Abstract
Site-specific homing endonucleases are capable of inducing gene conversion via homologous recombination. Reprogramming their cleavage specificities allows the targeting of specific biological sites for gene correction or conversion. We used computational protein design to alter the cleavage specificity of I-MsoI for three contiguous base pair substitutions, resulting in an endonuclease whose activity and specificity for its new site rival that of wild-type I-MsoI for the original site. Concerted design for all simultaneous substitutions was more successful than a modular approach against individual substitutions, highlighting the importance of context-dependent redesign and optimization of protein–DNA interactions. We then used computational design based on the crystal structure of the designed complex, which revealed significant unanticipated shifts in DNA conformation, to create an endonuclease that specifically cleaves a site with four contiguous base pair substitutions. Our results demonstrate that specificity switches for multiple concerted base pair substitutions can be computationally designed, and that iteration between design and structure determination provides a route to large scale reprogramming of specificity.
INTRODUCTION
Homing endonuclease genes (HEGs) are mobile genetic elements found throughout the microbial universe. They are typically associated with self-splicing intervening sequences (IS; introns or inteins) that are capable of invading and persisting in host genomes, due in part to the site-specific DNA cleavage activity of the rare-cutting homing endonucleases that they encode (1). Cleavage of a DNA site by the homing endonuclease results in copying of the HEG and the surrounding IS into the host genome through double-strand break repair via homologous recombination (2). These properties and functions of homing endonucleases form the basis of new targeted genetic applications, including corrective gene therapy (3). Delivery or expression of a HEG, along with a DNA repair template that is homologous to the DNA sequence surrounding the enzyme’s target, results in the repair or modification of the recipient allele for distances up to one kilobase on either side of the endonuclease cleavage site (4).
The potential sites of cleavage for these applications are primarily limited by the specificities (both natural and engineered) of available homing endonucleases. Multiple techniques can be used to generate homing endonuclease variants that display novel and specific cleavage activities, including mutagenic library selection and structure-based computational design (5–11). These methods currently produce changes in specificity for a relatively small number of contiguous base pairs (one to three) that are then combined to access more distant target sites. If these redesigned regions are not adjacent or overlapping, they can be readily combined in a modular fashion to yield enzymes capable of cleaving new targets differing from the original wild-type site at many base pairs (12), allowing the repair or conversion of novel specific gene loci in vivo (3,13,14). However, the extent to which separately optimized clusters of interactions that involve adjacent base pair substitutions and mutations at the same amino acid positions can be combined has yet to be determined. Furthermore, while high-throughput selection has yielded large numbers of new specificities, the extent to which computational methods can be used to rationally predict and design broad changes in specificity is as yet unknown.
To explore the feasibility of using structure-based computational methods to design novel specificity at multiple adjacent base pairs within a homing endonuclease recognition site, we employed a computational protein design approach (6,15) to redesign I-MsoI (16) to specifically cleave a DNA sequence harboring three consecutive base pair changes relative to the wild-type site. To investigate the modularity of designed interactions at adjacent and overlapping positions, we compared the results of a concerted design for the entire three base pair cluster to the results of individual design for each single base pair substitution. The designed endonucleases were characterized and compared by assaying relative DNA cleavage efficiencies and specificities in vitro, and by X-ray crystallography of each protein–DNA complex. Finally, starting from the crystal structure of the triple base pair switch, we designed a further change in specificity, illustrating the power of iterating between computational design and experimental structure determination.
MATERIALS AND METHODS
Computational design of specificity
The computational methodology for the prediction and redesign of homing endonuclease specificity has been described previously (6,11). A starting model was built using the atomic coordinates from the crystal structure of the wild-type I-MsoI endonuclease in complex with its un-cleaved native DNA recognition site [pdb code 1M5X (16); Supplementary Data]. Nucleotide substitutions were modeled by superimposing the ideal coordinates of new nucleotides onto the backbone atoms of crystallographic nucleotides. The side chain conformations of all amino acids in the vicinity of the substituted nucleotides were allowed to reconfigure according to the Rosetta physics-based full-atom energy function. New combinations of amino acid identities were searched at those amino acid positions that were capable of directly contacting the substituted nucleotides. Positions were considered to be capable of contact if an arginine side chain at that position could be placed within 3.6 Å of any nucleotide base atom. Water-mediated contacts between protein and DNA were also searched by modeling water molecules attached to the major groove atoms of nucleotide bases. During the design for three simultaneous base pair substitutions, small shifts in the protein backbone were modeled using a loop-closure algorithm (17,18). The binding energies of all complexes were calculated by subtracting the energy of the bound complex from the sum of the energies of the separated protein and DNA.
