Abstract
Mrr superfamily of homologous genes in microbial genomes restricts modified DNA in vivo. However, their biochemical properties in vitro have remained obscure. Here, we report the experimental characterization of MspJI, a remote homolog of Escherichia coli’s Mrr and show it is a DNA modification-dependent restriction endonuclease. Our results suggest MspJI recognizes mCNNR (R = G/A) sites and cleaves DNA at fixed distances (N12/N16) away from the modified cytosine at the 3′ side (or N9/N13 from R). Besides 5-methylcytosine, MspJI also recognizes 5-hydroxymethylcytosine but is blocked by 5-glucosylhydroxymethylcytosine. Several other close homologs of MspJI show similar modification-dependent endonuclease activity and display substrate preferences different from MspJI. A unique feature of these modification-dependent enzymes is that they are able to extract small DNA fragments containing modified sites on genomic DNA, for example ∼32 bp around symmetrically methylated CG sites and ∼31 bp around methylated CNG sites. The digested fragments can be directly selected for high-throughput sequencing to map the location of the modification on the genomic DNA. The MspJI enzyme family, with their different recognition specificities and cleavage properties, provides a basis on which many future methods can build to decode the epigenomes of different organisms.
INTRODUCTION
Restriction-modification (RM) systems are widely present in prokaryotic genomes (1). They typically consist of restriction endonucleases (REs), which protect the hosts from invading DNA (e.g. bacteriophages) by cleaving DNA at defined sites, and DNA methyltransferases, which protect host DNA from being degraded by methylating the cognate RE sites. Although RM systems are effective at restricting foreign DNA, bacteriophage species sometimes modify their own DNA, thus acquiring resistance to cleavage by most conventional REs (2). For example, some bacteriophage genomes are fully cytosine methylated (3). Among many specificities characterized so far, only a handful of REs recognize sites with modified bases. The early observation that maintenance of foreign methyltransferase genes in E. coli induces an SOS response led to the discovery of the McrA, McrBC and Mrr systems (4,5). McrA recognizes CmCGG containing DNA (6,7) and McrBC recognizes pairs of (A/G)mC separated by 40–3000 bp (8). Mrr has been shown to restrict some cytosine-methylated or adenine-methylated DNA (9). Other examples which recognize sequence specific modified sites include DpnI (GmATC) (10), GlaI (GmCGmC) (11) and BisI (GmCNGC) (11). The presence of these methylation-dependent REs allows the hosts to defend against bacteriophages with modified DNA.
Previously, E. coli Mrr was shown to restrict some 5-methylcytosine (5mC) and N6-methyladenine containing DNA in vivo (9). However, no simple consensus sequence has been determined for its recognition site, nor has the in vitro endonuclease activity been observed for the recombinant E. coli Mrr (9). This opens the possibility that the E. coli Mrr achieves its biological role by a mechanism other than simple endonucleolytic cleavage. There have been reports which suggested Mrr’s involvement in the bacterial SOS response to high-pressure stress (12). A BLAST search using E. coli Mrr as the initial query reveals the existence of Mrr homologs in many bacterial species (13,14). However, other than E. coli Mrr, none of these homologs has been biochemically characterized and their function remains elusive.
Conserved domain analysis (15) suggests the existence of at least two domains in a typical mrr homolog: a conserved C-terminal domain resembling a catalytic endonuclease region and a less conserved N-terminal domain presumably responsible for the DNA recognition and binding activity. Putative catalytic motifs such as (D/E)…(D/E/Q)xK are conserved inside the C-terminal domain (14).
