Abstract
Recombineering is employed to modify large DNA clones such as fosmids, BACs and PACs. Subtle and seamless modifications can be achieved using counter-selection strategies in which a donor cassette carrying both positive and negative markers inserted in the target clone is replaced by the desired sequence change. We are applying counter-selection recombineering to modify bacmid bMON14272, a recombinant baculoviral genome, as we wish to engineer the virus into a therapeutically useful gene delivery vector with cell targeting characteristics. Initial attempts to replace gp64 with Fusion (F) genes from other baculoviruses resulted in many rearranged clones in which the counter-selection cassette had been deleted. Bacmid bMON14272 contains nine highly homologous regions (hrs) and deletions were mapped to recombination between hr pairs. Recombineering modifications were attempted to decrease intramolecular recombination and/or increase recombineering efficiency. Of these only the use of longer homology arms on the donor molecule proved effective permitting seamless modification. bMON14272, because of the presence of the hr sequences, can be considered equivalent to a highly repetitive BAC and, as such, the optimized method detailed here should prove useful to others applying counter-selection recombineering to modify BACs or PACs containing similar regions of significant repeating homologies.
INTRODUCTION
Recombineering is a homologous recombination (HR)-based genetic engineering technique mediated by transient, regulated expression of phage-encoded recombinases (1–3). Using tools developed from lambda Red (4) or Rec E/T (3) recombineering is commonly employed to modify large, low-copy number replicons, such as BACs, PACs or fosmids, and, less frequently, intermediate and multicopy plasmids. Sequence manipulations can range from point mutations to modifications in the kilobasepair range. Recombineering capacity can be supplied via a defective, chromosomally integrated lambda prophage, in Escherichia coli strains such as EL250 and EL350 (5) and derivatives (6), or from a variety of plasmid systems (7). An attractive feature of the procedure is that, in contrast to RecA, phage-encoded recombinases mediate efficient HR via relatively short (∼50 bp) sequences enabling a donor molecule with appropriate terminal homology sequences to be built easily by standard PCR.
In its simplest single-step guise recombineering involves transient induction of phage recombinase activities in bacteria containing the circular target followed by their transformation with linear, donor dsDNA molecules containing (i) terminal sequences shared with the target, (ii) the desired target sequence modification and (iii) an appropriate selection marker. Following HR the selection marker may be left in or, if flanked by appropriate sites, removed by site-specific recombinases, such as Cre or Flp, leaving a post-excision scar. An alternative approach, counter-selection recombineering, has been developed that, although more complex to perform, has the advantage of seamless modification generating recombinants with the desired sequence change only. Counter-selection strategies are, however, limited to low-copy number targets because the intrinsic inefficiency of the method makes it difficult otherwise to select negatively for desired recombinants. Several such methods, utilizing different counter-selection cassettes, have been described (6,8–10). We reported previously (8) an approach utilizing a counter-selection cassette, the RT-cassette, containing positive, tetA(C), and negative, rpsL+, markers generated by PCR from the plasmid pBAC-RT (11). The RT-cassette is inserted into the target by an initial recombineering step, using positive selection with tetracycline (Tc), and then subsequently replaced seamlessly with another cassette, containing the desired sequence, in a second recombineering step applying negative selection. When expressed in streptomycin (Sm) resistant (SmR) rpsL− host, such as EL250 or EL350, the wild-type rpsL+ gene expressed from the RT-cassette results in a dominant Sm sensitive (SmS) phenotype thereby providing the negative marker. Desired recombinants lacking the RT-cassette will be SmR revertants and selectable with Sm.
There are a number of methodological points to consider when performing recombineering. One is that the transient ‘hyper-recombination’ (hyperRec) state produced in the bacterium by phage recombinase induction may cause resident clones, particularly large PACs or BACs containing stretches of repeating homologous sequences, to undergo intramolecular rearrangements (12). If performing simple, positive-selection recombineering on such a potentially unstable clone then it is possible to prevent significant rearrangement and obtain intact, desired recombinants by attenuating the hyperRec state by simply decreasing the Red-induction time (12). However, when employing counter-selection such clone instability will be far more problematic as negative selection will actively select all deletion-bearing clones lacking the counter-selection cassette. A second consideration applies when combining recombineering with subsequent Cre recombinase-mediated modification to, for example, remove a marker sequence. Numerous near consensus loxP sites, so-called cryptic loxP sites, are distributed throughout mammalian genomes many of which are good recognition sites for Cre (13,14). The recombineering strain EL350 (5) contains an arabinose-inducible cre gene and is commonly used for combining recombineering with subsequent Cre-mediated marker removal. However, it has been demonstrated (14) that the cre gene in EL350 is leaky and, as such, if EL350 is used to propagate a BAC or PAC containing cryptic loxP sites then the resident clone, especially if based in a vector, such as pBeloBac, containing a consensus loxP site, may suffer Cre-mediated single-strand nicking leading to replication failure, marker loss and poor bacterial growth.
We are applying recombineering to develop the baculovirus Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) as a gene therapy (GT) vector for liver fibrotic disease. Although AcMNPV was originally developed as a vector for heterologous protein expression in insect cells (15) it has, more recently, received attention as a possible GT vector as it is also capable of transducing and delivering a functional expression cassette to a range of mammalian cells (16,17). In contrast to single-step, positive-selection recombineering, which has been employed by others to either selectively delete baculoviral genes (e.g. 18–20) or to improve its characteristics as a protein expression vector (21,22), we have begun to seamlessly modify the AcMNPV bacmid bMON14272 (23) by utilizing our counter-selection recombineering protocol (8). In the first instance we aim to replace the gp64 gene, encoding the promiscuous envelope fusion protein GP64, with a range of genes encoding the F (Fusion) proteins from the group II Alphabaculoviruses (24,25). These F proteins can functionally replace GP64 in insect cell entry, budding and endosomal escape (26,27) but F-pseudotyped AcMNPV is apparently unable to transduce mammalian cells (28–30). We aim to decorate, via recombineering, the surface of F-pseudotyped AcMNPV with targeting peptides to provide selective liver cell transduction. In the course of these experiments we determined that bMON14272 was unstable during the hyperRec phase resulting, after negative selection, in an unacceptably high background of RT-cassette-lacking deletion clones. Extensive restriction mapping and PCR analyses of a subset of these clones established that the instability was due to intramolecular recombination between pairs of the homologous regions (hrs) present in the genome that are thought to play roles in DNA replication and/or to act as transcriptional enhancers (31,32). As we wished to seamlessly modify the genome we explored whether protocol modifications could increase significantly the numbers of intact, final recombinant clones above the deletion-bearing background. These modifications included attempting to minimize the potential for Cre-mediated clone damage, via cryptic loxP sites identified in the bacmid sequence, by creating a non-Cre, non-Flp producing recombineering strain that was still compatible with use of the RT-cassette; co-transforming bacmid and donor dsDNA into Red-induced bacteria; and even inducing recombinase activity after donor DNA transformation. Of all the modifications attempted only the inclusion of longer terminal homology arms to the donor molecule designed to replace the RT-cassette proved effective permitting seamless modification of the bMON14272 bacmid following negative selection. Incorporation of a commercial in vitro recombination-based cloning system facilitated construction of donor DNA molecules equipped with such longer homology arms. Optimization of our protocol to permit seamless modification of the AcMNPV bacmid should not only prove useful to workers wishing to seamlessly modify this viral replicon but also to others planning to apply counter-selection recombineering to modify BACs or PACs containing similar regions of significant repeating homologies.