For the individual base pair substitutions at positions ±8 and ±7, an algorithm was employed that directly optimizes the specificity of designed amino acids for the target DNA target site sequence (19,11). The energies of interaction between the protein and DNA (‘affinities’) were computed for the target DNA site as well as for alternative DNA site sequences at the substituted base pairs. Using a genetic algorithm (19), a population of randomized amino acid identities at positions in contact with the substituted nucleotide positions was evolved in silico by enriching for combinations that maximized the discrimination between the target and alternative DNA sites. To excessive loss of affinity, amino acid combinations were disfavored if their affinities were more than 5–10 energy units worse than the best affinity found over all amino acid combinations. The optimal energy threshold for this criterion was estimated by recovery analysis of wild-type and previously-designed (6) interactions (data not shown). The specificities of all design models were calculated as a Boltzmann occupancy of the target complex, versus a partition function consisting of all competing single base pair variant sites (20).
Protein production and purification
Genes for the homing endonuclease designs were assembled by PCR from oligonucleotides, based on a DNAWorks (21) assembly that was codon-optimized for expression in Escherichia coli. 6X-His-tagged proteins were expressed in E. coli BL21-pLysS cells from a pET15 vector by auto-induction (22) at 18–22°C for 24 h. Proteins were purified by nickel affinity fast-performance liquid chromatography (FPLC). Protein purity and identity were verified by polyacrylamide gel electrophoresis (PAGE) and liquid chromatography mass spectrometry (LCMS), and their concentrations were determined by dividing absorbance at 280 nm by their predicted extinction coefficients (5500*Trp + 1490*Tyr + 125*Cys M−1cm−1) (23). For crystallography, I-MsoI designs contained within the pET-24 vector were transformed into BL-21(DE3)pLysS E. coli cells (Invitrogen). Single colonies were then inoculated into 5 ml cultures (LB containing kanamycin and chloramphenicol) that were again grown overnight. Cultures were added to 1l LB media containing 0.5% glucose to repress basal expression. At an optical density of 0.6 AU600, cells were collected by centrifugation and transferred to LB media containing 1 mM IPTG to induce expression. Cells expressed I-MsoI overnight while shaking at 16°C.
In vitro characterization of endonuclease activity
The relative cleavage activities and specificities of wild- type and designed endonucleases were determined by incubating serial dilutions of each enzyme with a constant amount of plasmid DNA. The plasmid substrate contained two I-MsoI cleavage sites, one wild type and one containing designed base pair substitutions. To preserve symmetry, palindromic base pair substitutions were incorporated into both the left (−) and right (+) half-sites of the substituted recognition sites. The plasmid substrates were created by temperature-annealing phosphorylated oligonucleotides into duplexes corresponding to wild type and designed cleavage sites. These sticky-ended duplexes were ligated into two different locations of a plasmid of length 3308 bp, originally obtained from Doyon et al. (7). The substrates were pre-linearized by digestion with the restriction endonuclease XbaI. The sizes of linear DNA fragments resulting from digestion by the endonucleases were as follows: of size 3308 bp (no cleavage), 2766 bp (wild-type site cleaved but not designed site), 2174 bp (designed but not wild-type), 1632 bp (wild-type and designed), 1134 bp (designed), 542 bp (wild-type), where the site whose cleavage results in each product is indicated in parentheses. Plasmid DNA substrates (50–200 ng) were incubated with varying concentrations of endonuclease in 20 mM Tris pH 8.0, 100 mM NaCl, 10 mM MgCl2 for 1 h at 37°C. The reactions were quenched by adding 10 mM EDTA and 1% SDS and incubating for 10 min at 60 °C. The DNA products were separated by agarose gel electrophoresis, visualized by staining with ethidium bromide and quantified by measuring spectral density using the program ImageJ (http://rsbweb.nih.gov/ij/). These data were fit to a sigmoid function to estimate the concentrations that corresponded to half-maximal cleavage of each target site (EC50).
Crystallization
Protein samples were further purified by size exclusion chromatography using a 150 mM NaCl, 0.02% sodium azide, 50 mM Tris pH 8.0 buffer with a flow rate of 1 ml/min on the Superdex75 16/60 column (120 ml volume). Resulting fractions were analyzed by electrophoresis using a 12.5% SDS denaturing polyacrylamide gel. Fractions containing the purified protein were pooled and concentrated from 15 to 1.5 ml with a final concentration of 440 µM. Crystal trays were set using a grid varying pH (6.6, 7.3, 7.8, 8.1, 8.5 and 9.2) and PEG 400 (v/v 18, 20, 22 and 24%). Each reservoir also contained 5 mM CaCl2, 20 mM NaCl. DNA was resuspended and annealed at 92°C for 2 min and then added to protein in a 2 : 1 concentration. Three 1 µl hanging drops of dimer protein concentration 180, 135 and 90 µM were added to each well. Crystals were left to grow at 18°C for 4 days. The crystals were looped and placed in a cryogenic solution containing 170 mM NaCl, 5 mM CaCl2 and 25% v/v PEG 400.
