Abstract
Transcripts of NANOG and OCT4 have been recently identified in human t(4;11) leukemia and in a model system expressing both t(4;11) fusion proteins. Moreover, downstream target genes of NANOG/OCT4/SOX2 were shown to be transcriptionally activated. However, the NANOG1 gene belongs to a gene family, including a gene tandem duplication (named NANOG2 or NANOGP1) and several pseudogenes (NANOGP2-P11). Thus, it was unclear which of the NANOG family members were transcribed in t(4;11) leukemia cells. 5′-RACE experiments revealed novel 5′-exons of NANOG1 and NANOG2, which could give rise to the expression of two different NANOG1 and three different NANOG2 protein variants. Moreover, a novel PCR-based method was established that allows distinguishing between transcripts deriving from NANOG1, NANOG2 and all other NANOG pseudogenes (P2–P11). By applying this method, we were able to demonstrate that human hematopoietic stem cells and different leukemic cells transcribe NANOG2. Furthermore, we functionally tested NANOG1 and NANOG2 protein variants by recombinant expression in 293 cells. These studies revealed that NANOG1 and NANOG2 protein variants are functionally equivalent and activate a regulatory circuit that activates specific stem cell genes. Therefore, we pose the hypothesis that the transcriptional activation of NANOG2 represents a ‘gain-of-stem cell function’ in acute leukemia.
INTRODUCTION
The NANOG protein—in combination with OCT4 and SOX2—was shown to be sufficient to establish an embryonic stem cell program. Since the discovery of NANOG in 2003 (1,2), NANOG has drawn very much attention, and the ‘core NANOG network’ has been unraveled for human and murine embyronic stem (ES) cells by ChIP-on-Chip experiments (3,4). Stem cell functions of the core NANOG network are maintained by the help of the Polycomb repressor complex II (PRC II: SUZ12, EED, EZH2) which specifically silences genes coding for transcription factors necessary for the development of all three germ layers and neuronal development (5).
Tumor research has been widely improved by the concept of cancer-initiating cells (6). Cancer-initiating cells provide features of stem cells, however, different tumors seem to use different pathways to obtain stemness (7), and little is known about the molecular mechanisms that are required to establish this unique cell population.
Recently, we have discovered that NANOG transcription was significantly enhanced (16-fold) in murine fibroblast when stably transfected with expression constructs coding for the MLL•AF4 and AF4•MLL fusion genes, deriving from the chromosomal translocation t(4;11)(q21;q23) (8). This genetic aberration is associated with high-risk acute lymphoblastic leukemia and very poor outcome. Subsequent analyses of the core NANOG network provided first evidence that NANOG downstream targets were indeed transcriptionally activated, while NANOG/PRCII-repressed genes were transcriptionally silenced. To verify this unusual finding, leukemic cells deriving from adult and pediatric t(4;11) patients were investigated and revealed the same transcriptional profile (8). Thus, it seems that the population of t(4;11) leukemia cells—or at least a small fraction thereof—is able to turn on a stem cell program similar to the core NANOG network identified in ES cells.
A precise analysis of NANOG transcription, however, is hampered by the fact that NANOG1 is transcribed along with several retroposed pseudogenes of the NANOG family, also demonstrating that NANOG2 (alias NANOGP1) is a tandem duplication of NANOG1 (9). The comparison of human and chimpanzee genome sequences has revealed that NANOG2 retained its intronic sequences, while NANOGP2 to P11 are dispersed, intronless and reverse transcribed integrants (10). Transcription of NANOG2 and NANOG pseudogenes (NANOGP2, P4, P5, P7 and P8) has been demonstrated for a large variety of different solid tumors (11,12). Thus, transcripts deriving from NANOG1, NANOG2 or these pseudogene copies can only be distinguished by cloning and sequencing the resulting PCR amplimers. Based on their specific mutation spectrum (missense, frame shift or deletion) their origin can be elucidated.
Therefore, we started a detailed investigation of the NANOG gene family and their transcriptional properties, using MLL-rearranged leukemic cells that seem to be capable of re-activating a NANOG-dependent stem cell program, probably as part of their pathological properties.
MATERIAL AND METHODS
Cell culture
SEM, RS4;11, KOPN8 and NOMO1 cells were cultured in RPMI 1640 containing 10% fetal calf serum. NTERA-2, HEK-293 and HEK-293T cells were maintained in DMEM supplemented with 10% fetal calf serum (and 5% horse serum for NTERA-2 cells). All media were supplemented with 1% l-glutamine and 1% Pen/Strep.
Total RNA extraction and reverse transcription
RNA was prepared from NTERA-2, SEM, RS4;11, KOPN-8 and NOMO-1 cell lines using the Qiagen RNeasy Mini Kit (Qiagen, Germany). The protocol was slightly changed to extract only cytosolic RNA (in order to prevent genomic DNA contamination for pseudogene analysis). One microgram RNA was reverse transcribed using hexamer primers in a total volume of 25 µl. Final cDNA synthesis was diluted to 50 µl using sterile water. In addition, all isolated RNAs were directly tested in PCR reactions to exclude any contamination with genomic DNA (data not shown). Five microliters of each cDNA was used as template in 50 µl PCR reactions throughout all experiments.
NANOG-specific RT–PCR experiments
Initial RT–PCR experiments were carried out by using three different primer sets (a–c). Two of these pair sets (a and b) specifically bind to transcripts deriving from NANOG1, NANOG2 and all other NANOG pseudogenes (P2–P11). Primer set c binds to the 5′-flanking UTR of NANOG1 and NANOG2 (one mismatch) and to an internal exon. Primer sets were as follows: set a (5′-gatcagatctAACATGAGTGTGGATCCAGCTTGTC-3′; 5′-ggaattcTCACACGTCTTCAGGTTGCATGTTC-3′) results in a 938-bp PCR amplimer; set b (5′-GCCTCCAGCAGATGCAAGAAC-3′; 5′-GCAGGAGAATTTGGCTGGAAC-3′) produces a 418-bp PCR amplimer; set c (5′-ATTATAAATCTAGAGACTCC-3′; 5′-TTGTTTGCCTTTGGGACTGGT-3′) results in a 444-bp PCR amplimer. The NANOG2-specific primer set d (5′-GTTAATGTGGTTACAAAAC GTGAC-3′; 5′-GCCACCTCTTAGATTTCATTCTCTGGTTCTGG-3′) should produce a 351-bp PCR amplimer. Finally, a NANOGP8-specific primer set e (5′-CAAAGCTTGCCTTGCTTTGAAGA-3′; 5′-CTGGTGGTAGGAAGAGTAAAGG-3′) resulted in a 525-bp amplimer. All amplifications were carried out with a denaturation step at 94°C for 2 min followed by 35 cycles with the following profile: 30 s at 94°C, 30 s at 58°C (45°C for set c; 59°C for set e) and 30 s at 72°C; a final extension step at 72°C for 5 min was performed for all reactions. QPCR experiments were performed by using the NANOG1-specific oligonucleotides 5′-CCTTCAGCAAAGAACAAAGCTTC-3′ and 5′-TGTCTATCCCTCCTCCCAGGTAG-3′ (hybridizing to NANOG1 exon 1b and exon 2) and NANOG1/2-specific oligonucleotides 5′-CACCTATGCCTGTGATTTGTGG-3′ and 5′-TTGTTTGCCTTTGGGACTGG-3′ (hydridizing to NANOG1/2 exons 3 and 4).
