Abstract
Differences in the substrate specificity of mammalian family X DNA polymerases are proposed to partly depend on a loop (loop 1) upstream of the polymerase active site. To examine if this is the case in DNA polymerase λ (pol λ), here we characterize a variant of the human polymerase in which nine residues of loop 1 are replaced with four residues from the equivalent position in pol β. Crystal structures of the mutant enzyme bound to gapped DNA with and without a correct dNTP reveal that the change in loop 1 does not affect the overall structure of the protein. Consistent with these structural data, the mutant enzyme has relatively normal catalytic efficiency for correct incorporation, and it efficiently participates in non-homologous end joining of double-strand DNA breaks. However, DNA junctions recovered from end-joining reactions are more diverse than normal, and the mutant enzyme is substantially less accurate than wild-type pol λ in three different biochemical assays. Comparisons of the binary and ternary complex crystal structures of mutant and wild-type pol λ suggest that loop 1 modulates pol λ’s fidelity by controlling dNTP-induced movements of the template strand and the primer-terminal 3′-OH as the enzyme transitions from an inactive to an active conformation.
INTRODUCTION
Family X polymerases are small, single-subunit enzymes devoid of 3′- to 5′-exonuclease activity that fill short gaps during DNA repair. Mammalian cells contain four family X polymerases, pol β, pol λ, pol µ and terminal deoxynucleotidyl transferase (TdT). Pols β and λ are involved in base excision repair (BER) (1–5), whereas TdT, pol µ and pol λ contribute to repairing double-strand DNA breaks via non-homologous end joining (NHEJ) (6,7). TdT is a template-independent polymerase that randomly adds nucleotides to the ends of broken DNA to generate N-regions during V(D)J recombination (8–10). Pol µ can also perform limited template-independent synthesis that contributes to junctional diversity during V(D)J recombination occurring at different developmental stages (11,12). In addition, pol µ is unique among family X members in having the ability to fill short gaps by template-dependent extension of primers that lack their template strand partner (13,14). Pol λ is a template-dependent polymerase that is also implicated in V(D)J recombination (15). Moreover, pol µ and pol λ, but not TdT, are implicated in general NHEJ of DNA damage-induced double-strand breaks that can have variety of different end configurations (16,17).
Given their important but somewhat diverse biological functions, it is of interest to understand the determinants of the substrate specificities of the four mammalian family X polymerases. X-ray crystal structures reveal that all four enzymes share a similar structure overall, yet each enzyme individually has small but important structural features that are likely to be relevant to their different substrate specificities (18). One such structural element is loop 1. Loop 1 was first described in TdT, where it lies between β sheets 3 and 4 of the palm subdomain. In the TdT structure, loop 1 is in a position proposed to preclude binding of the template strand, thereby limiting TdT to template-independent synthesis (19,20). In pol µ, which is a template-dependent polymerase that nonetheless can conduct limited template-independent synthesis, loop 1 is similar in length to loop 1 in TdT. Thus, when pol µ is conducting template-independent synthesis, loop 1 may play a similar function as in TdT. Consistent with this idea, pol µ that lacks loop 1 has strongly reduced ability to catalyze template-independent synthesis and template-dependent extension of a primer terminus lacking its template partner, yet it retains the ability to perform more classical, short gap-filling synthesis either alone or with its partners in NHEJ (13,21). Thus loop 1 in pol µ is an important determinant of substrate specificity.
In this study, we extend previous efforts to examine the function of loop 1 in pol µ (13,21) and TdT (20) to the analogous loop 1 in pol λ. As dNTP binding induces pol λ to transition from an inactive to an active conformation, β sheets β3 and β4 partially unravel to form loop 1, a nine-residue loop that repositions as the DNA template strand assumes its active conformation. Motivated by the fact that shortening loop 1 in pol µ was informative regarding its substrate specificity in NHEJ, here we decided to take the same approach with pol λ, by characterizing a pol λ derivative whose loop 1 was shorter than normal. In deciding how to shorten loop 1, we took advantage of the fact that in its family X sibling, pol β, strands β3 and β4 are shorter than in pol λ and their conformation does not change during catalytic cycling. Moreover, not only are the nine residues of loop 1 conserved in pol λ from different organisms (Figure 1A), but certain amino acids in this region of pol λ align with those in pol β (Figure 1B), and in a manner that suggests that five residues in pol λ might be deleted without inducing major structural changes. The goal in studying this variant enzyme is to better understand the nature of the catalytic cycle for template-dependent DNA synthesis by pol λ, both mechanistically, and also in the context of its biological role in NHEJ. Toward these goals, we compare several biochemical properties of the mutant enzyme to those of wild-type (wt) pol λ, when operating alone and when participating in the more complex NHEJ reaction. We solve binary and ternary complex crystal structures of the mutant enzyme, and compare them with each other and to their wt counterparts. The results demonstrate that loop 1 is not essential for catalytic activity, but it is important for the fidelity of DNA synthesis and the accuracy of NHEJ. We present a model for involvement of loop 1 in modulating fidelity by controlling the position of the template strand and the 3′-O of the primer terminal nucleotide, as pol λ cycles from an inactive to an active conformation.
Figure 1.
Loop 1 is conserved in pol λ from different species. (A) Sequence alignment of pol λ loop 1 region from different species. Identical residues are in gray background except for K472, which is in red background. Conserved and non-conserved residues are in green and black background, respectively. Human (Hs) pol λ sequence is aligned with sequences of pol λ from: rhesus monkey, Macaca mulatta (Mcm); cattle, Bos taurus (Bt); horse, Equus caballus (Eqc); dog, Canis lupus familiaris (Clf); rat, Rattus norvegicus (Rn); mouse, Mus musculus (Mum); red jungle fowl, Gallus gallus (Gg); western clawed frog, Xenopus (Silurana) tropicalis (Xst); zebrafish, Danio rerio (Dr); human pol β, pol µ and TdT. (B) The sequence of loop 1 in pol λ (nine residues in green background) is replaced with the corresponding sequence in pol β (four residues in yellow background).
MATERIALS AND METHODS
Materials
The human full-length wt pol λ, full-length pol λ with deletion of loop1 (designated pol λ DL) and its 39-kDa domain were expressed in Escherichia coli and purified as described (22). Oligonucleotides were from Oligos Etc. (Wilsonville, OR, USA) and radioactive nucleotides were from Perkin Elmer (Boston, MA, USA).
