIR) with a succinimide attached to the SH group of Cys-39. Owing to the instability of sulfide bonds in MALDI-MS/MS, a fragment ion corresponding to the non-modified peptide (y14, HHNQPYCGIAPYIR in Figure 2A) was observed. The sequence of the cross-linked peptide was confirmed by the detection of fragment ions in the lower m/z part of the spectrum (see Figure 2A). The sequence of the cross-linked SmD2 peptide was confirmed by its y4 fragment ion ([PVNK+H]+, Figure 2A), thus revealing Lys-88 as the actual cross-linked amino acid. Several other lysines in the immediate vicinity were also found to be crosslinked (SGKGKKKSKPVNK, positions 80–92 of Sm D2, cross-linked with Lys-83 and Lys-85–87) by manual inspection of the corresponding product ion spectra (data not shown).
Figure 2.
MALDI-MS/MS analysis of cross-linked peptides derived from reconstituted [U1 snRNP-SPN1] complexes. (A) MALDI product-ion spectrum of a peptide–peptide cross-link at m/z 2557.3 derived from the cross-linked proteins U1-70K–SmD2 after in-gel trypsinization (Figure 1B, band I). The sequences of the cross-linked peptides (U1-70K, HHNQPYCGIAPYIR; Sm-D2, SKPVNK) with their specific y-type ions are listed within the spectrum. (B) MALDI product-ion spectrum of a peptide–peptide cross-link at m/z 1499.7 derived from band II in Figure 1B. MS analysis identified SPN1 cross-linked to SmB/B′. The cross-linked sequences from SPN1 (SGYCVNR) and from SmB/B′ (IKPK) are given as insets within the spectrum. Sequences were determined according to the y-type fragment ions.
Accordingly, the specific fragmentation pattern of this particular cross-linked species was then used to scan for further peptide–peptide cross-links in product ion spectra derived from bands II and III (Figure 2B), expected to bear cross-linked SPN1. Indeed, a peptide–peptide cross-link at m/z 1499.7 corresponding to a tryptic peptide derived from SPN1 (positions 145–151, SGYCVNR, 797.4 Da) cross-linked via sulfo-MBS (199.1 Da) to an SmB/B′ tryptic peptide (positions 51–54, IKPK, M.W.:484.3 Da) was detected (Figure 2B). Note that the measured precursor mass again corresponds to that of the cross-link after addition of a water molecule (+18 Da) by the hydrolytic cleavage of the succinimide ring (which still leaves the cross-link intact, see above). A product ion spectrum of the cross-link precursor without the addition of water could also be obtained, but showed weak precursor and fragment ion intensities (data not shown).
The cross-linked band III in the SDS-PAGE (Figure 1B, lane 3) was analyzed in the same way and found to contain a protein–protein cross-link between SPN1 and SmD3. MSMS analysis showed that the cross-linking site on SPN1 encompassed the above peptide SGYCVNR with Cys-148 cross-linked. The cross-linking peptide in SmD3 was found to be SMoxKNKNQGSGAGR (positions 85–97, 1349.6 Da) with Lys-87 or -89 cross-linked (data not shown).
Taken together, two novel protein–protein interactions of SPN1 and proteins of the Sm ring have been identified, namely between SPN1 (145–151, Cys-148) and Sm B/B′ (Lys-52) as well as SmD3 (Lys-87 or Lys-89).
Protein–RNA contacts in the U1 snRNP–SPN1 complex
A wealth of data is available that show that SPN1 binds the hypermethylated terminal 5′ cap [m3GpppAUA (TMG)] of U1 snRNA in the U1 snRNP (32,37,38). The fact that SPN1 can be cross-linked to Sm proteins in native U1 snRNP raises the intriguing question of whether SPN1 might also form additional contacts to U1 snRNA apart from its interaction with the 5′ cap of U1 snRNP and, if so, which region(s) of SPN1 (besides the TMG) is/are involved.
To investigate potential protein–RNA contacts between SPN1 and U1 snRNP on a molecular level, we first applied Electrospray ionization (ESI) MS with multiple reaction monitoring (MRM). This method has been proven to be fast and highly sensitive for the specific detection of peptide-RNA oligonucleotides derived from UV irradiated samples (11). It is thus ideally suited to rapidly obtain an overview about potential cross-links in UV irradiated particles. This particular method is based on (i) precursor ion scanning experiments in the negative mode of the MS for detection of phosphate groups containing species (–79 Da), i.e. putative peptide-RNA cross-links derived after hydrolysis of UV irradiated particles with endoproteinases and nucleases and (ii) MRM triggered fragmentation of the peptide-RNA cross-linked species in the positive mode for unambiguous sequence determination of the cross-linked peptide and RNA moiety.