5′-RACE experiments
All 5′-RACE experiments were performed by using the Invitrogen RACE Kit according to the manufacturer’s instructions. Briefly, 5 µg of extracted RNA was used for the initial dephosphorylation step and a subsequent decapping step resulting in a 5′-phosphate only at bona fide mRNA molecules. Next, a ligation with an 44-nt long RNA oligonucleotide was performed, leading to 5′-tagged RNA molecules. Then, a first strand cDNA synthesis was performed using a NANOG-specific oligonucleotide (5′-GTTGCTCCACATTGGAAGG-3′) that specifically binds to the final exon. After completion of first strand cDNA synthesis, two consecutive PCR reactions were performed to amplify specifically NANOG1 and NANOG2 transcripts. For the first PCR the specific oligonucleotides were used (5′-CGACTGGAGCACGAGGACACTGA-3′; 5′-CACCAGGCATCCCTGCGTCAG-3) in combination with a touch-down PCR protocol: 10 cycles with 30 s at 94°C and annealing and elongation for 3 min at 68–64.4°C (–0.4°C per cycle); this was followed by 25 cycles with 30 s at 94°C, 30 s at 64°C and 3 min at 68°C. An aliquot of the resulting amplification products were used for a nested PCR reaction using the oligonucleotides 5′-GGACACTGACATGGACTGAAGGAGTA-3′ and 5′-GCCACCTCTTAGATTTCATTCTCTGGTTCTGG-3′ in combination with a second PCR program (35 cycles with 30 s at 94°C, 30 s at 64°C and 90 s at 68°C). Reactions were loaded on a 2% agarose gel and the different PCR amplimers were cut out, eluted from the gel and cloned into the pCR®4-TOPO® vector (TOPO TA Cloning® Kit For Sequencing, Invitrogen, UK). Subsequent sequence analyses using an universal T7 oligonucleotide (5′-TAATACGACTCACTATAGGG-3′) revealed different mRNA species deriving from the NANOG1 and NANOG2 genes, respectively.
Establishment of NANOG1- and NANOG2-specific PCR reactions
NANOG1-specific oligonucleotides were 5′-CACCCACACGAGATGG-3′ (specific for novel exon 2) and 5′-CAGAAGACATTTGCAAGG-3′ (specific for novel exon 3) and produce PCR amplimers of 274 bp. Amplification conditions for NANOG1 were 45 cycles with 30 s at 94°C, 30 s at 56°C and 30 s at 72°C. Oligonucleotides specifically binding to NANOG1 and NANOG2 were 5′-CCTTCAGCAAAGAACAAAGCTTC-3′ (specific for novel exon 1b) and 5′-CATCTCAGCAGAAGACATTTGCAAGG-3′ (specific for novel exon 3) and produce PCR amplimers of 195 bp (NANOG2) and 356 bp (NANOG1). In case of exon 1b* usage, a 314-bp amplimer of NANOG2 was obtained. Amplification conditions for NANOG1/2-specific transcripts were 26 cycles with 30 s at 94°C, 30 s at 65–54°C (–0.5°C per cycle) and 90 s at 72°C; this was followed by 14 cycles with 30 s at 94°C, 30 s at 54°C and 90 s at 72°C. A final extension step at 72°C for 2 min was performed. These conditions were also used to analyze a large variety of tissue cDNAs, biopsy material isolated from acute leukemia patients and several control samples. The identity of PCR-amplified NANOG1/2-specific transcripts was verified by DNA sequencing analyses.
Cloning and expression of NANOG1/2 protein variants
Identified splice variants of NANOG1 (A, Bb) and NANOG2 (D1, D2c and E) were amplified using specific oligonucleotides exhibiting appropriate restriction recognition sites for further cloning into the pEXPR-IBA10-3 vector (IBA BioTAGnology, Germany), resulting in a Strep-tag fusion to all 3′-ends of the amplified NANOG1 and NANOG2 open reading frames. Successfully cloned open reading frames were verified by sequence analyses. Subsequently, plasmids (25 µg per 15 cm petri dishes) coding for different NANOG1 and NANOG2 protein variants were lipotransfected into 293T cells and grown for 2 days. Transfection efficiencies were monitored by co-transfecting pEGFP plasmids (25 µg per 15 cm petri dishes). GFP-positive cells were always in the range of 60–70% transfected cells. Cells were harvested and lysed for at least 1 h in a cell lysis buffer containing 150 mM NaCl, 20 mM HEPES pH 7.5, 1% TritonX-100, 0.4 mM EDTA and a phosphatase/protease inhibitor mix. The soluble fraction was added to Strep-Tactin Superflow material and the Strep-tagged proteins were purified according to the manufacturer’s instruction (IBA BioTAGnology, Germany). An aliquot of the eluted protein fractions (10 µl) were used to separate the recombinant proteins on SDS page and blotted for subsequent western blot experiments.
Western blot experiments
30 × 106 SEM and 1 × 106 NTERA cells were lysed in the above mentioned lysis buffer. Soluble fractions (S), pellet fractions (P) or Strep-tag affinity purified recombinant proteins were mixed in 2× Lämmli buffer and separated on a 12% SDS–PAGE. After blotting onto a PVDF membrane, transferred proteins were incubated in blocking solution (5% skim milk in 0.2% Triton X-100/PBS) for 1 h. The C-terminal NANOG mouse antibody (clone 2C11, Abnova) was diluted 1:1000 in blocking solution and incubated over night. The membrane was washed three times with 0.2% Triton X-100/PBS and developed by using the ECLTM Western Blotting Analysis System (GE Healthcare).