Specific activity
Reaction mixtures (40 µl) contained 50 mM Tris (pH 7.5), 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml bovine serum albumin (BSA) and either 2.5 mM MgCl2, 1µg of activated DNA (Amersham Biosciences Corp. Piscataway, NJ, USA), 50 µM dNTPs, 5 µCi of α 32P dCTP and 50 nM full-length wt pol λ or pol λ DL or 0.5 mM MnCl2, 0.75 µg of pol(dA)/olido(dT) (Amersham Biosciences Corp.), 10 µM dTTP, 5 µCi of α 32P dTTP and 10 nM wt pol λ or pol λ DL. Reactions were incubated in 37°C, and two 5 µl aliquots were removed after 5, 10, and 15 min of incubation and mixed with 45 µl of 16 mM EDTA, followed by precipitation of the synthesized DNA with 0.5 ml of ice-cold 10% trichloroacetic acid (TCA) for 15 min on ice. The precipitated DNA was collected by vacuum filtration through Whatman GF/C glass fiber filters and the amount of radioactivity bound to the filter was measured in a Beckman Coulter LS 6500 Scintillation Counter.
Primer extension reactions
Measurements were performed with T5g, 36-mer oligonucleotide, 5′-CTCCGTCGTCCGGCACGTAATACTGCTCAGTCGTAC, as template primed at a 1.2:1 molar ratio with 5′ 32P-labeled P17 primer: 5′-GTACGACTGAGCAGTAT. Reaction mixtures (20 µl) contained 50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 1mM dithiothreitol, 2 µg of BSA, 4% glycerol and 50 µM each of dATP, dGTP, dCTP and dTTP, 200 nM DNA and 10, 50, and 150 nM wt or pol λ DL. The reactions were incubated in 37°C. Aliquots (10 µl) were removed at 10 and 20 min and mixed with equal volume of 99% formamide, 5 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue. DNA products were analyzed by electrophoresis in 12% denaturing polyacrylamide gels.
Kinetic analysis of nucleotide insertion
DNA substrates were prepared by hybridizing a 32P-5′-end-labeled 18-nt primer (P18, 5′-AGCAGGGTAGCCCACTGC) and a 17-nt downstream primer (DP17, 5′-GTGGACGTTCGGTGGAG) with a phosphate on the 5′-end to a 32-mer template (TT36, 5′-CTCCACCGAACGTCCACTGCAGTGGGCTACCCTGCT) to create a 1-nt gap substrate. Reaction mixtures (10 µl) contained 50 mM Tris, pH 7.5, 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml BSA, 2.5 mM MgCl2, 200 nM DNA and 4 nM full-length wt pol λ or pol λ DL. Reactions were initiated by adding dATP at one of nine concentrations (0.05, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20 µM) and incubated at 37°C for 3 min. To measure misincorporation, reaction mixtures contained 50 or 100 nM wt pol λ or 5 or 7 nM pol λ DL. Reactions were initiated by adding dGTP at one of eight concentrations (1, 2, 5,10, 25, 50, 100 or 150 µM to reactions with wt pol λ or 2, 5, 10, 20, 50, 100, 150 or 200 µM to reactions with pol λ DL) and incubated at 37°C for 6 min. After adding an equal volume of 99% formamide, 5 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue, products were resolved on a 12% denaturing polyacrylamide gel and quantified by phosphor screen autoradiography. The data were fit to the Michaelis–Menten equation using non-linear regression.
Processivity
Measurements were performed with the same DNA substrate as used in the primer extension reactions. Reactions (20 µl) contained 50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 1mM dithiothreitol, 2 µg of BSA, 4% glycerol and 50 µM each of dATP, dGTP, dCTP and dTTP, 200 nM DNA and 2 nM wt pol λ or pol λ DL. The reactions were incubated in 37°C. Aliquots (5 µl) were removed at 3, 6 and 9 min and mixed with 5 µl 99% formamide, 5 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue. DNA products were analyzed by electrophoresis in 12% denaturing polyacrylamide gels. Product bands were quantified by phosphorimagry, and the probability of terminating processive synthesis at any given site was expressed as the ratio of the amount of product at that site to the amount of product at that site plus all longer length products (23).
NHEJ reactions
To generate internally labeled plasmid substrates, plasmid pRZ56 was linearized with MluI and then 10- and 11-base 5′-overhangs were formed by controlled 3′-resection with T4 DNA polymerase in the presence of dTTP (24). Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end joining in CV-1 cells (24). An unlabeled 13-mer (5′pGCCGGACGCGACG) and a 5′-end-labeled 14-mer (5′*pCGAGGAACGCGACG) were ligated into the 10- and 11-base overhangs, respectively, and the plasmid was purified by agarose gel electrophoresis and electroeluted (25). End-joining reactions contained 3.2 mg/ml extract (Promega in vitro transcription grade), 50 mM triethanolamine-NaOH (pH 7.5), 1 mM Mg(OAc)2, 40 mM KOAc, 0.5 mM dithiothreitol, 1 mM ATP, 50 µM of each dNTP, 50 µg/ml BSA, 1 µg/ml substrate and 5 µg/ml XRCC4/DNA ligase IV complex (Trevigen). In some cases, extracts were immnuodepleted with antibodies against pol λ (gift of Luis Blanco), as described (17). Following incubation for 6 h at 37°C, DNA was deproteinized, cut with BstXI and AvaI, and analyzed by denaturing gel electrophoresis and phosphorimagry (17). Aliquots of some reactions were transfected into DH5α bacteria, clones were isolated and the plasmid repair joints sequenced.
Forward mutation assay
This assay score errors generated in the LacZ α-complementation gene in M13mp2 during synthesis to fill a 407-nt gap. Reaction mixtures (25 µl) contained 2 nM M13mp2 gapped DNA substrate, 50 mM Tris–HCl (pH 8.5), 2.5 mM MgCl2, 1mM dithiothreitol, 2 µg of BSA, 4% glycerol and 50 µM each of dATP, dGTP, dCTP and dTTP. Polymerization reactions were initiated by adding full-length wt (300 nM) or DL (50 nM or 150 nM) pol λ, and were incubated at 37°C for 1 h, and terminated by adding EDTA to 15 mM. Reaction products were analyzed by agarose gel electrophoresis as described (26). Correct synthesis produces M13mp2 DNA that yields dark blue phage plaques upon introduction into an E. coli α-complementation strain and plating on indicator plates. Errors are scored as light blue or colorless mutant phage plaques. DNA from independent mutant clones was sequenced to define the lacZ mutation. Since most of the mutant clones generated by pol λ contained both phenotypically detectable and silent changes, the error rates are described as the number of observed mutations divided by the number of nucleotides sequenced.