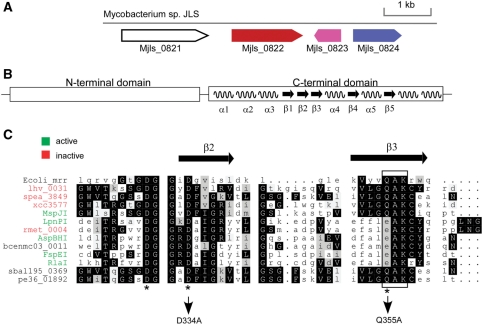
Our analysis of REBASE (1) identified a fairly typical-looking RM system in the genome of Mycobacterium sp. JLS, which comprises a predicted cytosine DNA methyltransferase (Mjls0824), an endonuclease (Mjls0822) and a predicted vsr gene (Mjls0823) (Figure 1A). During the course of this work, we have renamed Mjls0822 as MspJI, Mjls0824 as M.MspJIP and Mjls0823 as V.MspJIP in REBASE. Surprisingly, sequence analysis revealed the presence of an Mrr-like catalytic domain at the C-terminus of MspJI (Figure 1B), suggesting the possibility of its activity on modified DNA. However, if this is true, it was unexpected that MspJI would be associated with a DNA methyltransferase, as their combined action would be detrimental to the host genomic DNA. A BLAST search against GenBank using MspJI as the initial query retrieved more than 10 close homologs for which there is significant similarity covering most of the sequence length (Figure 1C). One of the notable differences between the E. coli Mrr gene and the MspJI family is their sequence length, while the E. coli Mrr is 304 amino acids (aa) long, MspJI is 456 aa and most of its close homologs are >400 aa.
Figure 1.
Genomic and sequence features of the MspJI RM System. (A) The genomic segment of Mycobacterium sp. JLS encoding the MspJI RM system. NCBI annotations for open reading frames are: Mjls0821, putative helicase; Mjls0822 (MspJI), restriction endonuclease; Mjls0823 (V.MspJIP), DNA mismatch endonuclease vsr; Mjls0824 (M.MspJI), DNA cytosine methyltransferase. Color coding used the REBASE coloring scheme. (B) Schematic domain structure of MspJI subfamily. Secondary structure elements are predicted by the PROMALS webserver (18) and shown for the C-terminal domain. (C) Multiple sequence alignment of the catalytic motif in the MspJI subfamily. The E. coli Mrr protein is also included as reference. The black box highlights the conserved catalytic motif (Q/E)AK. The REBASE names of active enzymes and their corresponding GenBank IDs are: FspEI, YP_001509600; LpnPI, YP_095265; RlaI, ZP_03168528; AspBHI, YP_931859. For others, see Supplementary Table S2 for a complete list of GenBank IDs. lhv_0031, YP_001576608; spea_3849, YP_001503694; xcc3577, AAM42847; rmet_0004, YP_582159.
Here, we describe our initial biochemical characterization of MspJI as a modification-dependent endonuclease. Results accumulated during the course of MspJI studies further suggest that some members in the Mrr family, if not the majority, may have endonuclease activity toward modified DNA (5).
MATERIALS AND METHODS
All enzymes, plasmids, bacterial strains, if not otherwise specified, are from New England Biolabs Inc.
Cloning, protein expression and purification
The gene for MspJI was codon optimized using in-house software for optimal expression in E. coli. Cassettes of synthetic DNA (∼500 bp) were first assembled using overlapping PCR from oligonucleotide DNA and then joined by USER cloning (22). Synthetic DNA encoding MspJI was then ligated into pTXB1 with an N-terminal His-tag and expressed in a dcm− E. coli strain T7 Express (C2566). Clones were grown in LB-Amp to OD600 0.6–0.8 and induced with a final concentration of 0.5 mM IPTG. Induced cultures were then grown overnight at 25°C and stored as frozen cell pellet at −20°C. Re-suspended cell pellet was sonicated and cleared lysate was collected after centrifugation. Purification was carried out on an AKTA FPLC machine (GE Healthcare). MspJI was first purified on a HiTrap Heparin HP column, then a HisTrap HP column, and a final HiTrap SP column. The endonuclease activities of the MspJI fractions were assayed on regular λ DNA, which is partially dcm methylated. The final product ran as a single band on SDS–PAGE. The final concentration of MspJI after purification was estimated to be 0.8 mg ml−1.
Endonuclease assay and DNA substrates
All digestion reactions were carried out in standard NEB buffer 4 (50 mM potassium acetate, 20 mM Tris–acetate, 10 mM magnesium acetate, 1 mM dithiothreitol, pH 7.9 at 25°C). pBR322(dcm+) was from NEB and pBR322(dcm−) was prepared using the dam−/dcm− E. coli strain. Methylated pBR322(dcm−) was prepared using different recombinant methyltransferases (NEB) according to manufacturer's protocol.
