Abstract
Eukaryotic cells use numerous pathways to regulate RNA production, localization and stability. Several of these pathways are controlled by ribonucleases. The essential ribonuclease, Dis3, plays important roles in distinct RNA metabolic pathways. Despite much progress in understanding general characteristics of the Dis3 enzyme in vitro and in vivo, much less is known about the contributions of Dis3 domains to its activities, subcellular localization and protein–protein interactions. To address these gaps, we constructed a set of Drosophila melanogaster Dis3 (dDis3) mutants and assessed their enzymatic activity in vitro and their localizations and interactions in S2 tissue culture cells. We show that the dDis3 N-terminus is sufficient for endoribonuclease activity in vitro and that proper N-terminal domain structure is critical for activity of the full-length polypeptide. We find that the dDis3 N-terminus also contributes to its subcellular distribution, and is necessary and sufficient for interactions with core exosome proteins. Finally, dDis3 interaction with dRrp6 and dImportin-α3 is independent of core interactions and occurs though two different regions. Taken together, our data suggest that the dDis3 N-terminus is a dynamic and complex hub for RNA metabolism and exosome interactions.
INTRODUCTION
Ribonucleases (RNases) degrade or process RNA by endonucleolytic or exonucleolytic mechanisms. These activities are vital to cellular function, as reduction of RNase activities can severely impact cell growth, division and viability. Moreover, RNases are evolutionarily conserved in archaea, eubacteria and eukaryotes and hence represent a crucial and fundamental feature of all living cells.
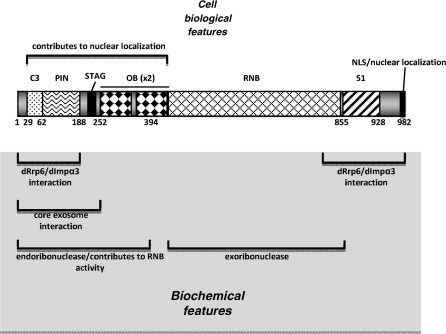
Dis3 is an essential, conserved RNase that possesses 3′ → 5′ exo- and endoribonuclease activites (1,2). A homolog of the eubacterial RNase II/R, Dis3 has been implicated in the processing and degradation of ribosomal RNAs [rRNA (3–8)], messenger RNAs [mRNA (5,9)] and transfer RNAs [tRNA (10,11)]. The major structural difference between RNase II/R and Dis3 is an ∼300 amino acid N-terminal extension that contains multiple bioinformatically identified domains [(2), Figure 1A]. These include a conserved set of three cysteine residues that resemble an iron–sulfur cluster motif [referred to here as C3 (7,12)], a PIN endoribonuclease domain (6–8), and, in the Drosophila melanogaster protein, a motif with homolgy to the cohesin protein STAG (12). Both RNase II/R and Dis3 harbor two N-terminal oligonucleotide-binding (OB) fold domains, an RNB exoribonuclease domain and a C-terminal S1 RNA-binding domain (2). Most Dis3 homologs have a C-terminal extension that contains additional, uncharacterized variant sequences and, in the Drosophila homolog, a nuclear localization sequence [NLS, (12)]. Few of these domains have been unequivocally demonstrated to have functional relevance to Dis3 activity, localization or interactions.
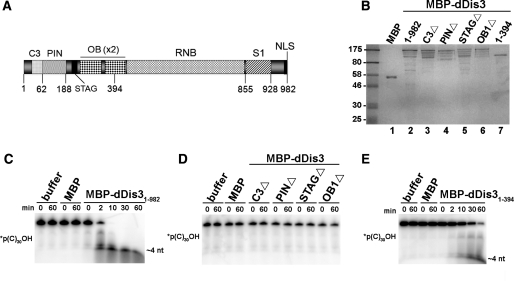
Figure 1.
N-terminal dDis3 domains are necessary and sufficient for in vitro ribonuclease activity. (A) Schematic of full-length D. melanogaster Dis3. (B) Recombinant MBP-Dis3 proteins used in in vitro ribonuclease assays. Dis3 proteins were purified and quantified as described in ‘Materials and Methods’ section. Full-length proteins (200 ng) were loaded onto the gel, as well as pre-stained protein marker (New England BioLabs). Molecular weight standards are labeled on the left side of the gel. (C) MBP-dDis31–982 degrades a 5′-end-labeled polyC RNA substrate. RNA was incubated alone (buffer) or with recombinant protein for the times indicated in each figure. Composition of the RNAs is depicted on the left side of each gel (asterisk represents the position of the label); this notation is used hereafter. The smallest reaction product is marked on the right side of each gel. (D) dDis3 N-terminal internal domain deletion mutants are ribonucleolytically inactive. Expression of these mutants was low, so they were used at 5 nM in this assay. As a comparison, full-length MBP-dDis3 at the same concentration (5 nM) is active (Supplementary Figure S1). (E) MBP-dDis31–394 cleaves polyC RNA. Data shown is representative of at least two independent experiments.
Analyses regarding Dis3 structure and function have been primarily done in yeast cells and/or using recombinant yeast polypeptides. Thus, it is not known if the majority of observed domain functions are conserved in multicellular eukaryotes. Studies of Saccharomyces cerevisiae Dis3 have revealed that mutations in the N-terminal C3 domain impede cell growth, but for unknown reasons (7). Mutations to conserved residues in the PIN or RNB domains result in loss of endoribonuclease or exoribonuclease activities, respectively (5–8,11). These analyses indicate the PIN and RNB domains of yeast Dis3 contain ribonuclease active sites, although it is not known how other domains in the protein contribute to these activities. Consistent with these observations, the RNB domains of Drosophila Dis3 and bacterial homologs RNase II/R alone contain exoribonuclease activity, thus Dis3 exoribonuclease activity is conserved (13–15). However, it is not known if PIN endoribonuclease activity is conserved in Dis3 homologs. Finally, N-terminal domains appear to be important for localization of Drosophila Dis3 (12), but the relationship of the Dis3 N-terminus to its subcellular distribution is largely unknown.
