Abstract
We have shown that ent-kaurenoic acid oxidase, a member of the CYP88A subfamily of cytochrome P450 enzymes, catalyzes the three steps of the gibberellin biosynthetic pathway from ent-kaurenoic acid to GA12. A gibberellin-responsive barley mutant, grd5, accumulates ent-kaurenoic acid in developing grains. Three independent grd5 mutants contain mutations in a gene encoding a member of the CYP88A subfamily of cytochrome P450 enzymes, defined by the maize Dwarf3 protein. Mutation of the Dwarf3 gene gives rise to a gibberellin-responsive dwarf phenotype, but the lesion in the gibberellin biosynthesis pathway has not been identified. Arabidopsis thaliana has two CYP88A genes, both of which are expressed. Yeast strains expressing cDNAs encoding each of the two Arabidopsis and the barley CYP88A enzymes catalyze the three steps of the GA biosynthesis pathway from ent-kaurenoic acid to GA12. Sequence comparison suggests that the maize Dwarf3 locus also encodes ent-kaurenoic acid oxidase.
Gibberellins (GAs) are an important class of plant hormone involved in the regulation of processes from seed germination through the development and reproduction of plants (1). GA biosynthesis is catalyzed by three classes of enzymes: terpene cyclases catalyze the synthesis of ent-kaurene from geranylgeranyl diphosphate; cytochrome P450 monooxygenases catalyze the steps of the pathway from ent-kaurene to GA12; and soluble dioxygenases catalyze the final steps of the pathway.
Genes encoding enzymes for the two terpene cyclase-catalyzed steps, copalyl diphosphate synthase and kaurene synthase, have been isolated from a number of species (2, 3). Similarly, a wide range of 2-oxoglutarate-dependent dioxygenases encoding GA 7-oxidase, GA 20-oxidase, GA 2β, 3β-hydroxylase, GA 3β-hydroxylase, and GA 2-oxidase activities have been isolated (2, 4–6). GA3 from Arabidopsis thaliana is the only gene encoding a cytochrome P450-mediated step of GA biosynthesis isolated (7), although the maize Dwarf3 gene is likely also to be involved in GA biosynthesis, and it encodes a member of the CYP88A subfamily of cytochrome P450 enzymes (8). GA3 encodes the cytochrome P450 CYP701A3 (7) and has been shown to catalyze the three-step oxidation of ent-kaurene to ent-kaurenoic acid (KA) (9). In line with current nomenclature for GA biosynthesis enzymes and the corresponding genes (10), we will refer to GA3 as AtKO1. The genes encoding the enzymes catalyzing the conversion of KA to ent-7α-hydroxy-kaurenoic acid (7OH-KA) and 7OH-KA to GA12-aldehyde have not been isolated, but it is known that these reactions are likely to be catalyzed by cytochrome P450s requiring O2 and NADPH for activity (11).
The maize Dwarf3 gene encodes the cytochrome P450 CYP88A1 (8). The dwarf3 mutant is a GA-responsive dwarf (12); however, the reaction catalyzed by CYP88A1 has not been defined (2). It is likely that Dwarf3 is a GA biosynthetic enzyme, and therefore it may catalyze one of the steps between KA and GA12. We have described the isolation of a cDNA-encoding CYP88A2 from pumpkin (13) but were unable to demonstrate an enzyme activity for this P450 when the cDNA was expressed in yeast. The CYP88A subfamily of P450 is not related closely to the CYP701A (AtKO1) subfamily (Fig. 1). The CYP701A subfamily belongs to the plant group A P450s (14), whereas the CYP88A subfamily groups with P450s from other organisms (15). Among the plant P450s closely related to CYP88A are CYP85, CYP90A, and CYP90B, which catalyze steps in the brassinosteroid biosynthesis pathway (15–17).
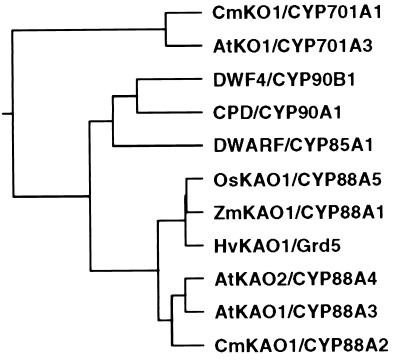
Figure 1.