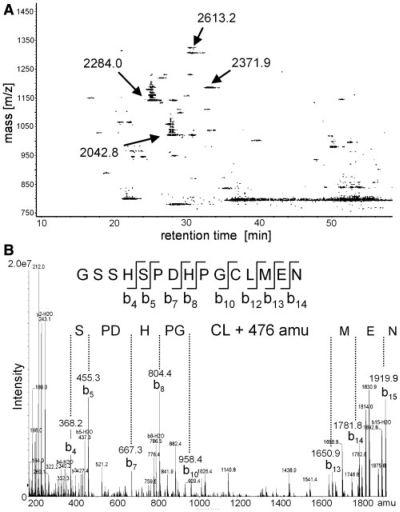
Figure 3A shows the precursor ion-scanning experiments of UV irradiated [U1 snRNP–SPN1] after hydrolysis of the protein moiety with trypsin and the RNA moiety with ribonucleases A and T1. Shown are the masses of those species over LC elution time that produce PO−3 ions in the negative mode upon fragmentation in the MS. The precursor-ion-scanning experiments revealed several putatively cross-linked species that had not been observed in earlier UV cross-linking experiments with U1 snRNPs (11). The most prominent ones are observed at m/z values of 2042.8 Da., 2284.0 Da, 2371.9 Da and 2613.2 Da, respectively (Figure 3A). These species were selected for MRM-triggered product ion-scan experiments to acquire sequence information on the cross-linked peptides. An example is shown in Figure 3B for the precursor with the molecular mass of 2042.8 Da. The corresponding peptide GSSHSPDHPGCLMEN (as identified by product ion spectra) represents the C-terminal 15 residues of SPN1 (positions 346–360) with an ‘extra mass’ of 476 Da. MRM-triggered product ion experiments of the other species also revealed the C-terminus of SPN1 (2284.8 Da, IKGSSHSPDHPGCL- MEN, positions 344–360, data not shown) and the C-terminus with a cross-linked adenosine (2371.9 Da, GSSHSPDHPGCLMEN + A (329 Da); 2613.2 Da, IKGSSHSPDHPGCLMEN + A). Surprisingly, all sequencing results revealed an extra mass of 476 Da that does not match with the mass of any (modified) mono- or dinucleotide.
Figure 3.
Detection and structural analysis of [U1 snRNA-SPN1] cross-links by LC/MS/MS. (A) Precursor-ion-scanning experiments for m/z 79 (PO−3) in negative-ion mode to determine the molecular weight of putative cross-linked species (see text and ‘Materials and Methods’ section for details). Observed m/z values of PO−3-generating precursors are plotted against their retention time. The corresponding molecular weights listed (MW = 2042.8 Da, 2284.0 Da, 2371.9 Da and 2613.9 Da) are selected in a second experiment for MRM-triggered MSMS analysis in positive-ion mode (see text and ‘Materials and Methods’ section for details). (B) MRM-triggered product-ion scan of a cross-linked precursor (MW = 2042.3 Da) derived from SPN1. MRM was triggered by the monitoring production of specific RNA-base fragment ions, i.e. [M + nH]n+ → [{111, 112, 135, 151} + H]+. The sequence of the cross-linked peptide (GSSHSPDHPGCLMEN) with the corresponding fragment ions is given within the deconvoluted spectrum. The deciphered sequence shows an extra mass of 476 Da that cannot be explained by any combination of RNA mono- and/or dinucleotides.
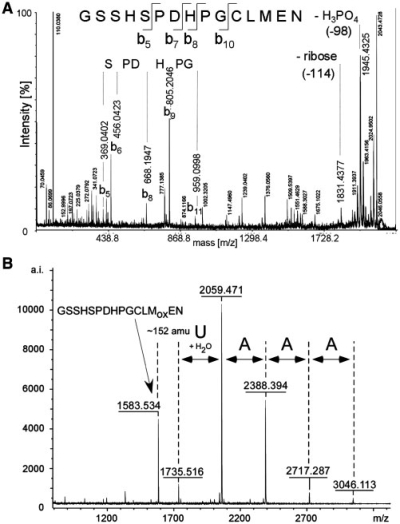
Further investigation of these cross-links by ESI-MS was not possible. MS3 experiments of a fragment ion containing the extra mass gave no further result about the nature of the extra mass (data not shown). We therefore applied matrix assisted laser desorption/ionization (MALDI)-MSMS as alternative fragmentation strategy in order to investigate whether the extra mass of 476 Da belonged to a cross-linked RNA moiety. We performed MALDI product ion-scan experiments (MSMS) on the cross-linked precursor ([M+H]+ m/z 2043.8) that was purified under semi-preparative conditions for that purpose [according to (46)]. Product ion-scan analysis revealed a characteristic neutral loss of H3PO4 (98 Da) and ribose (114 Da) under high energy collision-induced decay (CID) conditions (Figure 4A), demonstrating that the extra mass must include a ribophosphate moiety.
Figure 4.