Immunohistological experiments
HeLa cells were lipotransfected with pEXPr-IBA10-3 vectors (10 µg for 10 cm petri dishes) containing open reading frames for different NANOG1 and NANOG2 protein variants. After 24 h cells were spread out on cover glass and incubated for additional 24 h. The cells were fixed with 100% Methanol (–20°C) for 20 min at −20°C. Cells were incubated with blocking solution [10% goat serum, 0.1% Triton X-100 in PBS (PBST)] for 30 min. Methanol was removed by drying the cells and subsequently rehydrating them with PBS. The C-terminal NANOG rabbit antibody (H-155, Santa Cruz, USA) was diluted 1:300 in blocking solution containing 1% goat serum and incubated over night. Cells were washed three times for 5 min with 0.1% PBST and incubated with FITC-conjugated anti-rabbit antibody (111-095-003, Dianova, USA) which was diluted 1:150 in blocking solution containing 1% goat serum. Cells were washed three times with 0.1% PBST and treated for 1 min with 0.1 µg/ml DAPI solution. After a second washing step, cells were embedded in 0.1% DABCO/Mowiol solution. All pictures were taken with the Axio Observer Z1 (ZEISS), the Digital CCD Camera C4742-80-12AG (Hamamatsu ORCA-ER) and the Volocity software package.
Microarray analysis
Gene expression analysis was performed using the Affymetrix HG-U133® Plus 2.0 oligonucleotide microarrays. Two µg of purified RNA from transfected cell lines expressing either GFP (mock-control) or the variants NANOG1, NANOG2D2 and NANOG2E were converted by reverse transcription into double-stranded cDNA (Roche Applied Science) and then purified using the GeneChip Sample Cleanup module (Affymetrix). Then, labeled cRNA was generated using the Microarray RNA target synthesis kit (Roche Applied Science) and an in vitro transcription labeling nucleotide mixture (Affymetrix). The cRNA was then purified using the GeneChip Sample Cleanup module (Affymetrix) and quantified using the NanoDrop spectrophotometer. Eleven micrograms of labeled cRNA were fragmented. Hybridization, washing, staining and scanning protocols were performed on Affymetrix GeneChip instruments (Hybridization Oven 640, Fluidics Station 450Dx, Scanner GCS3000Dx, respectively), following the manufacturer’s instructions. The obtained data were stringently screened by comparing against mock-control, by their P-values, log2-fold changes and signal intensities to obtain only very robust data sets. Presented genes were at least 4-fold deregulated.
RESULTS
Transcriptional properties of NANOG gene family members in MLL-rearranged and control cell lines
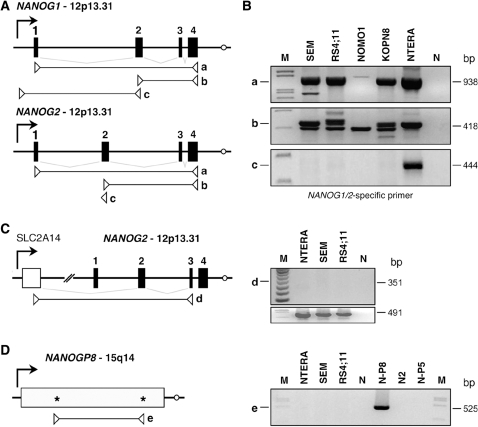
Transcripts of the NANOG gene/pseudogene family were monitored by RT–PCR experiments using different primer combinations. As shown in Figure 1A, three different primer sets (a–c) were used to analyze cDNAs obtained from four different MLL-rearranged cell lines (t(4;11) cell lines: SEM, RS4;11; the t(9;11) cell line NOMO1; the t(11;19) cell line KOPN8. The NTERA-2 (human embryonic carcinoma) cell line served as positive control. By using the exon-specific primer sets a and b, PCR amplimers were obtained for all investigated cell lines (Figure 1B, panels a and b). The forward primer of set c binds to exclusively the 5′-UTR of the NANOG1 gene, demonstrating that NANOG1 transcripts were not present in the investigated MLL-rearranged cell lines (Figure 1B, panel c). Thus, we concluded that (i) NANOG1 transcripts can be distinguished from all other family members, and (ii) that all investigated cell lines are only able to transcribe RNA species deriving from NANOG2 or other NANOG pseudogenes.
Figure 1.
NANOG transcription in MLL-rearranged and Teratocarcinoma cell lines. (A) Gene structures of NANOG1 and NANOG2. Primer sets a–c were indicated for both genes. Forward primer of primer set c binds specifically to the 5′-NTR of NANOG1, and thus, enable to monitor transcripts that derive only from the NANOG1 gene. (B) RT–PCR analysis of four different MLL-rearranged cell lines (SEM, RS4;11, NOMO1, KOPN8). The NTERA-2 cell line represents a human embryonic carcinoma cell line and served as positive control for all experiments. M: DNA size marker. N: water control. (C) Validation of transcripts starting in the upstream located SLC2A14 gene. M, DNA size marker. N: water control. (D) Validation of NANOGP8 transcription. Specific oligonucleotides that bind only to the NANOGP8 open reading frame, we were not able to detect NANOGP8 transcripts in the investigated cells lines. M: DNA size marker. N: water control. N-P8, N2, N-P5: cloned NANOGP8, NANOG2 and NANOGP5 cDNAs served as positive controls.
Subsequent sequence analysis of 60 individual clones that derived from NANOG amplimers of the SEM cell line revealed a first glimpse on the spectrum of activated NANOG genes/pseudogenes. Comparison to available NANOG sequences (NCBI database) demonstrated the following genes/pseudogenes to be transcribed in this MLL-rearranged leukemia cell line: NANOG2 (27%), NANOGP2 (2%), NANOGP4 (55%) and NANOGP5 (16%).
Since Booth and Holland reported for the NANOG2 gene an unusual splice event involving a cryptic exon located about 20 000-bp upstream of the NANOG2 gene, we analyzed this possibility by using specific primers (set d) in an RT–PCR experiment. As shown in Figure 1C, neither NTERA-2 cells nor the two analyzed t(4;11) cell lines SEM and RS4;11 seem to use this upstream exon that is located in intron 1 of the SLC2A14 gene (9), transcribing in the opposite direction relative to the orientation of the NANOG2 gene.
Since NANOGP8 transcripts were not identified in the 60 clones that derived from SEM cells, we wanted to rule out that NANOGP8 is indeed not transcribed in the investigated cell lines. Corresponding RT–PCR experiments were conducted with cDNA from NTERA-2 cells and both t(4;11) cell lines. Cloned NANOG2, NANOGP8 and NANOGP5 cDNAs were used as positive control and to demonstrate specificity of the applied primer sequences (set e). As shown in Figure 1D, the investigated cell lines does not seem to transcribe NANOGP8.
