Abstract
DNA telomeric repeats in mammalian cells are transcribed to guanine-rich RNA sequences, which adopt parallel-stranded G-quadruplexes with a propeller-like fold. The successful crystallization and structure analysis of a bimolecular human telomeric RNA G-quadruplex, folded into the same crystalline environment as an equivalent DNA oligonucleotide sequence, is reported here. The structural basis of the increased stability of RNA telomeric quadruplexes over DNA ones and their preference for parallel topologies is described here. Our findings suggest that the 2′-OH hydroxyl groups in the RNA quadruplex play a significant role in redefining hydration structure in the grooves and the hydrogen bonding networks. The preference for specific nucleotides to populate the C3′-endo sugar pucker domain is accommodated by alterations in the phosphate backbone, which leads to greater stability through enhanced hydrogen bonding networks. Molecular dynamics simulations on the DNA and RNA quadruplexes are consistent with these findings. The computations, based on the native crystal structure, provide an explanation for RNA G-quadruplex ligand binding selectivity for a group of naphthalene diimide ligands as compared to the DNA G-quadruplex.
INTRODUCTION
G-quadruplexes are non-canonical nucleic-acid structures with unusually high stability. This stability is derived in part from the stacking together of G-quartets, which are planar arrangements of four guanines held together by eight hydrogen bonds. These G-quartets stack through π–π interactions to form stable quadruplex motifs (1,2). The recently identified non-coding telomeric RNA’s are composed of extended tandem r(UUAGGG) repeats, transcribed from telomere DNA sequences located at the terminal ends of chromosomes (3–7). The G-rich telomeric repeat-containing RNA sequences (TERRA/telRNA) have been shown to form stable parallel-stranded G-quadruplex structures in solution (8–10), analogous to their single-stranded DNA counterparts. TERRA molecules have regulatory roles in telomere maintenance and other regulatory functions in both yeast and mammalian cells (3–7). They were recently shown to directly associate with two core proteins of the Shelterin complex—telomere repeat factors 1 and 2 (TRF1 and TRF2) (11). The Shelterin complex is an important multimeric complex involved in telomere maintenance, and is located proximal to the 3′-end of the chromosome (12). Composed of several proteins, it forms a tight complex with double-stranded G-rich telomeric DNA, and is directly involved in recruitment of the enzyme telomerase to single-stranded 3′-ends (12). Additionally, G-rich TERRA sequences have the potential to directly interfere with telomere extension and maintenance: by hybridizing to the complimentary single-stranded C-rich telomeric DNA. This is transiently exposed during the replication process, thereby interfering with the replication machinery; by hybridizing to the C-rich template region of the RNA subunit of telomerase (hTR), it can directly disrupt telomerase function.
The current interest in G-rich telomeric DNA folded into quadruplex structures stems from its attractiveness as an anti-cancer target, linked to the ribonucleoprotein telomerase, an enzyme that maintains chromosomal integrity and is up-regulated in >85% of various human cancers types (13). Telomerase catalytic function requires hybridization between its RNA template sequence and the single-stranded 3′-end of telomeric DNA; this association can be impeded by the stabilization of this DNA into a higher order-structure. The hybridization equilibrium can be shifted by addition of small-molecule ligands acting as stabilizers of these higher-order structures. By inhibiting substrate binding, telomerase is down-regulated, so interfering with overall telomere maintenance. Selective molecules that bind and topologically trap G-quadruplex structures can prevent other important telomere regulatory proteins such as POT1 from binding single-stranded DNA, resulting in telomere attrition and eventually senescence or cell death (14–16). Critically, the extreme ends of mammalian chromosomes terminate with a single-stranded 3′ overhang of ∼120–150 nt (17). With no complimentary C-rich strand, this region is free to form G-quadruplex structures and influence overall genomic stability, cellular division and cellular replicative lifespan (18). Thus, the overhang has been extensively studied as a therapeutic target. TERRA molecules have the same sequence as the repeats of single-stranded telomeric DNA, and are similarly free to self-associate and form higher order G-quartet based quadruplex motifs. An understanding of ligand selectivity between RNA and DNA quadruplexes, and thus of RNA quadruplex architecture, will be important for the future design of selective telomere targeting agents.
We report here the first crystal structure of a quadruplex formed from human telomeric RNA (TERRA). This provides fine detail of RNA quadruplex folding, such as water structure, groove widths and specific hydroxyl group interactions, as well as being a basis for the design of selective RNA quadruplex-targeted ligands. The chosen sequence contains two G3 runs, linked by a UUA sequence and flanked on the 5′- and 3′-ends by uracil and adenine bases. Previous crystallographic analysis of the DNA sequence d(UBrAGGGUBrTAGGGT) revealed a folded, parallel-stranded quadruplex topology, with the two strands associating to form a bimolecular quadruplex (19). We hypothesized that an analogous RNA sequence r(UBrAGGGUUAGGGU) should also crystallize; this was synthesized and indeed has crystallized. This RNA structure displays the same overall topology and structural arrangement as its DNA analogue, with the expected C3′-endo sugar puckers for many of the ribose sugars, albeit with a modified hydration structure. The influence of the hydroxyl groups on the ribose sugars in this context will be described in detail, particularly in the light of recent modelling and mass spectrometry studies which have identified several quadruplex-binding ligands with DNA versus RNA selectivity (20).
