Abstract
HOXA9-mediated up-regulation of miR-155 was noted during an array-based analysis of microRNA expression in Hoxa9−/−bone marrow (BM) cells. HOXA9 induction of miR-155 was confirmed in these samples, as well as in wild-type versus Hoxa9-deficient marrow, using northern analysis and qRT–PCR. Infection of wild-type BM with HOXA9 expressing or GFP+ control virus further confirmed HOXA9-mediated regulation of miR-155. miR-155 expression paralleled Hoxa9 mRNA expression in fractionated BM progenitors, being highest in the stem cell enriched pools. HOXA9 capacity to induce myeloid colony formation was blunted in miR-155-deficient BM cells, indicating that miR-155 is a downstream mediator of HOXA9 function in blood cells. Pu.1, an important regulator of myelopoiesis, was identified as a putative down stream target for miR-155. Although miR-155 was shown to down-regulate the Pu.1 protein, HOXA9 did not appear to modulate Pu.1 expression in murine BM cells.
INTRODUCTION
Hoxa9 is expressed in numerous tissues during development including rib (1), limb (2), motor neuron progenitors (3), reproductive tract (4) and mammary gland (5). Hoxa9 is also expressed in normal adult bone marrow (BM) (6,7) and loss of Hoxa9 leads to multiple relatively mild defects in normal hematopoiesis (8–10). HOXA9 is often up-regulated in acute myeloid leukemias (AMLs; 11,12), and in an analysis of 6817 genes, HOXA9 was the most highly correlated with treatment failure in AML patients (13). Forced expression of HOXA9 in murine BM cells in culture results in immortalization of a myeloid progenitor cell (14,15), whereas transplantation of HOXA9-infected BM cells results in the eventual induction of AML (16). More recently, we have shown that HOXA9 regulates its DNA binding partner Meis1, and that deletion of one Meis1 allele in Hoxa9 deficient mice results in profound Pre/pro-B cell defects (17).
Despite the broad expression of Hoxa9 and other Hox genes, relatively little is known about how the HOX proteins function. An important advance was the discovery that many HOX proteins gain DNA binding specificity by forming complexes with the PBX (18,19), MEIS1 (20) and PREP1 (21) proteins. Although HOXA9 is capable of binding DNA alone (22), it forms cooperative DNA binding complexes with MEIS1 alone (20), as well as in a triple complex with PBX proteins (23,24). Despite these apparent advances and the publication of the HOXA9 transcriptome (25), only a few direct downstream targets for HOXA9 have been confirmed (26–28).
Following the early report that microRNAs (miRs) regulate aspects of hematopoietic lineage commitment (29), several studies have enumerated miR expression in normal BM fractions as well as in various leukemias (30,31). These studies have revealed that miRs play diverse roles in hematopoietic lineage decisions as well as in several types of leukemias. However, the regulation of miR expression in BM remains relatively poorly understood. Since the HOX proteins are thought to function as DNA binding transcription factors, as part of ongoing studies on the mechanism of action of the HOXA9 protein, we used an array analysis to identify downstream miR gene targets in murine BM cells. This analysis showed that HOXA9 up-regulates miR-155. HOXA9-mediated induction of myeloid colonies was impaired in miR-155-deficient mice, suggesting that miR-155 mediates some HOXA9 function(s) in BM cells. Although miR-155 was shown to down-regulate the hematopoietic modulator, Pu.1, HOXA9 did not alter Pu.1 protein levels, indicating that miR-155 functions in other pathways as a downstream mediator of HOXA9 action.
MATERIALS AND METHODS
Mice
Hoxa9-deficient mice have been described previously (10). miR-155-deficient mice were provided by Jackson Laboratories (Bar Harbor, MA, USA) and were re-derived from animals developed by Thai et al. (32).
Retroviral constructs and virus production
MSCV-HOXA9-IRES-EGFP (MIG-HOXA9) and MSCV-HOXA9NS-IRES-EGFP, encoding a DNA binding mutant (MIG-HOXA9NS) retroviral constructs were described previously (28). A hairpin expression construct for miR-155 was constructed by PCR amplification of the murine miR-155 sequence and 150 nt of 5′ and 3′ genomic sequence, followed by cloning into the MDH retroviral vector (29). High-titer retrovirus was obtained by transfecting Phoenix amphotropic packaging cells using Metafectene transfection reagent (Biontex Laboratories, Munich, Germany). Virus-containing medium was harvested at 48 and 72 h after transfection, filtered through a 0.45-µm filter, and used without storing or freezing.