Based on these initial experiments and on the fact, that only NANOG1, NANOG2 and the pseudogene NANOGP8 are per se able to be translated into NANOG proteins, we concluded that NTERA-2, the t(4;11) cell lines and t(4;11) patient cells most likely use NANOG1 or NANOG2 protein to activate the downstream core NANOG network.
RACE experiments revealed an extended gene structure for NANOG1 and NANOG2
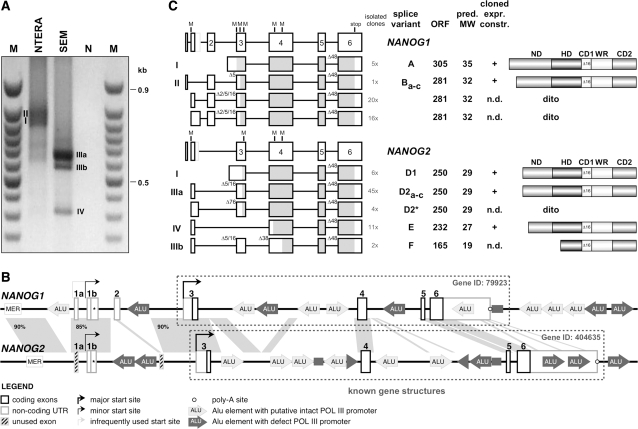
Based on the obtained experimental results, 5′-RACE experiments were conducted with cDNA obtained from NTERA-2 and the SEM cell line by using an anchored oligonucleotide in the final exon of the NANOG1/2 gene. As shown in Figure 2A, several but different PCR bands were obtained for both investigated cell lines. Therefore, all RACE–PCR amplimers were cut out from the gel and cloned. Subsequent sequence analyses revealed the presence of NANOG1 and NANOG2 transcripts in NTERA-2 cells, and NANOG2 transcripts in SEM cells. The large number of analyzed clones allowed us to identify complex splice patterns in transcripts deriving from both NANOG genes. In addition, the presence of novel 5′-exons for both NANOG genes revealed extended gene structures. As shown in Figure 2B, the NANOG1 gene exhibits three additional exons, named exon 1a, 1b/1b* and 2, while the NANOG2 gene exhibits an additional exon 1b/1b*. Surprisingly, exon 2 seems to be absent, however, a careful analysis of genomic sequences revealed that there exists an NANOG2 exon 2 remnant, missing most of its 3′-portion due to an ALU integration event. Based on these findings, we propose an revised exon/intron structure for both NANOG genes which is depicted in Figure 2B. The yet known gene structures for both genes are indicated by dotted frames. It is important to note that an earlier report (9) already described exon 1b in transcripts deriving from the NANOG1 gene.
Figure 2.
RACE experiments reveal splice variations of NANOG1 and NANOG2 and novel upstream exons. (A) 5′-RACE experiment. NTERA-2 and SEM cDNA was analyzed by using an anchored primer in the final NANOG exon. I–IV: bands cut out from the gel, cloned and sequenced. M: 50-bp ladder; N: water control. (B) Extended gene structure for NANOG1 and NANOG2. Homologous regions between both genes are depicted by grey areas. Repetitive DNA elements are indicated. Potentially transcriptional active ALU-elements are shown in light grey, while promoter-mutated ALU-elements are shown in dark grey. MER: MER-repeat. Arrows: indicate transcription initiation sites. All exons are numbered according to the nomenclature used throughout the manuscript. (C) Splice variants of NANOG1 and NANOG2. All cloned RACE amplimers were sequenced and revealed the additional NANOG1 exons 1a, 1b, 1b*, 2 and NANOG2 exons 1b, 1b*. Open reading frame analyses (marked in grey) of all cloned splice variants revealed putative open reading frames (ORFs) coding for two NANOG1 (splice variants A and B) and three NANOG2 protein (splice variants D, E and F). The number of isolated cDNA clones is indicated. Length of ORFs, predicted molecular weight (MW) and putative protein structures are depicted for all NANOG1/2 mRNA variants.
Detailed overview about the novel NANOG1 and NANOG2 gene structures
NANOG2 most likely represents a tandem duplication of the NANOG1 gene located at 12p13.31. Both genes are separated by about 80 kb, a genomic region coding for the SLC2A14 gene. Based on our results, the NANOG1 gene exhibits two alternatively used first exons (1a and 1b/1b*) that both splice to the consecutive exons 2–6. For the NANOG2 gene, we identified only transcripts containing exon 1b/1b* sequences, although the presence of a NANOG2 exon 1a could be identified in the genomic DNA.
Both genes are highly saturated with repetitive ALU elements. All ALU integration events are unique for both genes, indicating that both NANOG gene copies diverged during the evolution. Although both NANOG genes seem to be quite diverse, the regions upstream and surrounding NANOG1/2 exons 1a/b are highly conserved (90 and 85% identity), similar to the region upstream of NANOG1/2 exons 3 that display 90% identical nucleotides. All other intronic sequences are much less conserved and show very little homology. This indicated that the regions used as transcriptional start sites of both NANOG genes have been conserved during evolution.
Alternative splice variants of NANOG1 and NANOG2
A series of different splice variants for NANOG1- (n = 7; named A and Ba–c) and NANOG2-derived transcripts (n = 7; named D1, D2a-c, D2*, E and F) were cloned (Figure 2C). All these variant mRNAs can be potentially translated into 2 different NANOG1 (predicted MW: 32 and 35 kDa) and three different NANOG2 proteins (predicted MW: 19, 27 and 29 kDa). Splice variant A is derived from transcripts starting upstream of NANOG1 exon 3 and uses the first AUG start codon of exon 3; this transcript encodes an open reading frame with the potential to produce a protein with a predicted molecular weight (MW) of 35 kDa. Splice variants B come in many different flavors, because exons 1a, 1b or 1b* are used for splice processes to consecutive exons 2–6 of NANOG1. Moreover, splice processes between exons 2 and 3 result in exon 2 sequences fused to nt +3, +6 or +17 of exon 3, thereby skipping 2, 5 or 16 nt. All these different mRNAs lead to shorter open reading frames with the potential to produce a protein with a predicted MW of 32 kDa. In a few of these NANOG1 transcripts, alternative splicing is leading to the skipping of the first 48 nt of exon 6. This results in the loss of 16 amino acids of the CD1 domain which has previously been described (13).