Crystallography
Crystals of the 39-kDa pol λ DL in complex with oligos T11 (5′-CGGCAGTACTG), PB (5′-CAGTAC) and DT (5′-GCCG) for a binary complex or oligos T11T (5′-CGGCAATACTG), PT (5′-CAGTA), DT and ddTTP for a ternary complex were grown using the hanging drop method. The protein/DNA solution contained 100 mM NaCl and 10 mM MgCl2. Binary complex crystals grew in 50 mM sodium cacodylate, pH 6.5, 12% 2-propanol, 100 mM ammonium acetate and 15 mM magnesium acetate. Ternary complex crystals grew in 50 mM sodium cacodylate, pH 5.5, 21% 2-propanol and 200 mM sodium citrate. Crystals were transferred in four steps to a solution containing 50 mM sodium cacodylate, pH 6.5, 12% 2-propanol, 100 mM ammonium acetate, 15 mM magnesium acetate, 100 mM NaCl and 25% ethylene glycol (binary complex crystals) or 50 mM sodium cacodylate, pH 5.5, 21% 2-propanol, 10 mM magnesium chloride, 100 mM NaCl, 200 mM sodium citrate and 25% ethylene glycol (ternary complex crystals) and flash cooled. Data collection for both structures were performed at −178°C on a Saturn92 CCD area detector mounted on a MicroMax-007HF (Rigaku Corporation) rotating anode generator equipped with Varimax HF mirrors. All data were processed using the HKL2000 data processing software (27). The structures were solved by molecular replacement using ternary or binary structures of pol λ wt (PDB codes 1XSN and 1SXL, respectively) as search models and using MOLREP (28). Refinement was carried out using CNS (29) and phenix (30), and model building with O (31) and Coot (32). The quality of the model was assessed using Molprobity (33) and found to have good stereochemistry (Table 5).
Table 5.
Summary of crystallographic data
| PDB ID | 3MGH (binary complex) | 3MG1 (ternary complex) |
|---|---|---|
| Unit cell dimensions (Å) | 95.529 × 190.931 × 58.724 | 56.188 × 63.315 × 140.477 |
| Space group | P21212 | P212121 |
| Number of observations | 327 225 | 122 110 |
| Unique reflections | 41 236 | 29 412 |
| Rsym (%) (last shell) | 14.7 (63.8) | 11.6 (64.6) |
| I/σI (last shell) | 10.1 (2.4) | 14.0 (2.4) |
| Completeness (%) (last shell) | 95.6 (88.8) | 99.8 (99.7) |
| Refinement statistics | ||
| Resolution (Å) | 2.40 | 2.60 |
| Rcryst (%) | 20.82 | 20.14 |
| Rfree (%) | 26.01 | 27.06 |
| Number of complexes in asymmetric unit | 2 | 1 |
| No. of atoms | ||
| Protein atoms | 4972 | 2440 |
| Water | 358 | 158 |
| Ions | 4 | 2 |
| Heteroatoms | 853 | 453 |
| Mean B value (Å2) | 45.05 | 39.83 |
| RMS deviation from ideal values | ||
| Bond length (Å) | 0.007 | 0.003 |
| Bond angle (°) | 1.192 | 0.736 |
| Ramachandran statistics Residues in | ||
| Favored regions (%) | 96.20 | 96.85 |
| Allowed regions (%) | 99.84 | 99.68 |
RESULTS
Construction of pol λ DL
The nine amino acids of loop 1 in human pol λ (463-SQEENGQQQ-471) were replaced with the four-residue turn between β3 and β4 in human pol β (Figure 1B). The resulting polymerase (pol λ DL) was expressed in E. coli and purified as described (34), in two different forms. One was the full-length protein containing the BRCT domain that is required for use in the NHEJ reactions (35). The other form was the 39-kDa protein that lacks the BRCT domain but contains both the polymerase and 8-kDa dRP lyase domains. This protein was used for crystallography.
Catalytic activity of pol λ loop DL
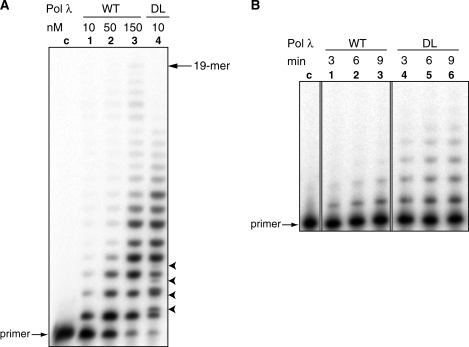
The catalytic activity of pol λ DL was first measured using activated calf thymus DNA as a generic primer-template. The specific activity of pol λ DL was 3-fold higher than that of wt pol λ (1800 versus 560 pmol/nmol/min). Because manganese has been suggested to be the physiologically relevant metal for pol λ (36), we also measured the specific activity of the mutant enzyme in the presence of manganese as the activator. We performed this measurement as described by Blanca et al. (36), using poly(dA)/oligo(dT) as the DNA substrate. The specific activity of pol λ DL was 4-fold higher than that of the wt pol λ (2800 versus 700 pmol/nmol/min). Next, equivalent amounts (10 nM) of the two enzymes were compared for the ability to extend a primer hybridized to an oligonucleotide template (Figure 2A). Pol λ DL (Lane 4) reproducibly extended more primers and generated longer products than did wt pol λ (Lane 1). A 15-fold higher concentration of wt pol λ (Lane 3) was required to extend most of the original primer and to obtain a DNA product ladder similar to that observed with pol λ DL (Lane 4).
Figure 2.
Catalytic activity of pol λ DL. (A) Products of primer extension reactions by wt pol λ and pol λ DL. Lane c: control reaction without enzyme; Lane 1–3, reactions with 10, 50 and 150 nM pol λ wt, respectively; and Lane 4, 10 nM pol λ DL. All reactions were incubated for 10 min in 37°C. The position of the 19-mer full-length product is indicated. Arrowheads point to the double bands in Lane 4 (pol λ DL) suggesting products of nucleotide misincorporation. (B) Products of primer extension reactions under single hit conditions; Lane c, control without enzyme; Lanes 1–3 with wt pol λ; and Lanes 4–6 with pol λ DL after 3-, 6- and 9-min incubation, respective.
Processivity of pol λ DL
The experiment in Figure 2A was performed under conditions that allow primers to be extended multiple times. To measure the processivity of a single cycle of synthesis, we decreased the ratio of enzyme to DNA substrate to ensure that each primer was extended only once. Under these conditions, as expected based on an earlier study (37), wt pol λ is largely distributive (Figure 2B, Lanes 1–3), terminating synthesis after each incorporation with ∼90% probability. However, pol λ DL is more processive, as illustrated by the ability to generate somewhat longer product chains (Figure 2B) and by lower termination probabilities following each of five incorporation events (∼60%).
Steady-state kinetics analysis of dNTP insertion
Next, we determined steady-state parameters for single nucleotide incorporation into a 1-nt gap. For correct incorporation of dATP opposite template T, the apparent Km and kcat of pol λ DL differed by <2-fold from the values for wt pol λ (Table 1), such that the catalytic efficiency (kcat/Km) of the two enzymes differed by only 3-fold.