DNA oligos with or without internal methylated cytosine were synthesized either in-house or at Integrated DNA Technologies. In the digestion series presented in Figure 3, ∼1 µM double-stranded annealed oligos were digested by 1.6 µM MspJI in the presence of 1.5 µM double-stranded DNA activator in a 10 µl volume. All reactions were incubated at 37°C.
Figure 3.
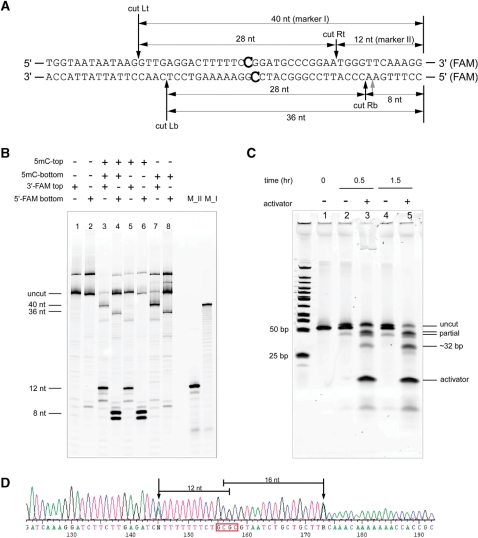
Determination of the MspJI cleavage sites. (A) Oligonucleotides used in the cleavage assay and cleavage sites (designated Rt, Rb, Lt, Lb). The 3′ end of the top strand or the 5′ end of the bottom strand is labeled with FAM, respectively. The wobbling cut is labeled as grey arrow. (B) Determination of cleavage sites. Lane 1, no methylation, top strand labeled; lane 2, no methylation, bottom strand labeled; lane 3, fully methylated, top strand labeled; lane 4, fully methylated, bottom strand labeled; lane 5, top strand methylated, top strand labeled; lane 6, top strand methylated, bottom strand labeled; lane 7, bottom strand methylated, top strand labeled; lane 8, bottom strand methylated, bottom strand labeled. As a control, markers (M_I,M_II) are run on the right-hand side of the gel. (C) Digestion of the oligo substrate shown under native conditions.1 µl of the ds DNA oligonucleotide (10 µM) was mixed with 1 µl of MspJI (0.8 µg µl−1) in the presence or absence of a DNA activator (1 µM final) in 10 µl reaction volume. The reactions were incubated at 37°C for different time points. Five microliters of the reaction mix was run on a 20% polyacrylamide TBE gel and visualized by SYBR GOLD staining. (D) A representative run-off sequencing trace around a methylated HhaI site on the pBR322 DNA. The distances between the cleavage sites, which can be inferred from run-off peaks, to the methylated cytosine are shown.
RESULTS
Modification-dependent endonuclease activity of MspJI
The codon optimized MspJI gene was synthesized by oligonucleotide assembly and USER™ ligation (‘Materials and Methods’ section). During the cloning process, we noticed that transformation of the plasmid, which contains the MspJI gene, resulted in dramatic decrease in efficiency in the dcm+ E. coli host ER1992 but not in a dcm− strain T7 Express. These results indicate that the presence of methylated dcm sites (CmCWGG) may adversely interfere with the expression of MspJI. Recombinant MspJI with an N-terminal His-tag was then expressed in the dcm− strain T7 Express and purified to apparent homogeneity (‘Materials and Methods’ section).
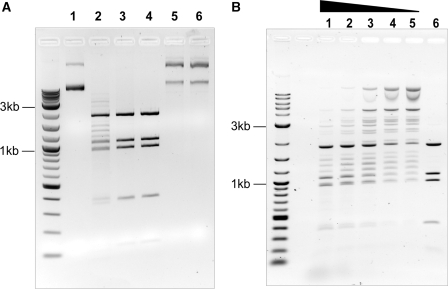
The in vitro activity of MspJI was assessed on a variety of methylated and non-methylated DNA substrates, as shown in Figure 2A for the dcm methylated plasmid DNA pBR322. MspJI shows clear endonuclease activity (lane 1 and 2) and this endonuclease activity is DNA methylation dependent, as MspJI does not act on pBR322 without dcm modification (Figure 2A, lane 5 and 6). By using a methylation insensitive restriction enzyme BstNI (CC↓WGG) in a double-digestion assay, cleavage sites on pBR322(dcm+) by MspJI can be deduced to be at or close to the dcm sites (Figure 2A, lanes 2–4).