MATERIALS AND METHODS
Crystallization and data collection
The oligonucleotide sequence, r(UBrAGGGUUAGGGU), was purchased from Eurogentec (Belgium) and used without further purification. Mass spectrometry showed it to comprise a single species. A stock solution was prepared by dissolving the RNA in RNAse-free water to a final single-strand concentration of 3 mM. For crystallization trials, the RNA solution was further diluted to 1.5 mM (ssRNA) and annealed in 30 mM KCl and 50 mM potassium cacodylate (pH 6.5) by heating to 90°C for 5 min, followed by cooling to room temperature overnight. RNA quadruplex crystals were grown by the hanging drop vapour diffusion method. One microlitre of the annealed RNA quadruplex was mixed with 1 µL reagent solution, composed of 15% MPD, 150 mM NaCl, 50 mM sodium cacodylate (pH 6.5) and 5 mM spermine. The hanging drop was equilibrated against a well containing 15% MPD. Crystals appeared overnight and were left to grow for 1 week at 12°C. A dataset was collected at 105 K on a single flash-frozen crystal, on an Oxford Diffraction Xcalibur NovaT X-ray diffractometer. The data were processed and scaled using CrysalisPro (Oxford Diffraction) and Scala (from the CCP4 suite) (21). The crystals were assigned to space group P3121, with unit cell dimensions a = b = 57.58 Å, c = 38.38 Å, α = β = 90°, γ = 120°.
Structure determination and refinement
The structure was solved by molecular replacement using the Phaser program (22) (CCP4), using the bimolecular DNA quadruplex structure, PDB 1K8P, as a model, and refined using Refmac5 (23). Data collection and refinement statistics are shown in Table 1. The RNA model was generated by modification of the DNA structure (addition of hydroxyl groups at the C2′ position and deletion of methyl groups from the C5 position of thymine). The three guanine quartets, bromine atoms and K+ ions could be clearly seen in the initial σA-weighted 2Fo–Fc electron density maps, while the initial Fo–Fc difference electron density maps showed residual density for the bromine atoms and the propeller loop of strand A. The RNA model was initially built into maps calculated from low-resolution data, and extended and refined into maps calculated with progressively higher-resolution data, through iterative cycles of manual adjustment with maximum-likelihood restrained refinement, using Coot (24) and Refmac5 (23). Electron density for the hydroxyl groups could also be clearly seen in the initial σA-weighted 2Fo–Fc electron density maps, allowing these features to be accurately placed at an early stage of the solution process, and ensuring the sugar puckers and backbone could be modelled accurately. A translation/libration/screw (TLS) motion determination (25) approach was used in the final stages of refinement in an attempt to fit the propeller loop of strand B (residues U18, U19, A20) into density. The RNA model was divided into four groups (two per strand) for the TLS refinement. Although the TLS refinement improved the accuracy of the model, the majority of the UUA loop of strand B could not be fully fitted into density. The occupancy of this loop has therefore been set to 0% and has been assigned geometry based on the A-strand linking loop (U6, U7, A8). The phosphate group (atoms P, OP1 and OP2) for residue U18 could be fitted into density and has been given full occupancy in the model. The final model (including solvent molecules) was refined using data between 11.69 and 2.20 Å, with final R and Rfree values of 21.6 and 23.1% respectively.
Table 1.
Data collection and refinement statistics for the telomeric RNA quadruplex crystal structure
| Data collection | |
| Sequence | r(UBrAGGGUUAGGGU) |
| Space group | P3121 |
| Unit cell dimensions: | |
| a, b, c (Å) | 57.58, 57.58, 38.38 |
| α, β, γ (°) | 90.00, 90.00, 120.00 |
| Resolution (Å) | 49.87–2.20 |
| Rint (%) overall | 5.0 (24.0) |
| I/σ | 43.38 (4.54) |
| Completeness (%) | 99.6 (97.3) |
| Redundancy | 6.3 (6.0) |
| Refinement | |
| Resolution (Å) | 11.69–2.20 |
| Reflections | 3899 |
| Rwork/Rfree (%) | 21.6/23.1 |
| No. of atoms | 520 |
| Ions | 2 |
| Water | 45 |
| Overall B-factor (Å2) | 24.26 |
| RMS deviations | |
| Bond-lengths (Å) | 0.013 |
| Bond-angles (°) | 0.859 |
| PDB ID | 3IBK |
Values in brackets refer to the highest resolution shell, 2.28–2.20 Å.
Modelling
In order to explore the effects of dynamics on structural stability and inter- and intra- molecular interactions, multiple multinanosecond molecular dynamics (MD) simulations were performed on the crystal structure of this 12-mer RNA quadruplex, sequence UBrAGGGUUAGGGU. The X-ray structure has a vertical column of consecutive K+ ions within the central core, sandwiched equidistant between G-quartets, which was retained for the simulations. Explicit solvent (TIP3P; 20 829 waters) and counter ions (20 K+ ions) were added such that the total charge was zero. Energy minimization (steepest descent algorithm) was run for 5000 steps, followed by a 500-ps equilibration phase during which the quadruplex was harmonically restrained with a force constant of 1000 kJ mol−1 nm−2 at 300 K, which was gradually relaxed until no restraints were applied. During the entire process, the ions and the solvent were allowed to equilibrate. The final production run was carried out without any restraint on the system using the GROMACS program, version 4.0 (26) (http://www.gromacs.org) with the AMBER ff99 and parmBsc0 forcefield (27) that has been ported into the GROMACS MD suite (28). Additional details of modelling methods can be found in the Supplementary Data.
The specificity of ligand interactions with the RNA quadruplex was examined by means of three additional MD simulations with bound ligands (Supplementary Figure S1). Molecular models of the naphthalene diimide series of quadruplex-stabilizing ligands (29) were constructed; minimized and partial charges calculated semi-empirically using the MOPAC program (30) as implemented in the Insight II suite software (http://www.accelrys.com). Prior to docking, terminal uracil and adenine residues were removed in order to fully expose the flat planar quartet to the ligand. Several uracil and adenine terminal 3′ and 5′ residues are involved in crystal packing interactions but are not integral to the stabilization of this quadruplex arrangement. Of the two uracils at the 3-ends, one was not observed while the second is observed stacking through crystal packing with the loop of a symmetry related quadruplex. The ligand structures were minimized and docked on the 3′ surface of the quadruplex structure using the AFFINITY docking program (http://www.accelrys.com) employing the grid docking method available with AFFINITY. This approach has been previously validated in our studies on the rational design of quadruplex ligands (31,32). The G-quartets were frozen during the entire docking procedures since they have been shown to be exceptionally stable and structurally rigid, as confirmed by several experimental and simulation studies (19,33–42). The automated docking process identifies and ranks positions based on interaction energies within the binding site. The final conformation of the complexes were then subjected to a further 500 steps of unrestrained molecular mechanics minimization (steepest descent algorithm). The force-field parameters for ligands were generated using the AMBER force field ff99 (43) and subsequently ported to GROMACS format using amb2gmx perlscript (44). The three production runs were each performed for 20 ns.