A tree showing the relationship between CYP88A, ent-kaurene oxidase (KO), and brassinosteroid biosynthesis P450 proteins. CYP88A proteins are CYP88A3 (AtKAO1), CYP88A4 (AtKAO2), Grd5 (HvKAO1), Dwarf3 (ZmKAO1) (8), CYP88A2 (CmKAO) (13), and CY88A5 (OsKAO1; GenBank accession no. AP000616). KO sequences are CYP701A3 (AtKO1) (7) and CYP701A1 (CmKO1) (13), and brassinosteroid biosynthesis genes are CYP90B1 (DWF4) (15), CYP90A1 (CPD) (17), and CYP85A1 (DWARF) (16). The tree was produced from a ClustalW alignment by using the KITSCH program on Bionavigator (http://www.bionavigator.com).
Cytochrome P450 proteins are membrane-bound monooxygenases that probably are ubiquitous to all living organisms (18), catalyzing many reactions of secondary metabolism. Cytochrome P450s require at least an NADPH-cytochrome P450 reductase for activity. The method of choice for defining the activity of cytochrome P450s is expression in yeast, because it has an endogenous NADPH-cytochrome P450 reductase to support the activity of the expressed P450 protein. We have used the G1315 yeast strain previously to express functional AtKO1 (9) but were unsuccessful with the same yeast strain in detecting activity of the putative pumpkin KO, CmKO1 (13). An explanation for this lack of activity may be that the yeast cytochrome P450 reductase in G1315 cannot interact with CmKO1. An improved expression system uses yeast strains that have been engineered to express NADPH-cytochrome P450 reductases from Arabidopsis (19, 20). These yeast strains have been used to define the activity of a number of plant cytochrome P450s successfully (16, 19).
In this paper we report the identification of a dwarf mutant of barley that accumulates KA in developing grains. Three independent mutations at this locus were associated with alterations in a barley CYP88A-related sequence. Arabidopsis has two genes encoding CYP88A proteins; both are expressed in all plant parts examined. An improved yeast expression system is used to demonstrate that the two CYP88A proteins of Arabidopsis and the CYP88A protein of barley catalyze the three steps of the GA biosynthesis pathway from KA to GA12. We have named this enzyme ent-kaurenoic acid oxidase (KAO).
Materials and Methods
Content of KA, 7OH-KA, and GA12 in Developing Barley Grains.
Plants of Himalaya barley (wild type) and a representative dwarf mutant at the grd5 locus, M574 (homozygous stocks that had been through three backcrossing generations), were grown in a glasshouse of the Canberra phytotron at 18°C (day) and 13°C (night) with a 16-h photoperiod. Heads were tagged at awn emergence (about 2 d before anthesis) and harvested 1 to 5 weeks after anthesis. Developing grains from the two central floret rows of each head (excluding smaller grains at the base and tip) were detached and frozen on dry ice. Ten grains from each sample were homogenized in 20 ml MeOH, and then standards consisting of 60,300-dpm ent[17-14C]kaurenoic acid [47 mCi⋅mmol−1 (1 Ci = 37 GBq)], 25 ng 7β-hydroxy[17,17-2H2]kaurenoic acid, and 6,630-dpm [17-14C]GA12 (56 mCi⋅mmol−1) were added. All standards were provided by L. N. Mander (Australian National University). The samples were extracted overnight at 4°C with stirring and centrifuged at 1,000 × g, and the supernatant was decanted into a round-bottom flask. The pellet was re-extracted with another 10 ml of MeOH for 6 h and centrifuged as above, and the supernatant was combined with the first. Samples were dried by rotary evaporation, dissolved in 5 ml of hexane, and stored at −20°C overnight. Fractionation of extracts on silica columns was based on an unpublished procedure of S. Croker and J. R. Lenton (Institute of Arable Crops Research Long Ashton Research Station, University of Bristol). Silica columns (1.5 g) were made in plastic cartridges with a frit placed on top and washed with 2× 5-ml aliquots of hexane. Each sample was loaded onto a column and washed with a further 2× 5 ml aliquots of hexane. The initial load and the subsequent hexane washes were individually collected. This pattern of 3× 5 ml elutions was