Although Dis3 has been shown to function independently in vitro, it was initially co-purified in a multiprotein complex (where it was termed Rrp44) of exoribonucleases called the exosome (3). The original exosome core, thought to assemble and function in the nucleus and cytoplasm, contains RNase PH subunits Rrp41/Ski6, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3, and S1 domain subunits Rrp4, Rrp40 and Csl4 (16,17). In S. cerevisiae, the N-terminal PIN domain of Dis3 is responsible for interactions with these proteins (8). N-terminal domains also appear to be important for Drosophila Dis3 interactions with the exosome (12), although the exact interacting domain has not been identified.
The exosome core associates with additional proteins, including the RNase D homolog Rrp6, and the nuclear exosome cofactor Rrp47/Lrp1. However, Rrp6 and Rrp47 function independently of the exosome core as well (18–20). In this regard, there is significant biochemical evidence that exosome polypeptides assemble into multiple distinct complexes, several of which lack many subunits, including Dis3 (21,22). Dis3 itself is found in complexes independent of the exosome (23–26). The exact number of Dis3 complexes, the site(s) of their assembly and disassembly, and the domains of Dis3 that mediate specific protein–protein interactions remain largely unknown.
In this work, we focus on the contributions of D. melanogaster Dis3 N-terminal domains to functions in vitro and in vivo. First, we examine if the N-terminal endoribonuclease activity reported for S. cerevisiae Dis3 is conserved in the Drosophila Dis3 enzyme. We also use truncation and point mutants to explore the contributions of the dDis3 N-terminus to its subcellular distribution, and interactions with core exosome subunits, dRrp6 and dImportin-α3. Our study reveals novel features of the dDis3 N-terminus that are functionally relevant and hence of general and broad importance to exosome-mediated RNA metabolic pathways and mechanisms.
MATERIALS AND METHODS
Molecular cloning
All plasmids were constructed using basic molecular cloning techniques. All constructs were screened by digestion with restriction endonucleases and sequenced to confirm the absence of errors. The following description details how constructs were specifically generated. dDis3 fragments used in the cell-based analysis were PCR amplified from the full-length ORF using primers shown in Supplementary Table S1. The 5′ primer has a unique BglII site and the 3′ primer has an in-frame FLAG (DYKDDDDK) tag followed by a stop codon and a unique SalI site. This PCR product was digested with BglII and SalI and cloned into the BamHI and SalI sites of pRmHa3 to obtain metallothionein (Mtn) promoter driven dDis3 gene fragments. These constructs were transiently transfected into S2 cells using CELLFectin (Invitrogen), tested for copper-inducible expression, and then established as stable cell lines as described previously (27,28). Mutant dDis3 genes were PCR amplified from DNA templates as previously described (12). For in vitro experiments, wild-type or mutant dDis3 genes were cloned into pMAL-c2 to create maltose binding protein (MBP) fusions.
Expression and purification of recombinant proteins
MBP, full-length MBP-dDis3, or mutant MBP-dDis3 constructs were transformed into Escherichia coli. Expression of recombinant proteins was induced with 1 mM IPTG (Denville Scientific) in 500 ml cultures, either overnight at 20°C (MBP, MBP-dDis31–982, MBP-dDis31–394), or for 2 h at 37°C (MBP-dDis3C3Δ, MBP-dDis3PINΔ, MBP-dDis3STAGΔ, MBP-dDis3OB1Δ). Cells were resuspended in buffer containing 1 mg/ml lysozyme (Sigma), 20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1× protease inhibitor cocktail (Roche), 0.1 mM PMSF (Sigma) and 1 mM EDTA (domain deletion mutants). Sonication was used to lyse the cells. Cells harboring MBP-dDis31–394 were further lysed in 1% Triton. Recombinant proteins were purified over amylose resin (New England BioLabs). Columns were washed with 80 ml of wash buffer (20 mM Tris–HCl, pH 7.5, 100 mM NaCl, and for some full-length aliquots and the domain deletion mutants, 1 mM EDTA). Recombinant proteins were eluted from the column with buffer containing 20 mM Tris–HCl, pH 7.5, 100 mM NaCl and 50 mM maltose. Eluants were dialyzed into buffer containing 20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10% glycerol, and for domain deletion mutants, 1 mM EDTA. All pure protein preparations were run out on 12% SDS–polyacrylamide gels, and visualized by Coomassie staining. QuantityOne software was used to determine protein concentrations based upon comparisons to a BSA standard curve.
Preparation of RNA substrates
RNA substrates utilized in the ribonuclease assays included polyA (30 nt, Dharmacon), polyC (30 nt, Dharmacon), polyU (32 nt) and an RNA containing all four nucleotides, polyN (31 nt; 5′GCGUCUUUACGGUGCUUAAAACAAAACAAAA3′). T4 polynucleotide kinase (New England BioLabs) was used to label RNAs at the 5′-end with γ32ATP (Perkin-Elmer). Unincorporated radiolabeled nucleotides were removed with NucAwayTM spin columns per manufacturer’s recommendations (Ambion). Circular RNA substrates were generated by incubating 5′-end-labeled RNAs with T4 RNA ligase 1 (New England BioLabs) for 15 min at 37°C. Ligation was terminated by boiling the reactions for 2 min. For 3′ end-labeling, 3′CMP was first incubated with γ32ATP (Perkin-Elmer) and T4 polynucleotide kinase (New England BioLabs) to generate radiolabeled pCp. RNAs and [32P]pCp were then incubated with T4 RNA ligase 1 (New England BioLabs) for 5 h at room temperature to generate 3′-end-labeled RNAs. Unincorporated nucleotides were removed by NucAwayTM spin columns (Ambion).