BM cell culture and retroviral transduction
BM cells from wild-type, Hoxa9-deficient mice, or miR-155-deficient animals were harvested 4 days after intraperitoneal injection with 150 mg/kg 5-fluorouracil and cultured in prestimulatory media consisting of Dulbecco’s modified Eagle medium (DMEM) supplemented with 20% heat-inactivated FCS, 100 ng/ml recombinant murine (rm) SCF, 50 ng/ml rm IL-6 and 10 ng/ml rm IL-3 (StemCell Technologies, Vancouver, BC, Canada) for 48 h. Mouse BM cells were infected with retrovirus on two consecutive days by spinoculation in the presence of 4 µg/ml polybrene (Sigma, St Louis, MO, USA).
Analysis of miRs regulated by HOXA9 in murine BM cells
Total RNA from GFP+ Hoxa9−/−BM cells, infected with retrovirus expressing HOXA9-IRES-GFP, HOXA9NS-IRES-GFP or MIG vector alone, was harvested in lysis buffer (Ambion, #1931). Control and test miRs were labeled with Cy5 (Alexa Fluor 647, Invitrogen #A32757; Ambion labeling kit #1562) and hybridized to an Ambion miR array containing 397 unique miRs (Ambion #1566V1), according to the manufacturer's instructions. Microarrays were analyzed using GenePix software. miR analysis was performed in triplicate on two separate sets of infected BM samples. All values are means ± SD. A P < 0.05 denoted the presence of a statistically significant difference. Statistical analyses were performed using a two-tailed Student t-test.
Quantitative real-time RT–PCR and northern blotting analysis
GFP+-sorted BM cells from the different MIG-, MIG-HOXA9- or MIG-HOXA9NS-infected cells were resuspended in lysis buffer (#1931, Ambion). BM progenitors were sorted into long-term hematopoietic stem cells (HSCs), short-term HSCs and early progenitor pools through multichannel sorting (33). Equal amounts of total RNA were reverse transcribed to cDNA using Superscript II polymerase (Invitrogen). All PCR reactions were performed in an ABI Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA) using FAM (Applied Biosystems) as described (7). qRT–PCR was used to measure murine miR-155 or U6 RNA levels by using Applied Biosystems kits (Applied Biosystems). Northern blotting was performed using antisense miR-155 or U6 probes. Statistical analyses were performed using Student’s t-test.
Fluorescence-activated cell sorting analysis
Fluorescence-activated cell sorting (FACS) sorting of primitive hematopoietic pools was performed following the methods developed in the Weissman laboratory (33). For the immunophenotyping of primitive marrow populations, the femurs and tibia of 15-day old mice were flushed with Hanks Balanced Salt Solution (Life Technologies, Bethesda, MD, USA) supplemented with 2% fetal calf serum (FCS) and 2 mM ethylenediaminetetraacetic acid. Then the BM cells were incubated with labeled antibodies for 30 min on ice, washed with cold-PBS twice and analyzed by FACS on Becton Dickinson FACScan using Cellquest software (Becton Dickinson, San Jose, CA, USA). Antibodies used for BM cell staining included phycoerythrin (PE)-conjugated Mac-1 (#553311), PE-Cy7-conjugated-Gr-1 (#552985), PE-conjugated B220 (#553089), fluorescein isothiocyanate (FITC)-conjugated IgM, PE-conjugated CD71 (#553267), PE-Cy7-conjugated Ter119 (#557853), FITC-conjugated CD41 (#553848), PE-conjugated CD61 (#553347), PE-conjugated Sca1 (#553108) and PE-Cy7-conjugated cKit (#558163), all of which were purchased from BD Pharmingen (BD Biosciences, San Diego, CA, USA).
Myeloid colony assays
CFU-myeloid colony assays (CFU-GM) were performed by culturing 2 × 103 BM cells in M3434 medium (StemCell Technologies). Colonies were enumerated 7 days after plating (28).