repeated by using solutions containing progressively higher concentrations of EtOAc consisting of EtOAc/hexane/HOAc 2.5:96.5:1 first, then 5:94:1, and finally 15:84:1. Radioactivity determined for each wash revealed that KA eluted with 5% (vol/vol) EtOAc, and GA12 eluted with 15% (vol/vol) EtOAc. Previous studies had indicated that 7OH-KA eluted with GA12. Pooled fractions were dried and dissolved in 2 ml of 100% MeOH, and then 0.5 ml of 2 mM HOAc was added. Sep-Pak C18 cartridges (WAT051910) were preconditioned with 5 ml of 100% MeOH and 5 ml of 2 mM HOAc. The samples were loaded and washed with successive 5-ml aliquots of 80% MeOH (4 ml 100% MeOH + 1 ml 2 mM HOAc), 90% MeOH (4.5 ml 100% MeOH + 0.5 ml 2 mM HOAc), and then 100% MeOH. For the KA sample, radioactivity eluted in the 80% and 90% washes, whereas for 7OH-KA/GA12, it eluted in 80% only. The appropriate fractions were pooled and dried by Speedivac in preparation for reverse-phase C18 HPLC. Samples were injected in 80% MeOH and eluted with an 80–100% gradient of solvent B (100% MeOH) in solvent A (10% MeOH in 2 mM HOAc) over 20 min, collecting 1-ml fractions with a flow rate of 1 ml/min. Based on radioactivity, KA eluted in fraction 21 and 7OH-KA and GA12 in fractions 8 and 9. The appropriate fractions were dissolved in a small volume of methanol, methylated with excess ethereal diazomethane, and dried in preparation for GC/MS. For KA analysis, a shallower than normal temperature ramp was used based on the data of D. Pearce (personal communication) indicating that a contaminating compound was not resolved from authentic KA under normal programs. The temperature profile was 60°C for 1.5 min, 60–150°C at 25°C/min, 150–230°C at 2°C/min, and 230–300°C at 70°C/min. The content of KA was determined by isotope dilution using the M+ and M2+ ions. The material for full-scan authentication of KA was purified in the same fashion as above, although no standards were added. The GC/MS program used the slow temperature ramp, and samples were injected with parafilm so that a Kovats' retention-index measurement could be made. An unlabeled KA standard also was injected to allow comparisons to be made. The samples were reinjected for analysis by multiple-ion monitoring so that better peak shapes and larger areas were obtained for the two samples.
The fractions containing 7OH-KA and GA12 were trimethylsilylated (7OH-KA) by using pyridine and N,O-bis(trimethylsilyl)trifluoroacetamide plus 1% trimethylchlorosilane (Alltech Associates) before analysis by GC/multiple-ion monitoring. The content of 7OH-KA was calculated from peak areas of the 314–316 and 404–406 ion pairs by using two arithmetic correction factors calculated by examining the percentage of 314 (or 404) compared with 316 (or 406) in 7β-OH[2H2]KA, and the percentage of 316 (or 406) in 314 (or 404) in unlabeled 7β-OH KA. The content of GA12 was calculated by using isotope dilution and the M+ and M2+ ions.
Yeast Expression.
Yeast expression constructs were made in the pYEDP60 vector (20). Oligonucleotide primers with restriction sites incorporated at the 5′ end were designed to amplify the putative protein-coding sequence of each P450-encoding cDNA. The PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced to ensure no mutations had occurred during the PCR amplification. The P450 cDNAs then were excised from pGEM-T easy by using the restriction sites introduced in the PCR primers and inserted in the sense orientation with respect to the GAL10-CYC1 promoter in the pYEDP60 polylinker. The expression clones in pYEDP60 were used to transform the WAT11 and WAT21 strains by using a LiCl method (21) and selected on SGI minimal-media plates (6.7 g/liter yeast-nitrogen base without amino acids, 20 g/liter glucose, 10 g/liter Bacto casamino acids, 20 mg/liter tryptophan, and 20 g/liter agar). Transformed yeasts were checked for expression of the P450 cDNA by RNA gel blots of RNA extracted from yeast cultures inoculated from single colonies into YPL liquid media (10 g/liter Bacto yeast extract/20 g/liter Bacto peptone/20 g/liter galactose).