Figure 2.
Modification-dependent endonuclease activity of MspJI. (A) Lane 1, pBR322(dcm+) DNA only; lane 2, pBR322(dcm+) + MspJI; lane 3, pBR322(dcm+) + MspJI + BstNI; lane 4, pBR322(dcm+) + BstNI only; Lane 5, pBR322(dcm−) DNA only; lane 6, pBR322(dcm−) + MspJI. All reactions were incubated at 37°C for 1 h and resolved on 1% agarose gel. (B) Effect of DNA activator on MspJI activity. From lane 1 to 5, each reaction contains 1 µg (0.35 pmol) pBR322(dcm+) and 1.6 pmol MspJI in 30 µl volume. DNA activator with methylated CCWGG sites is titrated from lane 1–4 (40, 20, 10, 5 pmol). Lane 5, pBR322 digestion using MspJI without DNA activator. Lane 6, pBR322 digestion using BstNI (CCWGG).
To determine the specificity of MspJI, pBR322(dcm−) was methylated in vitro using different DNA methyltransferases prior to MspJI digestion. MspJI shows apparent endonuclease activity on modified DNA including CmCGG (M.HpaII modification), AGmCT (M.AluI modification), GGmCC (M.HaeIII modification), GmCGC (M.HhaI modification) and mCCGG (M.MspI modification) (Supplementary Figure S1). We noticed that MspJI cleaves M.MspI modified DNA more efficiently than other modified DNA, suggesting the possibility of its dependence on the flanking nucleotides besides the modified cytosine. Two sets of synthetic oligonucleotide substrates were used to investigate the dependence of the flanking nucleotides: one with every possible combination of NmCGN (N=A/T/G/C) and the other with that of NCmCGGN. The detailed results and analysis on the oligo substrates will be published later in a separate paper (Cohen-Karni, D. et al., unpublished data). As a summary here, MspJI appears to recognize mCNNR (R=G or A) sites. Besides 5mC, MspJI shows no detectable endonuclease activity in vitro on DNA containing 6-methyladenine such as M.TaqI (TCGmA) or dam (GmATC) modified DNA, which is consistent with the observation that it can be stably maintained and expressed in a dam+ strain (T7 Express). Moreover, MspJI does not seem to act on N4-methylcytosine containing plasmid DNA or PCR DNA.
During the course of this work, there have been reports suggesting the existence of 5-hydroxymethylcytosine (5hmC) in addition to 5-methylcytosine (5mC) in mammalian genomes (16). MspJI’s activity was tested on variants of phage T4 genomic DNA which contain 5hmC (T4 gt DNA) or 5-glucosyl-hydroxymethylcytosine (T4 wt DNA). Our results indicate MspJI is able to cleave 5hmC sites but is blocked by glucosylation on the 5hmC (Figure S2 in SOM). Since the modified sites for MspJI cleavage are much denser on the T4 gt DNA than on the pBR322(dcm+) DNA, the digestion pattern on agarose gel appears as smear instead of discrete bands (Figure 2 and Supplementary Figure S2). Overall, it appears that MspJI specifically targets cytosine modification with 5-CH3 or 5-CH2OH addition on the pyrimidine ring.
MspJI endonuclease activity is stimulated by a double-stranded DNA activator
Initially, we observed that the MspJI cleavage efficiency on a DNA fragment with only one methylated site is much lower than that with two methylated sites on the same DNA fragment, which suggests that MspJI may require interaction with multiple recognition sites for cleavage, like many other Type II enzymes (17).
We then investigated the possibility of using a double-stranded DNA with methylated sites as an activator in trans. We used a 15-bp double-stranded DNA fragment with a dcm (CCWGG, W=A/T) site either with or without C5 modification (Supplementary Table S1). Since the distance from the methylated site to either end is less than the reach of MspJI for cleavage, MspJI should only be able to bind, but not cleave, the short DNA activator. As shown in Figure 2B, in a digestion containing fixed amounts of MspJI (1.6 pmol) and dcm− methylated pBR322 DNA (1 µg or 0.35 pmol), by increasing amounts of the methylated 15-nt long nucleotide DNA activator, the digestion reaction is driven closer to completion, as compared with BstNI digestion (Figure 2b). However, no stimulation effect is seen in the control experiment in which the 15-nt long nucleotide DNA fragment is unmethylated. A 30-bp cleavage-resistant DNA activator containing phosphothioate linkages at the cleavage sites also stimulates the reaction (Supplementary Table S1). These observations suggest that MspJI may interact with multiple sites, either in cis or in trans.