RESULTS
HOXA9 regulates multiple miRs in BM cells
We previously described the cDNA transcriptome of the HOXA9 protein (25). The initial impetus for this project was to build on these data by describing the possible downstream miR targets regulated by HOXA9. A differential microarray analysis was performed using an Ambion chip containing 397 miR probes with small RNA from Hoxa9-deficient BM cells retrovirally infected with an MSCV vector expressing HOXA9 plus GFP or a GFP vector control. Figure 1A shows the relative expression levels of the HOXA9 and HOXA9-NS proteins expressed from the retroviral vector, compared to the GFP control vector, in Hoxa9-deficient BM cells. HoxA9 significantly up-regulated 30 miRs but only down-regulated five miRs in murine BM cells (Table 1). Interestingly, 22 of the most highly expressed miRs in murine BM were not altered by HOXA9 expression. Among the putative miR targets regulated by HOXA9 identified in this analysis, we chose to focus on miR-155, which was up-regulated 1.56-fold (P = 0.001) in the array analysis, because of the published importance of this microRNA in hematopoiesis (34,35).
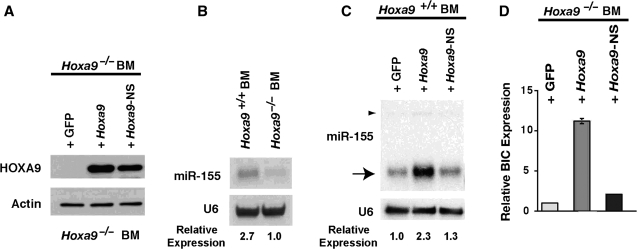
Figure 1.
HOXA9 up-regulates miR-155 and BIC mRNA. (A) Western blot showing HOXA9 or HOXA9-NS protein expressed from a GFP-expressing retroviral vector after infection of Hoxa9−/−BM cells. (B) Northern gel analysis showing that miR-155 was ∼2.7-fold higher in wild-type BM cells compared to Hoxa9-deficient cells. (C) Retroviral-mediated expression of HOXA9 caused a 2.3-fold increase in miR-155, while a DNA binding mutant (HOXA9NS) did not significantly change miR-155 levels. The arrowhead indicates the miR-155 primary hairpin form. (D) The primary hairpin miR-155 is generated from the BIC transcript (34). Forced expression of HOXA9 produced a 10-fold increase in BIC message, while the HOXA9NS mutant did not significantly increase BIC mRNA.
Table 1.
miRs regulated by HOXA9 in BM cells
| miRNA | Expression levels | Fold change | P-value |
|---|---|---|---|
| miRs down-regulated by HoxA9 | |||
| miR-15b | High | 0.77 ± 0.12 | 0.02 |
| miR-23a | High | 0.40 ± 0.06 | 0.01 |
| miR-25 | High | 0.55 ± 0.14 | 0.01 |
| miR-320 | High | 0.78 ± 0.02 | 0.01 |
| miR-342 | High | 0.48 ± 0.07 | 0.01 |
| miRs up-regulated by HoxA9 | |||
| miR-19b | High | 1.53 ± 0.10 | 0.001 |
| miR-106b | High | 1.43 ± 0.38 | 0.03 |
| miR-147 | High | 2.46 ± 0.93 | 0.03 |
| miR-155 | High | 1.56 ± 0.24 | 0.001 |
| miR-191 | High | 1.27 ± 0.18 | 0.014 |
| miR-297 | High | 2.21 ± 0.29 | 0.0002 |
| miR-122a | Medium–high | 2.67 ± 1.60 | 0.04 |
| miR-142-5p | Medium | 3.94 ± 2.30 | 0.05 |
| miR-302c-AS | Medium | 1.67 ± 0.34 | 0.003 |
| miR-124a-mm | Medium–low | 3.45 ± 2.60 | 0.05 |
| miR-142-3p | Medium–low | 3.98 ± 1.80 | 0.04 |
| miR-190 | Medium–low | 3.42 ± 0.11 | 0.01 |
| miR-211 | Medium–low | 3.50 ± 2.01 | 0.01 |
| miR-326 | Medium–low | 3.64 ± 2.10 | 0.03 |
| miR-525 | Medium–low | 5.07 ± 1.20 | 0.0002 |
| miR-34c | Low | 3.68 ± 1.51 | 0.01 |
| miR-98 | Low | 3.31 ± 1.21 | 0.02 |
| miR-181c | Low | 2.78 ± 1.23 | 0.03 |