Single yeast colonies were used to inoculate 10-ml YPG liquid cultures (10 g/liter Bacto yeast extract/20 g/liter Bacto peptone/20 g/liter glucose) in 50-ml polypropylene tubes. They were grown overnight with shaking at 28°C. Cells from 2.5 ml of culture were harvested and resuspended in 10 ml YPL liquid culture and grown overnight with shaking at 28°C. Assays essentially were as described (9) except they were carried out in a volume of 2 ml with 20 μg of substrate. The substrates tested were ent-kaurene, KA, 7OH-KA, and GA12-aldehyde. The substrates were all obtained from J. Metzger (Ohio State University, Columbus, OH). The assay mixture was incubated at 28°C with shaking in 50-ml polypropylene tubes for 6 h. The assay mixture was extracted once with 2 ml of hexane and twice with 2 ml of ethyl acetate. The pooled solvents were dried in a Speedivac before methylation and trimethysilation for GC/MS analysis as described above. Initial analyses were carried out in full-scan mode, and putative kaurenoids were identified by comparison to a library of spectra (22). To confirm that the products detected by GC/MS were authentic, samples and known standards were analyzed by using GC/multiple-ion monitoring to determine Kovats' retention indices and to measure relative ion abundances. This analysis was sufficient to authenticate the presence of 7OH-KA. Unequivocal confirmation of the presence of KA and GA12 was not possible because of the presence of contaminating compounds, thus the remaining portions of the extracts were fractionated by HPLC as described above. KA-methyl ester (Me) eluted in fraction 25 and GA12-Me eluted in fraction 16. This procedure removed the contaminating compounds and allowed the identification of KA and GA12.
Other Methods.
CYP88A3 and CYP88A4 cDNA clones were isolated from an Arabidopsis Landsberg erecta silique cDNA library (A. Koltunow, CSIRO Plant Industry, Adelaide, Australia) by screening with PCR-amplified fragments of the putative genes. A wheat expressed-sequence tag (EST) was identified in a database of wheat endosperm ESTs (23) with 74% deduced amino acid identity across 202 aa to the maize Dwarf3 protein. This EST was used as a probe to isolate the Grd5 cDNA from a library made from RNA of developing endosperm of Himalaya barley (S. Ali, CSIRO Plant Industry).
RNA was extracted from rosette leaf, cauline leaf, inflorescence stem, inflorescence (buds and mature flowers), and siliques of Arabidopsis ecotype C24 using TRIzol (Life Technologies, Melbourne, Australia). RNase protection assays were carried out by using the HybSpeed RPA kit (Ambion, Austin, TX) with antisense RNA probes hybridizing to bases 5–235 of the CYP88A3 mRNA and 1–300 of the CYP88A4 mRNA. Protected fragments separated on 8 M urea/5% acrylamide gels were visualized using a PhosphorImager.
Reverse transcription (RT)–PCR of Grd5 sequences from barley used RNA isolated from the endosperm of three dwarf mutants (2–3 weeks after anthesis) at the grd5 locus (M362, M574, and M594). Products were amplified, by using the Access RT-PCR kit (Promega) following the manufacturer's methods, purified, and sequenced in both strands.
Results
Barley grd5 Dwarf Mutant Accumulates KA.
GA-responsive dwarf mutants of Himalaya barley define six different genetic loci (grd1-6) on the basis of complementation grouping (ref. 24 and P.M.C., unpublished data). Analysis of GA biosynthetic intermediates in representatives of the different complementation groups showed that M574, a mutant at the grd5 locus, accumulated KA in developing grains. The GA1 content of M574 grains is only 15% of that of wild-type Himalaya barley (P.M.C., unpublished data). The KA content of developing Himalaya grains remains low (about 1 ng per grain) from 1 to 5 weeks after anthesis, whereas KA accumulates in the grd5 mutant M574, attaining levels of about 60 ng per grain. Grains of Himalaya and M574 were harvested at 2 and 4 weeks after anthesis, and KA, 7OH-KA, and GA12 were measured (Table 1). The results confirmed the accumulation of KA in developing grains of M574 and demonstrated that there is little if any change in 7OH-KA and GA12. The material identified as KA by GC/multiple ion monitoring showed an excellent match to authentic KA by full-scan GC/MS and GC/multiple-ion monitoring (Table 2).
Table 1.
Contents of KA, 7OH-KA, and GA12 in developing grains of Himalaya and grd5-M574
| Sample | Content, ng per grain
|
||
|---|---|---|---|
| KA | 7OH-KA | GA12 | |
| Himalaya | |||
| 2 wpa | 0.40 | 0.83 | 0.033 |
| 4 wpa | 0.31 | 0.45 | 0.023 |
| M574 | |||
| 2 wpa | 27.4 | 0.71 | 0.031 |
| 4 wpa | 49.4 | 1.68 | 0.028 |
Grains were harvested at 2 and 4 weeks postanthesis (wpa), and the contents of GA biosynthetic intermediates were determined.