Ribonuclease assays
RNase assays were adapted from previously published protocols (5–7). Labeled RNAs were incubated alone or with recombinant proteins at 37°C in buffer containing 10 mM Tris–HCl, pH 8.0, 75 mM KCl, 1 mM 2-mercaptoethanol and 40 µM MgCl2 for experiments in Figure 1 and Supplementary Figure S2. Assay buffer for experiments in Figures 2 and 3 contained 10 mM Tris–HCl, pH 8.0, 75 mM KCl, 1 mM 2-mercaptoethanol and 40 μM MnCl2. RNA concentrations were 120 nM for experiments in Figures 1 and 3, Supplementary Figures S1 and S2, and 20 nM for experiments in Figure 2. For all assays except those in Supplementary Figure S1, the concentration of full-length dDis3 and the 1–394 mutant was 60 nM. The concentration of domain deletion mutants was 5 nM. For time courses, a 10 µl aliquot of each reaction was removed at the designated time points and stopped with one reaction volume of formamide loading buffer. Reaction products were separated on 12.5%, 8 M urea denaturing polyacrylamide gels and visualized by autoradiography.
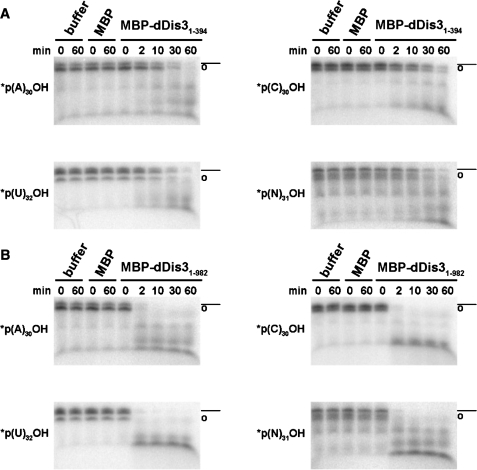
Figure 2.
The dDis3 N-terminus has endoribonuclease activity. MBP-dDis31–394 (A) and MBP-dDis31–982 (B) cleave circularized polyA, polyC, polyU and polyN RNA substrates. Some linear RNA is present due to inefficiency of the ligation reaction during preparation of circular substrates. Full-length linear RNA is marked as (dashed line) and RNA circles as (open circle). These symbols are used throughout. Data shown is representative of at least two independent experiments.
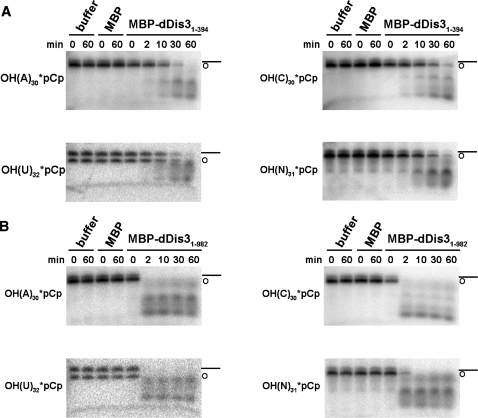
Figure 3.
dDis3 cleavage of 3′-end-labeled RNAs confirms endoribonuclease activity. MBP-dDis31–394 (A) and MBP-dDis31–982 (B) cleave 3′-end-labeled polyA, polyC, polyU and polyN RNA substrates. Circular RNAs were a by-product of the 3′-end-labeling procedure. Note that reaction products for both proteins are RNA fragments. Data shown is representative of at least two independent experiments.
Cell culture
Drosophila melanogaster S2 cell culture, including transient transfections and stable cell lines were established and maintained as previously described (12,20,27).
Immunochemistry
All immunoprecipitations and western blotting with anti-FLAG and anti-dDis3, -dRrp6, -dImportin-α3 and exosome antibodies were performed as previously described (12, 20, 27). Immunoprecipitation (IP) experiments were performed with RNase A and/or ethidium bromide to confirm that interactions were independent of nucleic acids. Indirect immunofluorescence experiments were also performed as detailed previously (27). Images of all dDis3 proteins were captured within 1 s exposure times, using a Zeiss Axioplan 2 microscope with a Hamamatsu Digital CCD camera.
RESULTS
The dDis3 N-terminus harbors an endoribonuclease activity
To examine the contributions of N-terminal domains to dDis3 RNase activity, we first purified full-length and domain mutant recombinant proteins as MBP-dDis3 fusions from E. coli (Figure 1B). We then assessed activity of the purified recombinant proteins using an in vitro system that we previously developed (13). Within 10 min, full-length MBP-dDis3 (MBP-dDis31–982) completely degraded a polycytidine (polyC) RNA (Figure 1C) as well as RNAs composed of polyadenine (polyA), polyuridine (polyU) and an RNA containing all four nucleotides (polyN; Supplementary Figures S1 and S2A). Conversely, proteins lacking the C3, PIN, STAG or OB1 domains displayed no activity within the duration of the time course (Figure 1D). Moreover, this effect was not substrate-specific, as these mutants did not degrade polyA, polyU, or polyN (Supplementary Figures S1 and S2B). As a control, MBP itself did not degrade any RNAs in any assay. This data suggests that N-terminal domains are important for maintaining the ribonuclease activity of full-length dDis3.
