Abstract
Unlike classical 2-oxoglutarate and iron-dependent dioxygenases, which include several nucleic acid modifiers, the structurally similar jumonji-related dioxygenase superfamily was only known to catalyze peptide modifications. Using comparative genomics methods, we predict that a family of jumonji-related enzymes catalyzes wybutosine hydroxylation/peroxidation at position 37 of eukaryotic tRNAPhe. Identification of this enzyme raised questions regarding the emergence of protein- and nucleic acid-modifying activities among jumonji-related domains. We addressed these with a natural classification of DSBH domains and reconstructed the precursor of the dioxygenases as a sugar-binding domain. This precursor gave rise to sugar epimerases and metal-binding sugar isomerases. The sugar isomerase active site was exapted for catalysis of oxygenation, with a radiation of these enzymes in bacteria, probably due to impetus from the primary oxygenation event in Earth’s history. 2-Oxoglutarate-dependent versions appear to have further expanded with rise of the tricarboxylic acid cycle. We identify previously under-appreciated aspects of their active site and multiple independent innovations of 2-oxoacid-binding basic residues among these superfamilies. We show that double-stranded β-helix dioxygenases diversified extensively in biosynthesis and modification of halogenated siderophores, antibiotics, peptide secondary metabolites and glycine-rich collagen-like proteins in bacteria. Jumonji-related domains diversified into three distinct lineages in bacterial secondary metabolism systems and these were precursors of the three major clades of eukaryotic enzymes. The specificity of wybutosine hydroxylase/peroxidase probably relates to the structural similarity of the modified moiety to the ancestral amino acid substrate of this superfamily.
INTRODUCTION
RNAs, especially transfer RNAs (tRNA), show diverse post-translational modifications of bases. To date, at least 90 distinct modifications have been described (1,2). These include simple substitutions (e.g. deamination of adenosine to inosine), addition of small functional groups to bases (such as a methyl group in 1-methylguanosine) and chemical transformations resulting in large complex bases (such as threonyl carbamoyladenosine, queuosine and wyosine and their further derivatives). A veritable biochemical menagerie of enzymes catalyzing these modifications has become apparent over the past 15 years through a combination of computational and experimental studies (1). The catalytic mechanisms and phyletic distributions of these enzymes have greatly contributed to our understanding of novel biochemical reactions and the evolutionary significance of RNA modifications, respectively. However, lacunae remain in terms of candidate enzymes and reaction mechanisms responsible for several of the more complex modifications. Modified bases are particularly prevalent at position 37 of tRNA, which is adjacent to the anticodon, and is known to stabilize mRNA–tRNA pairing and maintenance of reading frame (1,3). An important complex base derived from guanosine is wybutosine (yW) present at position 37 of tRNAPhe in eukaryotes. Precursors of yW, such as wyosine and its derivatives, are detected in the same position in tRNAPhe in archaea (4–6). In contrast, bacteria contain an adenosine at position 37 of tRNAPhe and may show a completely different modification at this position such as isopentenyladenosine and its derivatives (7). Biochemical studies in Saccharomyces cerevisiae have identified five proteins, Trm5, Tyw1, Tyw2, Tyw3 and Tyw4, in the biosynthetic pathway that convert a guanosine to yW (Figure 1; 8). Orthologs of some of these enzymes, such as Trm5, Tyw1, Tyw2 and Tyw3 are also present in archaea, which synthesize yW precursors such as 4-demethylwyosine and its archaeal-specific derivatives (5,6,9,10). This suggests that the basic pathway for yW biosynthesis was already present in the common archaeo-eukaryotic ancestor, and was further elaborated in eukaryotes. Though yW is an ancestral modification, it shows considerable variation in its distribution across eukaryotes. A particularly prevalent variant of yW is its hydroxy or hydroperoxide derivative that is observed in several eukaryotes like animals (including humans) and fungi such as Geotrichum candidum (6,11,12). Despite the complete characterization of the yW pathway in yeast, the enzyme/s catalyzing the hydroxylation or peroxidation step resulting in hydroxywybutosine and/or hydroperoxywybutosine has evaded detection (6).
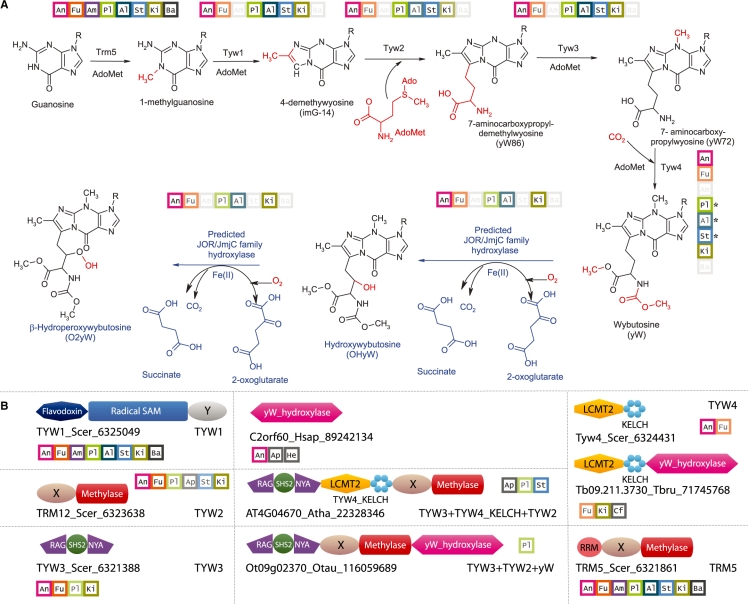
Figure 1.
Predicted hydroxy/hydroperoxy wybutosine biosynthesis pathway and domain architecture of key enzymes involved in its synthesis. (A) Shown above each enzyme of the reaction pathway, in colored squares, is the phyletic profile of the protein family. The color intensity of the outline is used to depict its distribution within a particular lineage. Thus, darkly colored squares are used when an enzyme is present in most members of that eukaryotic lineage. Lighter coloring is used for enzymes that are present only in some members of a lineage. When an enzyme is completely absent from a lineage, the square is not colored. The Tyw4 enzyme in plants, apicomplexans and stramenopiles is a specialized case and marked with an asterisk in the phyletic profile. In these cases, the kelch domain of TYW4 is found between Tyw3 and Tyw2. These also have a solo LCMT domain that in trees groups with LCMT1 and may substitute for the LCMT2 seen in Tyw4. The eukaryotes are divided into eight major groups and the expansion of the abbreviations within the boxes are as follows: An, animals; Cf, choanoflagellates; Fu, fungi; Am, Amoebozoa; Pl, plants; Al, Alveolates, Ap, Apicomplexa; St, Stramenopiles; Ki, Kinetoplastids and Heterolobosea; He, Heterolobosea only; Ba, the basal lineages diplomonads and parabasalids. We predict that the hydroperoxywybutosine reaction seen in some lineages is catalyzed by the same wybutosine hydroxylase. (B) Domain architectures from representative eukaryotes are labeled with the gene name, species abbreviation and Genbank index number separated by underscores. Domains are labeled with standard names. X and Y represent poorly characterized globular domains. The phyletic profile for the domain architecture is shown next to them in colored boxes. The abbreviations are the same as in (A).
Using sensitive computational analysis, we have recently described several novel nucleic acid-modifying enzymes that were predicted to catalyze hydroxylation of bases (13). Most of these belong to the clade of classical 2-oxoglutarate and iron-dependent dioxygenases (2OGFeDO) with the double-stranded β-helix fold (DSBH), which includes numerous enzymes acting on diverse substrates such as: amino acids and proteins (e.g. EGL-9, prolyl and lysyl hydroxylases), small molecules (e.g. clavaminate synthase, isopenicillin synthase and plant leukoanthocyanin hydroxylases), antibiotic precursors (e.g. halogenases in syringomycin and coronamic acid) and bases in RNA and DNA (such as the DNA-repair protein AlkB, TET/JBP and thymine-7-hydroxylases; 14–19). Subsequent experimental studies on some of the enzymes we identified have confirmed that indeed some of these mediate the hydroxylation of bases in DNA such as 5-methylcytosine, while others were predicted to perform similar modifications in RNA (13,20). Yet, none of these newly identified enzymes showed the phyletic patterns or other contextual features that made them candidates for the yW hydroxylase/peroxidase.
Hence, we undertook a new computational study of the yW biosynthesis pathway and identified an enzyme which belongs to a structurally related but distinct superfamily of dioxygenases, the jumonji-related (JOR/JmjC) dioxygenases (21–23), as the yW hydroxylase/peroxidase. This identification provided us with a molecular marker to infer the diversity of the position 37 of tRNAPhe modifications across eukaryotes. While these enzymes too utilize iron and 2-oxoglutarate as co-factors, in contrast to the catalytic diversity of the classical 2OGFeDO superfamily, the jumonji-related superfamily has been exclusively characterized as protein-modifying enzymes. They are known to catalyze the hydroxylation of different amino acids or the demethylation of ε-methyl-lysine (24). This observation raised the question of how and when the nucleic acid-modifying activity (i.e. yW hydroxylase/peroxidase) emerged in the midst of a superfamily of enzymes traditionally known to modify proteins. Another distinct group of proteins, frequently termed cupins, are also known to share a similar DSBH fold with the above two superfamilies. These are typified by the non-catalytic sugar-binding domain of the bacterial transcription factor AraC, the plant seed-storage proteins and enzymes such as the oxalate oxidase and sugar epimerases/isomerases (e.g. RmlC, mannose-6-phosphate isomerase and glucose-6-phosphate isomerase; 25). In the literature, there is rampant confusion in the nomenclature of these proteins with different overlapping groups termed cupins, JmjC or 2OGFeDO without a regard for their biochemistry (i.e. dependence on 2-oxoglutarate, or configuration of active site residues) or an objective analysis of their actual evolutionary relationships. This has hindered an evaluation of the emergence of multiple nucleic acid- and protein-modifying activities among these enzymes—e.g. questions pertaining to how many of these biopolymer-modifying activities emerged convergently as opposed to divergently and from what kind of precursors. Hence, we undertook a systematic higher order classification of the catalytic and small-molecule-binding DSBH fold proteins and objectively defined major evolutionary radiations within them. This provided a new understanding of the major catalytic innovations within the fold. We also clarified relationships within the jumonji-related superfamily and the higher order relationship of this superfamily to other members of this fold. We present evidence that enzymes belonging to all the distinct superfamilies of the DSBH fold underwent major radiations within a previously unappreciated array of bacterial biosynthetic systems that are involved in synthesis of hydroxylated and halogenated peptide–derived secondary metabolites. Based on this, we propose that peptide-modifying activities are likely to be the ancestral feature of the jumonji-related superfamily. Three distinct lineages of jumonji-like enzymes emerged in bacteria within the context of these biosynthetic systems and were transferred to eukaryotes, where they radiated further to acquire protein and RNA-modifying specificities.