Table 2.
Authentication of GA biosynthesis intermediates in the barley and yeast extracts
| Reference compound Putative compound | KRI | Characteristic ions, relative abundance as % of base peak | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Barley extracts | 316(M+) | 301 | 273 | 257 | 256 | 241 | 213 | |||
| KA-Me M574 | 2291 | 81 | 33 | 60 | 100 | n/a | 70 | 25 | ||
| KA-Me Standard | 2292 | 76 | 32 | 59 | 100 | 37 | 72 | 28 | ||
| Yeast extracts | 316(M+) | 301 | 273 | 257 | 256 | 241 | 213 | |||
| KAMe Standard | 2245 | 60 | 29 | 57 | 100 | 38 | 79 | 33 | ||
| KAMe CmKO1 | 2246 | 50 | 30 | 47 | 100 | 43 | 86 | 51 | ||
| Yeast extracts | 404(M+) | 389 | 314 | 299 | 255 | 254 | 239 | 223 | 185 | |
| 7OH-KA MeTMS Standard | 2367 | 15 | 7 | 100 | 24 | 40 | 34 | 23 | 10 | 13 |
| 7OH-KA MeTMS CYP88A3 | 2367 | 12 | 5 | 100 | 30 | 59 | 57 | 51 | 18 | 23 |
| 7OH-KA MeTMS CYP88A4 | 2367 | 23 | 9 | 100 | 30 | 47 | 41 | 34 | 22 | 19 |
| 7OH-KA MeTMS Grd5 | 2367 | 24 | 10 | 100 | 29 | 48 | 45 | 36 | 22 | 22 |
| Yeast extracts | 360(M+) | 328 | 300 | 285 | 241 | 240 | 225 | 185 | ||
| GA12Me Standard | 2324 | 2 | 24 | 100 | 19 | 25 | 20 | 17 | 8 | |
| GA12Me CYP88A3 | 2324 | 2 | 21 | 100 | 18 | 24 | 19 | 17 | 12 | |
| GA12Me CYP88A4 | 2324 | 1 | 22 | 100 | 20 | 26 | 20 | 18 | 11 | |
| GA12Me Grd5 | 2324 | 1 | 21 | 100 | 19 | 27 | 21 | 18 | 10 | |
Data shown for barley grd5 mutant M574, yeast expressing CmKO1 fed with ent-kaurene and yeast expressing CYP88A3, CYP88A4 and Grd5 fed with KA. Relative ion abundances for KA, 7OH-KA, and GA12 were calculated by GC/multiple ion monitoring and compared to authentic standards. KRI, Kovats' retention index.
grd5 Mutants Are Altered in Sequences of a CYP88A-Family Member.
The accumulation of KA in developing grains of M574 suggests this mutant has a defective gene encoding an enzyme that oxidizes KA. The possibility that the Grd5 locus might correspond to the Dwarf3 locus in maize (8) was investigated by cloning a Dwarf3-related cDNA from Himalaya barley and investigating whether there were alterations in this sequence in grd5 mutants. The Grd5 cDNA clone encoded a cytochrome P450 protein of 499 aa that was 80% identical in sequence to the maize Dwarf3 protein. Reverse transcription–PCR experiments on endosperm RNA of three independent grd5 mutants revealed that each had specific alterations to the wild-type sequence that would lead to nonconservative changes in the polypeptide (Fig. 2). We conclude that the barley Grd5 locus corresponds to the Dwarf3 locus in maize, and that this locus potentially encodes an enzyme that oxidizes KA.
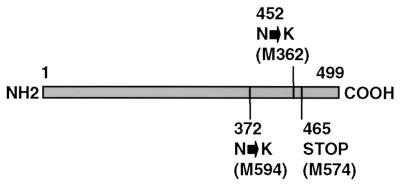
Figure 2.
Alterations in the Grd5 gene product in different grd5 mutants. Reverse transcription–PCR of endosperm RNA from different grd5 mutants (M362, M574, and M594) and sequence analysis of the products revealed amino acid substitutions at the positions indicated. A frameshift mutation in the M574 sequence causes an immediate termination codon.
Arabidopsis Expresses Two CYP88A Genes.