Table 1.
Steady-state analysis of single nucleotide incorporation by pol λ DL
| Enzyme | Km (µM) | kcat (1/s) | kcat/Km (/s/µM) |
|---|---|---|---|
| Incorporation of dATP | |||
| Pol λ wt | 0.39 ± 0.10 | 0.012 ± 0.004 | 3.0 × 10−2 ± 1.3 × 10−2 |
| Pol λ DL | 0.62 ± 0.126 | 0.0065 ± 0.001 | 1.0 × 10−2 ± 0.3 × 10−2 |
| Incorporation of dGTP | |||
| Pol λ wt | 7.4 ± 0.92 | 0.00014 ± 5.8 × 10−6 | 1.9 × 10−5 ± 2.5 × 10−6 |
| Pol λ DL | 28 ± 4.30 | 0.0064 ± 7.6 × 10−4 | 2.3 × 10−4 ± 0.4 × 10−4 |
The values for the kinetic constants are an average of 3–4 independent determinations.
Activity of pol λ DL in NHEJ
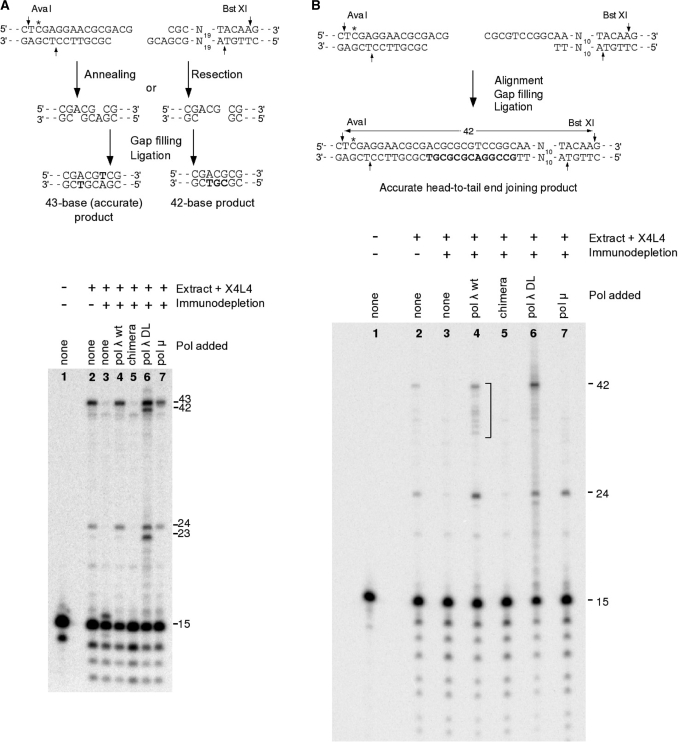
To determine if the deletion of loop 1 in pol λ affects its performance in NHEJ, we examined repair of a defined, site-specifically labeled double-strand break substrate bearing partially complementary overhangs. Repair was performed in nuclear extracts made from HeLa cells, which as described previously (17), were immunodepleted of pol λ and then supplemented with either wt pol λ or pol λ DL. Repair of this substrate proceeds predominantly by annealing of the terminal CG dinucleotides in the 3′-overhangs (Figure 3A), followed by single-base gap filling that is dependent on pol λ, Ku and XRCC4/DNA ligase IV, and finally ligation, resulting in 43-base (head-to-tail) and 24-base (head-to-head) products following AvaI/BstXI cleavage. Pol λ DL (Lane 6 in Figure 3A, bottom panel) was as effective as wt pol λ (Lane 4) in generating the diagnostic 43- and 24-mers. In addition, however, pol λ DL also generated substantial amounts of products that were 1-nt shorter (42- and 23-mer in Lane 6).
Figure 3.
Alignment-based gap filling by the pol λ DL mutant. Internally labeled (*) substrates bearing two 3′-overhangs (A) or one 3′- and one 5′-overhang (B) were incubated for 6 h in HeLa nuclear extracts that had been immunodepleted with either antibodies to pol λ (+), or preimmune antibodies (−), and supplemented with wild-type pol λ, pol λ /pol β chimera, pol λ DL or pol µ, as indicated. Samples were deproteinized, and DNA was cut with BstXI and AvaI and analyzed on denaturing gels. Schemes at top show mechanisms of formation of the major 43- and 42-base products. The 24- and 23-base products are formed by gap filling and ligation as shown in (A), except between the left-hand ends of two plasmid molecules.
These shorter products could arise by resection of one 3′-overhang to a blunt end, followed by 3-nt gap filling using the blunt end as primer and 3′-overhang on the opposite end of the break as template (Figure 3A, top panel). However, because these substrates were constructed by ligating 13- or 14-mer onto 10- or 11-base 5′-overhangs, there will always be some residual 5′-overhangs. Thus, the blunt end from which the putative synthesis on the 3′-overhang template is primed could also result from fill-in of such a 5′-overhang during end joining. To determine the relative efficiencies of pol λ and pol λ DL in catalyzing such fill-in reactions, a substrate was constructed with an oligomer ligated onto only one end of the plasmid, i.e. having a 3-base 3′-overhang at one end and a 10-base 5′-overhang at the other (Figure 3B). When this substrate was incubated in pol λ-supplemented extracts, a prominent 24-base head-to-head product was generated, as expected. However, in comparison to the predominant 43-base head-to-tail product that was generated from the substrate with two 3′-overhangs (Figure 3A), this new substrate yielded a broader distribution of less prominent products ranging from about 33 to 42 bases (bracketed bands in Lane 4 of Figure 3B, bottom panel). The 42-base product is consistent with continuous polymerization across both the 5′- and 3′-overhangs, while the shorter products may reflect varying degrees of trimming of the two overhangs prior to ligation.