MATERIALS AND METHODS
Structure similarity searches were conducted using the FSSP program (26), and structural alignments were made using the MUSTANG program (27). Protein structures were visualized and manipulated using the Swiss-PDB (28) and PyMol programs (http://www.pymol.org/). Sequence profile searches were performed against the NCBI non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda, MD, USA), and a locally compiled database of proteins from eukaryotes with completely or near-completely sequenced genomes derived from the genomes division of Genbank. PSI-BLAST searches were performed using an expectation value (E-value) of 0.01 as the threshold for inclusion in the position-specific scoring matrix generated by the program; searches were iterated until convergence (29). Profile-based HMM searches were performed using the newly released HMMER3 package (version beta 2) using the HMMSEARCH and JACKHMMER programs (30). Multiple alignments were constructed using the MUSCLE and Kalign programs (31,32), followed by manual correction based on PSI-BLAST high-scoring pairs, secondary structure predictions and information derived from existing structures. Protein secondary structure was predicted using a multiple alignment as the input for the JPRED2 program (33), which uses information extracted from a PSSM, HMM and the seed alignment itself. Pairwise comparisons of HMMs, using a single sequence or multiple alignment as query, against profiles of proteins in the PDB database were performed with the HHPRED program (34). Similarity-based clustering was performed using the BLASTCLUST program with empirically determined length and score threshold parameters (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html). Gene neighborhoods in prokaryotes were obtained by isolating conserved genes immediately upstream and downstream of the gene in question showing separation of <100 nt between gene termini. Neighborhoods were determined by searching NCBI PTT tables (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Genome) with a custom PERL script. Phylogenetic analysis was carried out using neighborhood-joining and minimum evolution-based methods with gamma-distributed rates and a JTT substitution matrix as implemented in the MEGA4 program (35). The shape parameter α was estimated empirically through a series of experimental trials that ensured recovery of the species tree within orthologous protein groups. Additionally, maximum likelihood trees were obtained first by using the Fasttree program (36). All large-scale procedures were carried out using the TASS software package (Anantharaman, V., Balaji, S., A.L., unpublished data).
RESULTS AND DISCUSSION
Identification of the candidate yW hydroxylase/peroxidase and the distribution of this modification across eukaryotes
To better understand the variation in position 37 modifications of tRNAPhe across eukaryotes, we systematically identified orthologs of the yeast yW biosynthesis enzymes (Figure 1; Supplementary Data) in completely sequenced eukaryotic genomes. We observed that these enzymatic domains often show fusions to each other in various lineages (8). For example, the enzymes Tyw3 and Tyw2 are combined in the same polypeptide in chlorophyte algae. The enzyme Tyw4, which catalyzes two distinct reactions, namely methylation of 7-(-amino-carboxypropyl)wyosine and fixation of carbon dioxide, comprises of two domains namely a Rossmann fold methyltransferase (LCMT2) and a C-terminal kelch-like β-propeller domain (37). This kelch-like β-propeller domain is also fused to Tyw3 and Tyw2 in plants, stramenopiles and apicomplexans (Figure 1). In Tyw4, from several ascomycetes (e.g. the yeast Yarrowia lipolytica), choanoflagellates and kinetoplastids, we found a previously unaccounted globular domain, which is not found in the Saccharomyces Tyw4, occurring C-terminal to the methyltransferase and β-propeller unit (Figure 1). A homologous domain was also found linked to the C-terminus of the Tyw3+Tyw2 unit in chlorophyte algae and as a standalone protein in animals and apicomplexans (Figure 1). These multiple fusions of the same domain with different enzymes of the yW biosynthesis pathway suggested that it might be a previously unrecognized module involved in the modification of yW.
To determine the affinities of this domain, we ran sequence profile searches with the PSI-BLAST and HMMER3 program against the non-redundant database with the representative from Gibberella (FG04298.1, gi:46116912; region 693–989). In addition to orthologous domains from other eukaryotes, these searches recovered within two iterations a series of JOR/JmjC domain proteins including versions such as the factor-inhibiting hypoxia-inducible factor 1 (FIH1) protein (gi: 27065810; e = 10−10), the HSP-associated protein 1 (HSBAP1; e = 10−15) and jumonji-related domain protein 5 (JmjD5; e = 10−15) from eukaryotes, and bacterial homologs (e.g. Myxococcus xanthus MXAN_4411; e = 10−18). Conversely, profile searches with RPS-BLAST using a position-specific score matrix, which we had previously prepared for the detection of JOR/JmjC profiles (38), recovered all representatives of the uncharacterized yW-associated domain with significant e-values (e < 10−6). Furthermore, a few of these representatives were also detected by the PFAM ‘cupin-4’ HMM with significant e-values. These observations indicated that the uncharacterized domain associated with the yW biosynthesis proteins was a member of the JOR/JmjC superfamily. We clustered all the sequences recovered in the first two iterations of the above PSI-BLAST search with the BLASTCLUST program (length threshold 0.4, score threshold 0.8) and observed that the majority of JOR/JmjC domains associated with yW synthesis enzymes formed a single cluster, distinct from other clusters of related proteins such as those typified by FIH1 (asparaginyl hydroxylase of the hypoxia-induced factor, HIF), HSPBAP1 and the bacterial representatives. The yW-associated cluster typically contained only a single representative in a given organism and was detectable in animals, choanoflagellates, fungi, chlorophyte algae, heterolobosea and kinetoplastids, suggesting that they comprised a distinct family of orthologous JOR/JmjC domains.
A multiple sequence alignment of this family revealed the conservation of all key catalytic active site motifs characteristic of JOR/JmjC-like 2-oxoglutarate-Fe(II)-dependent dioxygenases that are required for chelating Fe(II) (23,24): (i) the highly conserved HxD motif in strand-2 and (ii) a histidine in strand-7 (Figures 2 and 3). These features suggested that the yW-associated JOR/JmjC domain is catalytically active and catalyzes a dioxygenase reaction like other members of the clade. Given that it is strongly associated with other catalytic domains of the yW pathway and their phyletic pattern matches the currently characterized phyletic distribution of hydroxy-yW/peroxy-yW, we posit that this domain is the yW hydroxylase/peroxidase. Furthermore, a comparison of the within-group mean pairwise protein distances showed that the yW pathway-associated family of JOR/JmjC domains are clearly slower evolving than any of the other related families with comparable phyletic patterns (e.g. FIH1, JMJD7 and JMJD5). This pattern is indicative of these enzymes retaining the same substrate over evolution, such as a key conserved position in the tRNA. The relationship with peptide hydroxylases, such as the asparagine hydroxylase FIH1, detected in the above searches is also consistent with the predicted yW hydroxylase/peroxidase activity for this domain, given that the latter reaction occurs on a structurally comparable methionine-derived moiety in yW (Figure 1).
Figure 2.
Multiple sequence alignment of the JOR/JmjC superfamily. The alignment shows the key sequence features that distinguish the JOR/JmjC superfamily from other 2OGFeDO-related enzymes and also brings to attention features that distinguish the major clades of the JOR/JmjC superfamily. Additionally, it highlights conserved residues shared between the predicted yW hydroxylase and other related members, and lineage-specific residues conserved in the yW hydroxylases that may have a substrate binding role. Sequences are labeled using the gene name, species abbreviation and GenBank gi number and sequence identifiers. Clades and families of representative sequences are shown on the right. The predicted secondary structure is shown above the alignment and family-specific features are highlighted under each alignment block. The color code used to highlight conserved amino acids is listed in the lower panel and is based on 85% consensus. Numbers between alignment columns indicate the length of variable inserts. The species abbreviations are: Aexc, Asticcacaulis excentricus; BPsyn9, Synechococcus phage syn9; Bpse, Burkholderia pseudomallei; Bsub, Bacillus subtilis; Ccre, Caulobacter crescentus; Cjap, Cellvibrio japonicus; Ddis, Dictyostelium discoideum; Gzea, Gibberella zeae; Hsap, Homo sapiens; Lmaj, Leishmania major; Mmus, Mus musculus; Mxan, Myxococcus xanthus; Ncra, Neurospora crassa; Nvec, Nematostella vectensis; Otau, Ostreococcus tauri; Pfal, Plasmodium falciparum; Phum, Pediculus humanus; Pput, Pseudomonas putida; Saur, Stigmatella aurantiaca; Sdeg, Saccharophagus degradans; Spom, Schizosaccharomyces pombe.
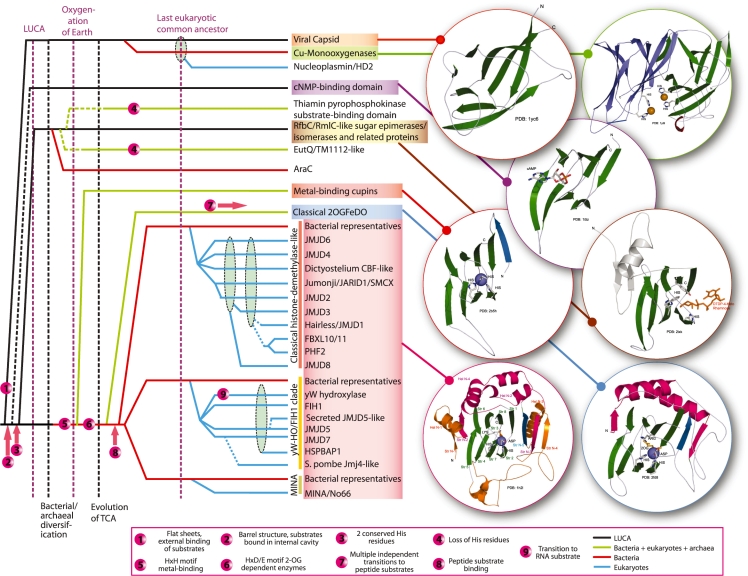
Figure 3.