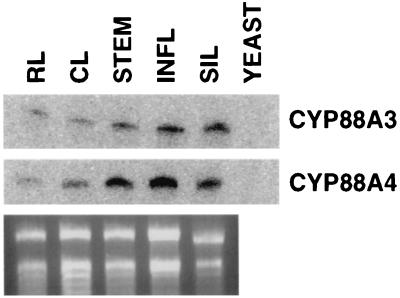
There are two genes likely to encode members of the CYP88A family in Arabidopsis: CYP88A3 and CYP88A4 (13). We isolated full-length cDNAs encoding CYP88A3 and CYP88A4 from the Landsberg erecta ecotype of Arabidopsis. The CYP88A3 gene is located on yUP8H12 (GenBank accession no. AC000098) on chromosome 1; CYP88A4 is located on section 181 of chromosome 2 (GenBank accession no. AC005700). The predicted proteins encoded by the CYP88A3 and CYP88A4 cDNAs are 491 and 490 aa long, respectively, and show 76% identity and 83% similarity to each other. Both the CYP88A3 and CYP88A4 mRNAs are expressed in all plant parts examined (Fig. 3), with higher expression in inflorescence stem, inflorescence, and silique tissue than leaf. There is some evidence of differential expression of the two genes; CYP88A4 mRNA is more abundant in stem and inflorescence than CYP88A3.
Figure 3.
Expression of CYP88A3 and CYP88A4 mRNA determined by ribonuclease protection assay. RNA samples prepared from rosette leaf (RL), cauline leaf (CL), inflorescence stem (STEM), stem tip including buds and mature flowers (INFL), and siliques (SIL). All RNA samples were from Arabidopsis ecotype C24. Yeast-negative control for the ribonuclease protection assay also is shown.
Arabidopsis and Barley CYP88As Catalyze the Oxidation of KA to ent-7α Kaurenoic Acid and GA12.
An improved yeast-expression system (20) was used to determine the function of the CYP88A P450s. The yeast strains WAT11 and WAT21 express the Arabidopsis cytochrome P450 reductases ATR1 and ATR2, respectively. Yeast strains WAT11 and WAT21 were transformed with the CYP88A3, CYP88A4, and Grd5 expression constructs and also with a CmKO1 expression construct encoding pumpkin KO (13).
WAT11 and WAT21 yeast strains expressing CmKO1, CYP88A3, CYP88A4, or Grd5 as well as untransformed yeast were incubated with ent-kaurene and KA. After incubation, the extracts from the yeast assays were analyzed by GC/MS, and putative kaurenoid peaks were identified by comparing their spectra to a library of spectra (22) and GC-retention times to those of authentic standards. Table 2 shows the results of this analysis for WAT21. WAT11 and WAT21 yeast strains gave the same products when expressing each of the P450 cDNAs. The WAT21 strains gave larger amounts of product in each case; therefore, further analysis was carried out by using the WAT21 strains.
Untransformed yeast did not metabolize either of the substrates to intermediates in the GA biosynthesis pathway, showing that the WAT11 and WAT21 yeast strains do not contain endogenous enzyme activities that can catalyze the steps of the GA biosynthesis pathway being examined. Yeast expressing CmKO1 produced a compound putatively identified as KA when incubated with ent-kaurene; this is the expected product of KO (9). The intermediates ent-kaurenol and ent-kaurenal were not detected. Yeast expressing CmKO1 did not produce any further intermediates of the GA pathway when incubated with KA. Yeast expressing CYP88A3, CYP88A4, or Grd5 did not oxidize ent-kaurene to KA, but when incubated with KA these yeast strains gave putative 7OH-KA and GA12 as products; GA12-aldehyde was not detected. To test whether the conversion of KA to GA12 was catalyzed enzymatically, untransformed WAT21, WAT21 expressing CmKO1, and WAT21 expressing CYP88A3 were incubated with 7OH-KA and GA12-aldehyde (Table 3). The CYP88A3-expressing yeast gave GA12 when incubated with either 7OH-KA or GA12-aldehyde. The untransformed WAT21 and CmKO1-expressing WAT21 did not produce any detectable GA12-aldehyde or GA12.
Table 3.