The NANOG2 gene gives rise to splice variants D–F. Splice variant D1 is similar to splice variant A of the NANOG2 gene. However, due to several missense mutations, the first two AUG start codons in exon 3 are absent, and thus, this splice variant encodes a protein with a predicted MW of 29 kDa. Splice variant D2 comprises transcripts that use either exon 1b or 1b*. Exon 1b is spliced to exon 3 by fusing exon 1b sequences again with nts +3, +6 or +17 of exon 3. Presumably due to the missing exon 2 in the NANOG2 gene structure, exon 1b* splice events fuse exon 1b* with nt +77 of exon 3; these different mRNAs all encode a NANOG2 protein variant with a predicted MW of 29 kDa. Splice variant E fuses exon 1b with exon 4, resulting in an open reading frame that can be translated into a 27 kDa protein. Finally, splice variant F is nearly identical to splice variant D2, however, exon 3 is fused to nt +39 of exon 4. This results in another open reading frame that potentially encodes a protein of 19 kDa. Moreover, all cloned NANOG2 splice variants exhibit the skipping of the first 48 nt of exon 6. The number of all cloned splice variants, the length of the potentially encoded open reading frames, the predicted MW and the protein domain structure of both NANOG genes are depicted in Figure 2C. All primary sequences and a complilation of all identified splice sites can be retrieved from the Supplementary Data (Supplementary Figures S1 and S2).
Western blot experiments revealed different NANOG protein variants
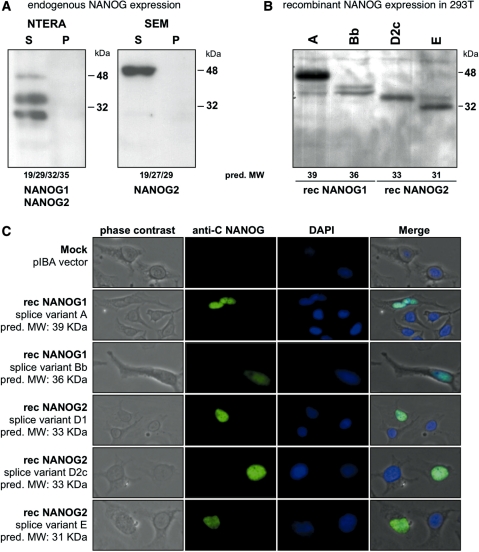
Western blot experiments with soluble cell lysates (S) of NTERA-2 cells revealed the expression of several protein bands. NTERA-2 cells transcribe both the NANOG1 and NANOG2 gene in a large variety of different splice variants. This results in weaker and stronger visible protein bands in the western blot experiments. Stronger protein bands seem to have a MW of 48, 35 and 29 kDa, respectively (see Figure 3A, left panel). SEM cells, shown to transcribe only the NANOG2 gene, expressed a strong 48 kDa and a weak 29 kDa protein variant (see Figure 3A, right panel).
Figure 3.
Western blot experiments, recombinant expression of NANOG1 and NANOG2 variants in 293T cells and immunohistological experiments. (A) Western blot experiments using an antiserum against the C-terminal portion of NANOG protein revealed the expression of NANOG1 and NANOG2 protein variants in NTERA-2 cells, and NANOG2 in SEM cells. (B) Cloned Strep-affinity-tagged splice variants of NANOG1 and NANOG2. Predicted molecular weight of all expression constructs is indicated. Recombinant expression of NANOG1 splice variants A and Bb and NANOG2 splice variants D2c and E in 293T cells were shown. Sizes of protein markers are indicated. (C) Immunohistological analysis of all recombinant NANOG1/2 protein variants expressed in Hela cells. Selected pictures from transfected cells were taken (20×). From left to right: phase contrast, anti-C NANOG antiserum, DAPI counter-staining and a merged picture is shown. All NANOG1/2 protein variants were localized within the nucleus of transfected cells.
In order to validate that the predicted MW of the different splice variants of both genes and migration behavior in SDS–PAGE are similar, we performed recombinant protein expression in 293T cells. Several splice variants were cloned into the pEXPR-IBA10-3 vector system (A, Bb, D1, D2c and E). This vector fuses a 4 kDa Strep-Tag to the C-terminus of all open reading frames. After transient transfection into 293T cells, all NANOG variants were affinity-purified and analyzed by western blot experiments. As shown in Figure 3B, two different NANOG1 and NANOG2 protein variants were successfully expressed in mammalian 293T cells. The NANOG1Bb, NANOG2D2c and NANOG2E protein variants migrated with their expected molecular weight (32 + 4 = 36, 29 + 4 = 33 and 27 + 4 = 31 kDa, respectively). By contrast, the NANOG1A variant migrated at ∼50 kDa. This indicated that the NANOG1A protein seems to be subjected to post-translational modifications (PTM). The recombinant NANOG1A protein was subjected to mass spectrometry and verified to be the Strep-tagged NANOG1 protein (Supplementary Figure S3). However, all our attempts to uncover the potential PTM failed so far. Therefore, we have no explanation for the observed shift of about +11 kDa (35 + 4 = 39) in SDS–PAGE.
Subcellular localization of NANOG1/2 protein variants
To further investigate the cellular distribution of different NANOG1/2 protein variants, we performed immunohistological experiments. As shown in Figure 3C, expression and cellular localization of the recombinant NANOG1/2 protein variants was monitored by using an antiserum raised against the C-terminal portion of NANOG in combination with DAPI staining. After transient transfection of all constructs into HeLa cells, the NANOG1/2 protein variants localized in the nucleus. Thus, it can be concluded that all different NANOG1/2 protein variants are per se able to translocate into the nucleus, where these protein variants are able to provide their specific function(s).