IMAC enrichment for the analysis of peptide–RNA oligonucleotide cross-links derived from UV-irradiated [U1 snRNP-SPN1] complexes. (A) MALDI-MSMS analysis of isolated cross-linked precursor (MW = 2042.3 Da). The fragmentation of the cross-link in the MALDI reveals that the extra mass is at least partly due to nucleic acids, as the loss of phosphate and ribose is seen clearly within the spectrum. Peptide-specific fragmentation is observed as well. (B) MALDI MS analysis of an enriched and semi-preparative purified cross-link species (see ‘Materials and Methods’ section) encompassing the cross-linked SPN1 tryptic fragment GSSHSPDHPGCLMoxEN. The MALDI MS spectrum shows mass signals for the non-cross-linked oxidized peptide and various corresponding cross-linked peptide-oligoribonucleotides. The differences in mass between the non-cross-linked and cross-linked species that correspond to nucleotides are listed; they reveal the cross-linked RNA moiety (AAAUp). The cross-link still has an extra mass of ∼152 Da that cannot be explained (see also text).
Additional information about the nature of the putatively cross-linked RNA was obtained by isolation of this cross-linked species under semi-preparative conditions combined with strategies to enrich it by immobilized metal-affinity chromatography [IMAC (14)]. Enrichment of cross-linked species by IMAC usually facilitates the detection of the larger hetero-conjugates in the mass spectrometer (7,9,14). MALDI-mass spectrometric analysis of an enriched sample containing the putatively cross-linked precursor in its methionine-oxidized form ([M+H]+ m/z 2059.5) demonstrates that this species is derived from a larger precursor with a cross-linked RNA moiety A3U ([M+H]+ m/z 3046.1, Figure 4B). Surprisingly, an additional signal corresponding to the non-cross-linked peptide moiety in its methionine-oxidized state ([M+H]+ m/z 1583.5 Da) was detected as well. This suggests a stepwise loss of nucleotides and that the occurrence of the non-cross-linked peptide is due to the acidic elution conditions of the cross-linked species from the IMAC beads, i.e. with phosphoric acid. The observed ladder-like pattern is due to acid hydrolysis of the larger cross-linked species. Importantly, the mass difference between m/z 2059.5 (corresponding to GSSHSPDHPGCLoxMEN plus 476 Da) and m/z 1735.5 exactly matches a single uridine nucleotide plus water and would therefore explain the loss of phosphate and ribose observed in the product-ion-scan experiments (Figure 4B). However, both cross-linked species still reveal a mass difference of ∼152 Da that cannot be easily explained, so that its nature remains unclear (see ‘Discussion’ section).
Despite this ambiguity, there are only two sequence stretches in U1 snRNA that harbor an oligonucleotide sequence with a nucleotide composition of three As and one U. These are positions 102–105 in stem-loop III (SL III, 5′AAAUp-3′) and positions 123–126 adjacent to the Sm site (5′-AUAAp-3′). Importantly, the observed loss of one uridine plus a water molecule in MALDI-MS argues that the 3′ terminus of the cross-linked oligonucleotide is uridine. Therefore, SL III of U1 snRNA must be the region cross-linked to SPN1.
Taken together, our protein–RNA cross-linking data demonstrate that the C-terminal region of SPN1 [presumably with C and/or L as the actual cross-linked amino acid(s)] is cross-linked to SL III of U1 snRNA.
DISCUSSION
In order to identify further interactions in [SPN1–U1 snRNP] complexes, we performed protein–protein and protein–RNA cross-linking studies. Using mass spectrometry, previously unknown sites of interaction between SPN1 and U1 Sm core proteins (i.e. the Sm proteins associated with Sm site RNA) and the U1 snRNA were identified at the molecular level. We identified a SPN1 peptide encompassing positions 145–151 (SGYCVNR) cross-linked to SmB and SmD3 and a peptide containing the 15 C-terminal residues of SPN1 cross-linked to SL III in U1 snRNA. The results provide valuable information on the site of the U1 snRNP that interacts with SPN1 and on the mutual orientation of the components of U1 snRNP. Our results can be integrated into the currently available 3D structure of U1 snRNP (23). This work is a further demonstration of the analytical power of mass spectrometry in protein–protein and protein–RNA cross-linking studies, and of the potential of its combination with other structural information, allowing one to draw conclusions about the spatial arrangements of components within macromolecular complexes.
Here we discuss the results in more detail in the context of (i) technical aspects of UV-induced protein–RNA cross-linking, (ii) interactions between SPN1, the Sm core and U1 snRNA, (iii) novel cross-linking sites in Sm proteins and (iv) location of SPN1-binding sites on the U1 snRNP.