A MD simulation of the equivalent 12-mer DNA quadruplex (PDB 1K8P; resolution 2.40 Å) was performed for 18 ns in order to highlight the differences between RNA and DNA quadruplexes. The simulation protocols were consistent for all systems. Trajectories were analysed using GROMACS and locally written scripts and the data visualized by means of the VMD program (45) with graphs plotted using the Xmgrace program (http://plasma-gate.weizmann.ac.il/Grace/).
RESULTS
Topological conservation between RNA and DNA G-quadruplexes
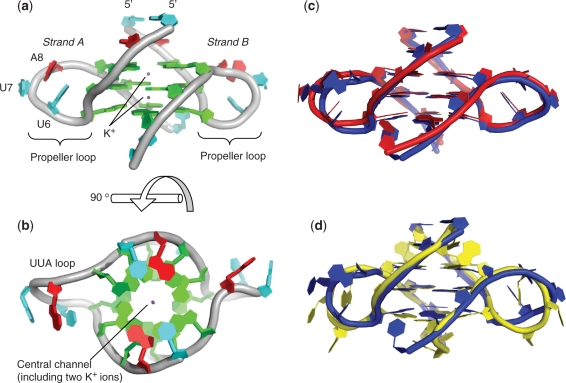
In the crystal packing arrangement for the P3121 space group, the asymmetric unit contains two RNA strands (A and B) of sequence r(UBrAGGGUUAGGGU). Each RNA strand contains two G-rich repeats, folding to form a parallel four-stranded bimolecular G-quadruplex. The intervening UUA nucleotides link the phosphate backbone through an external propeller-type arrangement, keeping the phosphate backbone strand associations parallel. The structure contains three stacked G-quartets, with two clearly identified K+ ions in the centre of the channel, positioned between the three G-quartets (Figure 1a and b) and coordinated in a bipyramidyl antiprismatic manner to the O6 atoms of each guanine. The overall quadruplex fold seen here is identical to that of the equivalent DNA quadruplex (19) (1K8P) and is in agreement with the NMR structure reported for an unmodified RNA sequence (10). A comparison of the root mean square (rms) deviations between the X-ray derived RNA quadruplex and the NMR derived structure is shown in Table 2. There is very little difference between the central G-quartets of the three structures (Figure 1c and d), however there are significant deviations between the structures when comparing the coordinates for the overall structures. These differences are primarily related to the diverse set of conformations adopted by the UUA loops of strand A and B for the NMR structure as reported in the ten deposited NMR-derived coordinate sets (10).
Figure 1.
Cartoon representation of a bimolecular quadruplex formed from human telomeric RNA contained within one asymmetric unit. The UUA loop for strand B is disordered, with the connecting backbone atoms modelled on the weakly observed residual electron density. The colouring scheme is based on nucleotide type, with guanines green, modified and non-modified uracils blue and adenines red. (a) Highlights the stacking of the UUA loops with the central uracil perpendicular to the stacked guanines. (b) shows the stacking of the uracils and adenines on to the G-quartets. (c) Overlay of the bimolecular RNA quadruplex (PDB 3IBK) (blue) with the equivalent bimolecular DNA quadruplex (PDB 1K8P) (red). (d) overlay of the bimolecular RNA quadruplex (PDB 3IBK) (blue) with the NMR model of the same sequence (PDB 2K8P, model 1) (yellow). The molecular graphics program PyMOL was used for structural alignments, calculation of rms deviations and visualization (http://www.pymol.org).
Table 2.
Comparison of the rms deviation between the two RNA models [2KBP—determined by NMR (model 1) and 3IBK determined by X-ray diffraction], and between the two crystal structures (1K8P–DNA quadruplex and 3IBK–RNA quadruplex)
| rmsd (Å) |
||
|---|---|---|
| Whole model | Quartets only | |
| 3IBK v 1K8P (DNA) | 0.990 (470 atoms) | 0.539 (232 atoms) |
| 3IBK v 2KBP (NMR) | 2.971 (473 atoms) | 1.011 (246 atoms) |
The rms deviations are generated from structural alignments, which can be seen in Figure 1c and d
Crystal packing interactions
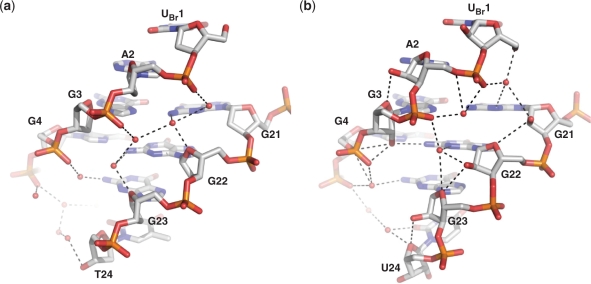
The two 5′ nts UBr1 and A2 associate together in the crystal lattice through a 2-fold crystallographic symmetry element forming two stacked UBrAUBr*A* tetrads, to generate a 5′ to 5′ stacked arrangement of two quadruplexes. The bromine atom attached at position C5 on the uracil base is a required modification, enhancing crystal diffraction quality by holding the dimer interface tightly together. The two UUA loops have different packing interactions, where the trinucleotide linking loop of strand A (U6, U7 and A8) is well ordered, folding as a stacked UAU loop, similar to strand A in the bimolecular DNA quadruplex (1K8P), whereas the UUA loop from strand B is not sufficiently ordered to model the position of the three nucleotides (Figure 1a and b). This overall lack of ordering in the B strand loop is not the result of an unfavourable crystal packing arrangement as space is available for folding into a conformation seen in the strand A loop.