Establishment of NANOG1- and NANOG2-specific RT–PCR conditions
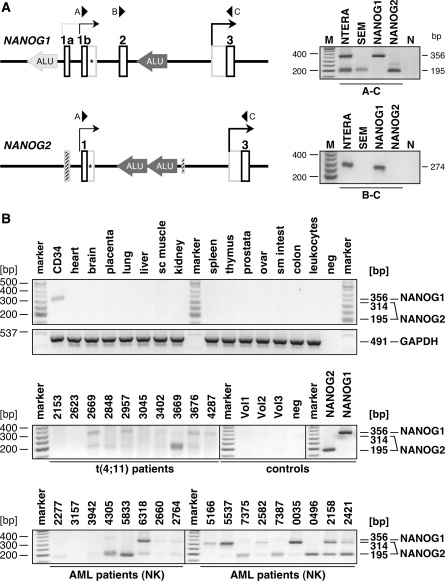
Since the novel gene structures of NANOG1 and NANOG2 exhibit additional exons that are absent in all known NANOG pseudogenes, they provide a perfect source to establish NANOG1- and NANOG2-specific PCR reactions. This may help to experimentally distinguish between bona fide NANOG gene transcripts and non-functional transcripts deriving from all other NANOG pseudogenes. In Figure 4 the first exons of the NANOG1 and NANOG2 gene are depicted. By using the specific primers A and C, we were able to establish specific conditions that specifically amplify only transcripts deriving from both NANOG genes. Since the NANOG1 gene encodes the additional exon 2, the resulting two different PCR amplimers indicate transcription of NANOG1 (356 bp) and NANOG2 (195 bp). In case of using the exons 1b* splice sites, the NANOG1-specific transcript will result in a 770-bp amplimer, while the NANOG2-specific transcript will result in a 314-bp amplimer. All experiments were controlled by using total RNA isolated from NANOG1Bb- and NANOG2D2c-transfected 293T cells (Figure 4, right upper panel). The results of these experiments clearly revealed that NTERA-2 cells transcribe both NANOG genes, while SEM cells exclusively transcribe the NANOG2 gene. The combination of primers B and C verified these results, since this PCR detected only transcripts deriving from the NANOG1 gene (274 bp amplimer). Thus, specific conditions were established that allow to distinguish between transcripts deriving from NANOG1 alone (B–C), NANOG1 and NANOG2 (A–C) from all other pseudogene transcripts of the NANOG gene family (Figure 1B, primer set a or b). From these experiments, we concluded that MLL-rearranged cell lines are able to transcribe the NANOG2 gene.
Figure 4.
NANOG1/2-specific RT–PCR experiments. (A) The two promoter regions of each NANOG genes are highlighted. Oligonucleotides A–C are indicated. Right upper panel: RT–PCR experiments using the primer combination A/C which allowed to monitor NANOG1- and NANOG2-derived transcripts. Right lower panel: RT–PCR experiments using the primer combination B/C, which allowed to monitor only NANOG1-derived transcripts. As internal control, cDNA from transfected cells were used (NANOG1 Bb and NANOG2 D2c) and analyzed under identical conditions. N: water control. (B) Tissue cDNA panel. By using the primer combination A/C, a tissue cDNA panel was tested. With the exception of CD34+ cells, all other investigated tissue cDNAs remained negative. M: DNA size marker; neg: water control. Middle panel: Screening of t(4;11) patients, healthy volunteers and positive controls. Seven out of ten analyzed t(4;11) patients transcribed the NANOG2 gene. Peripheral blood samples of healthy volunteers were negative. M: DNA size marker; neg: water control. Lower panel: Screening of AML patients with normal karyotype. Out of 20 samples, 16 were positive for NANOG2 transcription, while 5 out of 20 samples were also positive for NANOG1 transcripts. Only 17 out of 20 analyses are shown. M: DNA size marker.
By using the primer combination A–C, we investigated a tissue cDNA panel for NANOG transcripts. As shown in Figure 4B (upper panel), only CD34+ cells seem to express a 314-bp amplimer. This derives from the NANOG2 gene by using the exon 1b* splice site (Figure 4A). Thus CD34+ cells transcribe the NANOG2 but not the NANOG1 gene. All other tissues remained negative, as well as the investigated healthy individuals (n = 3). The same experiment was performed with leukemia biopsy material of t(4;11) patients and AML patients with normal karyotype (Figure 4B, middle and lower panels). These experiments revealed that leukemia patients weakly express predominantly NANOG2 (1b = 195 and 1b* = 314 bp), while some AML samples were also weakly transcribing NANOG1 (1b = 356 bp). Several bands were cut out from the gel and subjected to DNA sequencing analysis. In all cases, the appropriate NANOG1 or NANOG2 transcript could be confirmed. The weak amplimer production can be explained by two possibilities: (i) all cells produce only very few transcripts, or, (ii) only a minor fraction of the analyzed leukemia cells transcribe the NANOG genes. Based on immunohistochemistry experiments (Supplementary Figure S4), we assume that only a minor fraction of cells is expressing the NANOG or OCT4 protein. We also conducted QPCR experiments to analyze the absolute amount of transcripts produced by the two alternative start sites in the NANOG1 gene. For this purpose, PCR experiments were performed with total RNA isolated from NETRA-2 cells. The amount of transcripts between exon 1b/2 and exon 3/4 were measured. For quantification, we used a log-diluted NANOG1Bb expression plasmid (1–106 copies; Supplementary Figure S5). This allowed us to calculate the initiation events of transcripts starting upstream of exon 1b (10 000 transcripts in cDNA derived from 100 ng total RNA; assuming 15 fg total RNA per cells = 1 transcript in 600 cells) and exon 3 (490 000 transcripts in cDNA deriving from 100 ng total RNA; assuming 15 fg total RNA per cell = 1 transcript in 12 cells). Thus, the transcriptional start site upstream of NANOG1 exon 3 is about 50-fold more used than the start site upstream of exon 1b, assuming that both NANOG genes are equally transcribed. QPCR experiments could not be performed for the NANOG2 gene, because of the multitude of different-sized PCR bands in splice variants D2, D2*, E and F did not allow reliable quantification.