Data collection and refinement
Diffraction data were collected on an in house rotating anode generator, using a Saturn CCD area detector (Rigaku, Inc.). The crystals were maintained at cryological temperatures (72 K) and an X-ray wavelength of 1.54 angstroms was used. Exposure times were 3 to 7 seconds per frame. Images were recorded for 360° of crystal rotation, at 1° intervals. Diffraction images were analyzed by HKL2000 or CrystalClear 1.40r3 to determine the space group. Crystal structures were solved by molecular replacement using Phaser, followed by manual and automated refinement using Coot (24) and PHENIX (25), respectively. For molecular replacement, a modified I-MsoI [1M5X (16)] model was used where (i) waters were removed, (ii) target nucleotides were mutated and (iii) redesigned residues were mutated to alanine. Following molecular replacement and one round of rigid body refinement, redesigned residues were fit to observed electron density. Manual model adjustments, including movement of the phosphodiester backbone, within the electron density were performed using Coot. Finally, automated refinement of atomic positions and atomic displacement factors was performed using PHENIX. During refinement, structural adjustments were modeled using TLS motion determination (26). The Ramachandran statistics (% most favored/allowed/generously allowed/disallowed) for each of the new structures were: I-MsoI ‘GCG’ (0.85/0.15/0.01/0); I-MsoI ‘−8G’ (0.85/0.14/0.01/0); I-MsoI ‘−7C’ (0.87/0.13/0/0).
RESULTS
Computational design of specificity
The use of engineered homing endonucleases to target gene sequences depends on the practical ‘designability’ of available homing endonuclease scaffolds toward potential cleavage sites in a gene of interest. To identify new specificities that were both computationally predictable and therapeutically relevant, we predicted changes in specificity for all single- and double-base pair substitutions in the I-MsoI recognition site and then identified the most ‘designable’ sites in a gene sequence using a position weight matrix approach. This yielded a ranked list of hypothetically designable cleavage sites (Supplementary Table S1), from which therapeutically relevant changes in specificity could be chosen to examine the feasibility of computational design for gene targeting applications.
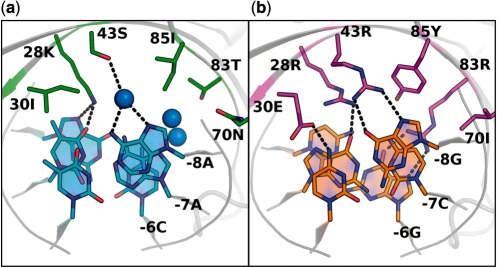
The site sequence GaAGgcgGTCGTGAGcagGgcagG (lower-case letters differ from native), which occurs in the human gene for fumaryl acetoacetate hydrolase (FAH), was chosen for further analysis due to its high rank. In a second round of computational design, we divided the DNA substitutions that occur within this target into separate clusters of contiguous changes, and then computationally searched for favorable interactions between each cluster and new combinations of amino acids at the surrounding residue positions (Supplementary Table S2). This resulted in favorable predictions for a specificity switch involving the three adjacent base pair substitutions {−8G, −7C, −6G}. The cluster of protein–DNA interactions in the region of these base pairs consists of a mixture of direct and water-mediated contacts to the DNA bases by six protein side chains (K28, I30, S43, N70, T83 and I85) in each identical subunit of the homodimeric endonuclease (Figure 1a). At these six amino acid positions, mutations were first optimized simultaneously to recognize the three bp cluster of altered base pairs (Table 1, ‘gcg’), and then were optimized separately for each single base pair substitution (‘−8g’, ‘−7c’, ‘6g’). The designed complexes were ranked based on their predicted binding energies and specificities, with particular emphasis placed on the latter criterion in order to identify designs with maximal specificity for their intended targets (Supplementary Figure S2). For example, in the case of design versus the ‘−8g’ and ‘gcg’ target sites, models of redesigned enzymes that harbor a glutamate at residue 30 were predicted to be more specific than those with glutamine (Supplementary Figure S1). Designs for the remaining two clusters of substitutions in the hypothetical FAH target site were also tested, despite the lack of a predicted change in specificity (Supplementary Table S2). Experimental characterization of these designed sequences showed little to no endonuclease activity on either wild-type or designed DNA substrates. Thus, the specificity measure is a useful criterion by which to predict the experimental outcome of computational designs.