Methodical considerations about MS-based detection of protein–RNA cross-links
Previous mass-spectrometric analysis of UV-induced cross-links between a protein or peptide and its RNA revealed that UV cross-linking is additive, i.e. the mass of the cross-linked product is the sum of the masses of the cross-linked peptide moiety and the cross-linked oligoribonucleotide (11–14,53). In the current study, we identified the C-terminal tail of SPN1 cross-linked to SL III of U1 snRNA (5′-A102AAU105p-3′) in a variety of MS experiments including precursor-ion scanning and MRM ESI mass spectrometry, as well as MALDI mass spectrometry after IMAC enrichment of cross-linked species. In this case, however, the entire mass of the cross-linked conjugate does not equal the sum of the masses of the cross-linked peptide (GSSHSPDHPGCLMEN, MW = 1566.6 Da) and RNA (AAAUp, MW = 1311.2 Da) moieties. We show that this cross-link harbors an additional mass of ∼152 Da (151.9572 Da as determined by MS on Orbitrap instrument, data not shown). Surprisingly, we have observed a similar phenomenon in other UV-irradiated RNP particles, where peptides that harbor a cysteine-residue are cross-linked to RNA. These were the U4/U6-specific protein 61K/hPrp31 bound to U4atac snRNA (H.U. and M. Wahl, unpublished results) and Sm proteins cross-linked to U1 and U2 snRNA (H.U. and F.R., manuscript in preparation). However, the exact chemical nature of the ‘extra mass’, and thus of this particular cross-link, still remains unclear as these cysteine-based cross-links seem to occur exclusively in native particles. A possible explanation for the crosslink derives from a study recently published by Chen et al. (54), who showed that modified guanosine base is reactive towards cysteine side chains. Indeed, the cross-linked stretch of U1 snRNA in SL III (5′-AAAUp-3′) contains a guanosine adjacent to the uracil (nucleotide position 106 in SL III, 5′102AAAUG-1063′). Moreover, in the crystal structure of the U4/U6-specific protein 61K/hPrp31 bound to U4atac snRNA the identified cross-linked cysteine in 61K/hPrp31 is in direct contact with a guanosine in the 5′ stem-loop of U4atac snRNA (M. Wahl, personal communication).
SPN1-U1 snRNA and Sm core interactions
Previous biochemical studies have shown that SPN1 interacts with the TMG of U1 snRNA/snRNP (32,38). Furthermore, UV cross-linking studies revealed that the TMG cross-links efficiently to SPN1, and that the cross-link cannot be inhibited by U1 snRNA/snRNP that lacks the 5′ TMG. Together, all these previous results suggest that no additional domain in U1 snRNA/snRNP contacts SPN1 except for the TMG. On the other hand, UV induced cross-linking has been demonstrated to monitor additional molecular interactions that are less strong and are located apart from the actual nucleotide-binding domain of a protein as it is for example shown in the protein–RNA complex consisting of 61K/hPrp31 and a 15.5K protein bound to the 5′ stem-loop of U4 and U4atac snRNA (55,56).
In the present study, we were not able to identify cross-linked residues or regions in SPN1 encompassing the TMG-binding site. This was probably due to the fact that the tryptophan residues 107 and 276 (that are crucial for the interaction with the TMG) are located in extremely large tryptic peptides and thus exhibit poor chromatographic and ionization yields, which hamper the detection of cross-linking within these regions by LC–MS/MS. Interestingly, we found the C-terminal residues of SPN1 to be cross-linked to SL III of U1 snRNA. The cross-linking is achieved by the last 15 residues of SPN1 and therefore SL III and the TMG-cap of the U snRNA must be separated by a distance that may be bridged by the TMG-binding domain and the C-terminus of SPN1 (see below and Figure 6C). The extreme C-terminus of SNP1 has also been recently shown to bind to CRM1 in a fully assembled export complex (36), suggesting a possible dual function of the C-terminal tail of SNP1, i.e. RNA interaction and binding to the export complex.
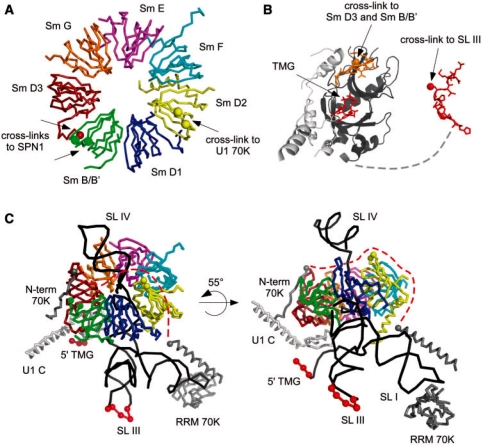
Figure 6.