Sugar puckering variation
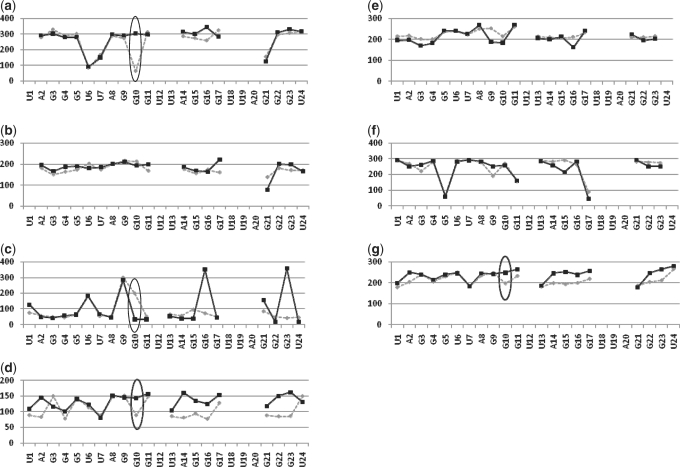
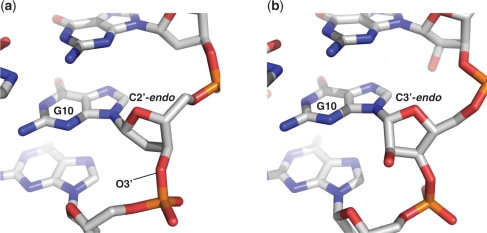
As expected for a RNA with 2′-OH groups, the majority of the RNA quadruplex ribose sugar puckers can be classified as being in the C3′-endo conformation, including the 5′ nts UBr1 and A2, and the central guanines of the G-quartets (Table 3). Uniform C3′-endo puckering is not observed throughout the structure however, as the ribose sugars of the loop residues and the guanines within the terminal G-quartets adopt a variety of pucker conformations. This mix of C3′-endo and C2′-endo puckering is consistent with the NMR model and with the DNA crystal structure (2KBP and 1K8P respectively), and was also observed in the r(UGGGGU) intermolecular parallel-stacked quadruplex refined to 0.61 Å resolution (46). The sugar puckers of the crystal structure reported here correspond well to those observed in the solution structure, especially for the nucleotides in the G-quartets (Table 3). Both solution and crystal structures have uniform C3′-endo puckers for the guanine residues of the central quartet (G4, G10, G16 and G22), and a mix of C2′-endo and C3′-endo puckers for the guanines of the outer G-quartets. Sugar pucker consistency between the RNA and DNA crystal structures is generally also observed, with the strand A UUA loop of both quadruplexes adopting the same pattern of sugar puckering (Table 3), implying that the loop geometry, base stacking and sugar puckers are strongly conserved in this stacked propeller loop conformation. There are though some significant differences between the RNA and DNA crystal structures, primarily for the sugar pucker conformations of non-loop residues, and with respect to intramolecular interactions (described below). Comparison of the backbone torsion angles of the RNA and DNA quadruplexes shows that, although the backbone conformations of these two structures to a large extent do correspond well with each other, there are some significant deviations which may explain how the two structures are able to adopt the same overall topology, despite differences in sugar puckers preference and available intramolecular interactions (Figure 2). An example is residue G10, which is situated within the plane of the central quartet in both DNA and RNA structures, with a C2′-endo sugar pucker for the DNA and a C3′-endo pucker for the RNA quadruplex. A detailed comparison of the backbone and glycosidic torsion angles of G10 reveals correlated differences in the backbone dihedral angles α, δ, γ and χ, suggesting that this difference in pucker is compensated for by changes in the torsion angles to allow the overall orientation of this residue to be conserved in the two structures (Figure 3).
Table 3.
Sugar pucker conformations for the 12-mer DNA crystal structure (1K8P), the 12-mer RNA crystal structure (3IBK) and the 12-mer NMR structure (2KBP, model 1)
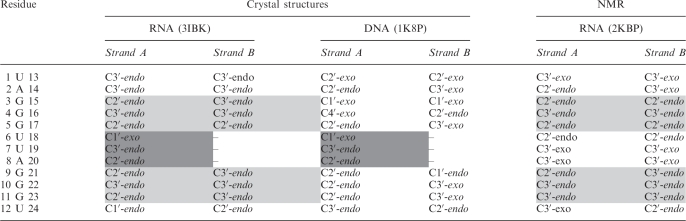 |
Linking U6 U7 and A8 residues for the B strands have been excluded for the crystal structures. Sugar puckers consistent between 3IBK and 2KBP are shaded light grey. Sugar puckers consistent between 3IBK and 1K8P are shaded dark grey. The sugar puckers are defined by the combined pseudorotation phase angle.
Figure 2.
Comparison of torsion angles in bimolecular quadruplex cyrstal structures. RNA (PDB 3IBK), solid black trace with squares; DNA (1K8P), grey dashed trace with diamonds. Dihedral angles are plotted separately; in order from (a) to (g)—alpha, beta, gamma, delta, epsilon, zeta, chi. Significant deviations between the two structures are circled (black). In all the graphs, x-axis shows residue and the y-axis shows torsion angle.
Figure 3.
Comparison of sugar puckering and backbone dihedral angles of residue G10 between DNA (a) and RNA (b) quadruplexes. The deoxyribose sugar of the DNA quadruplex adopts a C2′-endo conformation. A C2′-endo conformation for the RNA at this position would cause the C2′ hydroxyl group to sterically clash with the O3′ oxygen atom. The sugar of the G10 residue within the RNA structure therefore adopts a C3′-endo conformation (b), accompanied by alterations in backbone dihedral angles which preserves the positioning of the guanine base within the quartet.