Evolutionary history of the DSBH fold and structural features. On the left is a temporal diagram of the reconstructed evolutionary history of the DSBH fold. Individual superfamilies and families are listed to the right of the diagram, with solid horizontal lines tracing the inferred evolutionary depth of each family across several key evolutionary transition events shown as labeled vertical lines. Horizontal lines connecting to a dashed ellipse indicates the family descended from any one of the lineages bundled by the ellipse. Dashed horizontal lines represent an uncertainty in the inferred point of origin. Horizontal lines are color coded (see key) by observed phyletic distributions. Numbered circles illustrate the key transition events within the DSBH fold (see key). A more detailed classification of the DSBH fold is available in the Supplementary Data. On the right, the structures of some DSBH superfamilies are shown as cartoon representations with key structural, substrate binding and catalytic features. The conserved strands of the core of the DSBH fold are colored green, whereas the strand conserved in the metal-binding cupins, classical 2OGFeDO superfamily and JOR/JmjC is colored sky blue. Structural features conserved in classical 2OGFeDO and JOR/JmjC superfamilies are colored magenta, whereas those conserved in JOR/JmjC families are colored orange. In the classical 2OGFeDO and JOR/JmjC proteins, the 2-oxoglutarate is colored orange. The structure of the HIF1 JOR/JmjC contains the positions of the secondary structural annotations used in the text and multiple alignment. The proteins displayed include: Brome Mosaic Virus capsid (PDB: 1yc6), peptidylglycine alpha-hydroxylating monooxygenase (PDB: 1yi9), N-terminal domain of the catabolite gene activator protein (PDB: 1i5z), RmlC-like sugar epimerase (PDB: 2ixk), cysteine dioxygenase (PDB: 2b5h), AlkB-like 2OGFeDO (PDB: 2fd8) and FIH1-like JOR/JmjC (PDB: 1h2l).
Inference of phyletic patterns and functional implications of hydroxy-yW/peroxy-yW in eukaryotes
Although position 37 modifications of tRNA are universal across life, the enzymes catalyzing these modifications are very distinct in the three superkingdoms (39). Most position 37 methylations in bacteria appear to be catalyzed by the SPOUT superfamily methyltransferase TrmD (40–42). In contrast, comparable modifications in archaea and eukarya are catalyzed by the MJ883/Trm5-type Rossmann-fold methyltransferase (43). This modification is the first step in the synthesis of yW in tRNAPhe and is also potentially required to methylate the same position in other tRNAs. Interestingly, bacteria, which have an adenine at this position in the tRNAPhe, utilize the P-loop ATPase MiaA for isopentenylation, which presumably catalyzes a hemi-kinase reaction to activate isopentene (7) for synthesis of isopentenyladenosine. These observations imply that in spite of the need to modify this position for efficient maintenance of frame and mRNA–tRNA pairing, there is variability of the actual nucleotide at this position. The same trend continues in eukaryotes with respect to the nature of yW modification. Based on prediction of the yW hydroxylase/peroxidase, we can confidently infer that the hydroxylated version of this base is absent in the basal eukaryotes such as Giardia and Trichomonas. However, its presence in kinetoplastids and the heterolobosean amoeba Naegleria (Figure 1; Supplementary Data) suggests that it was present from relatively early in eukaryotic evolution. Further, it appears to have been lost in several lineages—in some cases entire lineages such as ciliates, multicellular plants and basidiomycete fungi appear to have lost the yW hydroxylase/peroxidase (Figure 1; Supplementary Data). In other cases, losses are restricted to certain terminal clades, including multiple independent losses among ascomycete yeasts and arthropods. In some of these cases, like certain arthropods and ciliates, this loss is accompanied by loss of the entire yW pathway except for the initial methylation catalyzed by Trm5 (Figure 1; Supplementary Data). Interestingly, retroelements use translational frame-shifting to express proteins such as the gag–pol fusion. This is achieved by as yet unclear viral mechanisms that deplete host yW-containing tRNAsPhe in infected cells (44,45). Given the variability in the distribution of yW and hydroxy-yW/peroxy-yW across eukaryotes, it is conceivable that some of these gene losses altering the base modification at position 37 of tRNAPhe might contribute to evasion of mechanisms by which retroelements target the base. The lack of hydroxy-yW has also been observed in neuroblastoma cells and reticulocytes, suggesting that alteration of this modification might also provide a means of regulating translation (44,45).
Higher order relationships of the DSBH fold and evolution of distinct active sites
The above-predicted yW hydroxylase is the first example of a JOR/JmjC domain inferred to modify RNA—all other experimentally characterized JOR/JmjC domains have been shown to modify amino acid side chains in peptides. This raised an interesting evolutionary question as to whether the yW hydroxylase arose secondarily from protein-modifying JOR/JmjC domains, or if it emerged from a larger group of hitherto uncharacterized RNA-modifying JOR/JmjC domains. The answer to this question is also intimately linked to the origin of these enzymes, their ancestral activities, and their relationship to the classical 2OGFeDO enzymes and those traditionally termed the cupins (25). However, there is considerable confusion among the existing classification schemes. For example, the SCOP database recognizes the common fold (DSBH fold) in these enzymatic groups but creates several distinct superfamilies such as ‘RmlC-like cupins’, ‘Clavaminate synthase-like’ and ‘Regulatory protein AraC-like’ (http://scop.mrc-lmb.cam.ac.uk/scop/). These are included with other superfamilies of non-enzymatic domains such as the cNMP-binding domains with no clear indication of their inter-relationships. While PFAM currently clubs all enzymatic versions under a ‘clan cupin’ (http://pfam.sanger.ac.uk/), SCOP to the contrary limits the term cupin to the RmlC-like superfamily, which is in line with the original definition of the cupin superfamily (25). Further, PFAM classifies certain JOR/JmjC-related proteins as a cupin family (i.e. cupin_4) and others separately in a JmjC family, and splits the various dioxygenases into multiple different families with no indication of their higher order relationships. Similarly, all the recently published phylogenetic classifications of JOR/JmjC domains have only considered the eukaryotic members (excluding the yW hydroxylase that we identify above), and none of the numerous bacterial representatives (24).
Hence, to delineate the higher order evolutionary relationships among the DSBH fold domains, we first recovered all known representatives in the PDB database by performing structure similarity searches with the DALI program. We then used the structural alignments generated by these searches and visually examined the structures to identify distinctive shared 3D features. The group of all-β strand domains with a shared core with β-strands arranged in a double helical pattern forms a distinctive structural category that might be defined as the classical DSBH fold. This core can be interpreted as comprising of eight β-strands units linked by short turns (Figure 3). This structurally distinct category includes all domains previously considered cupins, classical 2OGFeDOs and JOR/JmjC and also, cNMP-binding domains (including versions that bind heme, e.g. CooA or halogenated aromatic compounds, e.g. CrpK), the Cu(II)-dependent mono-oxygenases, the thiamin pyrophosphokinase substrate-binding domain, the AlcR-N domain the nucleophosmin/nucleoplasmin superfamily and viral capsid proteins of the all-β variety. Of these, pairwise structural comparisons suggest that the Cu(II)-dependent mono-oxygenases, nucleophosmin/nucleoplasmin superfamily and viral capsid proteins form a distinct clade that are characterized by flatter sheets. As a consequence, they have a limited cavity in the interior of the fold and all characterized members of this clade bind their peptide or nucleic acid substrates externally on the surface of the DSBH structure (Figure 3). Hence, even though the Cu(II)-dependent mono-oxygenases catalyze an ascorbate-dependent reaction comparable to the DSBH dioxygenases, their active site residues are on the exterior of the DSBH fold and is formed at the interface of duplicated DSBH units (Figure 3; 46). This indicates that a comparable oxygenase reaction emerged twice, independently in distinct clades of the DSBH fold. On the other hand, the domains traditionally considered cupins, classical 2OGFeDOs and JOR/JmjC and the cNMP- binding domains form a monophyletic clade unified by distinctive sequence signatures at the beginning of strands 4 and 6. The small residues at these positions result in the strands acquiring a more contiguous jelly-roll configuration, thereby forming a barrel-like structure. Consequently, this clade is characterized by a capacious internal cavity in the DSBH fold that accommodates the substrate binding site (Figure 3).
Within the above clade, the domains traditionally considered cupins (25), the classical 2OGFeDO superfamily, and the JOR/JmjC-like superfamily are further unified into a monophyletic clade by the presence of highly conserved histidines in strand-2 and strand-7. However, closer examination of the domains traditionally considered cupins (25) shows that the relationships between themselves and to other members of this monophyletic assemblage are more complicated. Of the cupins, the RmlC-like sugar epimerases contain the above- described conserved histidines but do not bind a metal; instead, they directly bind a sugar moiety via these residues (47). RmlC-like sugar epimerases also have a widespread distribution in all major bacterial and archaeal lineages suggesting that they were already present in the last universal common ancestor. The metal-binding cupins, the classical 2OGFeDOs and the JOR/JmjC share a further modification in the ancestral active site comprised of the two histidines—they possess a conserved residue downstream of the first ancestral conserved histidine, resulting in a signature of the form HXH or HXD (where X is any amino acid; Figure 3). This modification converts the active site into a metal-chelating configuration and the chelated ion (usually iron, zinc, nickel or manganese) is critical for binding oxygen and catalysis of the dioxygenase reaction. They also share a conserved strand that is anti-parallel to strand-1 of the core DSBH (Figure 3, strand N-5 in blue; henceforth structural elements N-terminal to the core eight strands shared by all members of the DSBH fold are labeled with an ‘N’ prefix and those to the C-terminus of the core with a ‘C’ prefix). Members of the classical 2OGFeDO superfamily and JOR/JmjC superfamily further share several unique features to the exclusion of the metal-binding cupins (Figure 3, in pink): (i) two N-terminal strands bracing the core DSBH—the first strand is on the side opposite to the mouth of the substrate-binding cavity and the second strand that stacks beside the N-terminal strand shared with the metal-binding cupins and contributes to the mouth of the substrate-binding cavity (Figure 3; strands N-2 and N-3). (ii) The straddling region connecting the first strand to the hairpin contains either a single long helix or a helical region interrupted by multiple loop-like regions. This region is also prone to independent insertions of distinct domains in members of both these superfamilies: e.g. the insertion of a cysteine-rich metal-chelating cluster in the metazoan TET proteins (classical 2OGFeDO superfamily) and the Bright and PHD domains in the Jumonji/ SMCX/JARID1 family (JOR/JmjC superfamily; 23). (iii) Strand N-5 is linked to the core of the DSBH via a further helical linker that might span across the length of the core DSBH (Figure 3, Helix N-4 in pink). (iv) The first pair of metal-chelating residues usually assumes the HXD signature in the 2OGFeDO and JOR/JmjC superfamilies as opposed to the HXH, the ancestral condition in the metal-chelating cupins (16). (v) Both these superfamilies are also united by their dependence on both 2-oxoglutarate and iron for catalysis. Thus, these two superfamilies form a distinct monophyletic clade to the exclusion of the metal-binding cupins with which they share a common metal-chelating active site.