Figure 1.
Amino acid base interactions in wild-type and designed complexes. The interactions between amino acid residues 28, 30, 43, 70, 83, 85 and DNA bases −8, −7, −6 are shown. Blue spheres are crystallographic water molecules. Dashed lines depict selected hydrogen-bonding interactions. (a) Wild-type I-MsoI interactions observed in the original crystal structure (pdb: 1M5X). (b) Predicted model of computationally designed interactions between novel amino acids and DNA bases for the I-MsoI ‘GCG’ design.
Table 1.
I-MsoI DNA cleavage sites
| Site name | Nucleotide changes (top strand) | DNA sequence (top strand) |
|---|---|---|
| ‘wt’ | – | GCAGAACGTCGTGAGACAGTTCCG |
| ‘−6g’ | −6G, +6C | GCAGAAgGTCGTGAGACcGTTCCG |
| ‘−7c’ | −7C | GCAGAcCGTCGTGAGACAGTTCCG |
| ‘−8g’ | −8G, +8C | GCAGgACGTCGTGAGACAGcTCCG |
| ‘gcg’ | −8G, −7C, −6G, +6C, +8C | GCAGgcgGTCGTGAGACcGcTCCG |
| ‘tgcg’ | −9T, −8G, −7C, −6G | GCAtgcgGTCGTGAGACAGTTCCG |
Base pair substitutions are indicated by lower-case, underlined letters. All cleavage sites were double-stranded duplexes and contained complementary substitutions in the bottom strands (not shown).
Novel specific cleavage of multiple adjacent base pairs
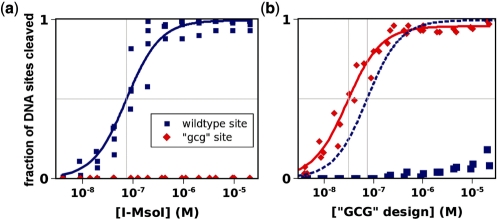
Upon expression and purification, the designed proteins displayed stabilities and yields comparable to that of the wild-type endonuclease. Table 2 shows the cleavage activities of the enzymes on the DNA target sites shown in Table 1. The wild-type endonuclease preferred its natural cleavage site over any of the altered sites, exhibiting 50% cleavage of the wild-type site at an endonuclease concentration of 74 nM. It cleaved the ‘−7c’ and ‘−8g’ sites at higher endonuclease concentrations (305 and 234 nM, respectively), but did not cleave the ‘−6g’ or ‘gcg’ sites at any endonuclease concentration up to 20 μM. This agreed qualitatively with the computed binding energies of the endonucleases for their target sites (Supplementary Figure S1). The endonuclease designed to cleave the ‘gcg’ cluster of three consecutive altered base pairs contained six amino acid mutations per domain in the homodimeric protein (Table 2, Figure 1b). This design cleaved its novel target site at a concentration lower than that at which the wild-type endonuclease cleaved the wild-type site (28.7 ± 2.2 versus 73.5 ± 8.4 nM, respectively, Figure 2 and Supplementary Data), and did not significantly cleave the wild-type site at any endonuclease concentration tested (up to 20 μM). Thus computational design resulted in a mutually-exclusive switch in specificity, with highly efficient cleavage of the significantly altered recognition sequence.
Table 2.
I-MsoI protein sequences and cleavage activities
| Protein | Amino acid sequence |
EC50 versus DNA target site (nM endonuclease) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 28 | 30 | 43 | 70 | 83 | 85 | ‘wt’ | ‘−8g’ | ‘−7c’ | ‘−6g’ | ‘gcg’ | |
| I-MsoI (wt) | Lys | Ile | Ser | Asn | Thr | Ile | 74 | 234 | 305 | >20 000 | >20 000 |
| I-Mso ‘GCG’ | Arg | Glu | Arg | Ile | Arg | Tyr | >20 000 | – | – | – | 29 |
| I-Mso ‘−8G’ | · | Glu | Arg | · | · | Tyr | >20 000 | 238 | – | – | – |
| I-Mso ‘−7C’ | Arg | · | Glu | Thr | · | Trp | >20 000 | – | ∼20 000 | – | – |
| I-Mso ‘−6G’ | Leu | · | · | · | Arg | · | ∼10 000 | – | – | 348 | – |
All amino acid mutations are shown for each designed protein. Amino acids in common with the I-MsoI ‘GCG’ design are underlined. Dots indicate no mutation relative to wild-type. On the right are relative cleavage efficiencies for selected combinations of endonuclease and DNA target site. EC50 indicates the concentration of the endonuclease at which half of the target site was cleaved under the conditions described in ‘Materials and Methods’. Dashes indicate no data.