Overall locations of protein–protein and protein–RNA cross-linking sites in Sm proteins, SPN1 and within the U1 particle. (A) Location of the protein–protein cross-linking sites in SmD2, SmD3, SmB/B′. The Sm proteins are depicted as a heptameric ring structure in various colors. Images were generated using PYMOL, and the coordinates of the respective proteins within the published U1 snRNP structure (22) available in the PDB (PDBid: 3CW1). No coordinates are available for loop 4 of SmD2, SmB/B′ or for the C-terminus of SmD3. Note that only coordinates from the α-C atoms were available in the PDB database. Yellow spheres mark the N- and C-terminal borders of loop 4 in SmD2, which was found to be cross-linked to U1-70K. Green spheres mark the borders of the corresponding loop region 4 in SmB/B′ protein, which was found to cross-linked to SNP1. The dark red sphere marks the last C-terminal amino acid in SmD3 for which coordinates were available. The subsequent amino acids in SmD3 were found to be cross-linked to SPN1. (B) Domain structures of SPN1 with cross-linking sites. Images were generated using PYMOL and the coordinates of SPN1 in the PDB (PDBid: 1XK5). SPN1 is shown with its co-crystallized cap-analogue structure (36,37). The orientation of the domains of SPN1 is as found in complex with CRM1 (PDBid: 3GJX) with the structured C-terminus of SPN1. The N-terminal IBB domain is colored in light grey, and the m3-Cap binding domain of SPN1 is shown in dark grey with the TMG cap in red superposed form (PDBid: 1XK5); the cross-linking sites to SmB/B′ and SmD3 are marked in orange with the actual cross-linked amino acid shown as an orange sphere. The cross-linking site to stem-loop III of U1 (SL III) is shown in red, and the actual cross-linked amino acid as a red sphere. The flexible region linking the central domain to the C-terminal residues is indicated as black dashed line. (C) Location of protein–protein and protein–RNA cross-linking sites identified on U1snRNPs (according to PDBid: 3CW1). The RNA is shown in black, with the area of cross-link in SL III and TMG are depicted as red spheres. The stem-loops I (SL I), SL III, and SL IV are indicated. Note that stem-loop II (SL II) has been truncated in order to obtain crystal structure (22). The Sm proteins are shown as in panel A using the same color code, whereas the U1-C and U1-70k are depicted in light and dark grey, respectively. The missing residues in U1-70k are indicated as dotted red line. Beginning and end point of cross-linked peptides are shown in colored spheres according to panel A.
It seems that cross-linking of SPN1 to the Sm proteins requires correct interaction of SPN1 with the TMG of the U1 snRNA, since no protein–protein cross-linking was observed when U1 snRNPs lacking the TMG were incubated with an excess of SPN1 and exposed to cross-linking conditions (data not shown). These results imply that the specific binding of SPN1 to the TMG is a prerequisite for the formation of additional contacts to the U1 snRNP.
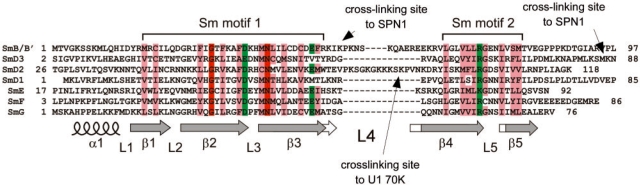
The second cross-link site in SPN1 was a peptide composed of residues 145–151 of SPN1, which cross-link to residues 51–54 of SmB/B′ [loop 4 according to Kambach et al. (18); see also Figure 5] and also to the C-terminal domain of SmD3 (residues 85–97; see Figure 5). SPN1 residues 145–151 are located within the TMG-binding domain of SPN1 adjacent to the actual TMG-binding site (see also Figure 6C), suggesting an additional structural and/or functional role of this domain. Since this site binds not only to SmB/B′ but also to SmD3, our data suggest a spatial proximity between the L4 loop region of SmB/B′ and the C-terminal region of SmD3. However, the structural analysis of the recombinant partial U1 snRNP indicates that precisely these regions in SmB/B′ and SmD3 are highly flexible, and are thus missing in the observed structure (22); however, this does not preclude being close, or adjacent, to one another.
Figure 5.
Location of protein–protein cross-linking sites identified in Sm proteins. Sequence alignment of the Sm1 and Sm2 motifs of the human Sm proteins B/B′, D2, D1, D3, E, F and G are according to Hermann et al. (15). The topology of the secondary structure was adapted from the crystal structures of SmB-D3 and SmD1-D2 protein complexes (18). Conserved amino acids are highlighted as follows: uncharged, hydrophobic residues (L, I, V, A, F, W, Y, C, M) are in light red; acidic and basic amino acids (D, E, R, K) are in green, and 100% conserved amino acids in dark red. Secondary structure elements in the Sm proteins are depicted as follows: α = helix; β = beta-sheet, L = loop. Arrows mark the cross-linked amino acids in the variable loop 4 of the protein sequences of SmD2 and SmB and in the C-terminal part of SmD3.
Interestingly, the site of cross-linking of SmD3 to SPN1 is located immediately adjacent to the first dimethyl RG dipeptide in SmD3 (57). Dimethylated RG repeats in the Sm proteins have been shown to be crucial for the interaction with Sm assembly factor SMN (58,59) as this particular protein interacts with dimethylarginine residues within RG dipeptide repeats of the Sm proteins B/B′, D3 and D1 (58,60).