MATERIALS AND METHODS
General molecular methods
Classical restriction enzyme (RE)-based genetic engineering techniques, preparation of media, etc were undertaken as described (33) unless otherwise stated. All PCRs were performed in volumes of 50 µl containing 15 pmol of each oligonucleotide (ODN) primer and 200 µM of each dNTP and were catalysed with a high-fidelity DNA polymerase (Phusion, NEB) using conditions designed to minimize mis-incorporation. ODN primers for generating recombineering cassettes were PAGE-purified and were from IDT (Leuven, Belgium). ODNs for sequencing and priming non-recombineering PCRs were desalted and from VH Bio (Gateshead, UK). Induction of lambda Red activities, preparation of electrocompetent E. coli cells and all subsequent recombineering steps were performed essentially as described (8) unless otherwise stated. Plasmids, bacmid and virus constructs were prefixed with lower-case p, b and v, respectively.
Identification of cryptic and secondary loxP sites
Cryptic loxP sites in the bMON14272 sequence were identified using the program loxpFinder at http://wilkie226.dmed.ed.ac.uk:8080/loxpFinder/ as described (14).
Generation of E. coli strain MW001
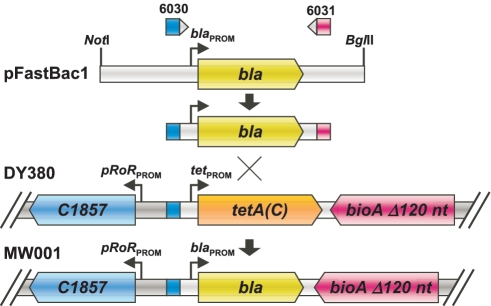
Strain MW001 was generated by PCR-amplification of a β-lactamase (bla) recombinering cassette (bla-cassette) using a gel-purified 3.2 kb NotI-BglII fragment of pFastBac1 (Invitrogen) (5 ng) as template and ODNs 6030-6031 (Supplementary Table S1), containing, respectively, 50- and 49-nt 5′ homology arms corresponding to sequences flanking the tetA(C) gene sequence in E. coli strain DY380 (5) (Figure 1). Non- (control) or Red-induced electrocompetent DY380 cells were electroporated with the bla-cassette (500 ng), recovered (SOC[–Mg] medium, 32°C, 220 r.p.m., 2.5 h), serially diluted (M9 salts) and aliquots (50 µl) spread on either selective (ampicillin [Amp], 50 µg/ml) or non-selective (for cell viability determination) LB-agar plates and incubated (32°C, 48 h). Five discrete Amp-resistant colonies were re-streaked on Amp-selective LB-agar plates and incubated further (32°C, 48 h). Colonies in which the bla-cassette had been inserted correctly into the host chromosome were identified by colony-PCR, using the insertion site-flanking ODNs 6032–6035 (Supplementary Table S1), and by direct sequencing of the resulting PCR products. A single colony was designated MW001 (Figure 1).
Figure 1.
Construction of E. coli strain MW001. A β-lactamase (bla) recombinering cassette, containing a bla gene (green directional box) and homology arms (blue and purple boxes), corresponding to sequences flanking the tetA(C) gene (orange directional box) sequence in E. coli strain DY380, was PCR-amplified from a pFastBac1 restriction fragment template with primers 6030 and 6031 and used as donor DNA to replace, via recombineering, the tetA(C) gene in DY380 generating strain MW001.
Construction of a dual promoter GFP reporter plasmid
A 1.4-kb fragment, containing CMVPROM-eGFP, released from pEGFP-N1 (Clontech) by sequential incubation with NsiI, T4 DNA Pol (NEB), to generate blunt ends, and, following enzyme inactivation, NotI, was ligated into pFastBac1, via the BstZ171 and NotI sites, to generate pMW002. A 127-bp region, containing the p10PROM sequence from Spodoptera exigua multiple nuclear polyhedrosis virus (SeMNPV), was PCR-amplified using bacmid SeBAC10 DNA (34) (10 ng) as template and ODNs 6028–6029 (Supplementary Table S1) that contained, in their respective 5′ ends, NheI and BshTI recognition sites. The resulting amplicon, following incubation with NheI, BshTI and gel-purification, was cloned into the corresponding sites of pMW002 generating pMW005 (Supplementary Figure S1).
Sequence confirmation of hrs of bMON14272
Because of potential discrepancies between the sequences of the hrs in the published AcMNPV genome [Acc. No L22858; (35)] and those of the bMON14272 base vector (23) the bMON14272 hrs 1, 1a, 2, 2a, 3, 4a, 4b, 4c and 5 were PCR-amplified, using respective ODN pairs 6193–6227, 6188–6194, 6189–6195, 6196–6197, 6183–6199, 6184–6200, 6190–6201, 6202–6203 and 6191–6204 (Supplementary Table S1), and sequenced.
Mapping of intramolecular recombination sites in deletion bacmids
Deletions in SmR bacmid clones that resulted following negative-selection recombineering were mapped by a combination of restriction fingerprinting, with ClaI, HindIII, NcoI and SpeI, and diagnostic PCRs designed to amplify across the predicted deleted region. Sequences of the resulting PCR products were aligned to and compared with the sequences of the bMON14272 hrs to identify precise break points.
Titration of Red-induction time
A 200-ml liquid culture (SOB[–Mg], Kanamycin [Kan], 50 µg/ml; Tc, 5 µg/ml), inoculated with 10 ml of an overnight mini-culture of MW001 harbouring bacmid bMW009 (see below), was incubated (32°C, 220 r.p.m.) and, at an OD600 of 0.8, aliquots (50 ml) transferred to each of three sterile, pre-warmed (42°C) 500 ml flasks. Red activities were induced at 42°C for 5, 10, or 20 min in a gently shaking water bath (100 r.p.m.) after which times these, and the original non-induced (0 min) culture, were chilled on ice (15 min). Aliquots (100 µl) of electrocompetent bacteria, prepared from each of the four cultures (0, 5, 10, 20 min induction), were mock-electroporated, recovered (SOC[–Mg] medium, 32°C, 220 r.p.m., 2.5 h), serially diluted (M9 salts) and aliquots (50 µl) spread on either selective (Kan, 50 µg/ml; Sm, 500 µg/ml) NSLB-agar (11) or non-selective (Kan, 50 µg/ml) LB-agar plates, incubated (32°C, 48 h), and the number of SmR colonies/106 viable cells determined.