| miR-200c | Low | 2.21 ± 0.62 | 0.02 |
| miR-301 | Low | 3.25 ± 1.88 | 0.04 |
| miR-383 | Low | 3.85 ± 1.63 | 0.04 |
| miR-422b-mm | Low | 2.24 ± 1.15 | 0.04 |
| miR-518b | Low | 4.64 ± 0.79 | 0.0002 |
| miR-524 | Low | 3.58 ± 1.14 | 0.0004 |
| miR-527 | Low | 3.29 ± 0.18 | 0.003 |
| miR-30e-3p | Very low | 3.81 ± 1.58 | 0.02 |
| miR-143 | Very low | 3.56 ± 2.04 | 0.01 |
| miR-204 | Very low | 4.13 ± 2.14 | 0.01 |
| miR-485-5p | Very low | 2.96 ± 1.25 | 0.05 |
| miR-518f | Very low | 4.98 ± 1.58 | 0.004 |
| Highly expressed miRs in BM, not regulated by HoxA9 | |||
| let-7a–7i | Very high | 0.99 ± 0.25 | 0.79 |
| miR-21 | Very high | 1.03 ± 0.13 | 0.67 |
| miR-93 | Very high | 0.97 ± 0.08 | 0.81 |
| miR-103 | Very high | 0.96 ± 0.45 | 0.71 |
| miR-106 | Very high | 0.96 ± 0.21 | 0.71 |
| miR-107 | Very high | 1.00 ± 0.24 | 0.90 |
| miR-210 | Very high | 1.04 ± 0.24 | 0.88 |
| miR-223 | Very high | 1.03 ± 0.22 | 0.90 |
| miR-494 | Very high | 1.00 ± 0.05 | 0.96 |
| miR-22 | High | 1.03 ± 0.25 | 0.85 |
| miR-23b | High | 0.97 ± 0.20 | 0.77 |
| miR-26a,b | High | 1.00 ± 0.28 | 0.95 |
| miR-29a | High | 1.01 ± 0.30 | 0.79 |
| miR-30a-5p | High | 1.04 ± 0.18 | 0.91 |
| miR-30c | High | 1.03 ± 0.38 | 0.92 |
| miR-31 | High | 1.01 ± 0.35 | 0.87 |
| miR-103b | High | 1.08 ± 0.28 | 0.75 |
| miR-181a | High | 1.06 ± 0.26 | 0.68 |
| miR-185 | High | 1.08 ± 0.34 | 0.79 |
| miR-198 | High | 0.96 ± 0.48 | 0.94 |
| miR-221 | High | 0.99 ± 0.10 | 0.88 |
| miR-513 | High | 1.00 ± 0.45 | 0.84 |
miR-155 is up-regulated by HOXA9
Northern blotting was initially used to confirm the array data for miR-155. Consistent with the array data, miR-155 was 2.7-fold higher in wild-type marrow than in Hoxa9-deficient BM cells (Figure 1B). Retroviral-mediated expression of HOXA9 in wild-type BM cells caused a 2.3-fold increase in miR-155, while a HOXA9-NS DNA binding mutant did not increase miR-155 significantly (Figure 1C). miR-155 is generated from the BIC non-coding transcript (34). In order to gain insight as to whether HOXA9 regulates miR-155 at the transcriptional level, we analyzed the effect of exogenous HOXA9 on BIC expression in BM cells. HOXA9 produced an ∼10-fold increase in BIC mRNA, whereas the HOXA9-NS mutant was only able to minimally increase BIC expression (Figure 1D).
miR-155 expression parallels Hoxa9 mRNA expression in BM progenitors and is highest in the stem cell pools
Because miR-155 has been reported to play a regulatory role in hematopoiesis, we analyzed miR-155 expression in fractionated HSC and progenitor pools. Consistent with both a regulatory role for HOXA9 and a function for miR-155 in hematopoiesis, miR155 expression was much higher in the long- and short-term HSC pools than in the other progenitor pools (Figure 2A). We have recently reported that HOXA9 expression is also highest in the long-term and short term HSC pools (17). Thus, miR-155 expression closely parallels Hoxa9 mRNA expression during early hematopoiesis. It should be noted that the levels of HOXA9 protein are too low to be detected in the small numbers of BM stem cells that can be obtained from FACS sorting.