Protein–protein interaction through variable loop regions of the Sm proteins
Phylogenetic and crystallographic studies initially demonstrated that the seven eukaryotic Sm proteins (B/B′, D1, D2, D3, E, F and G) share a common fold with two evolutionarily conserved motifs, Sm1 and Sm2 (15,16,18; Figure 5). Protein–RNA cross-linking (61) and crystallographic studies with Sm-like (LSm) proteins from Archaea (62,63) and Sm homologues from E. coli (64) with short oligoribonucleotides further demonstrated that the loop regions connecting β-strands 2 and 3 (loop L3) and 4 and 5 (loop L5), respectively, together with their adjacent amino acids from the respective β-strands, are in contact with the U-rich Sm RNA site.
Little is known about the structural and functional features of the variable loop regions of the Sm and LSm proteins that connect the Sm motifs, i.e. loop 4 and its adjacent β-strand regions. Indeed, this region of the Sm proteins (together with the C-terminal region of the proteins, see also below) exhibits the highest variability among the seven Sm proteins both in sequence and in size (Figure 5). We were able to show that these regions, in which the Sm proteins B and D2 are extended, are accessible to protein–protein cross-linkers, so that SmB is subsequently cross-linked to SPN1 and SmD2 to U1-70K (Figures 4 and 5).
Moreover, recent structural studies with the yeast Sm-like protein LSm3, which forms a homo-octameric ring, have shown that LSm3 is capable of binding to other LSm proteins (LSm2, 5 and 6) through interactions involving the variable region b3–L4–b4 (65). Our cross-linking data strongly support the assumption of a composite protein–protein interaction surface encompassing loop 4 and β-strands of the Sm motif, and they thus add another feature to the loop regions of the Sm fold, showing that these are involved in protein contacts via their largely flexible domains (Figure 5).
Overall localization of crosslinked proteins
Remarkable overall structural information, albeit at moderate to low resolution, has been obtained on native intact U1 snRNPs by electron cryomicroscopy (23) and on an in vitro assembled truncated, but active form of U1 snRNPs by X-ray crystallography (22). To date, only the coordinates of the protein C-alpha and RNA phosphorus atoms of U1 snRNP are available in the databank, thus preventing any detailed structural interpretation of our cross-linking experiments or a molecular docking approach. However, detailed structural information is available on the individual domains of SPN1 interacting with their respective binding partners, namely the N-terminal domain bound to importin β, the central TMG-cap binding domain in complex with a m3GpppG dinucleotide, and the C-terminal tail bound to CRM1 (36,37,41,43).
Besides the TMG-cap binding site, the central domain of SPN1 harbors the newly mapped interaction site for SmB/B′ and SmD3 (Figure 6B), which are in close proximity within the doughnut-like ring structure of the Sm proteins in the U1 snRNP (Figure 6A and C). The structural analysis of the recombinant partial U1 snRNP indicates that these regions in SmB/B` and SmD3 are highly flexible, and are thus missing in the reported structure [see above (22)]. Nevertheless, these loops could be arranged in a conformation that brings the interacting residues into close proximity to SPN1 (Figure 6C).
Structural information on the C-terminal tail of SPN1 is available for the last 12 residues bound to CRM1 (36). The absence of the residues connecting the TMG-binding domain and these C-terminal residues in the known structure indicates a high conformational flexibility, suggesting that additional interaction sites could be at various distances from the TMG-binding domain of SPN1 (Figure 6B). Interestingly, exactly these same C-terminal residues in SPN1 have been mapped as interaction partners for nucleotides in SL III of the U snRNA, its distance to the cap-binding site fulfilling the relaxed spatial criterion required for bridging the distance between the TMG and SL III (data not shown).
Taken together, our cross-linking data are in excellent agreement with the recent low-resolution U1 snRNP crystal structure (22), and the overall quaternary arrangement of its components, i.e. the orientation of the Sm proteins and in particular the location of the 5′ m3G cap and stem-loop III on the so-called ‘back’ side of U1 snRNP (Figure 6C). These data support earlier suggestions that structurally and functionally important regions of the U1 snRNP are located at one side of the U1 snRNP. Probing experiments with human U1 and U2 snRNPs bound to pre-mRNA in an early spliceosomal complex demonstrated that only the backside is accessible to HO radicals generated by the BABE reagent incorporated into U2 snRNA. On the basis of these results, it was suggested that this particular site of U1 snRNP is close to U2 snRNP when assembled into an early functional spliceosomal complex (49). On the basis of our crosslinking data, this side must be (at least partly) covered by SPN1 maybe as part of the large import machinery, which dissociates when it reaches the nucleus, to expose the back of the particle so that U1 snRNP can carry out its function in pre-mRNA splicing. Importantly, the location of SPN1 on this side also leaves enough freedom to accommodate the additional factors of the U snRNP import machinery around the N-terminal part of SPN1 and the C-terminal tails of the Sm proteins D3, B and D1.