Hydroxyl group interactions
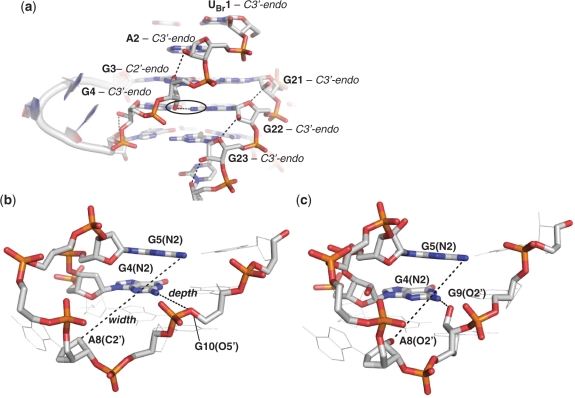
The RNA ribose sugars, with their 2′-OH groups, have the potential to make many more inter- and intra-molecular interactions than their DNA counterparts. In this quadruplex arrangement, the hydroxyl groups are seen to be preferentially forming intramolecular interactions with the quadruplex itself, rather than intermolecular contacts with solvent molecules. The resulting RNA quadruplex has fewer observed structured water molecules than the equivalently packed DNA quadruplex, and is also accompanied by an increase in intramolecular contacts. The 2′-OH hydroxyl groups are seen to form hydrogen bonds with a variety of acceptors within the RNA molecule, and interestingly, the type of intramolecular interaction appears to be dependent on the pucker conformation (Figure 4a). For example, within the quadruplex grooves, the hydroxyl groups of those sugars with a C3′-endo pucker are positioned such that they can form a hydrogen bond with the O4′ oxygen atom of the neighbouring residue. Alternatively, the hydroxyl groups of the sugars with a C2′-endo pucker preferentially hydrogen bond to the N2 amine group of the guanine base. For example, the hydroxyl group of residue G3, with a C2′-endo pucker, forms a hydrogen bond with the N2 amine group of G22 (circled black in Figure 4a). This interaction is also seen between the hydroxyl group of G9 and the N2 amine group of G4.
Figure 4.
(a) Intramolecular hydrogen bonding and sugar pucker conformation within the RNA quadruplex groove region in the crystal structure. The hydroxyl groups of residues with C3′-endo puckered sugars are seen to interact with the neighbouring O4′ atom, whereas the hydroxyl group of residue G3, which has a C2′-endo puckered sugar, is seen to interact with the N2 atom of a guanine base (circled). (b) DNA G-quadruplex and (c) RNA G-quadruplex loop dimensions (width and depth). The presence of 2′-OH groups reduces the dimensions of the loop grooves. The depth of the loop is the distance between N2 atom of guanine in the middle quartet to the first atom directly opposite it in the horizontal direction [DNA: G4(N2)-G10(O5′) – 6.40A and RNA: G4(N2)-G9(O2′) – 3.1 Å]. The width of the loop is the distance between N2 atom of guanine in the top quartet to the first atom which is diagonally opposite at the base of the loop [DNA:G5(N2)-A8(C2′) – 11.4 Å and RNA:G5(N2)-A8(O2′) – 9.8 Å].
MD simulations: stability and ligand binding
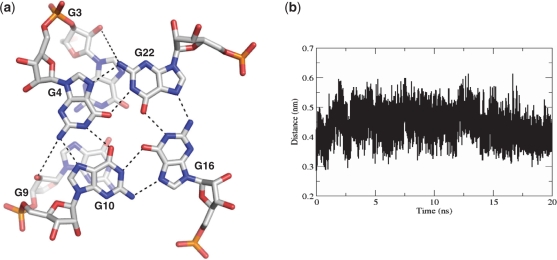
In order to assess the comparative conformational stability of the telomeric RNA quadruplex, a 20ns MD simulation was performed on both the RNA 12-mer and the DNA 12-mer quadruplex structures (PDB IDs 3IBK and 1K8P respectively). The root mean square deviations were monitored during the course of the simulations, as a measure of structural stability. The results of these simulations indicate that the 12-mer RNA quadruplex structure is more stable than the 12-mer DNA analogue, with rms deviations of 1.90 Å and 2.62 Å for the RNA and DNA respectively (Supplementary Figure S1). It is notable that the propeller topology of the UAU loop in the RNA quadruplex was maintained throughout the simulation, which was in contrast to the DNA simulation, during which the loops un-stack and open up. The C2′ hydroxyl groups can be seen during the course of the RNA MD simulation to interact with O3′ atoms (within the same residue), O4′ and O5′ atoms (of the adjacent residue in the backbone) and with N2 amine groups of the guanine bases (Supplementary Figure S2). In accordance with the crystal structure, the MD simulations show that the 2′-OH-N2 interactions are dependent upon the sugar adopting a C2′-endo pucker, and also show that this interaction is lost as the sugar conformation changes (Figure 5 and Supplementary Figure S3). The MD simulations of the RNA G-quadruplex further highlight the ability of the 2′-OH groups to make multiple interactions within the loop during the 20 ns of the simulation. These interactions impart rigidity to the overall sugar-phosphate backbone, and also explain the reduced flexibility observed within the UUA loops.
Figure 5.
C2′-endo 2′-OH–N2 interactions. (a) A 20-ns snapshot of interactions formed between 2′-OH and N2 atom of guanine residues when C2′-endo sugar puckering is present. (b) 2′-OH–N2 interactions calculated over 20 ns simulations. Over the course of the 20 ns simulations, the 2′-OH groups makes multiple interactions with O4′, O3′, O5′ and phosphate oxygen atoms. These interactions impart rigidity to the sugar phosphate backbone and are likely to contribute to the higher stability of RNA G-quadruplexes over DNA ones.