Putative products from WAT21 yeast strains expressing cytochrome P450s
| Substrate | Yeast
Strain
|
||||
|---|---|---|---|---|---|
| Untransformed | CmKO1 | CYP88A3 | CYP88A4 | Grd5 | |
| kaurene | none | KA | none | none | none |
| KA | none | none | 7OH-KA GA12 | 7OH-KA GA12 | 7OH-KA GA12 |
| 7OH-KA | none | none | GA12 | not tested | not tested |
| GA12-aldehyde | none | none | GA12 | not tested | not tested |
The identities of the putative reaction products shown in Table 3 were confirmed by comparison to authentic standards. To identify the KA and GA12 products, the extracts were purified by HPLC (Table 2). In all cases, both Kovats' retention index and relative ion abundances confirmed the identities of the putative products shown in Table 3. CYP88A3, CYP88A4, and Grd5 all catalyzed the conversion of KA to 7OH-KA and GA12 but did not oxidize ent-kaurene to KA. CmKO1 catalyzes the oxidation of ent-kaurene to KA but does not give any products with KA, 7OH-KA, or GA12-aldehyde.
Discussion
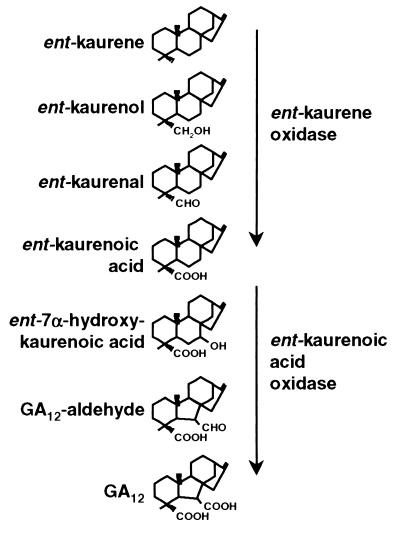
We have demonstrated that the CYP88A3, CYP88A4, and Grd5 genes from Arabidopsis and barley encode cytochrome P450 proteins that catalyze three steps of the GA biosynthesis pathway from KA to GA12 when expressed in yeast. Because the CYP88A cytochrome P450s catalyze the three oxidation steps from KA to GA12, we have named this enzyme KAO. In the barley grd5 mutant M574, which has a frameshift mutation in the Grd5 gene, KA accumulates in developing grains, providing confirmation that Grd5 catalyzes the oxidation of KA in vivo. Both 7OH-KA and GA12 were detected in the M574 mutant. M574 is a weak allele of grd5 in which GA1 is still detectable; the presence of 7OH-KA and GA12 may be due to residual activity in this allele. Our yeast expression studies show that all three KAO enzymes studied have the ability to metabolize KA to GA12. We have no direct evidence to confirm that KAO catalyzes all three steps in plants. The reactions from KA to GA12 are all at the C-7 position of the GA skeleton (Fig. 4). There are precedents in the GA pathway for enzymes carrying out multiple oxidations at the same carbon position. The cytochrome P450 AtKO1 (9) catalyzes three steps of oxidation at the C-19 position and the dioxygenases of the GA C-20 oxidase family carry out a three-step oxidation at the C-20 position (25, 26). Therefore, it is likely that the KAO enzymes do catalyze the three steps of the GA biosynthesis pathway from KA to GA12 in plants.
Figure 4.
Cytochrome P450-mediated steps of the GA biosynthesis pathway showing the steps catalyzed by KO and KAO.
There are two genes encoding KAO in Arabidopsis. In the yeast expression system, the activities of the two enzymes were the same. In plants, both AtKAO genes were found to be expressed in all aerial tissues examined. The lack of an equivalent mutant to grd5 in Arabidopsis can be explained therefore by gene redundancy. Determination of the Arabidopsis genome sequence has shown that much of the genome is duplicated (27). The high similarity between the two AtKAO genes suggests they have arisen by gene duplication, although they do not lie in areas that have been suggested to be major duplications. In barley and maize it seems likely that HvKAO1 and ZmKAO1 are the major KAO activities, because mutations in the genes encoding these enzymes give rise to GA-deficient dwarf plants. In pumpkin, the available evidence suggests that CmKAO1 (13) is a single-copy gene. The duplication of the AtKAO genes may be relatively recent.
The AtKO1 gene is activated by a basic leucine zipper-transcription factor from tobacco, RSG (28). The core binding site of RSG, CAANTTG, also is present in the promoters of AtKAO1 at −606 bp from the translation start and in AtKAO2 at −520 and −466 bp from the translation start so the AtKAO genes also may be regulated by RSG. The expression patterns of the AtKO1 (7) and the two AtKAO mRNAs are similar, suggesting that the P450-mediated steps of the GA pathway are regulated coordinately. RSG may be involved in the coordinate regulation of the P450-mediated steps of the pathway.