Recent reports have suggested that the N-terminus of Dis3 is important for enzymatic function because it contains an active site for endoribonuclease activity (6–8,29). To test directly if the dDis3 N-terminus alone harbors any ribonuclease activity, we purified a recombinant polypeptide in which MBP is fused to the first 394 amino acids of dDis3 (Figure 1B), and tested its activity. MBP-dDis31–394 degraded the polyC substrate (Figure 1E) as well as polyA, polyU and polyN (Supplementary Figure S2C), albeit much slower than the full-length protein (cf. Figures 1C and 1E). Consistent with this, an N-terminal fragment of S. cerevisiae Dis3 has RNase activity in vitro (6,7). These assays show that the dDis3 N-terminus contains an independent RNase activity.
To determine the type of RNase activity harbored by the N-terminus, we engineered substrates for endonucleolytic cleavage: circular polyA, polyC, polyU and polyN RNAs (marked with open circle symbols, Figure 2). As a control, RNAs marked as circles were not able to be dephosphorylated by calf intestine alkaline phosphatase, nor cleaved by the 5′→3′ exoribonuclease Xrn1, or by the 3′→5′ exoribonuclease RNase T (data not shown). In contrast, MBP-dDis31–394 cleaved each of the circular substrates (Figure 2A), suggesting that the dDis3 N-terminal activity is endoribonucleolytic. Similar to S. cerevisiae Dis3 (6,30), the ability to cleave circular substrates was retained in full-length dDis3 (Figure 2B). The full-length enzyme also cleaved the circular substrates more efficiently than the N-terminus alone.
Finally, we examined the activity of dDis3 on RNAs that were radioactively labeled at the 3′-end, under ionic conditions suggested to promote endoribonuclease activity (6–8). Specifically, manganese, and not magnesium, was added to the reaction buffers. Here, MBP-dDis31–394 cleavage of the RNAs produced multiple RNA fragments, which is again suggestive of endoribonuclease activity (Figure 3A). For full-length MBP-dDis3, we anticipated that we may not observe accumulation of product fragments. Rather, if the full-length enzyme utilized its RNB domain mediated 3′→5′ exoribonuclease activity to degrade the 3′-end-labeled substrates, we would expect accumulation of mononucleotides that would not be visible on our gels. Interestingly, full-length dDis3 cleavage of these RNAs also resulted in accumulation of RNA products of various sizes (Figure 3B), suggesting that full-length dDis3 uses endonuclease activity to cleave 3′-end-labeled RNAs in these assay conditions. Together, these data demonstrate that the N-terminus of dDis3 contains an independent endoribonucleolytic active site that is also functional in the context of the full-length enzyme.
Mutational analyses reveal N-terminal domain requirements for dDis3 localization
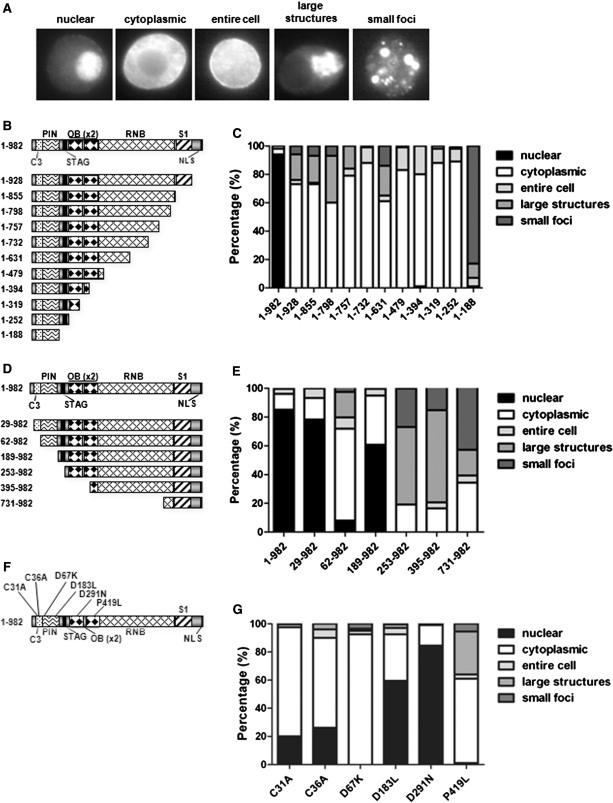
To determine the physiological relevance of dDis3 N-terminal domains, we analyzed the subcellular distribution and protein–protein interactions of multiple FLAG-tagged dDis3 mutant proteins. For cytological analyses, cells were scored for immunofluorescence staining predominantly in the nucleus, cytoplasm or throughout the entire cell. We also observed two minor types of staining that were classified as large structures or small foci, both of which appear in the cytoplasm (Figure 4A). As a control, we first examined the subcellular distribution of a set of C-terminally truncated polypeptides. These mutants, schematized in Figure 4B, were designed to remove the NLS as well as potentially functional domains as identified by the bioinformatic, structural and biochemical data from several Dis3/RNase homologs (12,14). Localization patterns of each mutant were quantified and are presented as graphs (Figures 4C) and fluorescence images (Supplementary Figure S3A). Consistent with previous reports (12), full-length dDis3 (dDis31–982) was predominantly nuclear (>90%). Conversely, all C-terminally truncated dDis3 mutants were predominantly cytoplasmic (Figure 4C). The dDis31–188 mutant was unique in that it accumulated within small cytoplasmic foci in ∼80% of cells. In the other ∼20%, dDis31–188 was found in large cytoplasmic structures. Together, these data confirm that loss of the dDis3 C-terminal NLS ablates nuclear accumulation of the protein.