Interestingly, the 42-base product was more prominent in the pol λ-depleted extracts that were supplemented with pol λ DL (Lane 6 in Figure 3B, bottom panel). This result suggests that pol λ DL is more efficient than the wt enzyme in filling in long gaps on aligned double strand break (DSB) ends, consistent with its intrinsically greater polymerization activity (Figure 2A) and processivity (Figure 2B) on simple primer-templates. As expected, a chimera consisting of pol λ with the catalytic domain of pol β (37) did not support any end joining of this substrate (Figure 3A and B). Extracts supplemented with pol µ yielded only the 24-base head-to-head product and no head-to-tail products at all, consistent with previous results indicating that it can efficiently fill only single nucleotide gaps on aligned DSB ends (17,38). Importantly, the pol λ DL yielded a broad smear of products surrounding the 42-mer (Figure 3B, Lane 6), consistent with errors made during gap filling. To test this possibility, we examined the sequences of gap filling products in more detail. DNA from pol λ DL-supplemented end-joining reactions was transfected into E. coli and 25 individual clones were sequenced (Figure 4). These sequences confirmed the broad heterogeneity of products. Some retained the 5′- and 3′-overhangs, while others contained deletions (of one or several nucleotides) and base substitutions. Four clones contained mixed sequences consistent with ligation of heteroduplexes generated by error-prone gap filling. These results suggest that gap filling by pol λ DL on aligned DSB ends during NHEJ in a HeLa cell nuclear extract is less accurate than is gap filling by wt pol λ. Consistent with these results is the observation that primer extension by pol λ DL generated additional bands (indicated by arrowheads in Figure 2A, Lane 4), likely products of incorrect incorporation, that were not observed among the reaction products of wt pol λ (Figure 2A, Lane 3).
Figure 4.
Repair joints formed in extracts depleted of pol λ and supplemented with pol λ DL. (A) Initial substrate. (B) Joints containing deletions (top strand) that were recovered in multiple clones. Bottom strand shows the initial overhangs. Bold letters indicate inferred gap filling. Dashes indicate bases that are present in the initial overhangs but missing in the repair product. Underlines indicate a 4-base microhomology. (C) Joints involving apparent ligation of heteroduplexes. At left, bottom strand shows overhangs of the initial substrate. Above it are two sequences that were recovered in a single clone, distinguishable by their abundance. Lower-case letters indicate apparent misinsertions. Diamond indicates apparent gap filling across the discontinuity in the template. Inferred ligated heteroduplexes are shown at right. (D) Additional sequences, each recovered in a single clone, including two with apparent untemplated insertions (overlines).
Error rates for stable misincorporation by pol λ DL
The heterogeneity of the products of the primer extension reactions (Figure 2A, Lane 4) and the NHEJ reactions (Figure 4) prompted further examination of the fidelity of DNA synthesis by pol λ DL. For this purpose, we used an assay that detects a broad range of different errors at a large number of sites as a polymerase copies a template encoding the LacZ α-complementation gene in gapped M13mp2 DNA (26). Both wt pol λ and pol λ DL completely filled the gap (data not shown). The DNA products were then introduced into E. coli cells, the cells were plated on indicator plates and the total number of dark blue M13 plaques (from correct synthesis) and light blue and/or colorless M13 plaques (containing DNA synthesis errors) were scored. Wild-type pol λ and pol λ DL generated mutants at frequencies of 21% and 54%, respectively (Table 2). A comparison of the DNA sequences of independent LacZ mutants revealed that, like wt pol λ, pol λ DL generated a variety of errors (Table 3) that were distributed throughout the LacZ sequence (data not shown). Amazingly, a total of 618 sequence changes were identified among only 50 LacZ mutants generated by pol λ DL, i.e. an average of 12 changes per mutant. This compares to 2.5 mutations per mutant generated by wt pol λ (39). When the sequence changes are used to calculate error rates for individual classes of events (see ‘Materials and Methods’ section), the results indicate that pol λ DL is 2- to 3-fold less accurate than wt pol λ for single-base insertion and single- and 2-base deletion errors (Table 2). Analysis of the specificity of single nucleotide deletions revealed that the largest rate increase is for deletions at non-iterated template sequences (6.6-fold, Table 3). Based on extensive previous data [reviewed in (40)], this specificity suggests that the majority of these single nucleotide deletions may have been initiated by nucleotide misinsertion followed by primer relocation to generate a deletion intermediate with an unpaired template nucleotide and a correct terminal base pair that facilitates extension. Pol λ DL also generates more complex, closely spaced substitution, substitution-deletion and substitution-addition errors at an increased frequency compared with wt pol λ (last line in Table 2). Finally and perhaps most tellingly, the largest effect of changing loop 1 in pol λ is the increase in the rates of single-base substitutions, by 13-fold overall, and by 7- to 31-fold for different base substitutions (Table 4). To confirm that the observed elevated substitution error rates of pol λ DL are independent of the nature of the DNA substrate (long versus short gap), we also measured single nucleotide misinsertion into a 1-nt gap DNA. For misinsertion of dGTP opposite template T, the catalytic efficiency of pol λ DL was 12-fold higher than that of wt pol λ, an increase that was due to a 46-fold higher kcat (Table 1). Collectively, these data demonstrate that deletion of loop 1 strongly increases the ability of pol λ to misincorporate nucleotides during DNA synthesis.
Table 2.
Fidelity of pol λ DL in the forward assay
| Pol λ DL | Pol λ wt | |
|---|---|---|
| Mutant frequency × 10−2 | 54a | 21b |
| Mutants sequenced | 50 | 103 |
| Nucleotides sequenced | 20 350 | 41 921 |
| Total changes | 618 | 253 |
| Classes | Error rate × 10−4 | |
|---|---|---|
| Base substitutions | 120 (252) | 9 (38) |
| 1-nt deletions | 120 (239) | 45 (190) |
| 2-nt deletions | 5.9 (12) | 2.1 (9) |
| 1-nt additions | 3.4 (7) | 1.6 (7) |
| Other changesc | (108) | (9) |
The number of occurrences is given in parenthesis.
aThe mutant frequency of pol λ DL represents one of seven independent determinations. The other six mutant frequencies were 43 × 10−2, 48 × 10−2 and 37 × 10−2 (reactions with 150 nM pol λ DL at pH 7.5), 39 × 10−2 and 50 × 10−2 (reactions with 50 nM pol λ DL at pH 8.5), 54 × 10−2 (reaction with 150 nM pol λ DL at pH 8.5).
bThe stated mutant frequency of pol λ wt, determined at the same time as the mutant frequency of pol λ DL, is identical to that reported earlier (39). The specificity of pol λ wt is from (39).
cClassified as ‘other changes’ are large deletions and complex errors involving substitution-deletion and substitution-addition errors.
Table 3.
Single nucleotide deletion error rates and the run length
| Runs | Length (nucleotide) | Pol λ DLa |
Pol λ wtb Error rate × 10−4 | |
|---|---|---|---|---|
| Observed | Error rate × 10−4 | |||
| One | 206 | 89 | 86 | 13 |
| Two | 114 | 91 | 160 | 77 |
| Three | 57 | 40 | 140 | 82 |
| Four/Five | 30 | 19 | 127 | 81 |
aThe error rates were calculated by dividing the number of observed deletions in the given category by the total number of template nucleotides present in runs of the listed lengths, among the 50 pol λ DL mutants sequenced [e.g. 89/(206 × 50) = 1-nt deletion rate at non-run sequences].
bRates for pol λ wt are from (39).