Figure 2.
Complete switch of activity and specificity for three novel adjacent base pairs by computational design of I-MsoI. The cleavage of either the wild-type site (blue) or the designed ‘gcg’ site (red) is plotted as a function of the endonuclease concentrations of wild-type I-MsoI (a) and the I-MsoI ‘GCG’ design (b). Data are densitometric measurements of ethidium bromide-stained agarose-electrophoresed DNA cleavage products. The data were fit to determine the endonuclease concentrations that correspond to half-maximal cleavage (EC50, gray lines). In (b), the best fit to the wild-type data in (a) is shown in dashed lines for comparison.
High specificity of designed interactions
We characterized the effect of mutations at three designed residues in I-MsoI ‘GCG’ in order to investigate the determinants of its high degree of specificity (Table 3). In agreement with qualitative predictions, the substitution of Glu30 with glutamine had little effect on the concentration at which the designed endonuclease cleaved its target, but resulted in considerable cleavage of the wild-type site at high endonuclease concentrations. This can be rationalized by considering that glutamate can only accept hydrogen bonds from the −8G:C base pair in the model, while glutamine can both accept and donate hydrogen bonds. However, the magnitude of this difference is underestimated by the computational prediction of binding energies (Supplementary Figure S1), indicating a need for training of the model to improve quantitative accuracy.
Table 3.
Cleavage of wild-type and ‘gcg’ sites by point mutants of the I-MsoI ‘GCG’ design
| Protein | Amino acid sequence |
EC50 versus site (nM endonuclease) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 28 | 30 | 43 | 70 | 83 | 85 | ‘wt’ | ‘gcg’ | ||
| I-MsoI (wt) | Lys | Ile | Ser | Asn | Thr | Ile | 74 | >20 000 | |
| I-Mso ‘GCG’ | Arg | Glu | Arg | Ile | Arg | Tyr | >20 000 | 29 | |
| I-Mso ‘GCG’−30Q | Arg | Gln | Arg | Ile | Arg | Tyr | 1319 | 34 | |
| I-Mso ‘GCG’−83T | Arg | Glu | Arg | Ile | · | Tyr | 2998 | 664 | |
| I-Mso ‘GCG’−43S | Arg | Glu | · | Ile | Arg | Tyr | (no expression in E. coli) | ||
This table is formatted as described for Table 2.
The reversion (to wild-type threonine) of Arg83, which makes contact to the −6G nucleotide in the design model, results in an increase in the concentration at which cleavage of the ‘gcg’ target site is observed, as well as cleavage of the wild-type site at particularly high concentrations. This confirms that Arg83 contributes to specificity, but that the remaining designed residues still contribute to specificity for the ‘gcg’ target site in its absence. Reversion (to wild-type serine) of Arg43, which makes contact with −8G in the design, was also attempted, but this protein was not expressible in E. coli.
We further characterized the specificity of the I-MsoI ‘GCG’ design by analyzing its ability to cleave every DNA site that contained a single base pair substitution within the designed three bp cluster (Table 4). As before, palindromic substitutions were introduced into both sides of the target site. The design displayed the highest specificity at position ±6, and at position ±8 only one other sequence (−8A/+8T) was cleaved at relevant concentrations (EC50 = 206 nM). The specificity of the design was lowest at position ±7, a property that was not reflected in the predictions. The designed Arg28 may interact with DNA more promiscuously than expected, or the interface may be flexible in this region in a manner that is not considered in the computational model. Also, the efficient cleavage of the −7T/+7A site suggests that the exclusion of a thymine at this position may require larger residues than Tyr85 or Ile70. However, the behavior of the single base pair ‘−7c’ design that contains Trp85 exhibits suboptimal activity, possibly due to insufficient room in the interface for this residue.
Table 4.