Determination of MspJI cleavage sites
To determine MspJI’s cleavage sites, a set of FAM-labeled synthetic oligonucleotide substrates was used in a digestion assay (Figure 3A). Double-stranded oligonucleotides with full-methylation and hemi-methylation either on the top or on the bottom strand were subject to MspJI digestion and resolved on a 7M urea 20% polyacrylamide denaturing gel with synthetic oligonucleotide markers, as shown in Figure 3B. Cleavage on either strand was monitored separately by using oligonucleotides labeled in the top or bottom strand. Figure 3A indicates the inferred cleavage sites and product sizes based on the results in Figure 3B.
On the hemi-methylated substrates, cleavage occurs on the 3′ side of the methylated cytosine and both strands are cleaved. For example, for a substrate with only top strand methylation, each strand is cut once at the 3′ side of the 5mC so that only shorter fragments are observed (lane 5 and 6, Figure 3B). On the top strand, cut Rt (Figure 3A) results in the labeled fragment of 12 bp (lane 5). The distance from cut Rt to the 5mC on the same strand is 12 bp. On the bottom strand, notice that cut Rb wobbles by one base and generates the labeled fragments of 8 or 7 bp (lane 6). The distance from cut Rb to the 5mC on the opposite strand is 16 or 17 bp. We have observed that the cleavage at 16 bp from 5mC is the major cut site. The same analysis applies to the substrate with bottom strand methylation where only longer fragments are seen (40 bp from cut Lt in lane 7 and 36 bp from cut Lb in lane 8, Figure 3B). Overall, these results suggest that MspJI preferably recognizes mCNNR(N9/N13–14).
On fully methylated substrates, MspJI appears to recognize each hemi-methylated site individually and cleave on both sides. As shown in lane 3 and 4 of Figure 3B, all four labeled fragments corresponding to the four cut locations are observed. Additionally, Figure 3C shows the MspJI digestion on the same fully methylated oligonucleotide but without FAM label, which was resolved on a 20% non-denaturing polyacrylamide gel. The gel was stained with Sybr Gold (GE) to visualize every cleaved fragment in double-stranded form. Partial cleavage products either at the left-hand side (∼40 bp) or at the right-hand side (∼45 bp) of the methylated site appeared as reaction intermediates while the complete cleavage product (∼32 bp considering both 5′-overhangs) on both sides accumulates as the reaction progresses (Figure 3C).
As a control, MspJI does not cleave non-methylated oligonucleotides with the same sequence (lanes 1 and 2 in Figure 3B). Although in this example the fully methylated site is palindromic, it appears that there may be no requirement for such symmetry since it is the half site that provides the directionality for each cleavage event.
Besides the oligos used in Figure 3A, we have used many other oligonucleotides as well as run-off sequencing of plasmid DNA digested with MspJI (Figure 3D) to confirm that the above conclusions are consistent on different sites. Note that in Figure 3D which shows a representative run-off sequencing trace, two run-off peaks correspond to the cleavage on either side of the fully methylated site.
MspJI digestion on genomic DNA samples
Genomes of many higher organisms are known to contain 5-methylcytosine modifications as an epigenetic marker. Most 5mCs exist in CpG dinucleotides or repetitive elements that are distributed unevenly on the genome. In Figure 4, we show MspJI digestion on a few representative genomic DNA samples from plant (Arabidopsis), human (Hela cells) and yeast. The digestion of the genomic DNA correlates with the presence of 5mC: plant and human DNAs result in smeared fragments on the gel while yeast DNA, which does not contain 5mC, remains intact.
Figure 4.