The JOR/JmjC domains are further distinguished from the classical 2OGFeDO domains by several sequence and structure features. These include: (i) an N-terminal strand–helix extension (strand N-1 and helix N-1, Figure 3). In several members of the JOR/JmjC family such as Jumonji, JARID1, SMCX and JMJD2, this region, in combination with the following strand (strand N-2, see above) has been erroneously defined as a distinct domain called the JmjN domain (48,49). However, it has no independent existence and merely represents a structural extension of the DSBH core. (ii) An additional strand-helix unit inserted into the hairpin formed by the strands N-3 and N-5. (iii) A helical hairpin with an extended connector loop is inserted between strands 4 and 5 of the core DSBH fold and packs against the side of the core barrel (this loop might bear additional inserts like a metal-cluster in the Jmjd2/KDM4 family; Table 1). (iv) A previously under-appreciated basic residue in strand 4 is conserved in most families (Figures 2 and 3). This residue forms a salt-bridge with the 2-oxoglutarate. In some JOR/JmjC families (namely CBF and Jmjd3), this basic residue has been substituted by lineage-specific conserved residues that are predicted to play a similar role (Table 1). In contrast, the classical 2OGFeDO superfamily has a highly conserved arginine on strand 8, which performs an equivalent role. This observation indicates that though the common ancestor of the JOR/JmjC and classical 2OGFeDO superfamilies already utilized 2-oxoglutarate, the positively charged residues for anchoring the cofactor emerged independently in the two superfamilies. Based on these features members of the ‘cupin-4’ group in PFAM (including the domain which we identify as yW-hydroxylase/yW-peroxidase and various bacterial proteins) are clearly a part of the JOR/JmjC superfamily. In conclusion, the originally defined cupin superfamily (25) is paraphyletic: the metal-binding cupins are closer to the clade uniting the JOR/JmjC and classical 2OGFeDO superfamilies than they are to the RmlC-like enzymes, which do not utilize metal (Figure 3; Supplementary Data). Several other DSBH domains have been classified separately from the ‘cupins’ in the SCOP database, but appear to be derivatives of RmlC-like enzymes. Of these, the sugar-binding DSBH domain found in the AraC-like transcription factors possess a similar structural configuration as the RmlC-like domains and like them binds sugars rather than metals. Consistent with this, they possess only one or two of the ancestral histidines in the active site. Some of them appear to be non-catalytic sugar-binding domains, while others might possess epimerase/isomerase activity like the RmlC-like enzymes which might be required for their transcription regulatory role. The thiamin pyrophosphokinase substrate-binding and AlcR-N domains also show the same configuration of strands as these sugar-binding domains; however, they appear to have lost the histidines. This suggests that they are also non-catalytic divergent representatives of the RmlC-like domains that have acquired specificity for other small molecules. Another comparable derivative is the EutQ-like family prototyped by the proteobacterial EutQ and the Thermotoga TM1112. They appear to retain the same binding pocket as other members of the RmlC-like enzymes but have lost the histidines. However, they possess a distinctive pair of conserved residues (aspartate and lysine) which could serve as an acid and base in catalyzing an as yet uncharacterized activity (Supplementary Data).
Table 1.
Phyletic distribution, relationships and shared sequence features of different JOR/JmjC families
| Family Name | Phyletic distribution | Additional comments | Clade synapomorphies |
|---|---|---|---|
| Histone demethylase clade | |||
| JMJD6a | An, Ch, Fu, Pl, My, Al, St, He, Kib | Conserved arginine just N-terminal to strand-1; sporadic fusions to F-box, AT-hook, Ankyrin repeats, C2, chromo and phospholipase domains. | (i) Two amino acid β bulge in connector between strands 2 and 3, (ii) Conserved asparagine residue in the middle of strand-3, (iii) HxxxN motif in strand-7 and (iv) Universally conserved C-terminal small residue (usually asparagine) displaced towards end of strand 8 in crystal structures. |
| JMJD4a | An, Ch, Fu, Pl, Ap, St, Ki | Conserved N[HR]NWhN motif in strand-8. | |
| Dictyostelium C-module binding factora | Fu, Am, He | Although the active site basic residue in strand-4 is lost, there is a highly conserved lysine residue before strand-8 that probably serves as an alternative active site residue; fusions to ZZ, CxxC, PHD, Bmb domains. | |
| Jumonji/JARID1/SMCX/KDM5a | An, Fu, My, Pl, Al, St, He | HxE in strand-2 catalytic site; Jumonji inactive; fusions to BRIGHT, PHD, SJA, C2H2 Zn-fingers and SANT domains. | |
| JMJD2/KDM4a | An, Ch, Fu, Plc, St, Ci, He | HxE in strand-2 catalytic site; A cysteine and histidine in insert between strand 4 and 5, and 2 cysteines after strand-8 form a Zn-binding cluster, Fused to PHD, TUDOR, C2H2, chromo, TAM/MBD and SET domains. | |
| JMJD3 (KDM6B)/UTX (KDM6A) | An, Ch, Fu, Ci | Basic residue in strand-4 is lost, there is a highly conserved lysine residue in strand-1 that probably serves as an alternative active site residue; HxE in strand-2 catalytic site; lacks asparagine of HxxxN motif, characteristic C-terminal domain with a C2C2 motif (treble clef fold); fused to N-terminal TPR repeats, expanded in Paramecium of which some inactive. | |
| Hairless/JMJD1 | An, Ba, Pl | Fused to an N-terminal ZZ finger, distinct N-terminal treble clef domain and CxC domains in plants. | |
| JMJD8 | An, Plc, St | Contain a HxH instead of a HxD in the catalytic site in strand-2. | |
| JHDM1/KDM2/PHF2/FBXL10 | An, Ch, Fu, St, Pl | HxxT instead of the HxxxN motif in strand-7; Fusions to CxxC, FBOX, LRR, PHD. | |
| Bacterial Histone-demethylase-like | Proteobacteria, Firmicutes, Bacteroidetes | See text and Figure 4 for gene neighborhoods and associations. | |
| yW hydroxylase/FIH1 clade | |||
| yW hydroxylase/peroxidasea | An, Ch, Fu, Plc, Ap, Ki, He | Conserved arginine in strand-N-5 and strand N-1; Fused to TYW3+TYW2, TYW4 and the kelch repeats of TYW4 in some species (Figure 1). | (i) Insert between strands N-3 and N-4, (ii) Highly conserved asparagine at beginning of strand-3 and (iii) Universally conserved C-terminal small residue (usually asparagine) in middle of strand 8 in structures. |
| FIH1a | An, Ch, My, Plc, St, He | Conserved HP motif in connector between strands 4 and 5; fusions to kelch repeats in Naegleria, and N-terminal P-loop sulfotransferase in Monosiga; expansion in Monosiga and stramenopiles. | |
| JMJD5a | An, Ch, Ba, Pl, My, Ap, St, He | Conserved glutamate in N-3 and aromatic between strands N-3 and N-4, DxxxP motif in Helix N-4; fusion to acetyltransferase in nematodes. | |
| Secreted JMD5-like family | An (lost in vertebrates, arthropods, nematodes) | Contains disulfide-bonding conserved cysteines and a unique N-terminal GHxxxhG-motif; three subfamilies each with lineage-specific expansions in various metazoans. | |
| JMJD7a | An, Ch, Fu, Pl, St, Ci, He | Conserved acidic N-terminal to strand-1; conserved asparagine in strand 1 shared with S. pombe Jmj4-like. | |
| Schizosaccharomyces pombe Jmj4-like | Fu, Ch, Plc, St, Ap | Conserved histidine in strands 3 and 8; Fused to TPR repeats and DNAJ in oomycetes; shares conserved asparagine in strand 1 with JMJD7. | |
| HSPBAP1 | An | Conserved histidine between strands 6 and 7; always a stand-alone DSBH domain. | |
| Bacterial yW hydroxylase/FIH1 | Widespread in bacteria, see supplement | See text and Figure 4 for gene neighborhoods and associations. | |
| MINA/No66 clade | |||
| MinA/No66a | An, Ch, Fu, Pl, My, Al, St, He, Kib | Expanded in Monosiga where it is fused to SH2, STY kinase, Ig-like domains, GCC repeat and cysteine containg extracellular domains. | Share a conserved C-terminal α-helical extension. |
| Bacterial MinA/No66-like/YxbC/YcfD | Wide-spread in bacteria, see supplement | Versions closer to eukaryotic homologs additionally share several conserved motifs, e.g. HxT motif in strand-8 and arginine between strands 6 and 7. |
aAncient eukaryotic JOR/JmjC lineages.
bSpecies abbreviations: Al, Alveolates; An, Animals; Ap, Apicomplexa; Ba, Basidiomycete fungi; Ch, Choanoflagellates; Ci, Ciliates; Fun, Fungi; He, Heterolobosea; Ki, Kinetoplastids; My, Mycetozoa; Pl, Plants; St, Stramenopiles.
cOnly present in chlorophyte algae.
Evolution of substrate preferences of the DSBH domains with internal binding sites
We then examined the nature of substrates of members of the DSBH clade that possessed internal active sites (see above; Figure 3) to infer ancestral substrate preferences. Majority of cNMP-binding domains appear to bind cyclic nucleotides, with the sugar moiety of the cNMP being bound inside the cavity of the DSBH fold (50). Of the remaining members of this clade, which are united by two conserved histidines the first to branch off are the RmlC-like epimerases and their non-catalytic relatives, like AraC that bind sugar moieties in their substrates (e.g. nucleotide diphosphate sugars) in the interior of the DSBH fold (16,47). The catalytic versions used at least one of the two histidines as both a general acid and base that alternatively abstracted and donated a proton while catalyzing epimerization. The metal-binding cupins were the next to branch off and eventually radiate into several distinct families. Some of these families retained the ancestral condition of a single DSBH domain, whereas the bicupin lineage underwent a duplication of the DSBH domain. Of the single domain versions, the archaeal-type glycolytic glucose-6-phosphate isomerase (G6PI; 51) and the type-II phosphomannose isomerase (PMI; 52) are widespread in bacteria and archaea (Supplementary Data), indicating that they are one of the most ancient lineages of metal-binding cupins. The bicupins, a family with a wide phyletic distribution comparable to the above single domain versions include sugar isomerases such as the type-I mannose-6-phosphate isomerases. Unlike the RmlC-like epimerases, these sugar ring-opening isomerases have been proposed to catalyze their reaction via a cis-enediol intermediate that requires the metal ion to stabilize the developing negative charge on the oxygen (53). Based on the crystal structures of the metal-binding cupin isomerases, the metal appears to be positioned to interact with the two cis oxygens on the enediol. On more than one occasion, metal-binding cupins related to G6PI/type-II PMI were recruited in polyketide biosynthesis pathways (e.g. TcmJ- and RemF-like families; Supplementary Data). We predict that in the cyclization of condensed poly-malonyl esters in the synthesis of the basic polyketide skeleton they are likely to catalyze a reaction similar to sugar isomerization probably via an enediol intermediate. These observations suggest that the metal-binding cupins also initially emerged as sugar-binding enzymes that utilized the chelated metal to catalyze a specific sugar isomerase reaction and this role was also widely adapted to polyketide/carbohydrate derived secondary metabolites.