Cleavage specificity of the ‘GCG’ design
| DNA cleavage site | EC50 (nM), I-MsoI ‘GCG’ | Predicted ΔEbinding |
|---|---|---|
| −8GCG/+6CGC (‘gcg’) | 29 | (0) |
| Alternative sites with palindromic single-base pair substitutions: | ||
| −8A/+8T | 206 | +2.4 |
| −8C/+8G | >1024 | +6.5 |
| −8T/+8A | >1024 | +2.8 |
| −7A/+7T | 310 | +8.6 |
| −7G/+7C | 68 | +6.7 |
| −7T/+7A | 24 | +4.5 |
| −6A/+6T | >1024 | +3.3 |
| −6C/+6G | >1024 | +2.4 |
| −6T/+6A | 507 | +1.8 |
Each indicated target site differs from the ‘gcg’ target site (Table 1) by corresponding single base pair changes on both sides of the palindromic target site. Top-stranded substitutions are indicated; complementary substitutions to the bottom strand are not shown. EC50 indicates the concentration of the endonuclease at which half of the target site was cleaved under the conditions described in ‘Materials and Methods’ section. The modeled binding energy is the predicted change in binding energy of the complex after repacking and minimizing the interface around each corresponding base pair substitution.
Design for individual base pair substitutions
In two out of three cases, the amino acid mutations that were predicted by computational design to alter the specificity of I-MsoI for individual base pair substitutions differed from those that were predicted by concerted design for the corresponding three base pair cluster. Each of these designs displayed a preference for its new target site (Table 2; Supplementary Figures S2 and S3) over the wild-type site. However, none of these proteins (which displayed 50% cleavage of their targets at 238 nM to 20 μM enzyme, respectively) were active at endonuclease concentrations as low as those observed for either the wild-type endonuclease vs. its wild-type target (EC50 = 74 nM), or the I-MsoI ‘GCG’ design vs. its ‘gcg’ target site (28 nM). The I-MsoI ‘−7C’ design in particular showed a significant increase in the enzyme concentration at which cleavage occurred, preventing precise estimation of EC50 values (Supplementary Figure S3). Subsequent characterization of a mutant of I-MsoI ‘−7C’ with Trp85→Tyr showed cleavage activity at slightly lower concentrations, but this was accompanied by lower specificity (Supplementary Figure S4). Thus, while in one case (I-MsoI ‘−8G’), the predicted mutations were completely complementary between the individual and concerted designs, the assembly of these individual designs to constitute a three bp change in specificity would be complicated by conflicting mutations at overlapping positions, as well as the poor outcome of the single-base pair I-MsoI ‘−7C’ design.
Crystallographic analysis and validation
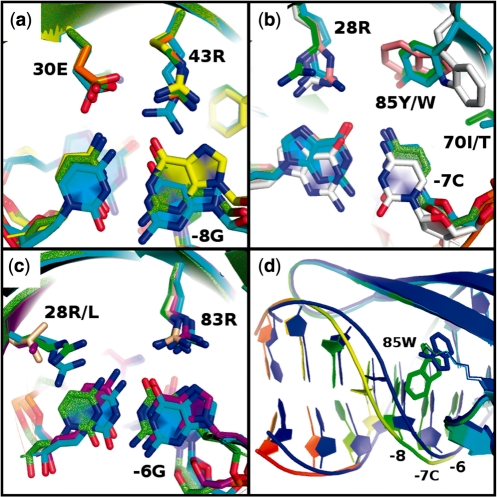
Crystal structures were determined for the I-MsoI ‘GCG’, I-MsoI ‘−8G’ and I-MsoI ‘−7C’ designs in complex with their designed recognition sequences (Supplementary Table S3). The structure of the designed I-MsoI ‘−6G’ complex was described previously (6). These structures show that the conformations and contacts adopted by most of the redesigned residues agree between the single- and triple-base pair redesigns, and were predicted accurately in the designed models (Figure 3). The triple-base pair I-MsoI ‘GCG’ design and the I-MsoI ‘−8G’ design both contain the designed residues Glu30, Arg43 and Tyr85. In both structures, Glu30 and Arg43 make direct contacts to nucleotides +8C and −8G, respectively (Figure 3a), while Tyr85 adopts the predicted position above −7C (Figure 3a and b). The designed Arg28 residue, which is common between the triple-base pair design and I-MsoI ‘−7C’, makes direct contact to the +7G nucleotide in both structures as predicted (Figure 3b). The designed Arg83 residue, which occurs in the triple-base pair design and in I-MsoI ‘−6G’, makes direct contact to the −6G nucleotide in both structures, also as predicted (Figure 3c).
Figure 3.
Comparison of designed and crystallographically observed interactions. (a–c) the crystal structure of the triple-base pair I-MsoI ‘GCG’ design (cyan) is aligned with the designed model (green) and with the crystal structures and designed models of each single-base pair design: (a) I-MsoI ‘−8G’ (X-ray: yellow, model: orange), (b) I-MsoI ‘−7C’ (X-ray: white, model: pink), (c) I-MsoI ‘−6G’ (X-ray: purple, model: beige). (d) A conformational shift in the DNA backbone is observed near Trp85 in the I-MsoI ‘−7C’ crystal structure (colored by increasing B-factor from light blue to red), compared to the designed model (dark blue).