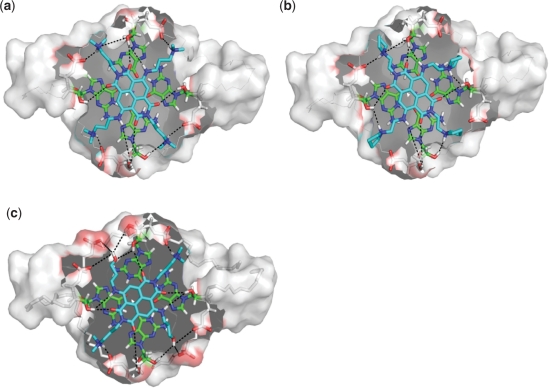
A recent study using electrospray–ionization mass spectrometry identified several small-molecule compounds with the ability to select between telomeric DNA and RNA quadruplexes (20). In order to rationalize these experimental findings, we have performed a series of MD simulations on the bimolecular RNA quadruplex structure (3IBK) bound with members of a group of three naphthalene diimide ligands. Details of the ligands used for the studies are shown in Supplementary Figure S4. MD simulations identified a ranking order for RNA quadruplex-ligand binding strength as follows: Ligand 3 > Ligand 1 > Ligand 2 that is in accord with the experimental data (20). The MD studies provide insight into the ligand binding mode, and can offer explanations for differences in binding affinity. The three naphthalene diimide compounds used for the present studies share a common core ring system, which is seen to stack effectively over the 3′ G-quartet. The 2′-OH groups of the RNA quadruplex make effective interactions with the carbonyl groups of the central chromophore, as well as with nitrogen groups within the side chain of the ligands. The interactions between the chromophore and 2′-OH groups at the apices of the G-quartets effectively locks the ligands in place and maximizes π–π stacking interactions (Figure 6). A total of six hydrogen bonds are formed between the ligand chromophore and 2′-OH groups. As the naphthalene diimide ring system is a common feature of these ligands, differences in quadruplex-ligand binding affinities are a result of differing side-chain functionalities. These side-chains interact with the quadruplex loops, rather than the G-quartets themselves, providing an explanation for the experimental data, which show that all three ligands do bind to DNA quadruplexes with high affinity, but only one ligand (Ligand 3) binds to the RNA quadruplex with comparable affinity (20). MD simulations of the RNA showed that the 2′-OH groups make multiple interactions within the loop, thereby reducing the depth and width of loop dimensions when compared to the equivalent DNA structure (Figure 4b and c). As a result, the RNA quadruplex is less amenable to bind ligands with side-chains terminating in bulky and/or inflexible functional groups. The lack of flexibility of the pyrrolidine group of Ligand 2, combined with the inaccessibility of the loop, makes this ligand the weakest binder in the series. The substitution of the pyrrolidine group by ––NHMe+2 (Ligand 1) results in a functional group with greater flexibility, but which is still ineffective in participating in stabilising 2′-OH interactions with the loop. Substitution of ––NHMe+2 with the –OH group (Ligand 3) increases functional group flexibility and also contributes towards hydrogen bonding interactions with the loop. An MD simulation snapshot of the ligand side-chain-loop interactions shows that a maximum of two hydrogen bonds per loop can be formed between Ligand 2 and the loop (Figure 6b). Additional flexibility of the –NHMe+2 group in Ligand 1 increases the ability of this molecule to pick up dynamic interactions during the simulation (Figure 6a). Replacement of the –NHMe+2 groups within two side chains having –OH groups (Ligand 3) increases the hydrogen-bonding ability of this ligand significantly, as the –OH group is seen to be involved in up to three hydrogen bonds (per side chain) with the quadruplex (Figure 6c).
Figure 6.
RNA quadruplex-ligand interactions studied by MD. Interactions between the side chain of ligands (a) Ligand 1, (b) Ligand 2 and (c) Ligand 3 and the 12-mer RNA G-quadruplex. The interactions are shown as dashed lines. The ligand (cyan) is stacked onto the 3′ G-quartet (carbons shown in green). Comparison of the surface charge distribution between the RNA and DNA bimolecular quadruplexes reveals areas of the RNA structure with increased polarity, which may be of significance for ligand binding (Supplementary Figure S5).
DISCUSSION
Similarity of topologies
The high quality of the diffraction data in this 2.20 Å resolution structure has enabled a detailed view of the RNA core structure to be obtained, with its stacked G-quartets, along with one of the two UUA loops, hydration structure and detail for all other ordered components in the crystal lattice. In contrast to the diverse folded topologies seen for human telomeric DNA quadruplex sequences, especially with changes in ionic environment, the RNA fold observed in this X-ray structure is consistent with the solution NMR data in K+ buffer (10) and in Na+ buffer, as confirmed by NMR, CD and MALDI-TOF MS (9). The striking similarity between the RNA and DNA structures is most apparent when they are overlaid (Figure 1c). This consistency in topology can be explained by the marked preference in RNA for an anti glycosidic bond angle, a consequence of steric constraints imposed by the C2′ hydroxyl groups (47); syn conformations for RNA nucleotides have been observed but only when contained within a very specific structural context, which includes quartet arrangements (48) for example with 8-bromo-guanine.
Correlations between sugar puckering and backbone geometries preserve G-quartet stacking
In stacked RNA structures, the ribose sugar puckers generally adopt the C3′-endo conformation, as seen in the present structure, where the majority of the sugars of the terminal residues have puckers in the C3′-endo conformation. However, for the stacked G-quartets, a more mixed puckering arrangement is observed, which is consistent with that observed in an intermolecular parallel stacked quadruplex of sequence r(UGGGGU) (46) albeit without the connecting loops. This mixing of ribose sugar puckers seen for the stacked guanines may reflect a conformational preference that is finely balanced between the requirements for stacking of the G-quartets and the steric hindrance caused by the addition of hydroxyl group at the C2′ position on the ribose sugar. This balance of sugar pucker between C3′-endo and C2′-endo may be shifted by the addition of the connecting propeller loops and constraints in geometry in the connecting backbones.