Moreover, cross-linking of the N-terminal peptide of U1-70K encompassing positions 33–46 to the Sm-D2 tryptic peptide encompassing positions 87–92 (loop 4, see above) is highly consistent with the proposed path of the N-terminus of U1 70K in the three-dimensional structure of Pomeranz Krummel et al. (22). In this structure, the path of the U1 70K N-terminus was traced by crystallization of seven SeMet derivatives of single methionine mutation at positions L9M, I19M, E32M, I41M, E49M, E61M, and I75M. In the electron density of the crystal structure, the thus substituted N-terminus is between I41(M) and E49(M) in contact to protein Sm D2. These data again prove the high specificity of the protein–protein cross-linking approach in order to decipher the spatial arrangements within molecular assemblies.
FUNDING
EURASNET YIP grant to H.U. within the 6th EU framework. Funding for open access charge: Dr Henning Urlaub Max-Planck-Institute for Biophysical Chemistry Bioanalytical Mass Spectrometry Group.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Monika Raabe and Uwe Plessmann for their excellent help in sample preparation and mass spectrometric analyses.
REFERENCES
- 1.Ihling C, Schmidt A, Kalkhof S, Schulz DM, Stingl C, Mechtler K, Haack M, Beck-Sickinger AG, Cooper DM, Sinz A. Isotope-labeled cross-linkers and Fourier transform ion cyclotron resonance mass spectrometry for structural analysis of a protein/peptide complex. J. Am. Soc. Mass Spectrom. 2006;17:1100–1113. doi: 10.1016/j.jasms.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 2.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell Proteomics. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nat. Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein–protein interactions. J. Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- 5.Yu ET, Hawkins A, Eaton J, Fabris D. MS3D structural elucidation of the HIV-1 packaging signal. Proc. Natl Acad. Sci. USA. 2008;105:12248–12253. doi: 10.1073/pnas.0800509105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyer H, Geyer R, Pingoud V. A novel strategy for the identification of protein–DNA contacts by photocrosslinking and mass spectrometry. Nucleic Acids Res. 2004;32:e132. doi: 10.1093/nar/gnh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen ON, Barofsky DF, Young MC, von Hippel PH, Swenson S, Seifried SE. Direct observation of UV-crosslinked protein-nucleic acid complexes by matrix-assisted laser desorption ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1993;7:496–501. doi: 10.1002/rcm.1290070619. [DOI] [PubMed] [Google Scholar]
- 9.Steen H, Petersen J, Mann M, Jensen ON. Mass spectrometric analysis of a UV-cross-linked protein-DNA complex: tryptophans 54 and 88 of E. coli SSB cross-link to DNA. Protein Sci. 2001;10:1989–2001. doi: 10.1110/ps.07601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiss A, Barre O, Michaud-Soret I, Forest E. Characterization of the DNA-binding site in the ferric uptake regulator protein from Escherichia coli by UV crosslinking and mass spectrometry. FEBS Lett. 2005;579:5454–5460. doi: 10.1016/j.febslet.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 11.Lenz C, Kuhn-Holsken E, Urlaub H. Detection of protein-RNA crosslinks by NanoLC-ESI-MS/MS using precursor ion scanning and multiple reaction monitoring (MRM) experiments. J. Am. Soc. Mass Spectrom. 2007;18:869–881. doi: 10.1016/j.jasms.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, Urlaub H, Wahl MC. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urlaub H, Hartmuth K, Kostka S, Grelle G, Luhrmann R. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA-protein contacts in native U1 and U4/U6.U5 snRNPs. J. Biol. Chem. 2000;275:41458–41468. doi: 10.1074/jbc.M007434200. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn-Holsken E, Dybkov O, Sander B, Luhrmann R, Urlaub H. Improved identification of enriched peptide RNA cross-links from ribonucleoprotein particles (RNPs) by mass spectrometry. Nucleic Acids Res. 2007;35:e95. doi: 10.1093/nar/gkm540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann H, Fabrizio P, Raker VA, Foulaki K, Hornig H, Brahms H, Luhrmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kambach C, Walke S, Nagai K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 1999;9:222–230. doi: 10.1016/s0959-440x(99)80032-3. [DOI] [PubMed] [Google Scholar]
- 18.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 19.Branlant C, Krol A, Ebel JP, Lazar E, Haendler B, Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Patel SB, Bellini M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 2008;36:6482–6493. doi: 10.1093/nar/gkn658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 25.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 26.Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell Biol. 2002;22:6533–6541. doi: 10.1128/MCB.22.18.6533-6541.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 28.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 29.Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- 30.Monecke T, Dickmanns A, Ficner R. Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1. Nucleic Acids Res. 2009;37:3865–3877. doi: 10.1093/nar/gkp249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouaikel J, Bujnicki JM, Tazi J, Bordonne R. Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 2003;31:4899–4909. doi: 10.1093/nar/gkg656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Luhrmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palacios I, Hetzer M, Adam SA, Mattaj IW. Nuclear import of U snRNPs requires importin beta. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum. Mol. Genet. 2002;11:1785–1795. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 36.Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324:1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 37.Strasser A, Dickmanns A, Luhrmann R, Ficner R. Structural basis for m3G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1. EMBO J. 2005;24:2235–2243. doi: 10.1038/sj.emboj.7600701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bahia D, Avino A, Darzynkiewicz E, Eritja R, Bach-Elias M. Trimethylguanosine nucleoside inhibits cross-linking between Snurportin 1 and m3G-CAPPED U1 snRNA. Nucleosides Nucleotides Nucleic Acids. 2006;25:909–923. doi: 10.1080/15257770600793901. [DOI] [PubMed] [Google Scholar]
- 39.Strasser A, Dickmanns A, Schmidt U, Penka E, Urlaub H, Sekine M, Luhrmann R, Ficner R. Purification, crystallization and preliminary crystallographic data of the m3G cap-binding domain of human snRNP import factor snurportin 1. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1628–1631. doi: 10.1107/S0907444904015380. [DOI] [PubMed] [Google Scholar]
- 40.Huber J, Dickmanns A, Luhrmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J. Biol. Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 42.Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Luhrmann R, Gorlich D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wohlwend D, Strasser A, Dickmanns A, Ficner R. Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus. J. Mol. Biol. 2007;374:1129–1138. doi: 10.1016/j.jmb.2007.09.065. [DOI] [PubMed] [Google Scholar]
- 44.Luhrmann R, Appel B, Bringmann P, Rinke J, Reuter R, Rothe S, Bald R. Isolation and characterization of rabbit anti-m3 2,2,7G antibodies. Nucleic Acids Res. 1982;10:7103–7113. doi: 10.1093/nar/10.22.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bringmann P, Luhrmann R. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J. 1986;5:3509–3516. doi: 10.1002/j.1460-2075.1986.tb04676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhn-Holsken E, Lenz C, Sander B, Luhrmann R, Urlaub H. Complete MALDI-ToF MS analysis of cross-linked peptide-RNA oligonucleotides derived from nonlabeled UV-irradiated ribonucleoprotein particles. RNA. 2005;11:1915–1930. doi: 10.1261/rna.2176605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelissen RL, Will CL, van Venrooij WJ, Luhrmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994;13:4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 49.Donmez G, Hartmuth K, Kastner B, Will CL, Luhrmann R. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol. Cell. 2007;25:399–411. doi: 10.1016/j.molcel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 50.Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA. 2009;15:153–175. doi: 10.1261/rna.1332609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hermanson GT. In Bioconjugate Techniques. 2008. The chemistry of reactive groups. 2nd edn. Elsevier. PART I. p. 184. [Google Scholar]
- 52.Kalkhof S, Sinz A. Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal. Bioanal. Chem. 2008;392:305–312. doi: 10.1007/s00216-008-2231-5. [DOI] [PubMed] [Google Scholar]
- 53.Meisenheimer KM, Meisenheimer PL, Koch TH. Nucleoprotein photo-cross-linking using halopyrimidine-substituted RNAs. Methods Enzymol. 2000;318:88–104. doi: 10.1016/s0076-6879(00)18046-2. [DOI] [PubMed] [Google Scholar]
- 54.Chen H-JC, Hsieh C-J, Shen L-C, Chang C-M. Characterization of DNA-protein cross-links induced by oxanine: cellular damage derived from nitric oxide and nitrous acid. Biochemistry. 2007;46:3952–3965. doi: 10.1021/bi0620398. [DOI] [PubMed] [Google Scholar]
- 55.Schultz A, Nottrott S, Hartmuth K, Luhrmann R. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J. Biol. Chem. 2006;281:28278–28286. doi: 10.1074/jbc.M603350200. [DOI] [PubMed] [Google Scholar]
- 56.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Luhrmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 57.Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- 58.Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J. Biol. Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 59.Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 60.Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urlaub H, Raker VA, Kostka S, Luhrmann R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001;20:187–196. doi: 10.1093/emboj/20.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thore S, Mayer C, Sauter C, Weeks S, Suck D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J. Biol. Chem. 2003;278:1239–1247. doi: 10.1074/jbc.M207685200. [DOI] [PubMed] [Google Scholar]
- 64.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naidoo N, Harrop SJ, Sobti M, Haynes PA, Szymczyna BR, Williamson JR, Curmi PM, Mabbutt BC. Crystal structure of Lsm3 octamer from Saccharomyces cerevisiae: implications for Lsm ring organisation and recruitment. J. Mol. Biol. 2008;377:1357–1371. doi: 10.1016/j.jmb.2008.01.007. [DOI] [PubMed] [Google Scholar]