Table 4.
Base substitution specificity of pol λ DL
| Base (n) | Mutation | Mispair | Number of occurrences | Error rate × 10−3 |
|
|---|---|---|---|---|---|
| Pol λ DL | Pol λ wt | ||||
| A (99) | A → G | A•dCMP | 51 | 10.3 | 0.69 |
| A → T | A•dAMP | 36 | 7.2 | 0.49 | |
| A → C | A•dGMP | 26 | 5.2 | 0.59 | |
| T (91) | T → C | T•dGMP | 42 | 9.2 | 1.40 |
| T → G | T•dCMP | 10 | 2.2 | ≤0.10 | |
| T → A | T•dTMP | 5 | 1.0 | ≤0.10 | |
| G (95) | G → A | G•dTMP | 35 | 7.4 | 0.30 |
| G → C | G•dGMP | 15 | 3.2 | 0.40 | |
| G → T | G•dAMP | 7 | 1.5 | ≤0.10 | |
| C (122) | C → T | C•dAMP | 15 | 2.5 | ≤0.08 |
| C → A | C•dTMP | 6 | 1.0 | ≤0.08 | |
| C → G | C•dCMP | 4 | 0.7 | ≤0.08 | |
Rates for wt pol λ wt are from (39).
X-ray crystal structures of pol λ DL
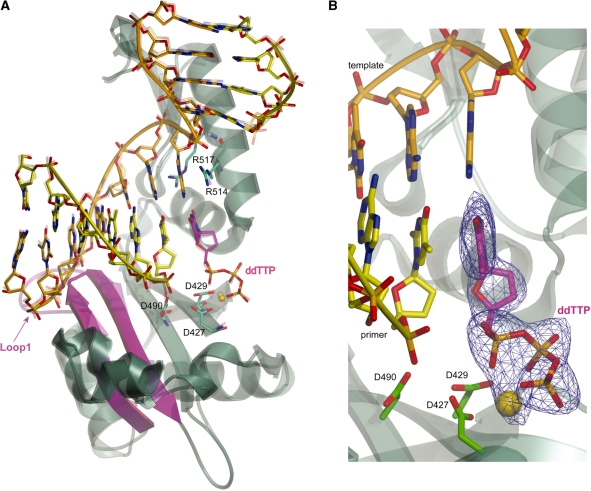
With the goal of understanding why the deletion of a loop positioned between the second and third template nucleotides upstream of the active site results in normal catalytic efficiency for correct incorporation, but reduces fidelity, we determined two crystal structures of pol λ DL. One was a ternary complex of pol λ DL bound to a 1-nt gapped DNA substrate and an incoming correct ddTTP (2.6 Å resolution; see ‘Materials and Methods’ section and Table 5). The single nucleotide gap substrate used here may be most relevant to pol λ’s role in BER. The substrates encountered by the enzyme during NHEJ may be more complex. The ternary complex structure is remarkably similar to that of the equivalent wt pol λ ternary complex (PDBID 1XSN; root mean square deviation (RMSD) 0.615 Å for 270 Cα atoms; Figure 5A and B), of course, with the exception of the loop 1 region. While β-strands 3 and 4 adopt a conformation similar to that observed in pol λ wt (22), the loop that connects them in the wt polymerase is absent (magenta in Figure 5A). This occurs without any structural perturbation of the DNA or any of the polymerase active site atoms, including the dNTP-binding Mg2+ ion observed in the wt structure. This is consistent with the nearly normal catalytic efficiency of pol λ DL.
Figure 5.
Superposition of pol λ wt and pol λ DL ternary complexes. (A) The structure of the pol λ DL ternary complex is shown (solid colors) in an overlay with the wt pol λ ternary complex [1XSN: transparent; (22)]. The three catalytic residues (D427, D429 and D490), R514 and R517 are shown in green, the magnesium atom is colored gold, the template strand is orange, the primer strand is yellow and the incoming nucleotide is magenta. Loop 1 in wt pol λ is shown in pink and the corresponding region in pol λ DL is shown in magenta. (B) Active site of pol λ DL showing density for the incoming dNTP and Mg2+ ion. A simulated annealing Fo-Fc omit density map is shown in blue, contoured at 3σ.
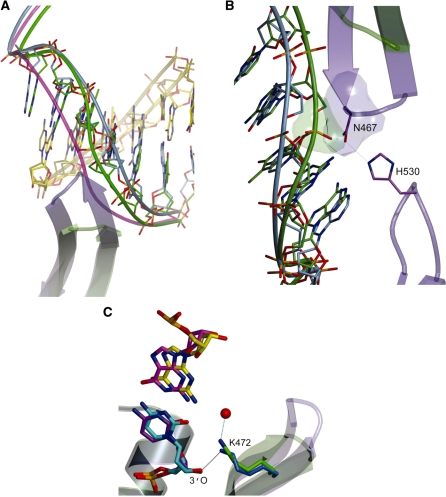
The catalytic cycle for correct incorporation by pol λ involves dNTP-induced repositioning of the DNA template strand, and changes in the positions of several amino acids that result in assembly of the polymerase active site (21). These changes are also believed to play an important role in ensuring the fidelity of synthesis (41). For these reasons, we attempted to understand why pol λ DL has reduced fidelity by solving a second structure, in this case a binary complex of pol λ DL bound to a 1-nt gapped DNA substrate but without a dNTP present (2.4 Å resolution; see Table 5). When the wt (1XSL) and pol λ DL mutant binary complex structures were overlaid (Figure 6A), there was good overall agreement between the two (RMSD 0.814 Å for 267 Cα atoms). Strikingly however, starting at the −1 nt in the template strand, and even more significantly for nucleotides −2 to −6, the template strand of pol λ DL upstream of the active site (green in Figure 6A) adopts a novel conformation that is intermediate between those observed in the wt ternary complex (magenta) and the wt binary complex (blue). In wt pol λ, the location of loop 1 in a binary complex prevents the template strand from assuming a catalytically competent position. Loop 1 and β strands 3 and 4 need to relocate to allow the template strand to occupy the position observed in the ternary complex (Figure 6A). In pol λ DL, the absence of loop 1, and perhaps more specifically the absence of N467 that is in loop 1 (Figure 6B), appears to allow the template to migrate toward the active conformation even in the absence of an incoming dNTP (Figure 6A and B). In addition, a comparison of the binary complexes of wt pol λ and pol λ DL reveals changes in K472 (Figure 6C) that could possibly be functionally significant. Based on the wt pol λ binary complex structure (1XSL, 34), K472 may contribute to maintaining an inactive conformation of the enzyme in the absence of a dNTP by forming a hydrogen bond with the 3′-O of the 3′ nucleotide of the primer (the distance between the NZ of K472 and the 3′-O is 2.8 Å). This may be less likely in pol λ DL because in the binary complex the NZ of K472 is 3.6 Å from the 3′-O, it is pointing away from the 3′-O and it is hydrogen bonding to a water molecule.