Eventually, the metal appears to have been exapted to bind a dioxygen molecule as it probably mimicked the two oxygens in the enediol intermediate of the sugar isomerases. This resulted in the emergence of dioxygenases, perhaps on more than one occasion among the metal-binding cupins. Evidence in support of this proposal emerges from the acireductone dioxygenase, which belongs to a widely distributed family of single domain metal-binding cupins, and is a key enzyme in the methionine salvage pathway. In catalyzing the oxidation of acireductone to 4-(methylthio)-2-oxobutanoate (a methionine precursor), it acts on a cis enediol moiety in acireductone that resembles the intermediate stabilized by the sugar isomerases. The 2-oxoglutarate co-factor used by both the 2OGFeDO and JOR/JmjC superfamilies is a derivative of sugar metabolism and could reflect a further development on the ring-opened sugar substrate. When taken together with the higher order relationships (see above; Figure 3), these observations suggest that the common ancestor of the entire clade of DSBH domains with internal binding sites might have bound sugars or their derivatives like nucleotides. However, with the emergence of the dioxygenase activity the substrate specificity appears to have greatly expanded. Representatives such as the acireductone dioxygenase and cysteine dioxygenase suggest that the initial acquisition of amino acid-related substrates occurred in the context of sulfur-containing amino acid salvage pathways. A further expansion to include peptides and amino acid derivatives occurred in the context of bacterial secondary metabolism. Evidence in support of this is offered by the identification of a novel metal-binding cupin domain fused to the N-terminus of a methionyl tRNA synthetase paralog (e.g. gi: 229076690 from Bacillus cereus; Supplementary Data). Based on the conserved gene neighborhoods, we predict that this enzyme catalyzes modification of a novel peptide-containing metabolite. A similar substrate expansion appears to have occurred within the classical 2OGFeDO superfamily, with the early emerging AlkB family acquiring specificity for bases (13,14). As with the metal-binding cupins, several lineages of the classical 2OGFeDO superfamily acquired preference for amino acid/peptide substrates in the context of bacterial secondary metabolism (see below; 14,54,55). In the JOR/JmjC superfamily, the only known substrates were peptides prior to the identification of the yW hydroxylase/peroxidase. Hence, deciphering the evolution substrate preferences of this superfamily needed a clearer understanding of their history, specifically in light of previously neglected bacterial members.
Evidence for major diversification events of the JOR/JmjC superfamily in bacteria followed by transfers to eukaryotes
The internal classification of the JOR/JmjC superfamily was developed using a combination of distance and maximum likelihood trees combined with detection of shared sequence and structure features (Table 1). Within eukaryotes 10 distinct families could be traced back to the common ancestor of the heterolobosean–kinetoplastid clade and the remaining eukaryotes. Interestingly, not a single member of this superfamily was identifiable in the two most basal eukaryotic lineages, namely parabasalids and diplomonads. The eukaryotic families could be further classified into three higher order clades: (i) the first and largest includes all families of well-characterized eukaryotic chromatin-modifying proteins [i.e. classical histone demethylases, namely the jumonji, Jmjd2, Jmjd4, and Dictyostelium C-module binding factor (Cbf) families] and the splicing factor U2AF65 lysyl hydroxylase Jmjd6 (56) (see Table 1 for synapomorphies). Histone demethylase versions of this clade are clearly distinguished from others by their multiple fusions to or insertions of various protein domains specifically linked to chromatin function. (ii) The second clade combines the newly identified yW hydroxylase, FIH1 (a protein asparaginyl hydroxylase), Schizosaccharomyces pombe Jmj4, JmjD5, JmjD7, a novel secreted family and HSPBAP1 families. This clade is distinguished by a conserved insert between strand 3 and 4 (Table 1). Of these, the S. pombe Jmj4 family appears to have emerged later in eukaryotic evolution and was secondarily lost in animals. In contrast, HSPBAP1 and the novel secreted family appear to have emerged only in metazoans (Table 1). The latter family shows certain shared features with JmjD5 and we predict that, unlike all other members of this clade, it hydroxylates residues on cell surface proteins. Fusions to chromatin-related domains are rare or non-existent in this clade. Among the intracellular versions, fusions of the yW-hydroxylases noted above and that of the nematode Jmjd5 to an acetyltransferase domain (C. elegans C06H2.3) are the only known examples. Interestingly, in the choanoflagellate Monosiga and several stramenopiles there is a lineage-specific expansion of an extracellular version of the FIH1 family, some of which are fused to a sulfotransferase domain. This sulfotransferase domain is also expanded in stramenopiles and found fused to or functionally linked to other 2OGFeDOs in several bacteria (see below). (iii) The third clade is comprised of a single family prototyped by the eukaryotic nucleolar protein MINA/No66. While all previously known eukaryotic members of this clade are intracellular proteins, we identified a lineage-specific expansion of this clade in Monosiga that comprises of receptor-like proteins with extracellular JOR/JmjC domains. All of these proteins combine a JOR/JmjC domain with one or more of several extracellular domains such as cysteine-rich GCC2/3 repeats, immunoglobulin, disintegrin or SUSHI domains and with intracellular SH2 or tyrosine kinase domains (Figure 4). These extracellular examples of the FIH1 and MINA appear to be the first examples of this superfamily that have been recruited for modifying cell surface proteins, probably as hydroxylases similar to leprecan and the prolyl hydroxylase. Further, the receptor-like MINA proteins could also function as sensors of redox conditions that signal via intracellular tyrosine phosphorylation pathways.
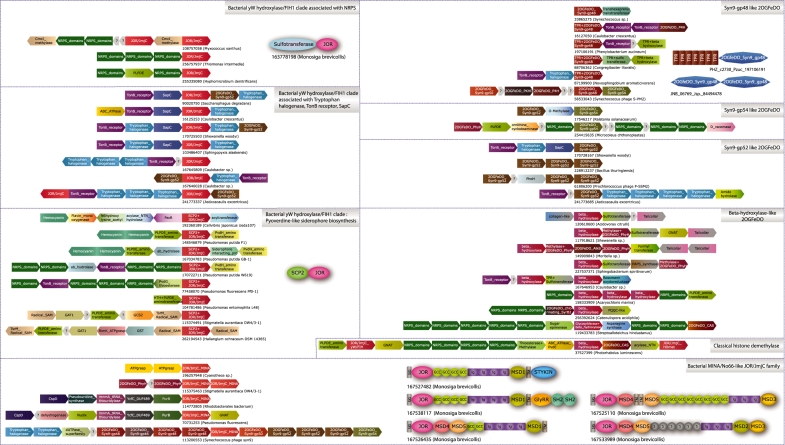
Figure 4.
Gene neighborhoods and domain architectures of various JOR/JmjC domains and classical 2OGFeDO containing systems described in this study. Genes are represented as arrows pointing from the 5′ to the 3′ end of the coding frame. Predicted operons are labeled with the gi and species name of the primary JOR/JmjC or 2OGFeDO enzyme in that context. Gene neighborhoods and domain architectures are arranged based on the primary JOR/JmjC or 2OGFeDO enzymes being described in a block. Standard domain names are used in domain architectures. MSD1-5 domains represent distinct Monosiga-specific domains that are predicted to be extracellular. MSD2, MSD4 and MSD5 also posses conserved cysteine residues. GlyRR denotes a distinct domain with glycine rich repeats that is also found in some prokaryotes in standalone form. S denotes sushi repeats. Note that in some antibiotic biosynthesis clusters, we only emphasize the genes in the immediate neighborhood of the JOR/JmjC or classical 2OGFeDO genes, as some of these are much larger gene arrays.
Upon addition of bacterial members to the phylogenetic analysis, we discovered that each of the above three clades had their own basal bacterial representatives within which the eukaryotic lineages were nested. Bacterial versions are sporadically, but widely distributed across the bacterial tree (Table 1) and also in certain bacteriophages (e.g. Synechococcus phage syn9 gp49, gi: 113200554, is a representative of the MINA clade; Table 1, Figure 3). This pattern suggests that the primary radiation of the JOR/JmjC family into three distinct clades happened in bacteria. This was followed by a transfer of at least one member from each of the three clades to the eukaryotes, prior to the divergence of the heterolobosean–kinetoplastid clade and the remaining eukaryotes. This scenario implies that after the acquisition from bacteria of ancestral members of the three clades of JOR/JmjC superfamily they extensively radiated within eukaryotes (Figure 3). This also meant that diversification of RNA-modifying yW hydroxylases and the peptide-modifying versions contained in the same clade (e.g. FIH1) happened in eukaryotes. To better understand the ancestral condition, we investigated potential functions and substrate specificities of bacterial members of the superfamily.
Gene neighborhoods and domain architectures of bacterial JOR/JmjC domains point to novel functional linkages
To investigate the roles of the bacterial JOR/JmjC superfamily proteins, we conducted a systematic survey of their gene neighborhoods and domain architectures. Such contextual analysis has greatly aided the decipherment of the function of uncharacterized proteins in terms of their biochemical partners and physical interactions (57–60). Unlike their eukaryotic counterparts, majority of the bacterial versions occur as single domain proteins. We analyzed the contextual information separately for the bacterial representatives of each of the major clades of JOR/JmjC proteins. A subset of the versions belonging to the clade that includes FIH1 and yW hydroxylase are encoded in neighborhoods along with genes for large multidomain non-ribosomal peptide synthetases in proteobacteria, bacteroidetes and cyanobacteria (Figure 4). In myxobacteria (e.g. Stigmatella and Haliangium), there are related gene neighborhoods that combine the JOR/JmjC gene with genes for distinct peptide synthetases of the ATP-grasp or COOH-NH2 ligase superfamilies (60). Additionally, most of these gene neighborhoods encode several proteins that could be involved in further peptide modifications. These include a methyltransferase closely related to CmcI, which is involved in methylating a hydroxyl group in the synthesis of cephamycin C (61), pyridoxal phosphate-dependent aminotransferases that synthesize novel amino acids, radical SAM-dependent enzymes and hemocyanin-like redox enzymes. One interesting group of these gene neighborhoods presents the JOR/JmjC gene embedded in the hypervariable part of the gene cluster required for the synthesis of the pyoverdine-like siderophores (Figure 4). These versions are distinguished by presence of a lipid/hydrophobic small-molecule-binding SCP2 domain fused to their N-termini. The remaining versions from diverse α- and γ-proteobacteria are part of a distinctive conserved gene neighborhood that might additionally encode one to three tryptophan halogenases, an outer-membrane receptor of the TonB-class and a gene prototyped by the Campylobacter fetus SapC (62). Given that the tryptophan halogenase homologs have an intact Rossmann-fold domain and a conserved lysine required to form a chloramine adduct, they are indeed likely to function as halogenating enzymes (63). Some versions of this gene neighborhood additionally encode a second predicted hydroxylase that belongs to the classical 2OGFeDO superfamily (Figure 4). The sporadic distribution of these gene neighborhoods and lack of congruence in the phylogeny of the JOR/JmjC protein encoded by them with species tree are suggestive of dissemination via lateral transfer (Supplementary Data).