Hydroxyl group interactions are linked to sugar pucker conformation, and appear to affect the hydration structure and overall stability of the RNA quadruplex
The sugar pucker arrangement in our structure for the guanine G3 stack has C2′-endo puckers at the ends and a C3′-endo for the central guanine, an arrangement consistent with the RNA quadruplex (PDB ID 2KBP) in NMR solution, but quite different from the two equivalent DNA quadruplex structures (PDB ID 1K8P and PDB ID 1KF1). The parallel-stranded quadruplex topology with a tightly folded propeller loop may impose additional steric constraints that need to be accommodated, such as inducing a C2′-endo conformation and being stabilized by additional 2′-OH – N2 hydrogen bonding. The modelling studies indicate that the propeller loops in the RNA have lower rms deviations over the time course of the simulation when compared to the modelling carried out on the 12-mer DNA, implying greater rigidity (Supplementary Figure S1). We infer that the additional hydrogen bonding arrangements derived from the 2′-OH groups are important contributors to stabilizing specific structural arrangements in this parallel-stranded topology.
The 2′-OH groups interact with a variety of hydrogen bond acceptors, including phosphate and backbone oxygens atoms (O3′, O4′ and O5′) and polar groups attached to the bases (such as the N2 amine group of the guanine base). With the additional hydroxyl groups, much of the internal hydrogen bonding of the RNA quadruplex is notably different to the equivalent DNA structure. In addition, these multiple intramolecular contacts of the RNA, which are observed both in the crystal structure and in our modelling studies (Supplementary Figure S2), would be expected to result in a reduction in associated water molecules, compared to the DNA (Figure 7). This data suggests to us that the extensive intramolecular contacts made by the C2′–OH groups makes a major contribution to the increased stability of the RNA quadruplexes over equivalent DNA structures. The overall loss of structural waters, the additionally constrained waters through hydrogen bonding and an increase in direct contacts through hydroxyl groups is consistent with recent biophysical studies (49) which showed the 24-mer telomeric RNA quadruplex (r(UUAGGG)4) to have a ΔTm value 12.6°C higher than the equivalent DNA quadruplex (in K+). In the same work, osmotic stress analysis showed RNA quadruplexes to have fewer associated water molecules when compared to the DNA ones (49). Additionally, recent CD analysis and melting studies (50) on various putative quadruplex-forming RNA sequences have shown a marked preference for these sequences to form parallel topologies, with higher melting temperatures in potassium when compared to their DNA equivalents. It is notable that the DNA quadruplex crystals diffracted to a significantly lower resolution compared to the RNA ones which also has overall considerably lower individual atomic temperature factors, implying greater ordering in the lattice and a higher rigidity.
Figure 7.
A comparison of water structure found within quadruplex groove regions. Left, DNA (1K8P); right, RNA (3IBK). Hydrogen-bonding interactions within the DNA quadruplex groove region are limited to intermolecular interactions with water molecules (a). In contrast, hydrogen bonding within the equivalent region of the RNA quadruplex includes both intramolecular and intermolecular interactions (b). With many of the hydrogen bonding groups of the RNA quadruplex involved in intramolecular contacts, there is an associated reduction in ordered water molecules within this structure, when compared to the equivalent DNA structure.
Modelling studies combined with structural data identify significant differences between RNA and DNA quadruplexes which may be useful in the design of selective quadruplex interacting ligands
Telomeric DNA quadruplexes have been extensively investigated as a target for anti-cancer therapies by the use of small molecule ligands (16), thus creating a wealth of biophysical and structural data accumulated over the last twelve years. This ligand/quadruplex data base will now have to be expanded to take into account an expanding diversity of targets generated by the discovery of TERRA sequences. The design of ligands that take into account DNA versus RNA quadruplex selectivity will have to be addressed. The studies presented here using differential analysis of dynamic interactions between ligands and RNA G-quadruplexes highlight some of the important structural features that should be considered when designing selective G-quadruplex stabilising ligands. For example, when comparing the naphthalene diimide ligands to DNA versus RNA selectivity, as well as binding affinities, the differences in association can be directly linked to the 2′-OH groups of the RNA. We observed that the 2′-OH groups make extensive interactions, both within the RNA molecule itself, by altering groove distances compared to the equivalent DNA structure, as well as with the ligands themselves, the 2′OH groups being directed upwards onto the 3′-quartet external surface (Figure 6). This highlights the importance of hydrogen bonding groups within quadruplex-binding small-molecule ligands.
Based on our observations, selectivity between DNA and RNA quadruplexes may be achievable through the appropriate selection of chromophores and side-chains. We therefore propose that, with the recent discovery of TERRA and telomeric RNA quadruplexes, it may now be necessary to screen potential telomeric DNA quadruplex stabilizing ligands in parallel with the equivalent RNA sequences, to assess this specificity. The application of structural data has played a major role in the understanding of interactions between ligands and DNA quadruplexes, particularly with the observations of large conformational changes in loop structures to accommodate diverse families of ligands. For example, the extensive loop modification induced by TmPyP4 binding to a parallel-stranded DNA quadruplex (51) may also be relevant to parallel-stranded RNA quadruplexes and may explain the observed similarity in binding affinity and stoichiometry of TmPyP4 to RNA and DNA human telomeric parallel-stranded quadruplexes (49). This similarity of loop structure for the connecting UUA loops in RNA and DNA in the crystal structures extends to almost identical backbone dihedral angles (Figure 2) even with the addition of the 2′-OH groups. The loops are stabilized in this arrangement with the inclusion of a U6(O2′)–A8(N7) hydrogen bond. However, the dimensions of the loops, as defined by width and depth, are significantly reduced with the addition of the 2′-OH groups (Figure 4b and c), which will affect the available space for ligand binding, at least in the absence of disruptions to the loop arrangement.