Figure 6.
The position of the template strand and K472 is altered in pol λ DL. (A) Overlay of a binary wt complex (1XSL, blue) with the binary pol λ DL structure (green) focusing on the DNA duplex around the active site. The template strand in the mutant adopts a conformation that is intermediate between the wt binary and ternary (1XSN, the template backbone is overlaid in magenta). The primer strands are shown in yellow (pol λ DL and wt are light and dark yellow, respectively). (B) The absence of N467 allows the template strand (green) to adopt the observed conformation. In the wt structure (blue), N467 hydrogen bonds with H530 (dotted line) and adopts a conformation that would clash with the conformation of the phosphate (green surface) bridging the −2 and −3 nt of the template strand observed in the mutant structure. The structure of the wt binary complex (blue) is overlaid for comparison.(C) Overlay of the wt (1XSL) and mutant binary complexes focusing on the position of K472. In the wt structure, this residue is hydrogen boding (black dashed line) to the 3′-O of the primer-terminal nucleotide (cyan), while in the mutant it appears to adopt a conformation in which it hydrogen bonds (cyan dashed line) to a water molecule (red).
DISCUSSION
The focus of this study, on the importance of loop 1 on pol λ’s substrate specificity, largely derives from previous studies indicating that loop 1 in the palm subdomain of TdT and pol µ is an important determinant of DNA substrate specificity during NHEJ of double-strand breaks in DNA. Loop 1 in TdT is long (20 residues) and has been suggested to preclude binding of the template strand and thereby contribute to its template-independent polymerization activity (19,20). Pol µ also contains a long loop 1, deletion of which abrogates its ability to catalyze template-independent synthesis as well as template-dependent extension of a primer terminus lacking its template partner during a reconstituted NHEJ reaction (13). Nonetheless, pol µ lacking loop 1 still retains the ability to perform more standard template-dependent gap-filling synthesis (e.g. when the primer is paired with its complementary template base), either alone or with its partners in NHEJ (13,21). Thus, loop 1 is not required for all template-dependent synthesis activity by pol µ, but it is an important determinant of the substrate specificity of pol µ. The present study leads to a similar general interpretation on the role of loop 1 in human pol λ, namely that loop 1 is not required for template-dependent synthesis, but it does have an important role in defining substrate specificity. Thus, wt pol λ and its loop 1 deletion derivative have similar kinetic constants for correct dNTP insertion (Table 1), similar specific activity with activated DNA, similar ability to extend a simple primer-template (Figure 2), and similar abilities to participate in NHEJ (Figure 3). If anything, the loop 1 deletion derivative performs slightly better than wt pol λ in the latter three assays. This may partly be a consequence of its slightly higher processivity (Figure 2B), a property suggesting that loop 1 may modulate movement of the template strand during catalytic cycling (see below), thus affecting translocation. However, in a manner generally akin to loop 1 in pol µ, loop 1 in pol λ also appears to have an important role in determining substrate specificity. This is because the loop 1 deletion enzyme is considerably less accurate than wt pol λ, as evidenced by the increased diversity of DNA products generated during NHEJ (Figures 3 and 4), by the increased rate of misinsertion of dGTP opposite template T (Table 1), and by the increased error rates for a variety of errors (Tables 3), especially all 12-single base–base mismatches (Table 4). The rate at which pol λ DL stably incorporates certain of these single base–base mismatches into duplex DNA approaches 1%. This rate is similar to the rate at which wt pol λ generates certain single-base deletions (39), an observation that motivated successful efforts to obtain crystal structures of pol λ bound in a catalytically competent conformation to misaligned template-primers (42). Thus, the high error rates in Table 4 suggest that it might be possible to obtain crystal structures of ternary complexes of pol λ DL protein containing mismatches in the nascent base pair binding pocket and/or at the primer terminus.
A previous study (43) showed that, when bound to Bacillus stearothermophilus DNA polymerase I large fragment (family A), the 12 single base–base mismatches are structurally diverse. It is therefore somewhat surprising that pol λ DL has increased error rates for all 12-single base–base mismatches (Table 4), which indicates that pol λ DL can accommodate, and relatively efficiently extend from, all 12 mispairs. Furthermore, the fact that modulation of base substitution fidelity by pol λ DL is not mispair specific is consistent with the idea that truncation of loop 1 likely does not alter the shape of the nascent base pair binding pocket [as, for example, is the case with mutator pol β R283K (44) or R517K pol λ (Bebenek,K., Garcia-Diaz,M. and Kunkel,T.A., unpublished data)]. Instead removal of loop 1 may allow the enzyme to more readily achieve the active conformation under less than ideal conditions, thus increasing a chance of misincorporation.
Clues to how the loop 1 deletion results in such a general loss of discrimination come from considering loop 1 interactions during the pol λ catalytic cycle. In a binary complex of wt pol λ bound to a gapped primer-template (1XSL), prior to binding of a correct dNTP (22), β3, β4 and the hairpin are stabilized by a network of hydrogen bonds. These include interactions between two residues of the hairpin: N467 and Q470 and H530 (2.6 Å) and E498 (2.9 Å), respectively. In addition β3, which is positioned parallel to the template strand, makes van der Waals contacts with the backbone of the template upstream of the primer terminal base pair. Upon dNTP binding, however, the switch from an inactive to active conformation involves partial unraveling of the β strands and displacement of the loop to permit the template strand to adopt its catalytic conformation. It is tempting to speculate that this loop is part of a system of checks and balances that controls insertion fidelity. The energetic penalty of relocating the loop would serve to preserve an inactive conformation, and would be generally only overcome upon binding of a correct nucleotide. Thus, the loop deletion, by lowering the energetic penalty of adopting an active conformation, eliminates one of the checks and facilitates the adoption of an active conformation, thus reducing the discrimination between correct/incorrect insertions. This would explain the general increase in base substitution mutagenesis, and would constitute a clear example of how actions ‘at a distance’ (i.e. modifications distant from the active site that influence active site function) can affect the fidelity of a polymerase.