Co-transformation of bacmid and donor DNA
For co-transformation studies, bacmid DNA (100 ng), isolated with a commercial kit (Qiagen plasmid midi kit) according to the manufacturer’s instructions, and appropriate linear donor dsDNA (500 ng) were mixed and electroporated into Red-induced electrocompetent MW001 (100 µl).
Post-transformation Red-induction
Non Red-induced electrocompetent MW001, harbouring an appropriate bacmid, were electroporated with linear donor dsDNA (500 ng) followed by the immediate addition of pre-warmed medium (SOC[–Mg], 2 ml, 42°C) and incubation (42°C, 10 min) prior to recovery (32°C, 220 r.p.m., 2.5 h).
Generation of linear recombineering cassettes
Two recombineering cassettes with ‘short’, defined as ∼50 bp, homology arms were generated by standard PCR. An RT-cassette, designed to replace the gp64 gene in bMON14272, was PCR-amplified as described (8) using ODNs 6005–6006 (Supplementary Table S1). A second cassette, containing the SeMNPV F gene, was generated by PCR-amplification using plasmid pSeBglII-H (36) as template (5 ng) and the primer pair 6009–6010 (Supplementary Table S1). Both amplicons contained ∼50-bp terminal sequences homologous to regions immediately 5′ and 3′ of the gp64 gene in bMON14272.
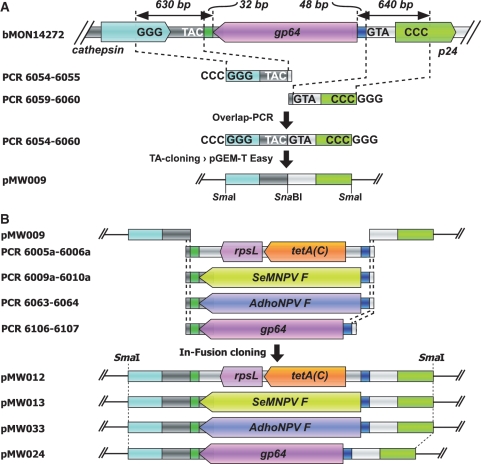
Recombineering cassettes equipped with ‘long’ (which we define here as 200 bp or longer) homology arms were created by an initial PCR, to generate the replacement sequence, followed by cloning of the resulting amplicon, via a commercial in vitro recombination system, into a construct previously fitted with the left and right homology arms and subsequent release of the homology arm-flanked cassette by RE digestion (Figure 2). To generate this construct, sequences in bMON14272, from 200 bp beyond the 5′ and 3′ ends of the gp64 gene, were scanned visually to identify either complete or partial sequences of RE recognition sites known to be absent from the two flanking sequences, from the various recombineering replacement cassettes and from the cloning vector pGEMT-Easy. Such a scan identified the SmaI half-sites GGG and CCC 630 bp and 640 bp from, respectively, the 3′ and 5′ ends of gp64 (Figure 2A). Next, 50-bp regions immediately flanking either end of the gp64 sequence were similarly scanned to identify half-sites that, when fused would create a blunt-cutting RE recognition site absent from the two ‘long’ homology arm sequences and also from pGEMT-Easy. This scan identified the SnaBI half-sites TAC and GTA 32 and 48 bp from, respectively, the 3′ and 5′ ends of gp64 (Figure 2A). Subsequently, the sequences between and including the partial SmaI and SnaBI recognition sites identified in the 5′ and 3′ flanking sequences of gp64 were PCR-amplified using bMON14272 DNA (5 ng) as template and the respective ODN pairs 6054–6055 and 6059–6060 (Figure 2A and Supplementary Table S1). The resulting amplicons, which now contained intact terminal SmaI sites created during the PCR, were subsequently fused, via short overlapping homologies introduced via the appropriate primers, in an Overlap-PCR (37), using ODNs 6054–6060, generating a single product, containing a unique SnaBI site at the fusion site, which was then cloned, via TA cloning, into pGEM-T Easy (Promega) to give pMW009 (Figure 2A). The RT-, SeMNPV F gene-, Adoxophyes honmai NPV (AdhoNPV) F gene- and gp64-cassettes were generated by PCR-amplification from, respectively, pBAC-RT (1 ng), pSeBglII-H (1 ng), bMON14272 (5 ng) and AdhoNPV DNA (5 ng) (38) using the respective ODN pairs 6005a–6006a, 6009a–6010a, 6063–6064 and 6106–6107 (Supplementary Table S1). The resulting amplicons were flanked with 31 and 47 bp sequences, corresponding to, respectively, the 3′ and 5′ flanking regions of gp64 as far out as the SnaBI half-sites, plus additional 15-bp termini equivalent to sequences either side of the SnaBI site in pMW009 (Figure 2B). These four cassettes were then cloned directly into SnaBI-linearized pMW009, via an in vitro recombination system (In-Fusion, Clontech) according to the manufacturer’s protocol, generating plasmids pMW012 (RT), pMW013 (SeMNPV F), pMW033 (AdhoNPV F) and pMW024 (gp64) (Figure 2B). Following DNA sequencing, each ‘long’ homology-arm equipped cassette was released from its respective construct via restriction with SmaI, gel-purified and quantified, by visualization against a DNA mass ladder (2-log ladder, NEB), prior to use in recombineering.
Figure 2.
Generation of ‘long’ homology arm negative-selection recombineering replacement cassettes via In-Fusion cloning. (A) PCR products, generated from template bMON14272 and primer pairs 6054–6055 and 6059–6060, containing 3′ and 5′ gp64 flanking sequences were fused in an Overlap-PCR with primer pair 6054–6060 to produce a single PCR product with central, unique SnaBI site and terminal SmaI sites that was cloned into pGEM-T Easy to create pMW009. (B) RT-cassette and replacement cassettes, containing coding sequences for SeMNPV F, AdhoNPV F and AcMNPV gp64 genes, were generated by PCR, with respective primer pairs 6005a–6006a, 6009a–6010a, 6063–6064 and 6106–6107, and cloned, via In-Fusion cloning, into pMW009 generating, respectively, pMW012, pMW013, pMW033 and pMW024.
Subsequent to the production, via In-Fusion cloning, of these ‘long’ homology-arm cassette-containing constructs an additional series of gp64 gene cassettes with differing, shorter homology-arm lengths were PCR-amplified from, as template, the gel-purified cassette (1 ng) previously excised from pMW024 using ODN pairs 6003–6258, 6259–6262 and 6256–6257 (Supplementary Table S1) generating, respectively, cassettes with homology-arms of 100, 200 and 400 bp. Amplicons were purified with a commercial system (Wizard DNA Purification, Promega) and visually quantified, as above, before being used directly in counter-selection recombineering.
Counter-selection recombineering
Briefly, non- (control) or Red-induced electrocompetent bMON14272-containing MW001 cells were electroporated with the ‘short’ homology arm RT-cassette (500 ng), recovered (SOC[–Mg] medium, 32°C, 220 r.p.m., 2.5 h), serially diluted (M9 salts) and aliquots (50 µl) spread on either selective (Kan, 50 µg/ml; Tc, 5 µg/ml) or non-selective (Kan, 50 µg/ml) LB-agar plates and incubated (32°C, 48 h). Five discrete KanR/TcR colonies were re-streaked on Kan+Tc-selective plates and incubated further (32°C, 48 h). A single bacmid in which the RT-cassette had been inserted correctly into the gp64 locus, identified by colony-PCR using the insertion site-flanking ODNs 6085–6101 (Supplementary Table S1) and restriction mapping, was designated bMW009 and used in all subsequent negative selection steps.