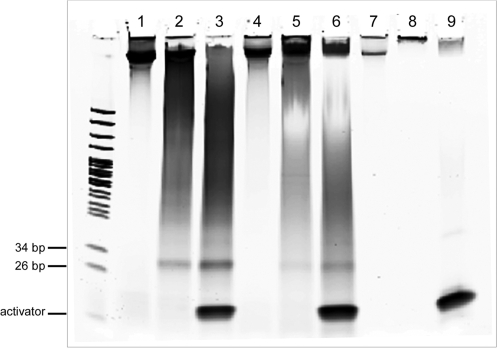
MspJI digestion on genomic DNAs. M, pBR322 MspI digest; lane 1, Hela genomic DNA; lane 2, Hela gDNA+MspJI; lane 3, Hela gDNA+MspJI+activator;4, Arabidopsis gDNA; lane 5, Arabidopsis gDNA+MspJI; lane 6, Arabidopsis gDNA+MspJI+activator; lane 7, yeast gDNA; lane 8, yeast gDNA+MspJI; lane 9, yeast gDNA+MspJI+activator. All digestion reactions contain 1.5 µg of genomic DNA, 1.5 µg of MspJI in 30 µl volume. When indicated, a final concentration of 0.8 µM of DNA activator was supplemented to stimulate MspJI digestion. All reactions were incubated overnight at 37°C.
In principle, every end of the fragments produced corresponds to a methylated CNNR site close to the cleavage site on the genome. Thus, by sequencing the ends of the MspJI digested fragments to a sufficient length and mapping them back to the reference genome, it is possible to generate a whole genome epigenomic map sampled at a subset of CpG sites (‘Discussion’ section). Moreover, as in the digestion of oligonucleotide substrate (Figure 3C), the cleavage of the fully methylated CpG sites in genomes leads to the appearance of a discrete band about 30 bp in size in the case of Arabidopsis and human genomic DNAs (lanes 3 and 6 in Figure 4). Direct sequencing of these fragments (after end polishing) should yield many 32–34 bp (considering wobbling cleavage) genomic reads with a CG or CNG in the middle. This would also provide a straightforward method to sequence the epigenome.
Members in the MspJI subfamily and its relatedness to E. coli Mrr
By using MspJI as the query sequence, a PSI–BLAST (13) search against GenBank retrieved >100 hits with significant sequence similarity, which we refer to as the Mrr superfamily. Most of the hits inside the superfamily are annotated as homologs of E. coli Mrr, or generally as RM genes. A distinguishing sequence feature of the Mrr superfamily is the presence of the conserved catalytic domain at the C-terminus, which harbors the D…(Q/E)×K metal-binding motif (Figure 1C) (14). The N-terminal domains, however, are much more divergent, both in sequence length and composition. Closer examination of the list reveals that ∼15 genes among the top hits have significant similarity to MspJI throughout the sequence length. We call these the MspJI subfamily. In Figure 1C, we show a partial multiple sequence alignment around the conserved catalytic motif inside the MspJI subfamily. The importance of the conserved catalytic motif is shown by the site-directed mutagenesis experiments, in which both D334A and Q355A mutations completely abolish the catalytic activity of MspJI (Figure 1C).
The predicted secondary structure elements of the MspJI subfamily were inferred simultaneously in the process of multiple sequence alignment by using the PROMALS web server (18) (Figure 1B). The secondary structure elements in the C-terminus are more reliably inferred than those in the N-terminus based on a higher degree of regional sequence conservation. The structure core of the catalytic C-terminal domain has three consecutive strands (β1β2β3 in Figure 1B), with the motif (Q/E)×K at the end of β3 and the conserved residue D at the beginning of β2, which remarkably resembles the structure of the FokI cleavage domain (Figure 1C) (19).
Inside the MspJI subfamily, we have chosen several other MspJI homologs for biochemical characterization. We found several of them also exhibit modification-dependent endonuclease activity (Supplementary Table S2). Our initial comparative analysis suggests these active enzymes have different substrate preferences. Detailed biochemical characterization of these modification-dependent enzymes and comparison with MspJI will be published elsewhere.
Surprisingly, examination of the genomic context of the members in the MspJI subfamily shows that about half of the members in the MspJI subfamily are located next to a DNA methyltransferase gene (Supplementary Table S2). Almost all of the neighboring DNA methyltransferases are predicted to be 5mC-specific. Among those MspJI homologs that are not associated with methyltransferases, some are often seen inside transposase islands, indicating the possibility of frequent horizontal gene transfer. For these, it is likely that the association with methyltransferases may have been lost.