Figure 4.
The N-terminus of dDis3 contributes to its subcellular distribution in Drosophila S2 cells. (A) Images are examples of possible major localization patterns of dDis3 mutant polypeptides. (B) Schematic of dDis3 C-terminally truncated mutants utilized as controls in this study. (C) dDis3 mutants lacking an NLS are predominantly cytoplasmic. Graph represents quantification of the distributions of full-length and mutant dDis3 polypeptides in S2 cells. For simplicity, localization patterns were grouped into the five categories listed (the nuclear category represents staining in the nucleus alone; cytoplasmic is general staining throughout the cytoplasm or at the plasma membrane; entire cell is staining in the nucleus and cytoplasm; large structures is staining in distinct large structures alone or staining in large structures and diffuse staining throughout the cytoplasm; small foci is staining in distinct foci alone, or staining in foci and diffuse staining throughout the cytoplasm). This method was used for all additional graphs. The ‘normal’ dDis3 distribution pattern is represented by the dDis31–982 bar, where full-length dDis3 is >90% nuclear. (D) Schematic of dDis3 N-terminally truncated mutants. (E) N-terminally truncated dDis3 mutants, despite harboring an NLS, are predominantly cytoplasmic. (F) Schematic of dDis3 N-terminal point mutants. (G) Point mutations to the N-terminus of dDis3 perturb its normal subcellular distribution pattern. Data shown was collected and averaged from two (point mutants) or three (truncations) independent experiments where at least 88 expressing cells were counted. Error bars are excluded for clarity. Relative expression levels of mutant constructs can be seen in Figure 5 and Supplementary Figure S4.
To expand upon this observation and improve our understanding of dDis3 domain contributions to nucleocytoplasmic distribution, we constructed a set of dDis3 N-terminal truncations (Figure 4D). All of these truncation mutants retain the C-terminal NLS and hence are predicted to be nuclear. In Figure 4E, we present a bar-graphical depiction of the localization of the mutants; cell images are in Supplementary Figure S3B. We observed no significant difference in the localization of dDis329–982 as compared to full-length dDis3. In contrast, a polypeptide lacking the C3 domain (dDis362–982) was predominantly (60%) cytoplasmic and <10% of cells had nuclear staining. When the first 188 amino acids were removed (dDis3189–982), we observed an unexpected reversal of localization, with 60% of the protein being found in the nucleus. The remaining constructs (dDis3253–982, dDis3395–982 and dDis3731–982) accumulated mainly in cytoplasmic foci and large structures. Thus, in addition to the C-terminal NLS, N-terminal domains appear to contribute to dDis3 subcellular localization.
To further investigate dDis3 localization, we engineered point mutations in dDis3 N-terminal domains and assessed their effects on subcellular distribution patterns (Figure 4F and G). We mutated two cysteine residues in the C3 domain (C31A, C36A), two active site aspartates in the PIN domain (D67K, D183L), a conserved residue in the OB1 domain (D291N); and a mutant that recapitulates the S. cerevisiae P463L mutation in the OB2 domain, described previously [(4), P419L]. Despite the presence of the NLS in all of these proteins, both the C31A and C36A mutants were only ∼20–30% nuclear. Disruption of the first active site residue in the PIN domain, D67K, elicited complete loss of nuclear localization. In contrast, nuclear accumulation of D183L (∼60%) and D291N (∼80%) mutants was comparable to wild-type levels. Finally, we observed complete loss of nuclear staining with the P419L mutant. These data show that specific changes to individual amino acids in the dDis3 N-terminus render the polypeptide incapable of maintaining its normal nucleocytoplasmic distribution pattern. Given the complex localization patterns of these mutants, it is possible that multiple, distinct N-terminal regions of dDis3 help regulate the nuclear targeting and/or retention of this enzyme in the cell.
dDis3 N-terminal domains are required for interactions with core exosome proteins and exosome co-factors
To ascertain the contributions of dDis3 domains to its protein–protein interactions, we used FLAG immunoprecipitation to recover full-length and mutant dDis3 polypeptides from whole cell extracts. We specifically examined the requirements for dDis3 interactions with core exosome proteins, exosome co-factors and the nuclear import protein dImportin-α3, all proteins previously shown to interact with dDis3 (12). The full-length dDis3 protein (dDis31–982) served as a positive control, and was able to co-precipitate core exosome proteins dRrp42, dRrp45, dRrp41, dRrp4, dRrp46, dCsl4 and dRrp40. dDis31–982 also co-precipitated the nuclear exosome cofactor dRrp47, as well as dRrp6 and dImportin-α3 (Figure 5A, lane 14; Figure 5B, lane 9). The negative control, beads incubated with extract from vector-harboring S2 cells, showed little or no background binding using any of these proteins (Figure 5A, lane 26; Figure 5B, lane 16). This confirms prior observations that Dis3 not only interacts with core exosome proteins and exosome co-factors, but also interacts with proteins involved in nucleocytoplasmic transport (3,4,12,23,24).
Figure 5.