Subsequently, negative selection recombineering was employed in attempts to replace the RT-cassette in bMW009 with each of the four linear replacement cassettes—the SeMNPV F gene-cassette, with either ‘short’ or ‘long’ homology arms, and the AdhoNPV F gene- and gp64 rescue-cassettes with ‘long’ arms. Following recovery and dilution as above, aliquots were spread on either selective (Kan, 50 µg/ml; Sm, 500 µg/ml) NSLB-agar or non-selective (Kan, 50 µg/ml) LB-agar (11) plates and incubated (32°C, 48 h). For each cassette several discrete SmR colonies were re-streaked to fresh Kan+Sm-selective plates, incubated further (32°C, 48 h) and subjected to colony-PCR, using ODNs 6085–6101, to identify clones in which the RT-cassette had either been successfully replaced with an F gene- or the gp64-cassette or had been removed via deletion. Bacmid DNAs in which the RT-cassette had been replaced were isolated and subjected to restriction mapping to confirm bacmid fidelity. Single bacmid clones, containing the correctly inserted SeMNPV F, AdhoNPV F or gp64 sequences were named, respectively, bMW010, bMW025 and bMW028 all of which subsequently received, via Tn7-mediated transposition (Bac-to-Bac, Invitrogen), the CMVPROMp10PROM-eGFP reporter sequence from pMW005 generating the respective bacmids bMW011, bMW036 and bMW033 from which viruses vMW011, vMW036 and vMW033 were produced.
Subsequently, negative selection recombineering was also undertaken on bMW009 with each of the additional gp64 gene cassettes with shorter homology-arms. Colony-PCR was employed on 10 randomly selected colonies to determine, for each homology-arm length, the proportion of colonies containing bacmids in which the gp64 gene sequence had successfully replaced the RT-cassette.
Confirmation of hr fidelity in F-containing recombinant bacmids
To confirm the presence and fidelity of the hr regions in the bMW010, bMW025 and bMW028 bacmids the hr1, hr1a, hr2, hr2a, hr3, hr4a, hr4b, hr4c and hr5 sequences were PCR-amplified and the lengths of the resulting products compared electrophoretically to equivalent PCR products amplified from bMON14272.
Virus generation
Sf9 insect cells were passaged (28°C) in shaking cultures (30 ml, 90 r.p.m.) using Sf-900II serum-free medium (Sf900II SFM, Invitrogen) supplemented with 2% fetal bovine serum (FBS). For virus generation, Sf9 cells (1.0 × 106) were transfected with bacmid DNA (∼1 µg), cultured (5 days) and an aliquot (500 µl) of the clarified (10 min, 1000 g), filter-sterilized (0.45 µm pore size) medium used to infect a new batch of Sf9 cells. At 72 h post-infection (p.i.) infected cells were inspected for GFP expression by fluorescence microscopy and titrated by end-point dilution assay. Amplified virus stocks were made by infecting Sf9 cells (1.0 × 106 cells/ml) in a shaking culture (30 ml, 90 r.p.m.) at a multiplicity of infection of 0.05 plaque-forming units (pfu)/cell. Cultures were harvested 5 days p.i., centrifuged and supernatants clarified, filter-sterilized as above, titres dertermined and stored (4°C).
Mammalian cell transduction
Saos-2, and both HeLa and HepG2 cells, were maintained (37°C and 5% CO2) in, respectively, Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% FBS, and in Eagle's Minimum Essential Medium (EMEM) supplemented with 1% nonessential amino acids (NEAA) and 10% FBS. Cells were seeded (1.0 × 105 cells/well) in 24-well plates and, after incubation (24 h), the medium was replaced with 500 µl of clarified, filter-sterilized baculovirus-containing Sf900II SFM resulting in virus: cell ratios of 50 (vMW011, vMW033) or 100 pfu/cell (vMW036). After 1 h incubation (28°C with gentle shaking) the medium was aspirated, the cells washed and fresh DMEM or EMEM added as appropriate. GFP expression was visualized in transduced cells by fluorescence microscopy 48 h post-transduction.
RESULTS AND DISCUSSION
Generation of MW001
We searched for primary and secondary cryptic loxP sequences using an online search tool (14). bMON14272 lacks a consensus loxP site but was found to contain one primary cryptic loxP site, located within the lef-11 gene, plus a number of secondary sites which were revealed as the consensus stringency used by the search tool was decreased. The primary cryptic loxP sequence identified (ATTAGCTCTTATATATTCTTTTATACGCTCAAAC) contained 10 mismatches in the 13-bp palindromic arms, 5 in the 18 conserved Cre contact points and none in the TATA sequences flanking the spacer region. Because the PBAD-driven cre gene exhibits leaky expression in EL350 (14) and because non-productive recombination between cryptic loxP sites can lead to DNA damage (14,39) we considered it prudent to undertake recombineering of bMON14272 in a non-cre expressing strain even though we did not establish whether the cryptic loxP sites identified were functional. Although EL250 (5) was a possible candidate this strain expresses Flp, another site-specific recombinase, from the same leaky AraC-PBAD promoter as in EL350, and thus might present with similar clone instability issues as EL350. Consequently, by replacing the tetA(C) gene in the non Cre-, non Flp-expressing strain DY380 (5) with bla we generated the RT-cassette-compatible strain MW001 (Figure 1) {F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZ M15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu) 7649 galU galK rspL nupG [λcI857 (cro-bioA) < > bla]} which we used for all recombineering procedures. MW001 is available from the non-profit plasmid repository Addgene.
Recombineering
Initially, a standard PCR-generated counter-selection RT-cassette (8,11), with ∼50-bp termini identical in sequence to regions immediately 5′ and 3′ of the gp64 gene in bMON14272, was used to replace gp64, via recombineering with positive (Tc) selection, generating recombinants with a frequency of ∼3 TcR colonies/106 viable cells. All isolated bacmid DNAs exhibited clone fidelity as determined by restriction mapping (data not shown) with one of these, bMW009, being used as the base RT-cassette-containing target for all subsequent negative selection manipulations.