As mentioned earlier, predicted gene neighborhoods encoding certain bacterial members of the classical histone demethylase clade show linkages to genes encoding multidomain non-ribosomal peptide synthetases (Figure 4). One such gene cluster from Photorhabdus has 12 linked genes, which includes, in addition to four modular peptide synthetase genes, three other distinct dioxygenase genes. Two of these encoded proteins belong to the clavaminate synthase (64) and β-hydroxylase families (54,65) of the classical 2OGFeDO superfamily and one of them a JOR/JmjC protein of the yW-hydroxylase/FIH1 clade. Additionally, this cluster also encodes genes for a GCN5-like acetyltransferase, diaminobutyrate aminotransferase, methyltransferase and a penicillin acylase-like enzyme (Figure 4). Similarly, the MINA/No66 clade also occurs in gene clusters along with genes for peptide synthetases, although in this case of the ATP-grasp superfamily (Figure 4) (60). Other MINA/No66-like proteins from cyanophages are encoded by a set of remarkable gene neighborhoods that consist of a tandem array of multiple dioxygenases (Figure 4). For example, in the Synechococcus phage Syn9 one of these gene clusters encodes 10 tandem dioxygenases including the MINA/No66 homolog, Syn9-gp49. The remaining nine dioxygenases belong to the classical 2OGFeDO superfamily. Analysis of these nine dioxygenases suggests that they are all not closely related. They belong to at least five distinct families, including one distinguished by a fusion to tetratricopeptide repeats (Figure 4; Supplementary Data). One of the 2OGFEDOs encoded by this gene cluster, namely Syn9-gp52, belongs to the same family as the second dioxygenase that is occasionally present in the above-described gene neighborhoods encoding JOR/JmjC proteins of FIH1/yW hydroxylase-containing clade (Figure 4). In addition to the 10 dioxygenases, this gene neighborhood contains two genes encoding tryptophan halogenases and a gene encoding a novel protein with a conserved cysteine and the same fold as the dUTPases (Figure 4).
A key theme emerging from all the above examples is the combination of multiple, distinct dioxygenases and tryptophan halogenase-type enzymes with other enzymes involved in peptide synthesis and modification in a single gene cluster. Importantly, the multiple dioxygenases in the same gene cluster often do not appear to have arisen solely through recent duplication. In many cases, they are distant homologs that might even belong to different superfamilies (i.e. the classical 2OGFeDO and JOR/JmjC; Figure 4) and appear to have been assembled into predicted operons due to selection for co-functionality in a common pathway. To explore this feature systematically, we first identified in complete genomes all members of the distinct families of dioxygenases represented in the above gene neighborhoods other than members of the JOR/JmjC superfamily with which we originally started. We then further investigated each of these dioxygenase families for conserved gene neighborhoods in order to capture additional functional associations. Interestingly, this search recovered several additional gene-neighborhood associations combining genes for dioxygenases and tryptophan halogenase-type enzymes. For example, certain Syn9-gp52-like genes occurred with tryptophan halogenase, SapC and TonB-like receptor genes, but independently of any JOR/JmjC superfamily members. In firmicutes, we found a conserved neighborhood comprised of a tandem array of three Syn9-gp52-like genes. Yet, others (e.g. versions related to the phage Syn9-gp52 and gp54 2OGFEDOs) occurred in conserved gene neighborhoods, which contained, in addition to genes encoding classical 2OGFeDOs, tryptophan halogenases and TonB-like receptors, genes for hexaprenyltranstransferases, sulfotransferases and non-ribosomal peptide synthetases. Two other families of 2OGFeDOs, whose genes frequently co-occur in gene-clusters with those encoding other dioxygenases, are bacterial cognates of the amino acid β-hydroxylases, which hydroxylate prolines, asparatates and asparagines (54,65), and the phytanoyl CoA hydroxylases (66). Multiple β-hydroxylases that co-occur in gene clusters for the synthesis of glycopeptide antibiotics such as bleomycin and tallysomycin have been characterized as enzymes that hydroxylate different amino acids or their derivatives in the peptide backbone of these molecules (67). Several uncharacterized gene neighborhoods that combine β-hydroxylase genes with non-ribosomal peptide synthesis systems (Figure 4; Supplementary Data) that we found in actinobacteria and rhizobia might represent other such novel peptide metabolites. We also uncovered previously uncharacterized conserved gene neighborhoods that combine a gene of the β-hydroxylase family, with those encoding phytanoyl CoA hydroxylases fused to methyltransferase domains and sulfotransferases that are related to those fused to Monosiga FIH1 family JOR/JmjC domains. Several of these predicted operons also encode PAPS synthetases, acetyltransferases AlkB-like dioxygenases and either or both of two types of non-enzymatic proteins (Figure 4): (i) a member of the bacteriophage tail–collar family prototyped by the phage T4 short tail–fiber protein (68). 2) Secreted glycine-rich peptides, some of which have a similar pattern of tri-peptide repeats as seen in animal collagen (e.g. Aave_1921, gi: 120610601 from Acidovorax).
Bacterial JOR/JmjC and 2OGFeDO domains proliferated in the context of peptide modification and secondary metabolite biosynthesis
The presence of multiple functionally linked dioxygenases and tryptophan halogenase-type enzymes in the above gene neighborhoods provided key evidence for uncovering their possible functions. In the well-studied biosynthetic pathway for the antibiotic cephamycin C, four dioxygenases of the classical 2OGFeDO superfamily act in tandem on a tripeptide to synthesize the final product (61,69). The tripeptide is synthesized first by ACV synthetase, a non-ribosomal peptide synthetase. The first dioxygenase is isopenicillin synthase that synthesizes isopenicillin from the tripeptide. The second dioxygenase, DAOC synthase, converts penicillin N to deacetoxycephalosporin via hydroxylation, while the third dioxygenase in this pathway converts this compound to deacetylcephalosporin via a further hydroxylation. The fourth hydroxylation is catalyzed by CmcJ resulting in an unmethylated precursor, which is then methylated to cephamycin C by CmcI. CmcJ was previously not considered as belonging to the 2OGFeDO superfamily (61); however, sequence profile searches reveal that it is indeed a classical 2OGFeDO that is only distantly related to the three other 2OGFeDO in this pathway. A similar pattern of multiple dioxygenases combined with non-ribosomal peptide synthesis systems are also seen in other antibiotic pathways such as those of the bleomycin family (see above). Halogenation of aromatic amino acids like tryptophan and tyrosine have been shown to be catalyzed by tryptophan halogenase-like proteins in the synthesis of antibiotic secondary metabolites like indolocarbazoles (e.g. rebeccamycin), pyrroindomycin B, pyrrolnitrin and chondrochloren (70–73). While being evolutionarily related, different Rossmann-fold halogenases have been found to show specificities for distinct positions of the indole or phenol rings or different halogens (i.e. Cl or Br). In a given metabolite, there could be multiple successive halogenations of different positions with the same or different type of halogen. Halogenated amino acids are usually further condensed or combined with long-chain fatty acids or glycosylated to generate terminal metabolites. Similarly, certain members of the phytanoyl-CoA dioxygenase family (66) of the classical 2OGFeDO superfamily (e.g. SyrB2, CmaB and HctB), lacking the acidic residue in the HXD/E signature, act as halogenases of amino acids such as l-allo-isoleucine and threonine and fatty acids such as hexanoic acid by forming a hypohalous acid through the primary dioxygenase reaction (15,17,74).
The template provided by the cephamycin C biosynthetic pathway and the above-described observations on secondary metabolite biosynthesis strongly implicates the bacterial JOR/JmjC, classical 2OGFeDOs and tryptophan halogenase-like enzymes that we recovered as co-occurring in gene neighborhoods (Figure 4) in synthesis of amino acid-derived secondary metabolites via successive halogenations and hydroxylations. The different types of non-ribosomal peptide synthetases present in some of these gene neighborhoods are likely to provide the initial peptide substrates on which the dioxygenases and halogenases catalyze additional modifications. The evidence suggests that some of these secondary metabolites are likely to be siderophores (Figure 4). In the case of the pyoverdine gene cluster the hypervariable segment, which encodes the JOR/JmjC protein, also usually encodes several distinct enzymes that differ from strain to strain, even within the same species. Thus, the products of this segment catalyze modifications that might primarily help in the diversification of the peptide and chromophore core derivative and consequently result in structurally distinct pyoverdines (75; Figure 4). This presumably helps bacteria prevent siderophore-stealing by environmental competitors. We found that the TonB-like receptor found in several of the conserved gene neighborhoods encoding JOR/JmjC proteins (Figure 4) is most closely related to siderophore-uptake receptors. Based on this, we predict that the conserved gene neighborhood is likely to be involved in synthesis of a previously unknown halogenated siderophore-like compound from amino acid derivatives. In this case, the tryptophan halogenases are likely to halogenate tryptophan, whereas the JOR/JmjC and classical 2OGFeDO proteins is likely to catalyze hydroxylations as they retain a typical HXD/E signature. Based on its conservation pattern (Supplementary Data), we also predict that the SapC protein found in these gene neighborhoods is likely to function as an as yet uncharacterized enzyme in the synthesis of the same metabolite. Similar gene neighborhoods with other modifying enzymes appear to be required for the synthesis of uncharacterized secondary metabolites through further modifications of the hydroxyl groups generated by the dioxygenases by methylation, sulfation and perhaps acylation (Figure 4). For predicted operons that combine the 2OGFeDOs of the β-hydroxylase and phytanoyl CoA hydroxylase families with sulfotransferases, we propose that the collagen-like or tail–collar domain proteins that are co-encoded with them are substrates of the former enzymes. As with eukaryotic cell surface and extracellular matrix proteins, we predict that these substrates are first hydroxylated by functionally linked dioxygenases and the hydroxyl group is then sulfated. This indicates that not only collagen-like proteins but also the hydroxylation system for them emerged first in the bacterial world.