Gene expression studies
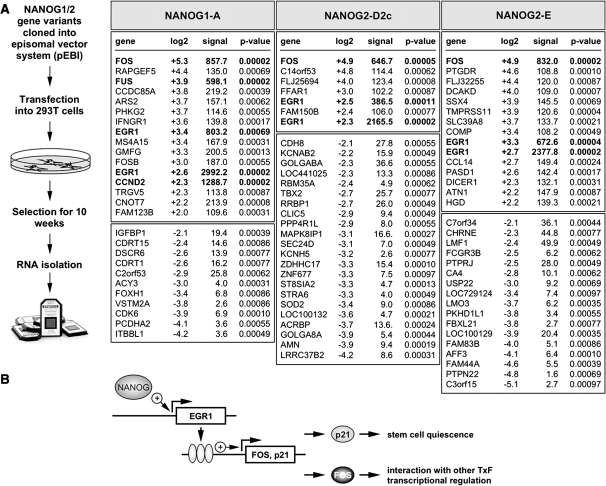
To understand the biological significance of the different NANOG1/2 protein variants, we used an episomal vector system (pEPI) to express three different NANOG protein variants (1A, 2D2 and 2E) in 293 cells. The pEPI-EGFP vector encodes an additional EGFP reporter gene and the neomycin resistance gene. An empty pEPI-EGFP vector expressing only the EGFP protein served as negative control and was kept under identical conditions. After 69 days of cell culture, about 60% of the different cell populations were green fluorescent and total RNA was prepared. These RNA samples were used to perform gene expression profiling experiments using available Affymetrix chips (Figure 5A). Subsequent analysis revealed a specific activation of very few genes in all three transfected cell populations. All investigated NANOG1/2 protein variants strongly induced FOS gene transcription (NANOG1A: 39-fold; NANOG2D2: 30-fold; NANOG2E: 30-fold). The FOS protein is able to bind directly to several other transcription factors or nuclear complexes (ATF2, BCL3, COBRA1, CREBBP, CSNK2A1, CSNK2A2, DDIT3, ETS2, GTF2E2, GTF2F1, JUN, MITF, NACA, NCOA1, NCOA6, NCOR2, PML, RPS6KA4, RUNX1, RELA, RUNX2, SMAD3, SPI1, TAF1, TBP, TCF1, TSC22D3, VDR, XBP1). The second gene that was transcriptionally activated was the EGR1 gene (5- to 6-fold). This zinc finger transcription factor directly interacts with several other nuclear factors (CEBPB, CREBBP, EP300, ERBB3, NAB1, NAB2, NFATC1, PITX1, PSMA3, RELA, TP53). Most importantly, EGR1 is able to transcriptionally upregulate the CDKN1A/p21 gene. The activation of the CDKN1A/p21 gene was recently shown to be a key step for cancer stem cell quiescence and maintenance (14). Thus, ectopic activation of different NANOG protein (variants) in non-ES cells may allow to activate a specific genetic program (Figure 5B). Based on recent findings, the transcriptional activation of p21 is p53-independent and one of the key features for tumor stem cells to aquire stem cell maintenance and drug resistance.
Figure 5.
Gene expression profiling experiment. (A) The cell line 293 was transfected with episomal pEPI-EGFP vector containing expression cassettes for NANOG1A, NANOG2D2c or NANOG2E. After 10 weeks of culture, GPF-positive cells were sorted and used for RNA isolation. These RNA samples were used for Affymetrix chip experiments, resulting in few deregulated genes shown in the right. White boxed areas: up-regulated genes. Light grey boxed areas: down-regulated genes. Common genes identified in all three experiments using the different NANOG1/2 protein variants are highlighted. (B) Mode of action for the investigated NANOG protein variants. NANOG protein is able to induce transcription of EGR1 (directly or indirectly). EGR1 protein is directly binding to the promoter regions of FOS and p21. P21 protein is necessary for stem cell maintenance, while the FOS transcription factor is necessary for cell proliferation.
DISCUSSION
NANOG is a homeodomain transcription factor that is—in conjunction with OCT4 and SOX2—responsible to establish a genetic circuit to maintain the stem cell compartment at the blastocyst stage of developing embryos (3,4). This important concept has recently been verified and extended by the genetic manipulation of differentiated cells with four different transgenes (Klf4, Oct4, Sox2 and c-Myc). Transient expression of these proteins led to the activation of the NANOG gene, and subsequently to a reprogramming of the manipulated cells into induced pluripotent stem cells (iPS) (15,16).
Many reports have demonstrated transcription of ‘NANOG’ in germ cells (17,18) and other human cancer tissues (11,12). In most of these reports, the authors were only able to distinguish between transcripts deriving from NANOG1, NANOG2 (10) or NANOG pseudogenes by cloning and sequencing the obtained PCR amplimers. However, only NANOG1-, NANOG2- and NANOGP8-derived transcripts are per se able to be translated into protein, while all other pseudogenes do not exhibit intact open reading frames (9).
This study tried to answer the question whether human cancer cells express either NANOG1 or the functional equivalents NANOG2 and NANOGP8, respectively. Expression of functional NANOG proteins may help to establish a ‘stem cell-like’ program which in turn could be a key event to establish and maintain cancer stem cells. To answer this important question and to validate our earlier finding, namely that t(4;11) leukemia cells transcriptionally upregulate NANOG transcripts (8), we started to investigate the transcriptional properties of NANOG genes and pseudogenes in MLL-rearranged cells lines.
First, the analysis of transcriptional properties of NANOG1 and NANOG2 in NTERA-2 and MLL-rearranged cell lines led to the discovery of novel 5′-exons which reflect on more extended gene structures for NANOG1 and NANOG2, respectively (Figure 2B). Several splice variants of both NANOG genes were cloned which putatively encode different NANOG1/2 protein variants (Figure 2C). The different NANOG protein variants display a shorter N-terminal domain (ND), without affecting the important homeobox domain (HD). The ND was shown to be dispensable for the function of human NANOG protein, because the transactivation domain is located in the C-terminal domain CD of human NANOG protein (19). Only the NANOG2F variant displayed a partial loss of the HD domain. Thus, all other NANOG1/2 protein variants seem to contain all necessary domains to exhibit similar functions. Noteworthy, the existence of exon 1b as part of the NANOG1 gene has been described earlier (9), however, the authors did not further investigated their own findings.
NTERA-2 cells transcribe several splice variants deriving from the NANOG1 and NANOG2 gene. By using an antiserum specific for the C-terminal portion of NANOG protein, different NANOG1 and NANOG2 protein variants were detected in western blot experiments. By contrast, the investigated t(4;11) cell line SEM predominantly expresses a 48 kDa protein. By using recombinant expression in HeLa cells, we could demonstrate that all tested NANOG1/2 protein variants translocated into the nucleus. This may indicate that all these different NANOG protein variants are able to function as transcription factors.
Based on the extended gene structures we established specific PCR conditions that allow to precisely distinguish between transcripts that derive from NANOG1 (B–C amplimer in Figure 4), NANOG2 (A–C amplimer in Figure 4) or all other NANOG pseudogene copies (primer sets a and b in in Figure 1B). Thus, the analysis of transcriptional properties in stem cells, cancer cells or any other cell line will become more accurate and reliable in the future. Because of the importance of this finding, we validated independently the existence of these NANOG1/2 upstream transcripts by RNase protection experiments (Supplementary Figure S6). These experiments validated all prior experiments, thus demonstrating that the novel upstream NANOG1/2 exons are existing and no cloning artifact. Therefore, the established PCR assay could be reliably used as read-out system for the transcriptional activity of both stem cell genes without detecting any other pseudogene.