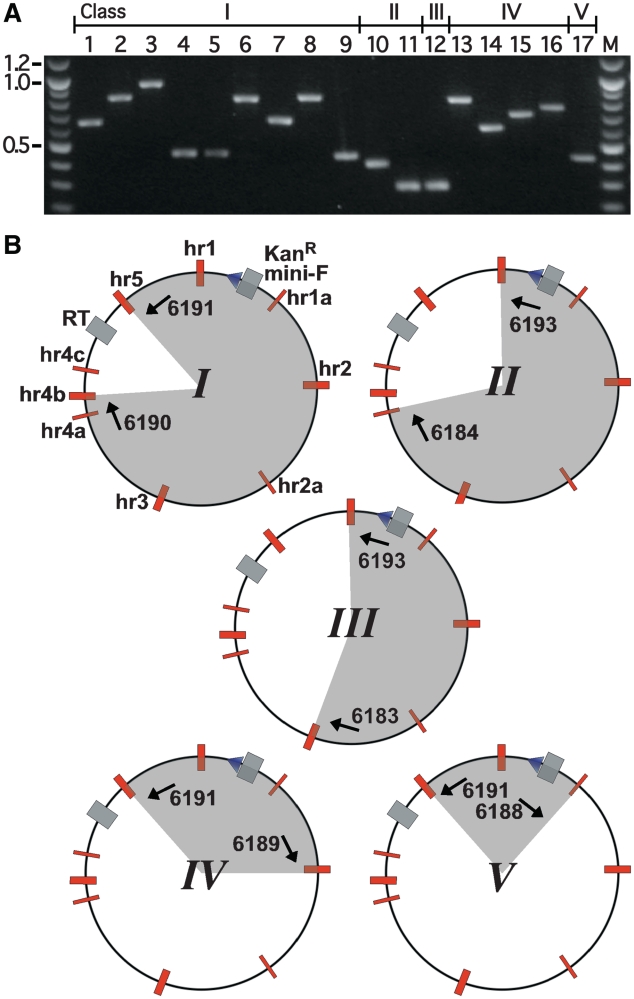
Having successfully replaced the gp64 sequence with an RT-cassette the intention was to replace this seamlessly with a series of F gene sequences. Our assumption was that this could be achieved via the negative-selection recombineering procedure, used successfully to seamlessly modify fosmid clones (8), using PCR-generated F gene-cassettes with standard 50-bp terminal homology arms. However, using the SeMNPV F gene-cassette with such ‘short’ terminal homologies resulted in large numbers of SmR colonies on both experimental plates and on control plates that had received Red-induced, mock-electroporated, i.e. no donor DNA, bacteria. Colony-PCR applied to colonies from both sets of plates, using ODNs that flanked the RT-cassette insertion site, failed to generate any products indicating the RT-cassette had, in all cases, been lost in a deletion event (data not shown). To investigate this further fine restriction mapping on bacmid DNAs isolated from 17 randomly selected colonies revealed the presence of five, I–V, distinct classes of deletion clone (Supplementary Figure S2 and data not shown). We reasoned that the hyperRec state, induced following Red-induction, might be leading to intramolecular recombination events producing autonomous episomal replicons lacking the RT-cassette, thereby restoring SmR to MW001, but, by necessity, retaining the mini-F ori and KanR gene. It also seemed plausible that such intramolecular recombination events would most likely occur between pairs of the nine hrs present in the AcMNPV genome. These hrs, considered to be involved in DNA replication and transcriptional enhancement (31,32), comprise between 1 and 9 imperfect palindromic repeats each of which contains a centrally located EcoRI sequence (35). Reexamination of the restriction maps supported this supposition and allowed, for each deletion class, the hr pair involved in the recombination to be provisionally assigned. PCR-amplification across the proposed deleted region generated products of the approximate expected sizes confirming, for all deletion classes, recombination had indeed occurred between the predicted hr pairs (Figure 3). For classes I, II and IV these products were not always the same size suggesting that, in these cases, recombination may have occurred between different combinations of the repeating palindromic sequences (Figure 3A). This was confirmed when these PCR products were subsequently sequenced (data not shown). The combined restriction mapping and PCR analyses data enabled the location of the deletion, and hr pair involved, to be defined for each deletion class identified (Figure 3B).
Figure 3.
Intramolecular recombination occurs between pairs of homologous regions in the baculoviral genome. (A) Electrophoretic separation of PCR products derived from amplification across regions predicted to be deleted in the 17 randomly selected clone DNAs, isolated following negative-selection recombineering of the RT-cassette-containing bMON14272-based clone bMW009 (lanes 1–17), using primer pairs 6190–6191 (lanes 1–9, Class I), 6184–6193 (lanes 10,11, Class II), 6183–6193 (lane 12, Class III), 6189–6191 (lanes 13–16, Class IV), 6188–6191 (lane 17, Class V). (B) Schematic representations of the deletion clone classes I–V illustrating (in the Class I deletion clone only) locations of homologous regions (hr) (red boxes), RT-cassette and mini-F ori (grey boxes), KanR gene (blue triangle). Annealing positions of primer pairs used to amplify across deleted regions and the remaining (grey segment) and deleted (white segment) segments in each clone class are also given.
Modifying negative-selection recombineering
Having established that the high number of background SmR clones would preclude the ready isolation of correctly engineered bMW009 we endeavoured to modify the negative-selection recombineering step in order to decrease the frequency of deletion clones produced and/or increase the HR efficiency between target and donor. Initially, as suggested by Narayanan (12), we attempted to simply attenuate the hyperRec state by decreasing the 42°C Red-induction time. However, attempting to replace the RT-cassette in bMW009 with the ‘short’ arm SeMNPV F gene-cassette still resulted in an unacceptably high SmR control background even when the induction time was reduced to 5 min (data not shown). Thus, it would appear that even the briefest Red-induction produces sufficient recombinase activity to mediate significant intramolecular rearrangements in bMW009 that, coupled with active selection of bacmids lacking the RT-cassette, leads to high numbers of SmR colonies harbouring deletion-bearing bacmids.
Next, we considered whether limiting the time the target bacmid DNA is exposed to the hyperRec state before a donor DNA is available as an alternative substrate for the phage recombinases might reduce intramolecular recombination and thus improve the chance of isolating desired recombinants under negative selection. Thus, as we are able to perform successful counter-selection recombineering by co-transforming fosmid (∼40 kb) target with ‘short’ homology arm-equipped linear donor (unpublished), we first investigated whether such co-transformation would work with the much larger (∼142 kb) bMON14272 target. We first determined that the transformation efficiency of MW001 with bMON14272 alone was approximately an order of magnitude lower (∼3 × 106 colonies/µg DNA) than we routinely achieve with the smaller fosmid clones (∼3 × 107 colonies/µg DNA). Subsequently, co-transformation of Red-induced MW001 with bMON14272 and either the ‘short’ or ‘long’ homology arm versions of the RT-cassette failed to generate any colonies under positive selection with Tc and Kan suggesting that the reduced transformation efficiency of the large bacmid target DNA (∼142 kb) lowers the overall recombineering efficiency to something that, at least in our hands, is impractically low. As might be expected from this result, large numbers of SmR/KanR colonies (data not shown) were still generated when Red-induced MW001 were either co-transformed with bMW009 and the ‘short’ homology arm SeMNPV F gene-cassette or simply transformed with bMW009 alone indicating that the presence of the linear donor molecule made little apparent difference to the propensity of the bacmid to undergo intramolecular recombination.