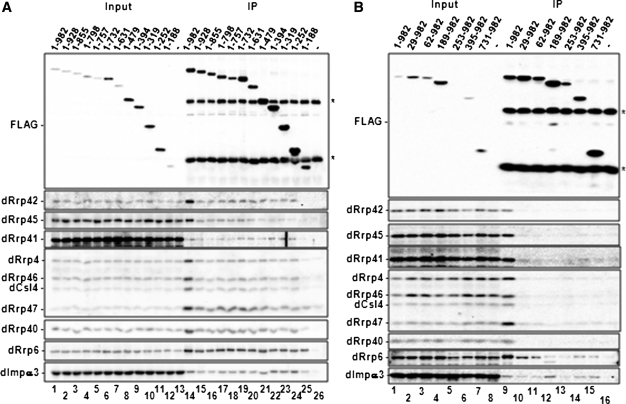
The dDis3 N-terminus is required for interactions with core exosome proteins. (A) dDis3 amino acids 1–252 contain the core exosome interacting region. Western blot analysis of dDis3 co-immunoprecipitation experiments are presented. Input, 2.5%;IP, 5%. Asterisks represent the heavy and light chains from the α-FLAG resin used to immunoprecipitate dDis3-FLAG constructs. Note that amino acids 1–188 retain interactions with dRrp47, dRrp6 and dImportin-α3, but not core proteins. (B) The dDis3 C-terminus interacts with dRrp6 and dImportin-α3, independently of core exosome proteins. Here, dDis3 amino acids 731–982 only co-precipitate dRrp6 and dImportin-α3. Schematics of all truncated polypeptides are shown in Figure 4.
The majority of dDis3 C-terminally truncated polypeptides (dDis31–928, dDis31–855, dDis31–798, dDis31–757, dDis31–732, dDis31–631, dDis31–479, dDis31–394, dDis31–319 and dDis31–252) also co-precipitated core exosome proteins, dRrp6, dRrp47 and dImportin-α3 (Figure 5A, lanes 15–24). However, the 1–188 fragment of dDis3 showed reduced binding to dRrp40, was severely compromised and/or deficient in co-precipitating dRrp45, dRrp41, dRrp4, dRrp46 and dCsl4, and did not co-precipitate dRrp42 (Figure 5A, lane 25). Despite this loss or reduction in core exosome binding, dDis31–188 still co-immunoprecipitated with dRrp6 and dRrp47. dImportin-α3 binding was observed as well, but modestly reduced. None of the C-terminally truncated mutants co-precipitate endogenous dDis3 (Supplementary Figure S4A). Based on these observations, the first 252 amino acids of the dDis3 N-terminus interacts with the exosome core, and the first 188 amino acids interact with dRrp6, dRrp47 and dImportin-α3.
Co-immunoprecipitation experiments with N-terminally truncated dDis3 mutants revealed that removal of the first 29 amino acids alone was sufficient to ablate dDis3 interaction with core subunits and dRrp47 (Figure 5B, lane 10). All additional N-terminal truncations (dDis362–982, dDis3189–982, dDis3253–982, dDis3395–982 and dDis3731–982) were unable to co-precipitate these proteins above background binding, confirming that the dDis3 N-terminus is required for these interactions (Figure 5B, cf. lanes 11–15 with lane 16). These mutants also did not recover endogenous dDis3 (Supplementary Figure S4B).
By comparison, all N-terminally truncated dDis3 polypeptides recovered dRrp6 and dImportin-α3, consistent with previous data supporting an interaction between the dDis3 C-terminus and these proteins (12). dDis3253–982 recovered qualitatively less dRrp6 and dImportin-α3, likely a consequence of its low level of expression (Figure 5B, FLAG panel, lanes 5 and 13). Thus, our data show that dRrp6 and dImportin-α3 interact with dDis3 N- and C-termini. Further, these observations suggest that dRrp6 and dImportin-α3 interactions with dDis3 are independent, as these proteins, but not core exosome proteins, bind to the dDis3 C-terminus.
DISCUSSION
In this work, we have performed a structure–function analysis of D. melanogaster Dis3. We show that N-terminal dDis3 protein domains contribute to its enzymatic activities, subcellular compartmentalization and interactions with the exosome core, dRrp6 and dImportin-α3 (Figure 6). Thus, our study confirms and extends upon data regarding yeast Dis3, which harbors two ribonuclease active sites, is proposed to possess the major activities associated with the exosome, and is involved in multiple RNA metabolic pathways (3–11,30–33). These findings help build a framework for understanding the conserved roles of Dis3 in RNA metabolism. Further, as Dis3 is a conserved endo- and exoribonuclease, these studies help us gain a better understanding of how RNases function in general.
Figure 6.
dDis3 functional regions identified in this work. (Top) dDis3 cell biological features include one N-terminal region that contributes to subcellular localization. The NLS was previously identified (12). (Bottom) dDis3 biochemical features include an N-terminal endoribonucleolytic active site and N-terminal domains that contribute to enzymatic activity overall. The N-terminus also interacts with core exosome proteins, dRrp47, dRrp6 and dImportin-α3. The dDis3 C-terminus contains a region for dRrp6 and Importin-α3 interactions as well. The exoribonuclease activity was previously identified (13).
Dis3 N-terminal endoribonuclese activity is conserved in metazoans
We show for the first time that the N-terminus of D. melanogaster Dis3 has an independent RNase activity. As the dDis3 N-terminus can cleave circular RNA substrates, we conclude that this activity is endoribonucleolytic. Although the dDis3 N-terminus itself has activity, it is less efficient at cleaving RNAs than the full-length enzyme. This is in contrast to studies of the S. cerevisiae Dis3, as its N-terminus alone typically has very robust endoribonuclease activity (6,7). The difference may lie in the composition of the N-terminal fragment we use here compared to those used in the yeast studies. Where our construct includes the C3, PIN and OB1 domains, the yeast constructs lack OB1. Interestingly, it has been suggested that the OB1 domain may regulate PIN activity (7). This is consistent with activity we see for dDis31–394; OB1 may reduce endoribonuclease activity.