The differences observed for K472 between the binary complexes of wt pol λ and pol λ DL described in ‘Results’ section are consistent with the possibility that K472 may be involved in modulating the activity and the fidelity of DNA synthesis by pol λ. K472 is conserved in family X polymerases (Figure 1), as a lysine in TdT and pol β, and as an arginine in pol µ (Arg387). Recently Andrade et al. (45) reported that substituting Arg387 with Ala resulted in a loss of template-independent synthesis, whereas a change to Lys increased this activity. Thus, Arg387 plays a key role in modulating template-independent synthesis by pol µ. Substituting the homologous lysine in TdT with arginine or alanine (20) also results in loss of template-independent activity, although the properties of the two TdT mutants are not identical. These results and the structures of TdT and pol µ led Andrade et al. (45) to propose that Arg387 stabilizes the position of the primer terminus and, through its interaction with the primer strand, controls the repositioning of the primer terminus upon dNTP incorporation to allow binding and incorporation of the subsequent dNTP during template-independent synthesis. According to their model, the repositioning of the primer is rate limiting for template-independent synthesis. Substituting Arg387 with Lys may allow the primer terminus to more readily adopt the catalytically active conformation. Our results suggest that K472 may help to modulate template-dependent synthesis. In the wt pol λ binary complex (1XSL), K472 is within H-bonding distance of the 3′-O of the primer terminal nucleotide. A hydrogen bond between K472 and the primer terminus that could stabilize the inactive conformation would need to be disrupted in order for the 3′-O to assume its catalytically competent position. A weakened interaction between K472 and the primer terminus, as might be the case in pol λ DL, would allow the 3′-O to more easily adopt a conformation that would support catalysis with an incorrect nucleotide bound, reducing the discrimination between correct and incorrect incorporation, as observed. Finally, the possible role of K472 is consistent with a quantum mechanical/molecular mechanical study of the catalytical mechanism of pol λ, indicating this lysine as one of the residues important for catalysis (46).
FUNDING
Division of Intramural Research of the National Institutes of Health, National Institute of Environmental Health Sciences (Project Z01 ES065070 to T.A.K, in parts); National Institutes of Health, National Cancer Institute (CA40615 to L.F.P., in parts). Funding for open access charge: Division of Intramural Research of the National Institutes of Health, National Institute of Environmental Health Sciences Project (Z01 ES065070 to T.A.K.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Bill Beard and Dr Michael Murray for helpful discussions and critical reading of the manuscript, and the NIEHS DNA Sequencing Facility for assistance in sequencing the M13mp2 LacZ α mutants.
REFERENCES
- 1.Wilson SH, Sobol RW, Beard WA, Horton JK, Prasad R, Vande Berg BJ. DNA polymerase beta and mammalian base excision repair. Cold Spring Harb. Symp. Quant. Biol. 2000;65:143–155. doi: 10.1101/sqb.2000.65.143. [DOI] [PubMed] [Google Scholar]
- 2.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 4.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 5.van Loon B, Hubscher U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc. Natl Acad. Sci. USA. 2009;106:18201–18206. doi: 10.1073/pnas.0907280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, Ramsden DA. Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol. Rev. 2004;200:156–164. doi: 10.1111/j.0105-2896.2004.00160.x. [DOI] [PubMed] [Google Scholar]
- 7.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 8.Bollum FJ. Terminal Deoxynucleotidyl Transferase. New York, NY: Academic Press; 1974. Inc. [Google Scholar]
- 9.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 10.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 11.Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003;19:203–211. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 12.Gozalbo-Lopez B, Andrade P, Terrados G, de Andres B, Serrano N, Cortegano I, Palacios B, Bernad A, Blanco L, Marcos MA, et al. A role for DNA polymerase mu in the emerging DJH rearrangements of the postgastrulation mouse embryo. Mol. Cell. Biol. 2009;29:1266–1275. doi: 10.1128/MCB.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Davis BJ, Havener JM, Ramsden DA. End-bridging is required for pol mu to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008;36:3085–3094. doi: 10.1093/nar/gkn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 18.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pedersen LC. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair. 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romain F, Barbosa I, Gouge J, Rougeon F, Delarue M. Conferring a template-dependent polymerase activity to terminal deoxynucleotidyltransferase by mutations in the Loop1 region. Nucleic Acids Res. 2009;37:4642–4656. doi: 10.1093/nar/gkp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juarez R, Ruiz JF, Nick McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34:4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat. Struct. Mol. Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 23.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 24.Bennett RA, Gu XY, Povirk LF. Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end-joining in CV-1 cells. Int. J. Radiat. Biol. 1996;70:623–636. doi: 10.1080/095530096144509. [DOI] [PubMed] [Google Scholar]
- 25.Povirk LF, Zhou RZ, Ramsden DA, Lees-Miller SP, Valerie K. Phosphorylation in the serine/threonine 2609–2647 cluster promotes but is not essential for DNA-dependent protein kinase-mediated nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res. 2007;35:3869–3878. doi: 10.1093/nar/gkm339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bebenek K, Kunkel TA. Analyzing the fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystollgr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol. Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Blanca G, Shevelev I, Ramadan K, Villani G, Spadari S, Hubscher U, Maga G. Human DNA polymerase lambda diverged in evolution from DNA polymerase beta toward specific Mn(++) dependence: a kinetic and thermodynamic study. Biochemistry. 2003;42:7467–7476. doi: 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Diaz M, Bebenek K, Larrea AA, Havener JM, Perera L, Krahn JM, Pedersen LC, Ramsden DA, Kunkel TA. Template strand scrunching during DNA gap repair synthesis by human polymerase lambda. Nat. Struct. Mol. Biol. 2009;16:967–972. doi: 10.1038/nsmb.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou RZ, Blanco L, Garcia-Diaz M, Bebenek K, Kunkel TA, Povirk LF. Tolerance for 8-oxoguanine but not thymine glycol in alignment-based gap filling of partially complementary double-strand break ends by DNA polymerase lambda in human nuclear extracts. Nucleic Acids Res. 2008;36:2895–2905. doi: 10.1093/nar/gkn126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J. Biol. Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 43.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 44.Osheroff WP, Beard WA, Yin S, Wilson SH, Kunkel TA. Minor groove interactions at the DNA polymerase beta active site modulate single-base deletion error rates. J. Biol. Chem. 2000;275:28033–28038. doi: 10.1074/jbc.M003462200. [DOI] [PubMed] [Google Scholar]
- 45.Andrade P, Martin MJ, Juarez R, Lopez de Saro F, Blanco L. Limited terminal transferase in human DNA polymerase mu defines the required balance between accuracy and efficiency in NHEJ. Proc. Natl Acad. Sci. USA. 2009;106:16203–16208. doi: 10.1073/pnas.0908492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cisneros GA, Perera L, Garcia-Diaz M, Bebenek K, Kunkel TA, Pedersen LG. Catalytic mechanism of human DNA polymerase lambda with Mg2+ and Mn2+ from ab initio quantum mechanical/molecular mechanical studies. DNA Repair. 2008;7:1824–1834. doi: 10.1016/j.dnarep.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]