Pursuing this strategy of reducing or eliminating unnecessary exposure of target DNA to recombinase activity one step further we next investigated whether inducing Red activities after electroporation with donor DNA duplex might prove more successful. Transformation of bMON14272-containing MW001 with either the ‘short’ or ‘long’ homology arm versions of the RT-cassette, followed by a 10 min induction at 42°C, generated, respectively, 0.03 and 0.09 TcR/KanR colonies/106 viable cells. For both these versions of the RT-cassette the recombineering efficiency was approximately two orders of magnitude lower compared to standard post-electroporation Red-induction. Transformation of bMW009-containing MW001 with the ‘short’ homology arm SeMNPV F gene-cassette, followed by Red-induction, again generated many background colonies (∼10 SmR/KanR colonies/106 viable cells) on both experimental and mock-electroporated, no donor DNA control plates. The reduced recombineering efficiency observed when Red-induction was performed after electroporation likely results from host RecBCD-mediated degradation of the linear donor molecule, due to the absence of the protective lambda Gam protein before Red-induction (40) combined with compromised cell recovery after electroporation. Whatever the reasons the results indicated post-electroporation Red-induction would not permit discrimination of intact, desired recombinants from deletion mutants under negative selection.
Increasing recombineering efficiency with ‘long’ homology arm cassettes
The lack of success in decreasing clone background after negative selection with the ‘short’ homology arm SeMNPV F gene-cassette led us to investigate the use of replacement cassettes with longer terminal homology arms on the basis that general recombineering efficiency between target and donor increases significantly with increasing lengths of homology (3,6) and that longer arms have been used previously to modify a Helicoverpa armigera NPV bacmid albeit, in this instance, only for less problematic positive selection strategies (41). To simplify the generation of these cassettes a construct, pMW009, containing ∼600-bp-long 5′ and 3′ homology arm sequences fused at a central blunt-cutting SnaBI site, was created to enable PCR-generated cassettes to be cloned via an in vitro recombination system and subsequently excised by simple restriction digestion (Figure 2). Although, in the case of pMW009, the cassette was excised via SmaI sites that were partially contained within the homology arms any RE recognition site(s) can be used as long as it (they) are absent from the complete ‘long’ homology arm cassette sequence. Alternatively, excision could be via rare-cutting RE sites, engineered into the end of each homology arm during their PCR-amplification, as the few additional non-homologous bases present at the donor DNA termini following RE digestion will have no effect on recombineering efficiency or fidelity of the final product. More importantly for efficient In-Fusion cloning is the design of the central RE site used for construct linearization. The simplest strategy, as employed for pMW009, is to locate two triplets either side of the target modification/insertion site that can create a blunt-cutting RE site during PCR-mediated fusion of the homology arms. However, in case such half-sites cannot be found, or one or both are further than ∼50 bp from the modification/insertion site, thus requiring very long ODN primers for cassette amplification, then a RE leaving a 5′ overhang can be used as long as the overhang sequence comes from the left side of the modification/insertion site. Use of a RE site that leaves a 3′ overhang will, because of the 3′–5′ exonuclease activity of the In-Fusion system, result in deletion of the overhang sequence from the final recombineered product.
To examine the effect of ‘long’ homology arms on positive selection recombineering, the RT-cassette excised from pMW012 (Figure 2B) was used to replace gp64 in bMON14272. In this case, compared to the ‘short’ homology arm-equipped RT-cassette, recombineering efficiency, as expected, increased significantly generating ∼60 TcR/KanR colonies/106 viable cells (data not shown). Next, we examined whether addition of longer homologies could increase recombineering efficiency sufficiently to enable desired recombinants to be isolated following negative selection. Numbers of SmR/KanR colonies obtained (per 106 viable cells) following transformation of bMW009-containing MW001 with the SeMNPV F-gene-, AdhoNPV F-gene- or gp64-cassettes excised from, respectively, pMW013, pMW033 and pMW024 (Figure 2B) were 2- to 3-fold higher than for control (no DNA) transformations suggesting this might be the case. PCR and restriction mapping analyses of bacmid DNAs isolated from a number of randomly selected SmR/KanR colonies confirmed that, in all three cases, the cassette had replaced the RT-cassette in approximately two out of five clones examined and there was no evidence of clone rearrangement (Supplementary Figure S3A and data not shown). Furthermore, for all clones in which the RT-cassette had been replaced successfully PCR analyses confirmed all nine hrs were present and of the correct length (Supplementary Figure S3B and data not shown). Thus, of all the methodological modifications attempted only the addition of longer terminal homologies to the replacement cassettes, thereby increasing the recombineering efficiency, proved successful in generating seamlessly modified, non-rearranged bMON14272 clones. It is unlikely that the increased recombineering efficiency observed with increased homology arm length correlates with any one factor and more likely represents the cumulative effects of several, for example, more efficient and rapid beta-mediated strand annealing, increased resistance of the donor molecule to host exonuclease activities, etc.
Finally, we examined, using a series of additional gp64 gene cassettes equipped with 100-, 200- and 400-bp-long terminal homologies, whether decreasing the homology-arm length from the 600 bp created via the In-Fusion method reduced significantly the proportion of picked colonies containing successfully recombineered clones. For each gp64 cassette, out of 10 randomly picked colonies, the RT-cassette was correctly replaced in two, three and five colonies for homology arm lengths of 100, 200 and 400 bp, respectively (data not shown). These data would indicate that, at least for the situation with bMON14272 described here, that homology arm lengths of at least 400 bp are required to enable facile identification of correctly engineered BAC clones from a screen of 5–10 colonies following negative-selection recombineering. Using terminal homologies of ∼600 bp, as we have done here to generate the In-Fusion-derived constructs, represents a practical compromise between gains achieved in recombineering efficiency and the ease with which the arms can be generated via PCR-amplification, fused and cloned via the In-Fusion procedure.