In our in vitro study, we also found that dDis3 cleaved all circular RNAs tested. Thus, dDis3 endoribonuclease activity, like its exoribonuclease activity (13), is not sequence-specific in vitro. In contrast, it is not known whether Dis3 endoribonuclease activity is substrate specific in vivo. To date, S. cerevisiae Dis3 endoribonuclease activity has only been linked to ribosomal RNA processing (6–8); other functions have not been tested. However, PIN endoribonuclease activities are not specific to rRNA processing pathways in multicellular eukaryotes. For example, the PIN domains of fly and human SMG6 have been linked to nonsense-mediated mRNA decay functions (34,35). Thus, it is possible that PIN-mediated endoribonuclease activity is a conserved mechanism utilized in the turnover and processing of different classes of RNAs.
Our in vitro analyses have shown that the dDis3 N-terminus not only possesses endoribonuclease activity, but N-terminal domains are also important for exoribonuclease activity in the full-length enzyme. Deletion of any N-terminal domain resulted in a complete loss of activity of full-length dDis3. It is possible that N-terminal domains are needed to maintain the stability of the protein, and loss of these domains results in an unstable, inactive protein, that would be rapidly turned over in the cell. However, this is unlikely, as proteins lacking these domains, when overexpressed in Drosophila S2 cells, are visible by western blotting [(12), Figure 5B], and hence are stable in vivo. Further, these particular domain deletion mutants still bind dRrp6 (12), so they have some normal protein–protein interactions. A more plausible explanation is that N-terminal domains are necessary structural elements that maintain the ribonucleolytically active conformation of the wild-type protein. When these domains are deleted, it is possible that remaining N-terminal sequences take on a dominant effect on the ribonuclease active sites, causing them to mis-fold or be blocked to substrate entry. Consistent with this, we have previously observed that recombinant MBP-dDis3 mutants truncated at the N-terminus (mutants 29–982, 62–982 and 189–982) retain RNB domain-mediated exoribonuclease activity (13). Similarly, S. cerevisiae Dis3 mutants lacking the first 241 amino acids are active in vitro (32). Thus, N-terminal domains appear to play multiple roles related to dDis3 enzymatic activity.
The dDis3 N-terminus is important for subcellular localization
We have shown that dDis3 subcellular localization is a consequence of a sensitive balance between N- and C-terminal sequences. In this regard, dDis3 N-terminal mutants containing the C-terminal NLS are not nuclear. This suggests several possibilities for the function of N-terminal domains in dDis3 subcellular localization. First, N-terminal domains could maintain the proper structure of the enzyme, such that the NLS is in a functional conformation. Consistent with this idea, N-terminal domains are also necessary, probably at the structural level, for RNase activity of the full-length enzyme. Alternatively, the N-terminus could contain an additional signaling or regulatory sequence that directs dDis3 localization, in conjuction with the NLS, to various subcellular compartments. Perhaps dDis3 shuttles in and out of the nucleus in an effort to degrade distinct classes of RNAs. S. cerevisiae Dis3 has been shown to participate in both the processing of ribosomal RNAs in the nucleus (3–8), as well as the degradation of mRNAs in the cytoplasm (5). It is unknown whether distinct pools of Dis3 proteins degrade these targets in each compartment, or if a single, shuttling pool of Dis3 proteins is responsible for the processing and/or turnover of both targets.
The presence of one N-terminal localization signal does not explain all of the results we observe with the N-terminal mutants. For example, mutations to either the C3 or OB2 domains disrupt proper subcellular distribution of dDis3. However, mutations to the PIN and OB1 domains have little effect on normal dDis3 localization. Based on these observations, it is possible that the dDis3 N-terminus contains multiple localization signaling sequences which may or may not be subject to several levels of regulation. It will be important to decipher the specific mechanisms by which dDis3 localization is directed in the cell in order to understand how and when Dis3 can function in different cellular locations.
The dDis3 N-terminus is responsible for interactions with core exosome proteins and exosome co-factors
Our interaction studies are consistent with work demonstrating that Dis3 N-terminal domains are responsible for core exosome interactions (8,12). We found that the interaction between the core and dDis3 is reduced to amino acids 1–252. Moreover, dDis329–982 does not interact with the core, indicating that the extreme N-terminal 28 amino acids are required for these interactions. Together, these data show that dDis3-core exosome interactions occur through a dDis3 region containing the C3, PIN and STAG domains. In contrast, the PIN domain alone is sufficient for S. cerevisiae Dis3 interactions with core exosome proteins (8). This could point to organismal differences in how Dis3 interacts with core proteins. However, the full-length S. cerevisiae Dis3-core interaction is more substantial than S. cerevisiae PIN-core interactions (8). It is reasonable that the domains surrounding PIN lend stability to the interactions in both organisms. Consistent with this, structural analysis of a S. cerevisiae Dis3 sub-complex containing Rrp41 and Rrp45 shows that regions of Dis3 outside of the PIN domain contact the core proteins (30). Although the interacting region of Dis3 differs slightly depending on the organism, it is clear from our studies and others that the N-terminus is important for Dis3-core exosome interactions.