We are now able to conclude that the six microsomal steps of the GA pathway from ent-kaurene to GA12 are likely to be catalyzed by the two cytochrome P450 enzymes, KO and KAO (Fig. 4). As both the Arabidopsis and barley CYP88A P450 enzymes are KAOs, it is likely that the other members of this P450 subfamily, including the maize Dwarf3 protein, have the same enzyme activity. The isolation of the genes encoding KAO means that the genes encoding the enzymes for all of the steps of the GA biosynthesis pathway from geranylgeranyl diphosphate to GA4 have now been isolated. Advances in methods for high throughput analysis of gene expression should reveal how each step in the GA biosynthesis pathway is regulated in plant developmental processes.
Acknowledgments
We thank Jane Pulford, Helen Adams, and Judy Radik for technical assistance, Dr. John Huppatz for useful discussions, and Professors Lew Mander and Jim Metzger for kaurenoid standards and substrates.
Abbreviations
- GA
gibberellin
- KA
ent-kaurenoic acid
- 7OH-KA
ent-7α-hydroxy-kaurenoic acid
- KO
ent-kaurene oxidase
- KAO
KA oxidase
- Me
methyl ester
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF318500, AF318501, and AF326277).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041588998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041588998
References
- 1.Hooley R. Plant Mol Biol. 1994;25:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- 2.Hedden P, Kamiya Y. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 3.Hedden P, Proebsting W M. Plant Physiol. 1999;119:365–370. doi: 10.1104/pp.119.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange T. Proc Natl Acad Sci USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange T, Robatzek S, Frisse A. Plant Cell. 1997;9:1459–1467. doi: 10.1105/tpc.9.8.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas S G, Phillips A L, Hedden P. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helliwell C A, Sheldon C C, Olive M R, Walker A R, Zeevaart J A D, Peacock W J, Dennis E S. Proc Natl Acad Sci USA. 1998;95:9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler R G, Helentjaris T. Plant Cell. 1995;7:1307–1317. doi: 10.1105/tpc.7.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helliwell C A, Poole A, Peacock W J, Dennis E S. Plant Physiol. 1999;119:507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coles J P, Phillips A L, Croker S J, García-Lepe R, Lewis M J, Hedden P. Plant J. 1999;17:547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- 11.MacMillan J. Nat Prod Rep. 1997;14:221–243. [Google Scholar]
- 12.Phinney B O, Spray C. In: Plant Growth Substances. Wareing P F, editor. New York: Academic; 1982. pp. 101–110. [Google Scholar]
- 13.Helliwell C A, Olive M R, Gebbie L, Forster R, Peacock W J, Dennis E S. Aust J Plant Physiol. 2000;27:1141–1149. [Google Scholar]
- 14.Durst F, Nelson D R. Drug Metab Drug Interact. 1995;12:189–206. doi: 10.1515/dmdi.1995.12.3-4.189. [DOI] [PubMed] [Google Scholar]
- 15.Choe S, Dilkes B P, Fujioka S, Takatsuto S, Sakurai A, Feldmann K A. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop G J, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones D G, Kamiya Y. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei G P, Nagy F, Schell J, Koncz C. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 18.Nelson D, Kamataki T, Waxman D, Guengerich P, Estabrook R W, Feyereisen R, Gonzalez F, Coon M, Gunsalus I, Gotoh O, et al. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 19.Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 20.Pompon D, Lauerat B, Bronine A, Urban P. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- 21.Cullin C, Pompon D. Gene. 1988;65:203–217. doi: 10.1016/0378-1119(88)90457-x. [DOI] [PubMed] [Google Scholar]
- 22.Gaskin P, MacMillan J. GC-MS of the Gibberellins and Related Compounds: Methodology and a Library of Spectra. Bristol, U.K.: Cantock's Enterprises; 1992. [Google Scholar]
- 23.Clarke B C, Hobbs M, Skylas D, Appels R. Funct Integr Genomics. 2000;1:44–55. doi: 10.1007/s101420000008. [DOI] [PubMed] [Google Scholar]
- 24.Chandler P M, Robertson M. Plant Physiol. 1999;120:623–632. doi: 10.1104/pp.120.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y-L, Li L, Wu K, Peeters A J M, Gage D A, Zeevaart J A D. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips A L, Ward D A, Uknes S, Appleforth N E J, Lange T, Huttly A K, Gaskin P, Graebe J E, Hedden P. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y. Plant Cell. 2000;12:901–915. doi: 10.1105/tpc.12.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]