As the majority of RNA quadruplexes studied to date exhibit a parallel topology (8–10,49,50), small molecule selectivity between RNA quadruplex types [e.g. TERRA quadruplex versus mRNA quadruplex (52,53)] offers a particular challenge in quadruplex targeting. This may be an important consideration, as ligands designed to stabilize mRNA quadruplexes (e.g for anti-cancer therapies) would need to be carefully designed to not bind to TERRA quadruplexes, as inadvertent stabilization of TERRA quadruplexes could potentially increase telomerase activity (by reducing the amount of hTR-template-RNA which is maintained in duplex form), although this is yet to be demonstrated experimentally. Sequence-dependent connective loop structures, and their accessible conformational diversity, may be the main avenue available for ligands to discriminate between the many similar parallel-stranded RNA structures, and we suggest therefore that this should be the main consideration in the design of inter- and intra- molecular RNA quadruplex selective ligands.
DATA BANK ACCESSION CODES
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with ID code 3IBK
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research UK (grant number C129/A10902); Research Studentship from the School of Pharmacy to G.C. Funding for open access charge: Cancer Research UK grant number C129/A10902.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Oxford Diffraction is thanked for the use of their data collection facilities.
REFERENCES
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge UK: Royal Society of Chemistry; 2006. [Google Scholar]
- 3.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 4.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 5.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 6.Schoeftner S, Blasco MA. A/‘higher order/’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Kaminaga K, Komiyama M. Human telomeric RNA in G-quadruplex structure. Nucleic Acids Symp. Ser. 2008:175–176. doi: 10.1093/nass/nrn089. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Kaminaga K, Komiyama M. G-quadruplex formation by human telomeric repeats-containing RNA in Na+ solution. J. Am. Chem. Soc. 2008;130:11179–11184. doi: 10.1021/ja8031532. [DOI] [PubMed] [Google Scholar]
- 10.Martadinata H, Phan AT. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 2009;131:2570–2578. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- 11.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 13.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 14.Stewart SA, Weinberg RA. Telomerase and human tumorigenesis. Semin. Cancer Biol. 2000;10:399–406. doi: 10.1006/scbi.2000.0339. [DOI] [PubMed] [Google Scholar]
- 15.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou J, Mergny J. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neidle S, Parkinson GN. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 19.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 20.Collie G, Reszka AP, Haider SM, Gabelica V, Parkinson GN, Neidle S. Selectivity in small molecule binding to human telomeric RNA and DNA quadruplexes. Chem. Commun. 2009:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- 21.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J. Appl.Crystallogr. 2006;39:109–111. [Google Scholar]
- 26.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 27.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys. J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuenca F, Greciano O, Gunaratnam M, Haider S, Munnur D, Nanjunda R, Wilson WD, Neidle S. Tri- and tetra-substituted naphthalene diimides as potent G-quadruplex ligands. Bioorg. Med. Chem. Lett. 2008;18:1668–1673. doi: 10.1016/j.bmcl.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 30.Stewart JJ. MOPAC: a semiempirical molecular orbital program. J. Comput. Aided Mol. Des. 1990;4:1–105. doi: 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- 31.Read M, Harrison RJ, Romagnoli B, Tanious FA, Gowan SH, Reszka AP, Wilson WD, Kelland LR, Neidle S. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl Acad. Sci. USA. 2001;98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore MJB, Schultes CM, Cuesta J, Cuenca F, Gunaratnam M, Tanious FA, Wilson WD, Neidle S. Trisubstituted acridines as G-quadruplex telomere targeting agents. Effects of extensions of the 3,6- and 9-side chains on quadruplex binding, telomerase activity, and cell proliferation. J. Med. Chem. 2006;49:582–599. doi: 10.1021/jm050555a. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 34.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefl R, Cheatham TE, Spacková N, Fadrná E, Berger I, Koca J, Sponer J. Formation pathways of a guanine-quadruplex DNA revealed by molecular dynamics and thermodynamic analysis of the substates. Biophys. J. 2003;85:1787–1804. doi: 10.1016/S0006-3495(03)74608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haider SM, Parkinson GN, Neidle S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 37.Fadrná E, Spacková N, Stefl R, Koca J, Cheatham TE, Sponer J. Molecular dynamics simulations of Guanine quadruplex loops: advances and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sponer J, Spacková N. Molecular dynamics simulations and their application to four-stranded DNA. Methods. 2007;43:278–290. doi: 10.1016/j.ymeth.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 44.Mobley DL, Chodera JD, Dill KA. On the use of orientational restraints and symmetry corrections in alchemical free energy calculations. J. Chem. Phys. 2006;125:084902. doi: 10.1063/1.2221683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Deng J, Xiong Y, Sundaralingam M. X-ray analysis of an RNA tetraplex (UGGGGU)(4) with divalent Sr(2+) ions at subatomic resolution (0.61 Å) Proc. Natl Acad. Sci. USA. 2001;98:13665–13670. doi: 10.1073/pnas.241374798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haschemeyer AE, Rich A. Nucleoside conformations: an analysis of steric barriers to rotation about the glycosidic bond. J. Mol. Biol. 1967;27:369–384. doi: 10.1016/0022-2836(67)90026-5. [DOI] [PubMed] [Google Scholar]
- 48.Pan B, Xiong Y, Shi K, Sundaralingam M. Crystal structure of a bulged RNA tetraplex at 1.1 Å resolution: implications for a novel binding site in RNA tetraplex. Structure. 2003;11:1423–1430. doi: 10.1016/j.str.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 49.Arora A, Maiti S. Differential biophysical behavior of human telomeric RNA and DNA quadruplex. J. Phys. Chem. 2009;113:10515–10520. doi: 10.1021/jp810638n. [DOI] [PubMed] [Google Scholar]
- 50.Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 51.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 52.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nature Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.