To investigate this a bit deeper, we analyzed the chromatin properties of both NANOG genes. This revealed open chromatin structures (H3K4me) in the promoter regions of both NANOG genes in NTERA2 cells, while the leukemia cell lines SEM and RS4;11 displayed open chromatin structures predominantly in the upstream region (region I and II) of the NANOG2 gene (Supplementary Figure S7). The existence of these novel upstream promoter regions was further validated by Luciferase reporter gene assays. These experiments demonstrated that both DNA sequences located directly 5′ to exon 1b function as promoter elements after transient transfection into NTERA2 cells (Supplementary Figure S8).
For the first time, the NANOG2 gene was shown to be transcribed in CD34+ cells (Figure 4B). This may indicate that the hematopoietic stem compartment may use the NANOG system to gain stem cell like properties. The same is true for most investigated leukemia samples that predominantly produced NANOG2D transcripts. We verified this finding by performing western blot experiments with biopsy material of acute myeloid leukemia patients (n = 10). We were able to validate NANOG protein expression in 4 out of 10 investigated leukemia patients (Supplementary Figure S9).
We further investigated the functional consequences of expressed NANOG1/2 proteins. For this purpose we used the NANOG1A, NANOG2D and NANNOG2E variants. These three NANOG variants were expressed for 69 days and remaining green fluorescent cells were used for RNA extraction. After substracting against GFP-expressing mock-transfected cells that were maintained in parallel, the specific expression profiles were obtained. Surprisingly, only very few genes were transcriptionally deregulated. Since we expressed the NANOG variants in 293 cells, we did not see an overlap with the known core NANOG network (3,4). However, the genes FOS and EGR1 were transcriptionally activated. EGR1 directly binds to the promoter region of the FOS and CDKN1A/p21 gene (20), and serves as gatekeeper for p53 expression (21). Beside the different functions in ES cells, ectopic NANOG protein expression may exhibit additional and yet unknown functions in non-ES cells that need to be investigated further.
In conclusion, we have identified additional exons reflecting on an extended gene structure of NANOG1 and NANOG2. Moreover, we have revealed complex splice patterns for both NANOG genes that are putatively translated in a large variety of different NANOG1/2 protein variants. We have also demonstrated that the NANOG2 gene is transcribed in CD34+ and different leukemia cells. Western blot experiments performed with t(4;11) cell lines demonstrated that the NANOG2 protein is produced, potentially helping to maintain stem cell functions. We also demonstrated functional equivalence for NANOG1 and NANOG2 protein variants in terms of their downstream target genes. Further studies are ongoing, aiming to understand the molecular consequences of NANOG2 expression in leukemic cells with MLL rearrangements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: BMBF (grant 01GS0875 to R.M.); Center of Excellence Frankfurt on Macromolecular Complexes (CEF-MC) funded by DFG (grant EXC 115 to R.M.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jennifer Merkens and Silvia Bracharz for their technical assistance and Torsten Pietsch for performing the immunohistochemistry experiments with SEM and RS4;11 cells. The authors are thankful to Thomas Burmeister, Lars Bullinger, Gesine Bug and Nicola Gökbuget who provided t(4;11) and AML biopsy samples, respectively.
REFERENCES
- 1.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 2.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 3.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 5.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–333. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udomsakdi C, Eaves CJ, Swolin B, Reid DS, Barnett MJ, Eaves AC. Rapid decline of chronic myeloid leukemic cells in long-term culture due to a defect at the leukemic stem cell level. Proc. Natl Acad. Sci. USA. 1992;89:6192–6196. doi: 10.1073/pnas.89.13.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chumsri S, Matsui W, Burger AM. Therapeutic implications of leukemic stem cell pathways. Clin. Cancer Res. 2007;13:6549–6554. doi: 10.1158/1078-0432.CCR-07-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaussmann A, Wenger T, Eberle I, Bursen A, Bracharz S, Herr I, Dingermann T, Marschalek R. Combined effects of the two reciprocal t(4;11) fusion proteins MLL-AF4 and AF4-MLL confer resistance to apoptosis, cell cycling capacity and growth transformation. Oncogene. 2007;26:3352–3363. doi: 10.1038/sj.onc.1210125. [DOI] [PubMed] [Google Scholar]
- 9.Booth HA, Holland PW. Eleven daughters of NANOG. Genomics. 2004;84:229–238. doi: 10.1016/j.ygeno.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Fairbanks DJ, Maughan PJ. Evolution of the NANOG pseudogene family in the human and chimpanzee genomes. BMC Evol. Biol. 2006;6:12. doi: 10.1186/1471-2148-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ezeh UI, Turek PJ, Reijo RA, Clark AT. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104:2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Wang X, Li M, Han J, Chen B, Wang B, Dai J. NANOGP8 is a retrogene expressed in cancers. FEBS J. 2006;273:1723–1730. doi: 10.1111/j.1742-4658.2006.05186.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim JS, Kim J, Kim BS, Chung HY, Lee YY, Park CS, Lee YS, Lee YH, Chung IY. Identification and functional characterization of an alternative splice variant within the fourth exon of human nanog. Exp. Mol. Med. 2005;37:601–607. doi: 10.1038/emm.2005.73. [DOI] [PubMed] [Google Scholar]
- 14.Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, Ronchini C, Ronzoni S, Muradore I, Monestiroli S, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 17.Hart AH, Hartley L, Parker K, Ibrahim M, Looijenga LH, Pauchnik M, Chow CW, Robb L. The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer. 2005;104:2092–2098. doi: 10.1002/cncr.21435. [DOI] [PubMed] [Google Scholar]
- 18.Hoei-Hansen CE, Almstrup K, Nielsen JE, Brask Sonne S, Graem N, Skakkebaek NE, Leffers H, Rajpert-De Meyts E. Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology. 2005;47:48–56. doi: 10.1111/j.1365-2559.2005.02182.x. [DOI] [PubMed] [Google Scholar]
- 19.Oh JH, Do HJ, Yang HM, Moon SY, Cha KY, Chung HM, Kim JH. Identification of a putative transactivation domain in human Nanog. Exp. Mol. Med. 2005;37:250–254. doi: 10.1038/emm.2005.33. [DOI] [PubMed] [Google Scholar]
- 20.Ragione FD, Cucciolla V, Criniti V, Indaco S, Borriello A, Zappia V. p21Cip1 gene expression is modulated by Egr1: a novel regulatory mechanism involved in the resveratrol antiproliferative effect. J. Biol. Chem. 2003;278:23360–23368. doi: 10.1074/jbc.M300771200. [DOI] [PubMed] [Google Scholar]
- 21.Krones-Herzig A, Adamson E, Mercola D. Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc. Natl Acad. Sci. USA. 2003;100:3233–3238. doi: 10.1073/pnas.2628034100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.