The remarkable cyanophage and firmicute gene neighborhoods with several tandem dioxygenases and halogenases are rather unprecedented and have not been observed before. Studies have shown that cyanobacteria produce a rich diversity of organo-halogen compounds such as nostocyclophanes, halogenated indole isonitriles and cryptophycins (76,77). Structures of these molecules indicate that their synthesis would require multiple halogenations and hydroxylation steps, but the biosynthetic mechanism for these unusual bioactive molecules remains largely unknown. We suggest that these phage-encoded gene clusters are probably a source for some these biosynthetic capabilities of the cyanobacteria. Furthermore, we also noted that homologs of several of these cyanophage dioxygenases are found in choanoflagellates, diatoms and chlorophyte algae, including lineage-specific expansions of certain representatives (e.g. of homologs of Syn9-gp54 in diatoms, Supplementary Data). It is possible that the above dioxygenases have a role in the synthesis of novel secondary metabolites even in these eukaryotes.
Thus, the weight of the evidence from the above observations indicates that the bacterial members of the JOR/JmjC superfamily primarily modify peptides in secondary metabolite biosynthesis. This, taken together with the predominance of peptide substrates among most characterized eukaryotic JOR/JmjC proteins, strongly supports a peptide-modifying role for the common ancestor of the JOR/JmjC superfamily in the context of bacterial secondary metabolism. Hence, the RNA-modifying activity of the yW hydroxylase is a secondary innovation, probably accommodated by the structural resemblance of the target moiety in yW to an amino acid substrate typical of JOR/JmjC enzymes (Figure 1). Furthermore, the secondary metabolite biosynthesis gene clusters also point to the large-scale independent recruitment of several families of the classical 2OGFeDO superfamily and some members of the cupin superfamily (16,55) (Figure 4, Supplementary Data). These include some representatives of originally nucleic acid-modifying families like AlkB (e.g. Moritella sp. gi: 149909845; Figure 4). These observations, together with our prediction that some bacterial β-hydroxylases probably modify collagen-like proteins, suggest that precursors of other eukaryotic peptide-modifying enzymes of the 2OGFeDO superfamily also emerged in the radiation of these enzymes in the bacterial peptide-modification systems. Certain evidence suggests that secondary recruitment for a RNA-related function could have occurred on a second occasion in the JOR/JmjC superfamily. Bacterial members of the MINA/No66 clade from a subset of proteobacteria are present in a conserved gene neighborhood that also encodes a pseudouridine synthase, the tRNA thiouridylase mnmA and the adenylosuccinate lyase PurB involved in purine biosynthesis (Figure 4). Hence, it could have potentially acquired a RNA-associated role. Eukaryotic MINA has been shown to localize to the nucleolus and is associated with pre-ribosomal complexes (78). It has also been shown to bind DNA (79) and is claimed to function as a histone demethylase (80,81). Nevertheless, in light of the possible RNA-related role of some bacterial MINA proteins and nucleolar localization of eukaryotic members, it remains to be seen if it might modify RNA or RNA-associated proteins.
General conclusions
The primary split in the DSBH fold appears to have been between the flattened versions that bind substrates externally and the more barrel-like forms that bind the metal co-factor and substrates internally (Figure 3). In the former clade, the viral capsid proteins are widely distributed across otherwise unrelated RNA and DNA viruses. The nucleoplasmin lineage is currently restricted to eukaryotes and the Cu(II)-dependent monooxygenases are of bacterial origin, with some late transfers to eukaryotes, e.g. in the metazoan clade (38,82). Previous studies on viral packaging ATPases, which function in a similar context as the capsid proteins, suggest that these are probably part of an early pre-cellular system for nucleic acid compartmentalization (83,84). In contrast, phyletic patterns of the remaining flattened DSBH representatives are suggestive of a much later derivation, perhaps from a viral capsid-like precursor. In the barrel-like DSBH clade, the cNMP-binding domains are encountered in archaea and bacterial along with distinct families of cyclic nucleotide-generating enzymes (21,85). RmlC-like sugar isomerases are found in all superkingdoms of life (16). However, the metal-binding cupin domains are relatively rare in archaea, show lower median counts, more restricted phyletic patterns, and are usually nested within larger clades of predominantly bacterial versions (16,21). In contrast, they are extremely widespread in bacteria. The classical 2OGFeDOs and the JOR/JmjC superfamilies are rarely or never observed in archaea (14,23). Together these observations suggest that the expansion of the metal-binding DSBH domains happened in the bacteria. The use of molecular oxygen by majority of DSBH metal-binding dioxygenases suggests that they probably rapidly diversified after cyanobacterial photosynthesis had made ambient oxygen available around 2.45 to 2.2 billion years ago (86,87; Figure 3). Furthermore, use of 2-oxoglutarate by two of the dioxygenase superfamilies suggests these superfamilies probably depended on a functional tricarboxylic acid cycle (TCA) for abundant supply of this metabolite. Hence, the expansion of aerobic metabolism in bacteria, resulting in an abundance of TCA-derived 2-oxoglutarate probably gave the impetus for the radiation of classical 2OGFeDOs and the JOR/JmjC superfamilies from a dioxygenase cupin-like precursor.
Another key finding in this work is that the major lineages of the cupin, JOR/JmjC and classical 2OGFeDO superfamilies had already radiated in the context of bacterial secondary metabolism prior to being transferred individually to eukaryotes. In eukaryotes representatives of many distinct families of DSBH domains were deployed as regulatory proteins that either non-catalytically bind peptides (e.g. the cupin CENP-C) or modify peptides in proteins such as chromatin components (e.g. histone demethylases, FIH1, and HIF) and extracellular matrix proteins [e.g. extracellular versions of FIH1 and MINA, leprecan, prolyl and lysyl hydroxylases of collagen (14,16)]. In this respect, a remarkable parallel is observed with the evolution of other regulatory peptide-modifying systems of eukaryotes. Precursors of the ubiquitin-conjugating systems (namely E1s and Ub-like proteins), amino acid conjugating ATP-grasp ligases (e.g. tyrosine tubulin ligase and polyglutamylases), GCN5-like acetyltransferase fold enzymes (histone acetylases and N-end rule amino acid ligases), methylases (e.g. protein arginine methylases), sulfotransferases (e.g. tyrosine sulfotransferase) and COOH–NH2 ligases (chromatin protein amino acid ligases) have emerged within bacterial amino acid and peptide-derived secondary metabolite biosynthesis systems (60,88–90). Thus, exaptation of enzymes that originally diversified in bacterial metabolic systems appears to have been a general theme in the origin of eukaryotic regulatory peptide modifications. It is conceivable that the elaboration of peptide modification systems in eukaryotes derived from bacterial precursors went hand-in-hand with proliferation of low complexity sequence extensions to globular domains in eukaryotes (e.g. histone tails and collagen-like tripeptide repeats; 91,92). These low-complexity sequences are either unstructured and solution-exposed, or have periodic repetitive patterns of accessible amino acids that potentially resembled the peptide substrates of the ancestral bacterial enzymes. Hence, these could have served as new substrate-niches that were colonized by these enzymes. Many of the peptide modification systems appear to be very early acquisitions from bacteria that contributed to the emergence of quintessentially eukaryotic features (38,60,89). But acquisition of peptide-modifying enzymes from bacteria appears to have continued throughout eukaryotic evolution. In this regard, our prediction that, in addition to modifying non-ribosomally synthesized peptides, some of these bacterial enzymes probably modify collagen-like proteins may point to a more direct recruitment of certain modification systems by eukaryotes. Thus, acquisition of bacterial collagen-like proteins and their modifying hydroxylases and sulfotransferases, might have contributed to major transitions in eukaryotes such as the origin of extracellular matrices and consequently multicellularity.