Whereas the crystal structures show that most designed interactions were correctly predicted, unprecedented shifts in the designed region of the interface occurred. In the structure of the I-MsoI ‘GCG’ complex, local rearrangement of the designed region resulted in a significant (1.4 Å) shift of the −8G:C base pair, which moved away from the protein (Figure 3a, cyan). This is accompanied by the extension of Glu30 and Arg43 to remain in specific contact with nucleotide −8G. In contrast, these shifts were not observed in the structure of the single-base pair I-MsoI ‘−8G’ design (Figure 3a, yellow). In the crystal structure of the I-MsoI ‘−7C’ design in complex, Trp85 juts outward toward the DNA backbone, rather than into the core of the interface as designed (Figure 3d). As a result, the neighboring DNA backbone shifted 2.4 Å away from the original wild-type position. This may explain the lower activity of this design.
Iterating between design and crystallography enables further switch in specificity
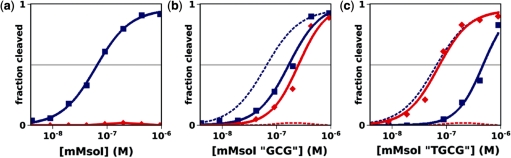
An important challenge in endonuclease engineering is to achieve specificity for genomic target sites which may differ by many base pairs from the original endonuclease target site. To investigate the utility of an iterative approach to structure-based computational design, we began with the crystal structure of the redesigned I-MsoI ‘GCG’ endonuclease in complex with its cognate DNA site ‘gcg’. Mutations were designed to alter the specificity for the adjacent base pair (wild-type: −9G:C) to allow cleavage of −9T:A (Table 1, ‘tgcg’). The I-MsoI ‘TGCG’ design contained eight predicted mutations (K28R, I30E, R32K, Q41Y, S43R, N70I, T83R, I85Y; additional mutations relative to the I-MsoI ‘GCG’ design are underlined). An additional requirement for most hypothetical gene targets is that specificity be alterable in an asymmetric fashion with regard to the two halves of the site. Therefore, these mutations were incorporated into the N-terminal domain of a monomerized construct of I-MsoI, referred to as mMsoI, which was previously created by engineering a peptide linker between the two domains of the wild-type homodimer (27). This resulted in the novel specific cleavage of a DNA target site containing four consecutive, asymmetric base pairs that could not be cleaved efficiently or selectively by the corresponding monomeric mMsoI ‘GCG’ endonuclease (Figure 4).
Figure 4.
Designed specific cleavage activity for an asymmetric four-base pair cluster. In vitro cleavage of wild-type (blue) and asymmetric ‘tgcg’ (red) DNA sites by monomerized I-MsoI (mMsoI) endonuclease designs. (a) wild-type mMsoI endonuclease, (b) N-terminal mMsoI ‘GCG’ design, (c) N-terminal mMsoI ‘TGCG’ design. Dashed lines in (b and c) represent the mMsoI trace from (a). Data are densitometric measurements of ethidium bromide-stained agarose-electrophoresed DNA cleavage products (Supplementary Figure S5).
DISCUSSION
Large changes in specificity by computational protein design
The ability to rationally design protein–DNA recognition is a critical test of our understanding, and could have considerable technological and medicinal value. Our results demonstrate the feasibility of using computational protein design to reprogram the target site specificity of homing endonucleases at multiple adjacent base pairs. The designed cleavage of a novel three- and four-base pair clusters represents a significant advance in computational design techniques, and could soon parallel the capabilities of the latest selection techniques for altering homing endonuclease specificity, which are combinatorially limited to simultaneously altering between three and six amino acids in a single library (7–10).
Concerted design of context dependent interactions
The relationship between the triple-base pair design I-MsoI ‘GCG’ and each of the single base pair designs provides insights into the specificities of homing endonucleases and how they can be reprogrammed. While it is feasible to computationally design single-base pair changes in specificity (6,11), the I-MsoI ‘GCG’ design shows that the simultaneous design of interactions between the protein and multiple adjacent base pair substitutions can be advantageous for introducing larger changes in specificity. This is because the physics-based modeling approach employed here is capable of capturing the context dependence of designed interactions, and optimizing the amino acid choices at positions that can interact with multiple adjacent base pairs. Thus, solutions found by concerted design for the three bp cluster differed from those yielded by design for individual base pair substitutions. For example, in the I-MsoI ‘GCG’ design, the −7C:G base pair is contacted by Arg43 and Tyr85 rather than Glu43 and Trp85 as in the case of the single-base pair change. A synergistic benefit of designing for concerted changes in specificity is evident in that the I-MsoI ‘GCG’ design cleaves its target site more efficiently than any of the designs for the single base pair substitutions, including I-MsoI ‘−8G’, which consists entirely of amino acid mutations that are also present in the triple-base pair design.