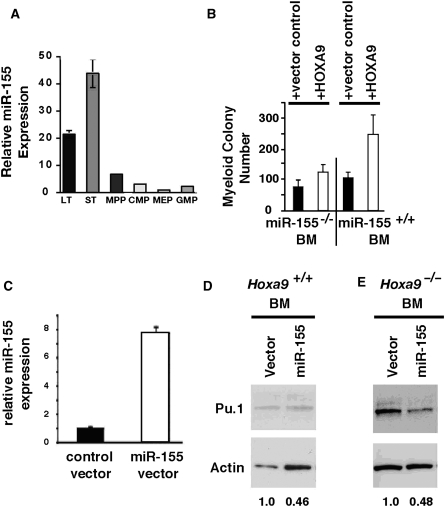
Figure 2.
miR-155 expression is highest in BM stem cell pools, miR-155 is downstream of Hoxa9, and miR-155 down-regulates Pu.1. (A) miR-155 was expressed predominantly within the stem cell-enriched pools in FACS-fractionated BM progenitors. LT, long-term HSC pool; ST, short-term HSC pool; MPP, multipotential progenitor pool; CMP, common myeloid progenitor pool; MEP, myeloid–erythroid progenitor pool; and GMP, granulocyte–monocyte progenitor pool. (B) HOXA9-mediated expansion of clonogenic myeloid progenitors is blunted in miR-155-deficient BM cells. 5-FU treated wild-type or miR-155−/−(C) BM cells were infected with HOXA9-GFP or GFP control retroviral vectors and then plated for myeloid colony forming capacity as described in ‘Materials and methods’ section (n = 4). There was a significant difference (P < 0.018) between HOXA9 induction of myeloid colonies in wild-type BM cells versus miR-155−/−cells. (C) qPCR analysis showing that retroviral-mediated expression of miR-155 in Hoxa9−/−BM cells produced an ∼8-fold increase in miR-155 RNA. (D and E) Retroviral-mediated miR-155 expression in either wild-type or Hoxa9-deficient cells, respectively, resulted in an ∼2-fold decrease in Pu.1 protein, by western analysis.
miR-155 is downstream of Hoxa9 in murine BM cells
To determine whether miR-155 mediates some of the biological properties of HOXA9, we used the fact that infection of murine BM cells with HOXA9 leads to a substantial expansion of myeloid progenitors as measured by colony formation (28). Retroviral-mediated expression of HOXA9 versus MIG control vector in wild-type murine BM compared to miR-155−/−BM was used to explore whether miR-155 was a downstream mediator of Hoxa9 (see Figure 1A for an example of HOXA9 expression generated using this system). HOXA9 induced a 2.6-fold increase in myeloid colony formation in wild-type BM cells (Figure 2B). In contrast, when HOXA9 was expressed in miR-155-deficient BM, there was a statistically significant decrease in induction of myeloid colonies (P < 0.018). These results demonstrate that the stimulatory effect of HOXA9 on colony growth was blunted in miR-155-deficient BM cells, strongly suggesting that at least some of the biological activity of HOXA9 in BM myeloid progenitor cells is mediated by miR-155.
miR-155 down-regulates Pu.1, but HOXA9 does not modulate Pu.1
After showing that HOXA9 up-regulated miR-155, and that miR-155 mediates at least some HOXA9 functionality, we used the available prediction programs to identify Pu.1 as a putative target for miR-155 (36). Pu.1 is an ETS family transcription factor that has been shown to play important regulatory roles in both normal myelopoiesis and leukemogenesis (37). To test the effects of miR-155 on Pu.1 expression, we generated a retroviral vector that expressed a miR-155 hairpin precursor. Although this vector produced an ∼600-fold increase in miR-155 in 293T cells (data not shown), miR-155 expression increased only 8-fold following viral infection of wild-type BM cells (Figure 2C). Infection of either wild-type or Hoxa9-deficient BM cells with this miR-155 expression vector resulted in an ∼50% decrease in Pu.1 protein (Figure 2D and E), a finding that has been reported by others since the inception of this work (38). These findings strongly suggested that HOXA9 might regulate Pu.1 via miR-155, and that this might be a significant window into understanding how the HOXA9 protein influences hematopoiesis. However, despite extensive analysis, we could not convincingly demonstrate a reproducible change in BM Pu.1 protein levels when HOXA9 was infected into either normal or Hoxa9-deficient cells (data not shown). We have concluded from these studies that the modulation of miR-155 by HOXA9 is not reflected by changes in Pu.1 expression.