In the example of LpnPI (Supplementary Table S2), it is close to a full RM system with both R and M genes in the genomic vicinity. The predicted specificity of the methyltransferase (M.LpnPI) is GGNCC. The host DNA of Legionella pneumophila strain Philadelphia 1 was tested and shown to be resistant to Sau96I digestion (G↓GNCC). Since the genome only has one putative C5-methyltransferase (20), it is likely that M.LpnPI is active and modifies GGNCC sites. Given the fact that we have observed that LpnPI is active on 5mC-containing DNA, it is likely that it will not cut modified GGNCC sites but act on other modified sites.
In the case of MspJI, we have expressed M.MspJI in E. coli but detected no methyltransferase activity on a number of DNA substrates. As a result, at least in the genome of Mycobacterium sp. JLS, the inactive M.MspJI should not interfere with the endonucleolytic activity of MspJI. Nevertheless, the high incidence of association between MspJI members and a DNA methyltransferase is unusual and calls for further investigation of their biochemical and physiological roles.
DISCUSSION
For a long time, genes in the Mrr superfamily have remained essentially uncharacterized as to their biochemical function. Although E. coli Mrr was shown to restrict certain epigenetic modifications in vivo, there seemed no simple consensus for its recognition sites (9). In this article, we characterize a remote homolog of E. coli Mrr, MspJI from a Mycobacterium sp., and show that it is a genuine endonuclease which recognizes cytosine residues modified at the C-5 position and cleaves at fixed distances away from the recognition site. The homology between MspJI and E. coli Mrr is weak and only revealed through multiple rounds of PSI–BLAST search. Following the finding of MspJI, we examined several other close homologs to MspJI, most of which also show modification-dependent endonuclease activity. Our experimental evidence lends strong support to the earlier proposal that genes in the Mrr family are endonucleases and recognize modified DNA (5,14). A notable difference between the MspJI subfamily and other homologs of E. coli Mrr appears to be their length. Further investigation is needed to characterize the in vitro activity of other E. coli Mrr homologs and compare the differences among the family members.
We have determined the preferred recognition site of MspJI to be mCNNR (R=G or A). Moreover, in our experiments we noticed that mCNNG provides a better site than mCNNA. These observations suggest that Mrr-like enzymes may be more promiscuous in their specificity by nature than typical restriction enzymes which recognize unmodified DNA. This may explain why it was difficult to infer a clear-cut recognition consensus for the E. coli Mrr. We reason that it may have to do with the different selection pressures associated with these two different types of enzymes. For a typical restriction enzyme, selection pressure includes not only the efficient cleavage against the target recognition sites to fight off invading DNA, but also minimal off-target cleavage which can be detrimental to the host DNA. However, for Mrr and many other modification-dependent nucleases, as long as the host DNA does not have the target modification sites, there would be little or no selection pressure to limit activity on non-target modification sites. If anything, the only selective pressure that might be present would be if there was a specific cytosine methyltransferase present elsewhere in the chromosome and the Mrr homolog would need to specifically avoiding recognizing those sequences. This is precisely the situation that is realized with the Mrr endonuclease in E. coli where it coexists with the dcm m5C methyltransferase. As a result, the co-existence of modification-dependent endonucleases such as the Mrr, McrA and McrBC superfamilies with the regular RM systems contribute to defining a delicate epigenetic landscape for each bacterial genome.
The association between MspJI and typical RM system elements, such as DNA methyltransferase genes and vsr genes, is another intriguing observation. On the one hand, the association does not seem to be incidental because a number of MspJI homologs have kept the association with the methyltransferases and the vsr genes. On the other hand, in some cases, the association seems dispensable. It is highly possible that MspJI-like genes may have originated from the conventional RM systems, for instance, one scenario could be that the R gene gradually acquires modification-dependent activity after losing its ability to recognize unmodified DNA. As a result, the M gene is no longer essential for the host survival and starts to accumulate inactivating mutations.