F-pseudotyped AcMNPV: virus generation and mammalian cell transduction
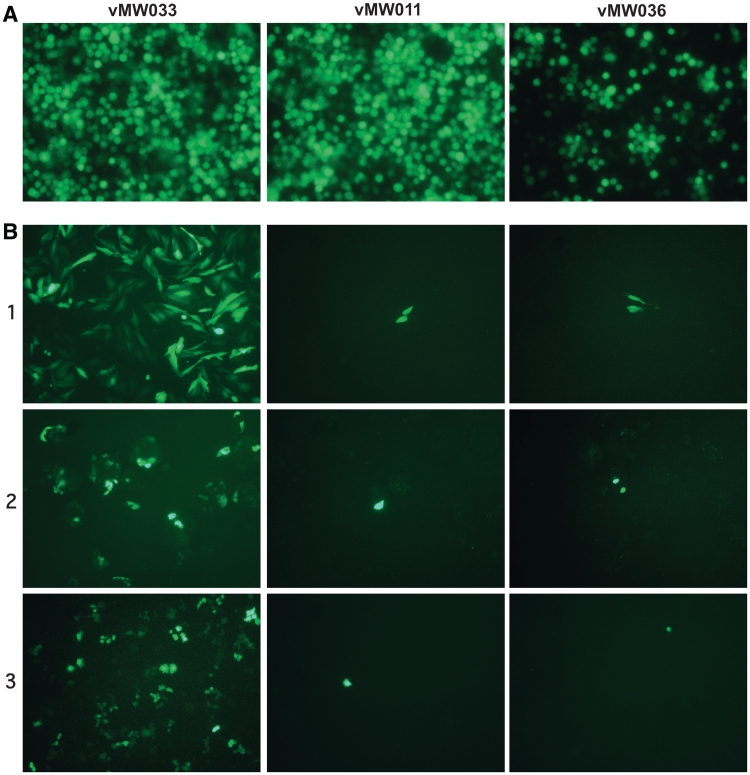
Following successful creation of seamlessly modified bacmids, and their fitment with the dual promoter GFP expression cassette, corresponding viruses were generated and amplified. The amplified viral titre for vMW033 (9.5 × 108 pfu/ml), in which the native gp64 gene was simply replaced with itself, was very similar to virus generated from unmodified bMON14272. In agreement with previous reports (26,27), titres for the SeMNPV F- (vMW011, 9.6 × 106 pfu/ml) and AdhoNPV F-pseudotyped (vMW036, 2.0 × 107 pfu/ml) viruses were lower. Nevertheless, both F-pseudotyped viruses were able to initiate and support multiple rounds of Sf9 infection, revealed by GFP expression in infected cells (Figure 4A and data not shown), confirming these F proteins are able to functionally replace GP64. While such functional homology has been demonstrated previously for SeMNPV F (26,27) this is the first time it has been demonstrated for the F protein from AdhoNPV. Furthermore, the strategy adopted here also differs from previous reports on F functionality in that we have replaced the gp64 coding sequence, in situ, with F gene sequences, thereby retaining the native genomic context and promoter elements associated with the gp64 locus, rather than introducing an F gene elsewhere on a Δgp64 background.
Figure 4.
SeMNPV F- or AdhoNPV F-pseudotyped AcMNPV supports viable insect cell infection but are not able to transduce mammalian cells. (A) Essentially all cells of an Sf9 culture infected with CMVPROM-p10PROM::eGFP cassette-containing AcMNPV in which the native gp64 gene was replaced either with itself (vMW033), the SeMNPV F (vMW011) or the AdhoNPV F gene (vMW036) exhibited GFP expression indicating SeMNPV F- and AdhoNPV F-pseudotyped AcMNPV both mediate viable insect cell infection. All cultures were x3 passage. (B) In all cases <0.01% of cultured Saos-2 (Row 1), HepG2 (Row 2) or HeLa (Row 3) cells exhibit GFP expression 48 h following incubation with either vMW011 (50 pfu/cell) or vMW036 (100 pfu/cell). For illustration purposes fields of view containing rare GFP-expressing cells have been chosen. In contrast, >90% of these cells exhibit GFP expression following incubation with the control gp64 rescue virus vMW033 48 h post-incubation. Images were taken with a Nikon Eclipse TE2000-S inverted microscope with FITC filter set (100× magnification).
The range of mammalian cells transduced by AcMNPV may hinder its development as a GT vehicle due to unwanted bystander effects. Interestingly, though F-pseudotyped AcMNPV is unable to transduce mammalian cells, the demonstration that F proteins mediate membrane fusion in an in vitro assay (27) and that naked gDNA from the group II Alphabaculovirus Helicoverpa armigera can deliver a reporter to mammalian cells when introduced directly into the cytoplasm (28) indicates this may result from a block at viral attachment or internalization rather than, for example, endosomal escape. Thus, our overall aim is to, first, replace gp64 with an F gene sequence that supports robust viral generation and propagation in insect cells but prevents indiscriminate transduction of mammalian cells, then, second, decorate the viral surface with peptides designed to promote targeting and transduction of selected cells. As a first step towards this we investigated whether either of our two AcMNPV F-pseudotypes would transduce, and deliver a GFP expression cassette, to any of the three cell lines, Saos-2, HepG2 and HeLa, all of which are readily transduced by native AcMNPV. Following incubation with either F-pseudotype less than 0.01% of cells of any line exhibited GFP expression (Figure 4B). Although the ratios of virus to cell used in these experiments were, at 50 and 100 pfu/cell for, respectively, the SeMNPV F- (vMW011) and AdhoNPV F- (vMW036) pseudotype, relatively low it is unlikely that significantly more cells would be transduced successfully if higher virus:cell ratios were used. In contrast, significant (>90%) of cells exhibited GFP expression after incubation (50 pfu/cell) with the gp64 rescue virus vMW033 (Figure 4B). These data confirm that Δgp64 AcMNPV, pseudotyped with either SeMNPV or AdhoNPV F, exhibits a functional block in mammalian cell transduction.
General applicability of extended homology arm counter-selection recombineering
Currently, we are applying our modified counter-selection protocol using extended homology arms to investigate whether F proteins from other members of the group II Alphabaculoviruses can functionally replace GP64. Furthermore, we have applied the same approach to seamlessly modify other targets in the bMON14272 replicon (Westenberg, unpublished). Because of the presence of the nine, relatively uniformly distributed hr sequences in AcMNPV, the bMON14272 replicon that we are endeavouring to modify can be considered equivalent to a single-copy BAC clone with extensive regions of repeating sequences. As we have demonstrated here with bMON14272, and others with repetitive BACs (12), such clones may exhibit instability and undergo rearrangements when subject to recombineering. This susceptibility to rearrange precludes easy isolation of intact clones containing desired sequence changes a situation that is exacerbated when applying negative selection strategies. BACs are commonly used as base clones for generation of targeting vectors for genome manipulation and recombineering is increasingly the method of choice for their sequence modification (42,43). If such a BAC contains repeating homologous regions then applying recombineering may prove problematic especially if the targeting vector requires subtle changes to be made, for example when creating knock-in style vectors, that can only be created seamlessly within the BAC if counter-selection approaches are employed. Although we acknowledge that the procedure to build replacement cassettes with longer homology arms is somewhat more involved than the standard PCR-based approach we have attempted to minimize that complexity by combining straightforward cloning with an in vitro HR system. That said, the significant increase in recombineering efficiency furnished by these extended homology arm sequences ensures that large, repeat-containing clones can now be modified seamlessly, via counter-selection recombineering, with desired recombinants being recovered without unwanted rearrangements. As such, the method described here should prove useful to others attempting to seamlessly modify repetitive BAC or PAC clones.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Wellcome Trust [WT078981]. Funding for open access charge: King’s College London—Wellcome Funded Authors membership agreement.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr M. Nakai (Tokyo University of Agriculture and Technology) for kindly providing AdhoNPV polyhedra.