DISCUSSION
Although the HOXA9 homeodomain protein appears to play important roles in numerous other tissues (1–5), we have focused in this study on its role(s) in normal hematopoiesis (29,39–43) and leukemia (40,44–46). In both blood and other tissues, despite the presence of a DNA binding domain and a presumption of transcriptional regulatory activity, the HOXA9 mechanism(s) of action remain relatively obscure. Although we have previously described the HOXA9 transcriptome (25), relatively few downstream targets have been authenticated. In this regard, we have previously reported that Pim1 is a direct downstream target (28). We also recently reported that HOXA9 can regulate its DNA binding partner Meis1, and that this process may be mediated by CREB1, which appears to be a direct HOXA9 target (17). Because of the dearth of well-understood mechanism to describe HOXA9 function, we undertook the current microarray-based survey of possible miR targets in BM cells. However, given our subsequent findings that the microarray survey data could be only partially confirmed by other methods used to assess miR expression (data not shown), we place only limited emphasis on the survey data. Nevertheless, some general trends were apparent; (i) there was a small class of miRs that were extremely highly expressed in BM, and none of these appeared to be modulated by HOXA9; (ii) the great majority of miRs interrogated were not highly expressed in BM cells; (iii) among the five miRs moderately down-regulated and the 30 moderately up-regulated by HOXA9, most of the changes detected by the relatively crude array strategy were modest.
Given the limitations of the array technology, we chose to focus on miR-155 based primarily on published evidence of a reported role in hematopoiesis and/or leukemogenesis (34–35,38,47–49). One study reported that transgenic mice in which miR-155 is strongly expressed within the B-cell lineage develop lymphomas (48). Another reported that miR-155 was over-expressed in some AMLs and that forced expression of miR-155 in murine BM induced a myeloproliferative disease (35). Since forced Nup98-HOXA9 expression in murine BM also produces a myeloproliferative state (50), while HOXA9 alone appears to immortalize a promyelocytic progenitor (14), the observation that HOXA9 up-regulates miR-155, and that HOXA9 and miR-155 expression was correlated in hematopoietic stem cells, suggested that this process might partially explain HOXA9 mechanism of action. Our data, showing that a DNA binding mutant form of HOXA9 is incapable of up-regulating miR-155, further supports a working model for HOXA9 functioning as a transcription factor to regulate miR155 and subsequent down stream targets. In support of this hypothesis, we have now also shown that miR-155 appears to act as a downstream mediator of HOXA9, since the proliferative effects of HOXA9 on BM progenitors is blunted in miR-155-deficient cells. In an attempt to discover a mechanism by which miR-155 might mediate the action of HOXA9, we used the existing prediction programs to identify Pu.1, an important regulatory protein for myelopoiesis and leukemogenesis (51), as a putative downstream target of miR-155. Our data confirmed this prediction by showing that forced expression of miR-155 does down-regulate Pu.1 protein. While this work was in progress, others also reported that Pu.1 is a direct target of miR-155 (38). Taken together, these data provided a strong working model for HOXA9 mechanism of action in hematopoiesis and leukemogenesis. We were therefore surprised that we were unable to demonstrate a regulatory role for HOXA9 on the Pu.1 protein. We posit that HOXA9 modulation of miR-155 regulates hematopoiesis by additional mechanistic pathways that remain to be described.
FUNDING
Research Service of the Department of Veterans Affairs and by National Institutes of Health (Grant #CA80029). Funding for open access charge: Northern California Institute for Research and Education.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
This article is dedicated in loving memory of Dr C.L., who passed away during the preparation of the article.