We had previously shown that the classical 2OGFeDOs had undergone considerable radiation in bacteria and phages to spawn several distinct nucleic acid-modifying enzymes, which were transferred to eukaryotes on several independent occasions, such as AlkB and multiple representatives of the Tet/JBP family (13,14). With prediction of the yW hydroxylase/peroxidase we present evidence for the first time that a member of the JOR/JmjC superfamily modifies nucleic acids. The findings presented here could potentially inspire further experimental test for some of the predictions and further investigations on this major class of proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online and also available at ftp://ftp.ncbi.nih.gov/pub/aravind/WYBUTOSINE/wybutosine.html
FUNDING
Intramural funds of the National Library of Medicine at the National Institutes of Health, USA. Funding for open access charge: National Institutes of Health, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 4.McCloskey JA, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, Stetter KO. Structure determination of a new fluorescent tricyclic nucleoside from archaebacterial tRNA. Nucleic Acids Res. 1987;15:683–693. doi: 10.1093/nar/15.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCloskey JA, Graham DE, Zhou S, Crain PF, Ibba M, Konisky J, Soll D, Olsen GJ. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbonavičius J, Droogmans L, Armengaud J, Grosjean H. Deciphering the Complex Enzymatic Pathway for Biosynthesis of Wyosine Derivatives in Anticodon of tRNAPhe. Austin, TX: Landes Bioscience; 2009. [Google Scholar]
- 7.Kaminska KH, Baraniak U, Boniecki M, Nowaczyk K, Czerwoniec A, Bujnicki JM. Structural bioinformatics analysis of enzymes involved in the biosynthesis pathway of the hypermodified nucleoside ms(2)io(6)A37 in tRNA. Proteins. 2008;70:1–18. doi: 10.1002/prot.21640. [DOI] [PubMed] [Google Scholar]
- 8.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J. Mol. Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Balaji S, Aravind L. The RAGNYA fold: a novel fold with multiple topological variants found in functionally diverse nucleic acid, nucleotide and peptide-binding proteins. Nucleic Acids Res. 2007;35:5658–5671. doi: 10.1093/nar/gkm558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasai H, Yamaizumi Z, Kuchino Y, Nishimura S. Isolation of hydroxy-Y base from rat liver tRNAPhe. Nucleic Acids Res. 1979;6:993–999. doi: 10.1093/nar/6.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakanishi K, Blobstein S, Funamizu M, Furutachi N, Van Lear G, Grunberger D, Lanks KW, Weinstein IB. Structure of the "peroxy-Y base" from liver tRNA Phe. Nat. New Biol. 1971;234:107–109. doi: 10.1038/newbio234107b0. [DOI] [PubMed] [Google Scholar]
- 13.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neidig ML, Brown CD, Light KM, Fujimori DG, Nolan EM, Price JC, Barr EW, Bollinger JM, Jr, Krebs C, Walsh CT, et al. CD and MCD of CytC3 and taurine dioxygenase: role of the facial triad in alpha-KG-dependent oxygenases. J. Am. Chem. Soc. 2007;129:14224–14231. doi: 10.1021/ja074557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Vaillancourt FH, Yin J, Walsh CT. SyrB2 in syringomycin E biosynthesis is a nonheme FeII alpha-ketoglutarate- and O2-dependent halogenase. Proc. Natl Acad. Sci. USA. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thornburg LD, Lai MT, Wishnok JS, Stubbe J. A non-heme iron protein with heme tendencies: an investigation of the substrate specificity of thymine hydroxylase. Biochemistry. 1993;32:14023–14033. doi: 10.1021/bi00213a036. [DOI] [PubMed] [Google Scholar]
- 19.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 20.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem. Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- 24.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 25.Dunwell JM, Khuri S, Gane PJ. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 2000;64:153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassmann T, Frings O, Sonnhammer EL. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009;37:858–865. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 34.Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Protein. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 36.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki Y, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res. 2009;37:2910–2925. doi: 10.1093/nar/gkp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 39.Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function, and Evolution. Austin, TX: Landes Bioscience; 2009. [Google Scholar]
- 40.Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. [PubMed] [Google Scholar]
- 41.Ahn HJ, Kim HW, Yoon HJ, Lee BI, Suh SW, Yang JK. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J. 2003;22:2593–2603. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J. Mol. Biol. 2003;333:931–949. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson BA, Kwon SY, Chamorro M, Oroszlan S, Hatfield DL, Lee BJ. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- 45.Carlson BA, Mushinski JF, Henderson DW, Kwon SY, Crain PF, Lee BJ, Hatfield DL. 1-Methylguanosine in place of Y base at position 37 in phenylalanine tRNA is responsible for its shiftiness in retroviral ribosomal frameshifting. Virology. 2001;279:130–135. doi: 10.1006/viro.2000.0692. [DOI] [PubMed] [Google Scholar]
- 46.Siebert X, Eipper BA, Mains RE, Prigge ST, Blackburn NJ, Amzel LM. The catalytic copper of peptidylglycine alpha-hydroxylating monooxygenase also plays a critical structural role. Biophys. J. 2005;89:3312–3319. doi: 10.1529/biophysj.105.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong C, Major LL, Srikannathasan V, Errey JC, Giraud MF, Lam JS, Graninger M, Messner P, McNeil MR, Field RA, et al. RmlC, a C3′ and C5′ carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation. J. Mol. Biol. 2007;365:146–159. doi: 10.1016/j.jmb.2006.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 49.Balciunas D, Ronne H. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 2000;25:274–276. doi: 10.1016/s0968-0004(00)01593-0. [DOI] [PubMed] [Google Scholar]
- 50.Kannan N, Wu J, Anand GS, Yooseph S, Neuwald AF, Venter JC, Taylor SS. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007;8:R264. doi: 10.1186/gb-2007-8-12-r264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berrisford JM, Akerboom J, Brouns S, Sedelnikova SE, Turnbull AP, van der Oost J, Salmon L, Hardre R, Murray IA, Blackburn GM, et al. The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis. J. Mol. Biol. 2004;343:649–657. doi: 10.1016/j.jmb.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 52.Wu B, Zhang Y, Zheng R, Guo C, Wang PG. Bifunctional phosphomannose isomerase/GDP-D-mannose pyrophosphorylase is the point of control for GDP-D-mannose biosynthesis in Helicobacter pylori. FEBS Lett. 2002;519:87–92. doi: 10.1016/s0014-5793(02)02717-5. [DOI] [PubMed] [Google Scholar]
- 53.Roux C, Lee JH, Jeffery CJ, Salmon L. Inhibition of type I and type II phosphomannose isomerases by the reaction intermediate analogue 5-phospho-D-arabinonohydroxamic acid supports a catalytic role for the metal cofactor. Biochemistry. 2004;43:2926–2934. doi: 10.1021/bi035688h. [DOI] [PubMed] [Google Scholar]
- 54.Korioth F, Gieffers C, Frey J. Cloning and characterization of the human gene encoding aspartyl beta-hydroxylase. Gene. 1994;150:395–399. doi: 10.1016/0378-1119(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 55.Rajavel M, Mitra A, Gopal B. Role of Bacillus subtilis BacB in the synthesis of bacilysin. J. Biol. Chem. 2009;284:31882–31892. doi: 10.1074/jbc.M109.014522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR, III, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 57.Maltsev N, Glass E, Sulakhe D, Rodriguez A, Syed MH, Bompada T, Zhang Y, D'Souza M. PUMA2–grid-based high-throughput analysis of genomes and metabolic pathways. Nucleic Acids Res. 2006;34:D369–D372. doi: 10.1093/nar/gkj095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huynen M, Snel B, Lathe W., III, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol. Biosyst. 2009;5:1636–1660. doi: 10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oster LM, Lester DR, Terwisscha van Scheltinga A, Svenda M, van Lun M, Genereux C, Andersson I. Insights into cephamycin biosynthesis: the crystal structure of CmcI from Streptomyces clavuligerus. J. Mol. Biol. 2006;358:546–558. doi: 10.1016/j.jmb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 62.Thompson SA, Shedd OL, Ray KC, Beins MH, Jorgensen JP, Blaser MJ. Campylobacter fetus surface layer proteins are transported by a type I secretion system. J. Bacteriol. 1998;180:6450–6458. doi: 10.1128/jb.180.24.6450-6458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT. Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochemistry. 2007;46:1284–1292. doi: 10.1021/bi0621213. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z, Ren J, Stammers DK, Baldwin JE, Harlos K, Schofield CJ. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat. Struct. Biol. 2000;7:127–133. doi: 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- 65.Mori H, Shibasaki T, Yano K, Ozaki A. Purification and cloning of a proline 3-hydroxylase, a novel enzyme which hydroxylates free L-proline to cis-3-hydroxy-L-proline. J. Bacteriol. 1997;179:5677–5683. doi: 10.1128/jb.179.18.5677-5683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherji M, Chien W, Kershaw NJ, Clifton IJ, Schofield CJ, Wierzbicki AS, Lloyd MD. Structure-function analysis of phytanoyl-CoA 2-hydroxylase mutations causing Refsum's disease. Hum. Mol. Genet. 2001;10:1971–1982. doi: 10.1093/hmg/10.18.1971. [DOI] [PubMed] [Google Scholar]
- 67.Tao M, Wang L, Wendt-Pienkowski E, George NP, Galm U, Zhang G, Coughlin JM, Shen B. The tallysomycin biosynthetic gene cluster from Streptoalloteichus hindustanus E465-94 ATCC 31158 unveiling new insights into the biosynthesis of the bleomycin family of antitumor antibiotics. Mol. Biosyst. 2007;3:60–74. doi: 10.1039/b615284h. [DOI] [PubMed] [Google Scholar]
- 68.Thomassen E, Gielen G, Schutz M, Schoehn G, Abrahams JP, Miller S, van Raaij MJ. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J. Mol. Biol. 2003;331:361–373. doi: 10.1016/s0022-2836(03)00755-1. [DOI] [PubMed] [Google Scholar]
- 69.Liras P, Demain AL. Chapter 16. Enzymology of beta-lactam compounds with cephem structure produced by actinomycete. Methods Enzymol. 2009;458:401–429. doi: 10.1016/S0076-6879(09)04816-2. [DOI] [PubMed] [Google Scholar]
- 70.Sanchez C, Zhu L, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc. Natl Acad. Sci. USA. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu X, De Laurentis W, Leang K, Herrmann J, Ihlefeld K, van Pee KH, Naismith JH. Structural insights into regioselectivity in the enzymatic chlorination of tryptophan. J. Mol. Biol. 2009;391:74–85. doi: 10.1016/j.jmb.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy CD. Recent developments in enzymatic chlorination. Nat. Prod. Rep. 2006;23:147–152. doi: 10.1039/b516588c. [DOI] [PubMed] [Google Scholar]
- 73.Buedenbender S, Rachid S, Muller R, Schulz GE. Structure and action of the myxobacterial chondrochloren halogenase CndH: a new variant of FAD-dependent halogenases. J. Mol. Biol. 2009;385:520–530. doi: 10.1016/j.jmb.2008.10.057. [DOI] [PubMed] [Google Scholar]
- 74.Ramaswamy AV, Sorrels CM, Gerwick WH. Cloning and biochemical characterization of the hectochlorin biosynthetic gene cluster from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2007;70:1977–1986. doi: 10.1021/np0704250. [DOI] [PubMed] [Google Scholar]
- 75.Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 2005;187:2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gribble GW. Naturally occurring organohalogen compounds—a comprehensive survey. Fortschr. Chem. Org. Naturst. 1996;68:1–423. [PubMed] [Google Scholar]
- 77.Gribble GW. The diversity of naturally occurring organobromine compounds. Chem. Soc. Rev. 1999;28:335–346. [Google Scholar]
- 78.Eilbracht J, Reichenzeller M, Hergt M, Schnolzer M, Heid H, Stohr M, Franke WW, Schmidt-Zachmann MS. NO66, a highly conserved dual location protein in the nucleolus and in a special type of synchronously replicating chromatin. Mol. Biol. Cell. 2004;15:1816–1832. doi: 10.1091/mbc.E03-08-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okamoto M, Van Stry M, Chung L, Koyanagi M, Sun X, Suzuki Y, Ohara O, Kitamura H, Hijikata A, Kubo M, et al. Mina, an Il4 repressor, controls T helper type 2 bias. Nat. Immunol. 2009;10:872–879. doi: 10.1038/ni.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010;29:68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Y, Chang Q, Zhang Y, Beezhold K, Rojanasakul Y, Zhao H, Castranova V, Shi X, Chen F. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle. 2009;8:2101–2109. doi: 10.4161/cc.8.13.8927. [DOI] [PubMed] [Google Scholar]
- 82.Iyer LM, Aravind L, Coon SL, Klein DC, Koonin EV. Evolution of cell-cell signaling in animals: did late horizontal gene transfer from bacteria have a role? Trends Genet. 2004;20:292–299. doi: 10.1016/j.tig.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 83.Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 84.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 85.Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 1999;289:729–745. doi: 10.1006/jmbi.1999.2827. [DOI] [PubMed] [Google Scholar]
- 86.Frei R, Gaucher C, Poulton SW, Canfield DE. Fluctuations in Precambrian atmospheric oxygenation recorded by chromium isotopes. Nature. 2009;461:250–253. doi: 10.1038/nature08266. [DOI] [PubMed] [Google Scholar]
- 87.Kopp RE, Kirschvink JL, Hilburn IA, Nash CZ. The Paleoproterozoic snowball Earth: a climate disaster triggered by the evolution of oxygenic photosynthesis. Proc. Natl Acad. Sci. USA. 2005;102:11131–11136. doi: 10.1073/pnas.0504878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 89.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 91.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 92.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]