REFERENCES
- 1.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 4.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 5.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 6.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muyrers JP, Zhang Y, Benes V, Testa G, Rientjes JM, Stewart AF. ET recombination: DNA engineering using homologous recombination in. E. coli. Methods Mol. Biol. 2004;256:107–121. doi: 10.1385/1-59259-753-X:107. [DOI] [PubMed] [Google Scholar]
- 8.Dolphin CT, Hope IA. Caenorhabditis elegans reporter fusion genes generated by seamless modification of large genomic DNA clones. Nucleic Acids Res. 2006;34:e72. doi: 10.1093/nar/gkl352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol. Biotechnol. 2009;42:110–116. doi: 10.1007/s12033-009-9142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong QN, Ng VC, Lin MC, Kung HF, Chan D, Huang JD. Efficient and seamless DNA recombineering using a thymidylate synthase A selection system in Escherichia coli. Nucleic Acids Res. 2005;33:e59. doi: 10.1093/nar/gni059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stavropoulos TA, Strathdee CA. Synergy between tetA and rpsL provides high-stringency positive and negative selection in bacterial artificial chromosome vectors. Genomics. 2001;72:99–104. doi: 10.1006/geno.2000.6481. [DOI] [PubMed] [Google Scholar]
- 12.Narayanan K. Intact recombineering of highly repetitive DNA requires reduced induction of recombination enzymes and improved host viability. Anal. Biochem. 2008;375:394–396. doi: 10.1016/j.ab.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 13.Thyagarajan B, Guimarães MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 14.Semprini S, Troup TJ, Kotelevtseva N, King K, Davis JR, Mullins LJ, Chapman KE, Dunbar DR, Mullins JJ. Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques. Nucleic Acids Res. 2007;35:1402–1410. doi: 10.1093/nar/gkl1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller LK. Baculoviruses as gene expression vectors. Annu. Rev. Microbiol. 1988;42:177–199. doi: 10.1146/annurev.mi.42.100188.001141. [DOI] [PubMed] [Google Scholar]
- 16.Hu YC. Baculoviral vectors for gene delivery: a review. Curr. Gene Ther. 2008;8:54–65. doi: 10.2174/156652308783688509. [DOI] [PubMed] [Google Scholar]
- 17.Huser A, Hofmann C. Baculovirus vectors: novel mammalian cell gene-delivery vehicles and their applications. Am. J. Pharmacogenomics. 2003;3:53–63. [PubMed] [Google Scholar]
- 18.Lin G, Blissard GW. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol. 2002;76:2770–2779. doi: 10.1128/JVI.76.6.2770-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanarsdal AL, Pearson MN, Rohrmann GF. Characterization of baculovirus constructs lacking either the Ac 101, Ac 142, or the Ac 144 open reading frame. Virology. 2007;367:187–195. doi: 10.1016/j.virol.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang M, Dai X, Theilmann DA. Autographa californica multiple nucleopolyhedrovirus EXON0 (ORF141) is required for efficient egress of nucleocapsids from the nucleus. J. Virol. 2007;81:9859–9869. doi: 10.1128/JVI.00588-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Chapman DA, Jones IM. Improving baculovirus recombination. Nucleic Acids Res. 2003;31:e6. doi: 10.1093/nar/gng006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noad RJ, Stewart M, Boyce M, Celma CC, Willison KR, Roy P. Multigene expression of protein complexes by iterative modification of genomic Bacmid DNA. BMC Mol. Biol. 2009;10:87. doi: 10.1186/1471-2199-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luckow VA, Lee SC, Barry GF, Olins PO. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson MN, Groten C, Rohrmann GF. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J. Virol. 2000;74:6126–6131. doi: 10.1128/jvi.74.13.6126-6131.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.IJkel WF, Westenberg M, Goldbach RW, Blissard GW, Vlak JM, Zuidema D. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology. 2000;275:30–41. doi: 10.1006/viro.2000.0483. [DOI] [PubMed] [Google Scholar]
- 26.Long G, Westenberg M, Wang H, Vlak JM, Hu Z. Function, oligomerization and N-linked glycosylation of the Helicoverpa armigera single nucleopolyhedrovirus envelope fusion protein. J. Gen. Virol. 2006;87:839–846. doi: 10.1099/vir.0.81592-0. [DOI] [PubMed] [Google Scholar]
- 27.Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 2002;76:5729–5736. doi: 10.1128/JVI.76.11.5729-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang C, Song J, Chen X. The GP64 protein of Autographa californica multiple nucleopolyhedrovirus rescues Helicoverpa armigera nucleopolyhedrovirus transduction in mammalian cells. J. Gen. Virol. 2005;86:1629–1635. doi: 10.1099/vir.0.80857-0. [DOI] [PubMed] [Google Scholar]
- 29.Westenberg M, Uijtdewilligen P, Vlak JM. Baculovirus envelope fusion proteins F and GP64 exploit distinct receptors to gain entry into cultured insect cells. J. Gen. Virol. 2007;88:3302–3306. doi: 10.1099/vir.0.83240-0. [DOI] [PubMed] [Google Scholar]
- 30.Yu IL, Lin YC, Robinson JH, Lung O. Transduction of vertebrate cells with Spodoptera exigua multiple nucleopolyhedrovirus F protein-pseudotyped gp64-null Autographa californica multiple nucleopolyhedrovirus. J. Gen. Virol. 2009;90:2282–2287. doi: 10.1099/vir.0.012138-0. [DOI] [PubMed] [Google Scholar]
- 31.Cochran MA, Faulkner P. Location of Homologous DNA Sequences Interspersed at Five Regions in the Baculovirus AcMNPV Genome. J Virol. 1983;45:961–970. doi: 10.1128/jvi.45.3.961-970.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson M, Bjornson R, Pearson G, Rohrmann G. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science. 1992;257:1382–1384. doi: 10.1126/science.1529337. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning; A Laboratory Manual. New York, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Pijlman GP, Dortmans JC, Vermeesch AM, Yang K, Martens DE, Goldbach RW, Vlak JM. Pivotal role of the non-hr origin of DNA replication in the genesis of defective interfering baculoviruses. J. Virol. 2002;76:5605–5611. doi: 10.1128/JVI.76.11.5605-5611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 36.IJkel WF, van Strien EA, Heldens JG, Broer R, Zuidema D, Goldbach RW, Vlak JM. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- 37.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 38.Nakai M, Goto C, Kang WK, Shikata M, Luque T, Kunimi Y. Genome sequence and organization of a nucleopolyhedrovirus isolated from the smaller tea tortrix, Adoxophyes honmai. Virology. 2003;316:171–183. doi: 10.1016/j.virol.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Abremski K, Wierzbicki A, Frommer B, Hoess RH. Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J. Biol. Chem. 1986;261:391–396. [PubMed] [Google Scholar]
- 40.Murphy KC. Lambda Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol. 1991;173:5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou S, Chen X, Wang H, Tao M, Hu Z. Efficient method to generate homologous recombinant baculovirus genomes in E. coli. Biotechniques. 2002;32:783–788. doi: 10.2144/02324st04. [DOI] [PubMed] [Google Scholar]
- 42.Cotta-de-Almeida V, Schonhoff S, Shibata T, Leiter A, Snapper SB. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 2003;13:2190–2194. doi: 10.1101/gr.1356503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Seed B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 2003;21:447–451. doi: 10.1038/nbt803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.