There are several other noteworthy observations gained from the immunoprecipitation data. First, dRrp40, dRrp47, dRrp6 and dImportin-α3 continue to interact with dDis31–188 despite the loss of core exosome binding. This could suggest that these proteins associate with dDis3 in a complex that is independent of the remaining subunits. Formation of this complex would be consistent with previously observed interactions between S. cerevisiae Rrp6 and Rrp47 (18,36). Notably, we also show that amino acids 731–982 of dDis3 interact with dRrp6 and dImportin-α3 (Figure 6). This occurs independently of any interactions with core exosome proteins and dRrp47. Thus, dRrp6 itself does not elicit indirect binding of the core proteins when it binds to either the dDis3 C-terminus, or the N-terminal region 1–188. This suggests that when core exosome proteins do bind dDis3, the interaction is direct, even in the presence of dRrp6. Direct interactions have been observed in the crystal structure of the S. cerevisiae Dis3–Rrp41–Rrp45 sub-complex (30). We speculate that these interaction profiles reflect different dDis3 complexes and/or different assemblages of polypeptides on distinct portions of the full-length protein. Moreover, this promotes a model that exosome subunits assemble into protein complexes that are independent of the core [(12,19,20,27); Kiss and Andrulis, in press].
Do dDis3 N-terminal domains link three different functions?
Our biochemical findings place additional importance on the roles of the Dis3 N-terminus. An examination of the schematic in Figure 6 shows that the N-terminus is a hub for endoribonuclease activity and interactions with core exosome proteins, dRrp6, dRrp47 and dImportin-α3. Additionally, the N-terminus is important for localization. As all of these functions are located at the N-terminus, it is possible that they are linked, providing a unique way to regulate dDis3 functions. For example, dDis3 ribonuclease activities could be regulated via interactions with core exosome proteins or exosome co-factors. In S. cerevisiae, it has already been shown that Dis3 interaction with core exosome proteins results in an increase or reduction in its ribonuclease activity depending on the substrate (5,30,31). Differential regulation of exo- and/or endoribonuclease activity could also occur with dependence on subcellular localization.
It is also possible that the N-terminus simply evolved in Dis3 proteins to mediate additional functions, like endoribonuclease activity, not possessed by its bacterial homologs. However, these N-terminal activities do not have to be interdependent. Comparison of our in vivo data sets suggests that proper dDis3 localization does not necessarily depend on interactions with core exosome proteins. For example, dDis329–982, which does not interact with the core exosome, is predominantly nuclear, consistent with the wild-type localization. Conversely, dDis31–252, which does interact with core exosome proteins, is predominantly cytoplasmic, and hence does not retain its normal subcellular distribution pattern. Further analysis will be required to determine exactly how the many functions of Dis3 are linked.
In sum, this study facilitates a broader advancement in aspects of RNase II/R enzymology, core exosome function and general RNA metabolic pathways. The goals now are to understand the mechanisms behind Dis3 complex assembly, disassembly, targeting, localization and substrate specificity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM072820 to E.D.A., T32HD007104 to M.M.). Funding for open access charge: National Institutes of Health R01.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephanie Davis for the initial characterization of the dDis3 truncations; Peter Harte, Alan Tartakoff, Edward Turk and members of the Andrulis lab for reviewing the manuscript; Amy Graham for technical support; Piet de Boer for microscope use; and Eckhard Jankowsky for generously providing the polyU and random RNA substrates.
REFERENCES
- 1.Kinoshita N, Goebl M, Yanagida M. The fission yeast dis3+ encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell Biol. 1991;11:5839–5847. doi: 10.1128/mcb.11.12.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki N, Noguchi E, Nakashima N, Oki M, Ohba T, Tartakoff A, Ohishi M, Nishimoto T. The Saccharomyces cerevisiae small GTPase, Gsp1p/Ran, is involved in 3′ processing of 7S-to-5.8S rRNA and in degradation of the excised 5′-A0 fragment of 35S pre-rRNA, both of which are carried out by the exosome. Genetics. 2001;158:613–625. doi: 10.1093/genetics/158.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 6.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 7.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNA Met in Saccharomyces cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham AC, Davis SM, Andrulis ED. Interdependent nucleocytoplasmic trafficking and interactions of Dis3 with Rrp6, the core exosome, and Importin-α3. Traffic. 2009;10:499–513. doi: 10.1111/j.1600-0854.2009.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mamolen M, Andrulis ED. Characterization of the Drosophila melanogaster Dis3 ribonuclease. Biochem. Biophys. Res. Commun. 2009;390:529–534. doi: 10.1016/j.bbrc.2009.09.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J. Mol. Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 15.Vincent HA, Deutscher MP. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J. Mol. Biol. 2009;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorentzen E, Basquin J, Conti E. Structural organization of the RNA-degrading exosome. Curr. Opin. Struct. Biol. 2008;18:709–713. doi: 10.1016/j.sbi.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham AC, Kiss DL, Andrulis ED. Core exosome-independent roles for Rrp6 in cell cycle progression. Mol. Biol. Cell. 2009;20:2242–2253. doi: 10.1091/mbc.E08-08-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding protein recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 22.Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. J. Biol. Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- 23.Noguchi E, Hayashi N, Azuma Y, Seki T, Nakamura M, Nakashima N, Yanagida M, He X, Mueller U, Sazer S, et al. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 1996;15:5595–5605. [PMC free article] [PubMed] [Google Scholar]
- 24.Shiomi T, Fukushima K, Suzuki N, Nakashima N, Noguchi E, Nishimoto T. Human Dis3p, which binds to either GTP- or GDP-Ran, complements Saccharomyces cerevisiae Dis3. J. Biochem. 1998;123:883–890. doi: 10.1093/oxfordjournals.jbchem.a022020. [DOI] [PubMed] [Google Scholar]
- 25.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 26.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 27.Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell. 2006;17:1399–1409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 29.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 2000;10:888–890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 30.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 32.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment, in the RNase II nuclease family. Mol. Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 33.Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS ONE. 2007;2:e317. doi: 10.1371/journal.pone.0000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 36.Hieronymus H, Yu MC, Silver PA. Genome wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.