DNA flexibility in the homing endonuclease interface
Crystallographic analyses demonstrate that novel specific interactions between protein and DNA can be successfully predicted using computational structure-based engineering. However, structural shifts in the interface, particularly of the bound DNA, can occur as a consequence of changes to protein and DNA sequence. Furthermore, these changes neither additive nor readily predictable using current modeling techniques. This reflects an inherent structural flexibility of the I-MsoI homing endonuclease interface that was not observed in previous studies of either I-MsoI (6,16) or its close relative I-CreI (12). That I-MsoI differentially cleaves DNA sequences with different intramolecular conformations also raises the possibility that indirect readout of sequence-dependent DNA structure (11,28) may be important throughout the homing endonuclease recognition site. This highlights the importance of accurately modeling significant shifts in DNA conformation for future efforts to predict and design the properties of protein–DNA interactions. Finally, the use of this new crystal structure to design high activity and specificity for additional changes in specificity illustrates the power of combining computational design and X-ray crystallography to generate novel cleavage specificities for genome engineering applications.
ACCESSION NUMBER
The crystal structures of the designed I-MsoI ‘GCG’, I-MsoI ‘-8G’, and I-MsoI ‘-7C’ complexes have been submitted to the RCSB Protein Data Bank, with the identifiers 3mip, 3mis, and 3ko2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health (GM084433 to D.B., GM049857 to B.L.S.); National Cancer Institute (CA133833 to B.L.S.); University of Washington Molecular Biophysics Training Grant (GM008268 to J.A. and G.K.T.); Foundation for the National Institutes of Health through the Gates Foundation Grand Challenges in Global Health Initiative; Division of Basic Sciences at the Fred Hutchinson Cancer Research Center; Howard Hughes Medical Institute. Funding for open access charge: US National Institutes of Health (GM084433 to D.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Andrew P. Leaver-Fay and others for their significant and ongoing contributions to the Rosetta macromolecular modeling suite, as well as Betty W. Shen, Jacob E. Corn, and Matthew C. Clifton for useful advice regarding crystallographic refinement.
REFERENCES
- 1.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 2.Belfort M, Perlman PS. Mechanisms of intron mobility. J. Biol. Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 3.Arnould S, Perez C, Cabaniols J, Smith J, Gouble A, Grizot S, Epinat J, Duclert A, Duchateau P, Pâques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C, Louvard D, Jaisser F. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol. Cell. Biol. 1998;18:1444–1448. doi: 10.1128/mcb.18.3.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Stoddard BL, Seligman LM. Isolation and characterization of new homing endonuclease specificities at individual target site positions. J. Mol. Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 6.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 8.Smith J, Grizot S, Arnould S, Duclert A, Epinat J, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat J, Stricher F, Petit A, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Wen F, Sun N, Zhao H. Directed evolution of homing endonuclease I-SceI with altered sequence specificity. Protein Engineering, Design and Selection. 2009;22:249–256. doi: 10.1093/protein/gzp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, Stoddard BL, Baker D. Exploitation of binding energy for catalysis and design. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redondo P, Prieto J, Muñoz IG, Alibés A, Stricher F, Serrano L, Cabaniols J, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 13.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao H, Smith J, Yang M, Jones S, Djukanovic V, Nicholson MG, West A, Bidney D, Carl Falco S, Jantz D, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61:176–187. doi: 10.1111/j.1365-313X.2009.04041.x. [DOI] [PubMed] [Google Scholar]
- 15.Havranek JJ, Duarte CM, Baker D. A simple physical model for the prediction and design of protein-DNA interactions. J. Mol. Biol. 2004;344:59–70. doi: 10.1016/j.jmb.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 16.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 17.Canutescu AA, Dunbrack RL. Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 2003;12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J. Mol. Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 19.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat. Struct. Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 20.Ashworth J, Baker D. Assessment of the optimization of affinity and specificity at protein-DNA interfaces. Nucleic Acids Res. 2009;37:e73. doi: 10.1093/nar/gkp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 26.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat RJ. Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael Gromiha M, Siebers JG, Selvaraj S, Kono H, Sarai A. Intermolecular and intramolecular readout mechanisms in protein-DNA recognition. J. Mol. Biol. 2004;337:285–294. doi: 10.1016/j.jmb.2004.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.