REFERENCES
- 1.Chen F, Capecchi MR. Targeted mutations in hoxa-9 and hoxb-9 reveal synergistic interactions. Dev. Biol. 1997;181:186–196. doi: 10.1006/dbio.1996.8440. [DOI] [PubMed] [Google Scholar]
- 2.Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Dierich A, Dolle P, Chambon P. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development. 1996;122:461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- 3.Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- 4.Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat. Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 5.Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc. Natl Acad. Sci. USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 7.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol. Cell. Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izon DJ, Rozenfeld S, Fong ST, Komuves L, Largman C, Lawrence HJ. Loss of function of the homeobox gene Hoxa-9 perturbs early T-cell development and induces apoptosis in primitive thymocytes. Blood. 1998;92:383–393. [PubMed] [Google Scholar]
- 9.Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- 11.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, Zeng C, Baron A, Hunger SP, Varella-Garcia M, et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, Buchberg AM, Largman C. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 13.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 14.Calvo KR, Sykes DB, Pasillas M, Kamps MP. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol. Cell. Biol. 2000;20:3274–3285. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischbach NA, Rozenfeld S, Shen W, Fong S, Chrobak D, Ginzinger D, Kogan SC, Radhakrishnan A, Le Beau MM, Largman C, et al. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 2005;105:1456–1466. doi: 10.1182/blood-2004-04-1583. [DOI] [PubMed] [Google Scholar]
- 16.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu YL, Fong S, Ferrell C, Largman C, Shen WF. HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol. Cell. Biol. 2009;29:5181–5192. doi: 10.1128/MCB.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 19.Mann RS, Affolter M. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 20.Shen WF, Montgomery JC, Rozenfeld S, Lawrence HJ, Buchberg A, Largman C. The Abd-B-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berthelsen J, Zappavigna V, Ferretti E, Malvilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen WF, Rozenfeld S, Lawrence HJ, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J. Biol. Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- 23.Shanmugam K, Green NC, Rambaldi I, Saragovi HU, Featherstone MS. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell. Biol. 1999;19:7577–7588. doi: 10.1128/mcb.19.11.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen WF, Rozenfeld S, Kwong A, Komuves L, Lawrence HJ, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorsam ST, Ferrell CM, Dorsam GP, Derynck MK, Vijapurkar U, Khodabakhsh D, Pau B, Bernstein H, Haqq CM, Largman C, et al. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood. 2004;103:1676–1684. doi: 10.1182/blood-2003-07-2202. [DOI] [PubMed] [Google Scholar]
- 26.Bei L, Lu Y, Eklund EA. HOXA9 activates transcription of the gene encoding gp91Phox during myeloid differentiation. J. Biol. Chem. 2005;280:12359–12370. doi: 10.1074/jbc.M408138200. [DOI] [PubMed] [Google Scholar]
- 27.Bruhl T, Urbich C, Aicher D, Acker-Palmer A, Zeiher AM, Dimmeler S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circ. Res. 2004;94:743–751. doi: 10.1161/01.RES.0000120861.27064.09. [DOI] [PubMed] [Google Scholar]
- 28.Hu YL, Passegue E, Fong S, Largman C, Lawrence HJ. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood. 2007;109:4732–4738. doi: 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 30.Fabbri M, Croce CM, Calin GA. MicroRNAs in the ontogeny of leukemias and lymphomas. Leuk. Lymphoma. 2009;50:160–170. doi: 10.1080/10428190802535114. [DOI] [PubMed] [Google Scholar]
- 31.Yendamuri S, Calin GA. The role of microRNA in human leukemia: a review. Leukemia. 2009;23:1257–1263. doi: 10.1038/leu.2008.382. [DOI] [PubMed] [Google Scholar]
- 32.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 33.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegue E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 34.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 37.Kastner P, Chan S. PU.1: a crucial and versatile player in hematopoiesis and leukemia. Int. J. Biochem. Cell Biol. 2008;40:22–27. doi: 10.1016/j.biocel.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 38.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp. Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr. Opin. Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 41.Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D, Chen Z, Zhang S. MicroRNAs play a role in the development of human hematopoietic stem cells. J. Cell. Biochem. 2008;104:805–817. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- 42.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur. J. Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 43.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, Garate L, Bandres E, Cordeu L, Aparicio O, Saez B, Navarro G, Vilas-Zornoza A, et al. Down-regulation of hsa-miR10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol. Cancer Res. 2008;6:1830–1840. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- 45.Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin. Oncol. 2006;33:167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turner M, Vigorito E. Regulation of B- and T-cell differentiation by a single microRNA. Biochem. Soc.Ttrans. 2008;36:531–533. doi: 10.1042/BST0360531. [DOI] [PubMed] [Google Scholar]
- 48.Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR, et al. Ship and C/EBPβ are targeted by miR155 in B cells of Eμ-miR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheedy FJ, O'Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann. Rheum. Dis. 2008;67(Suppl 3):iii50–iii55. doi: 10.1136/ard.2008.100289. [DOI] [PubMed] [Google Scholar]
- 50.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001;20:350–361. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU.1 (Spi-1) and C/EBPα regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]