As enzymatic reagents, the MspJI family should be especially useful in studies that aim at detecting the epigenetic status of DNA and mapping the locations of the modifications. The nature of the cleavage which takes place at fixed distances away from the methylated sites allows precise inferences of the methylated bases. In this regard, the fragments excised at a fully-methylated site would lend itself to analysis by some of the modern high throughput sequencing techniques. The biggest advantage of the method based on the MspJI-like enzymes, compared to the bisulfite sequencing method as the gold standard, is the convenience in pre-sequencing sample preparation and simplicity in post-sequencing data analysis. As an example, compared to some rather complicated algorithms designed to map bisulfite sequencing data, post-sequencing data analysis here would be much simpler: once the cleavage sites are located by mapping sequencing reads back to the reference genomes, the modified cytosines should be either down or upstream of the cleavage sites at 16 or 17 bases away.
In eukaryotic organisms, the most relevant epigenetic change is CpG or sometimes CHG methylation (21), both of which are recognized by MspJI. With the mCNNR specificities of MspJI, it is theoretically possible to introduce cleavage in the vicinity of up to 50% of the methylated CpG sites. Up to 25% of the genomic CpG sites can be directly interrogated by sequencing the 32-bp bands. A complicating factor is that if two substrate CpG sites are less than 16 bp apart, cleavages from MspJI bound to different half site may interfere with each other and produce fragments <32 bp. More studies are in progress to investigate the possibility of using MspJI in decoding epigenomes.
To our knowledge, MspJI and its homologs represent a unique group of modification-dependent endonucleases which extend our understanding of bacterial RM systems and may have practical application in manipulating modified nucleic acids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs; NLM (grant 5P41LM005800 to R.J.R.). Funding for open access charge: New England Biolabs Inc.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues E. Raleigh, T. Davis, H. Strimpel, S-Y. Xu, M. Sibley, R. Morgan, C. Pedamallu and members in the Division of Restriction Enzymes at NEB for their helpful suggestions and sharing of reagents. We thank Dr Janos Posfai in helping to analyze genomic sequence data. The genomic DNA of Legionella pneumophila strain Philadelphia 1 was a kind gift from Dr J.J. Russo (Columbia U.).
REFERENCES
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren RA. Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich M, Ehrlich K, Mayo JA. Unusual properties of the DNA from Xanthomonas phage XP-12 in which 5-methylcytosine completely replaces cytosine. Biochim. Biophys. Acta. 1975;395:109–119. doi: 10.1016/0005-2787(75)90149-5. [DOI] [PubMed] [Google Scholar]
- 4.Raleigh EA. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol. Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 5.Heitman J, Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J Bacteriol. 1987;169:3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulligan EA, Dunn JJ. Cloning, purification and initial characterization of E. coli McrA, a putative 5-methylcytosine-specific nuclease. Protein Expr. Purif. 2008;62:98–103. doi: 10.1016/j.pep.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 1992;225:327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 9.Waite-Rees PA, Keating CJ, Moran LS, Slatko BE, Hornstra LJ, Benner JS. Characterization and expression of the Escherichia coli Mrr restriction system. J. Bacteriol. 1991;173:5207–5219. doi: 10.1128/jb.173.16.5207-5219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacks S, Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J. Biol. Chem. 1975;250:4060–4066. [PubMed] [Google Scholar]
- 11.Tarasova GV, Nayakshina TN, Degtyarev SK. Substrate specificity of new methyl-directed DNA endonuclease GlaI. BMC Mol. Biol. 2008;9:7. doi: 10.1186/1471-2199-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aertsen A, Michiels CW. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Mol. Microbiol. 2005;58:1381–1391. doi: 10.1111/j.1365-2958.2005.04903.x. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bujnicki JM, Rychlewski L. Identification of a PD-(D/E)XK-like domain with a novel configuration of the endonuclease active site in the methyl-directed restriction enzyme Mrr and its homologs. Gene. 2001;267:183–191. doi: 10.1016/s0378-1119(01)00405-x. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 18.Pei J, Kim BH, Tang M, Grishin NV. PROMALS web server for accurate multiple protein sequence alignments. Nucleic Acids Res. 2007;35:W649–W652. doi: 10.1093/nar/gkm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bitinaite J, Rubino M, Varma KH, Schildkraut I, Vaisvila R, Vaiskunaite R. USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007;35:1992–2002. doi: 10.1093/nar/gkm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.